Abstract
DNA methylation (DNAm) is important in brain development, and potentially in schizophrenia. We characterized DNAm in prefrontal cortex from 335 non-psychiatric controls across the lifespan and 191 patients with schizophrenia, and identified widespread changes in the transition from prenatal to postnatal life. These DNAm changes manifest in the transcriptome, correlate strongly with a shifting cellular landscape, and overlap regions of genetic risk for schizophrenia. A quarter of published GWAS-suggestive loci (4,208/15,930, p<10−100) manifest as significant methylation quantitative trait loci (meQTLs), including 59.6% of GWAS-positive schizophrenia loci. We identified 2,104 CpGs that differ between schizophrenia patients and controls, enriched for genes related to development and neurodifferentiation. The schizophrenia-associated CpGs strongly correlate with changes related to the prenatal-postnatal transition and show slight enrichment for GWAS risk loci, while not corresponding to CpGs differentiating adolescence from later adult life. These data implicate an epigenetic component to the developmental origins of this disorder.
Keywords: brain development, DNA methylation, epigenetics, functional genomics
Introduction
DNA methylation (DNAm) plays an important role in epigenetic regulation of gene expression, orchestrating tissue differentiation and development during fetal life, childhood, and adolescence, and guiding functional activity in adulthood. Epigenetic control is especially important in human brain, where gene expression is extremely dynamic during fetal and infant life, and becomes progressively more stable at later periods of development1,2. Dysregulation of these precise and coordinated gene expression changes through epigenetic mechanisms may play a vital role in the pathogenesis of neurodevelopmental disorders, such as schizophrenia (SZ) and related conditions3–5. Pathologically, these epigenetic changes, acting through gene expression, could disturb the formation of essential brain circuits, fitting into one prevailing set of hypotheses for the causes of schizophrenia, namely the “neurodevelopmental” hypothesis6.
Exogenous factors have been associated with altering DNAm levels, both at specific loci and globally (averaged across all repeat elements), including changes in diet7, and exposure to cigarette smoking8 and arsenic9. Extensive research implicates environmental variables in the development of schizophrenia, especially during fetal and perinatal life, including maternal stress and infections, obstetric complications, and maternal nutrition during pregnancy6. For example, the Dutch famine of 1944–1945 led to a spike in the number of cases of schizophrenia two decades later10. Many of these factors have previously been associated with altering DNA methylation levels11,12. Lastly, several recent papers have explored the role of sequence variation on site- and region-specific DNA methylation13,14. The DNA sequence itself plays a large role in the maintenance of DNAm15, providing one potential mechanism, namely changes in DNAm, for the clinical associations of single nucleotide polymorphism (SNPs) from large genome-wide association studies (GWAS).
We generated DNA methylation (DNAm) data from postmortem dorsolateral prefrontal cortex (DLPFC) brain tissue from 526 individuals using the Illumina HumanMethylation450 (“450k”) microarray, extending earlier DNAm maps of human frontal cortex in normal subjects by twenty-fold increased genomic coverage (>485,000 versus 27,000 probes) in a much larger sample size (335 versus 108 normal subjects)2 and creating a more comprehensive landscape of epigenetic development in the human brain than previously available. While previous efforts to comprehensively measure DNA methylation across the epigenome using whole genome bisulfite sequencing have identified many important features of brain development16, we complement this work using a much larger sample at more continuous ages, albeit at lower genome-wide coverage, to obtain population-level spatial dynamics of DNA methylation across brain development.
Results
Widespread DNAm changes contrast fetal from postnatal life
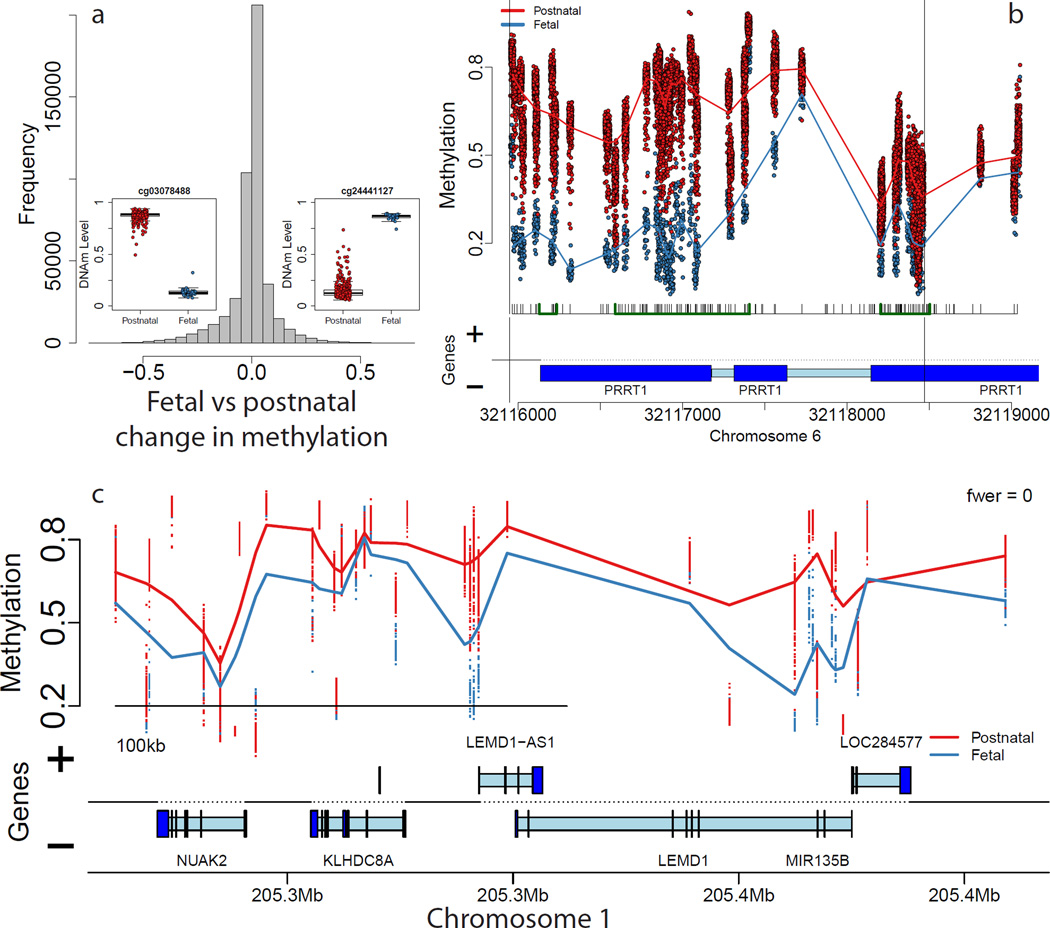
We obtained high quality DNAm data on 526 subjects from across the lifespan, including adults diagnosed with schizophrenia (Supplementary Table 1) using the Illumina 450k microarray (see Methods) after removing probes on the sex chromosomes and those containing known SNPs at the single base extension and target CpG sites, leaving 456,513 autosomal probes for analysis. We compared fetal (n=35) and post-natal (n=300) non-psychiatric control samples (including newborns and children) to identify changes in DNAm associated with the transition from the second fetal trimester to postnatal life, at varying spatial scales, adjusting for negative control probe factors to control for potential “batch” effects (see Methods). At the single probe level, the majority of assayed CpGs (N=231,415, 50.7%) were significantly differentially methylated (at Bonferroni-adjusted p-value, pbonf < 0.05, corresponding to p < 1.10×10−7, Online Table 1), suggesting a vastly different epigenetic landscape of the prefrontal cortex, during fetal compared with postnatal life (Figure 1A). While the Illumina 450k largely targets CpGs in and around gene promoter regions (islands, shores and shelves)17, almost every annotated gene (17,300/19,771 via UCSC knownGene hg19 table, 87.5%) contained at least one differentially methylated CpG within 5kb. These effects were further evident across the entire array - using principal component analysis, the first principal component, explaining 55.6% of variability in the data, strongly correlated with pre- versus post-natal life (Supplementary Figure 1).
Figure 1.
Differentially methylated loci comparing pre- and post-natal control subjects show large differences in DNA methylation. (A) Distribution of differences in DNAm across all individual CpGs/probes shows many sites with large changes in DNAm. Insets: examples of differentially methylated loci. (B) An example differentially methylated regions (DMRs) representing regional differences in DNAm levels. (C) An example methylation block representing long range changes. Proportion methylation is shown on the y-axis of the insets in panels B,C and the insets in panel A. Gene annotation panels in (B) and (C) are based on Ensembl annotation – dark blue represents exons and light blue represents introns.
These differentially methylated CpGs were classified into differentially methylated regions (DMRs) based on a “bump hunting” approach18 requiring at least 10% changes in DNAm at adjacent CpGs, resulting in 6,480 statistically significant DMRs (at family-wide error rate, FWER < 5%, Figure 1B, Supplementary Table 2, Supplementary Figure 2, median width=74bp) which overlap (within 5kb) 4,557 unique genes, which were strongly enriched for gene sets related to brain development and morphogenesis (Supplementary Table 3, Supplementary Text 1). We additionally identified 896 regions of long-range differential methylation (Figure 1C), termed “blocks”19, using an approach adapted to the Illumina 450k20 from whole genome bisulfite sequencing (WGBS) data (median width = 91.8kb, Supplementary Table 4). These blocks overlapped a combined 731 genes that were significantly enriched for 97 GO gene sets (at pbonf < 0.05, see Supplementary Table 5), largely related again to brain development, including neuron differentiation (109/1201 genes in set, p=6.99×10−18), generation of neurons (114/1308, p=2.23×10−17), and axonogenesis (65/569 genes, p=1.73×10−15). We further found significant overlap of differentially methylated blocks with previously identified blocks associated with cancer (791 of the 896 blocks; 88.3%, odds ratio of enrichment: 1.56, χ2 p-value = 3.63×10−86), including skewed directionality – fetal samples in these blocks were almost exclusively hypomethylated compared to adult samples, which mirrored the hypomethylated blocks associated with cancer (82.3% of overlapping blocks). These results support the idea that these blocks may represent more general developmental and/or proliferative phenomena21 but also link these long-range changes in DNAm to mechanisms underlying human brain development.
Many of these DNAm changes were further confirmed using WGBS data comparing one fetal sample to two adult NeuN+ and NeuN− samples from the study of Lister et al16 (see Supplementary Text 2) at all three spatial scales. Using Epigenome Roadmap data, many of the DNAm changes at birth were further associated with enhancer and repressive chromatin states in the adult DLPFC and suggest vastly different epigenetic landscapes of the prenatal versus postnatal human brain, consistent with our DNAm data (Supplementary Table 6, Supplementary Text 3). By matching these DNAm samples to publicly available gene expression data from the same donor, we confirmed that many of these DNA methylation changes associated with nearby gene expression levels, including at the CpG (71.0%, Supplementary Figure 3), DMR (87.3%, Supplementary Figure 4) and block (73.3%) spatial scales (Supplementary Text 4). These data, therefore, suggest a functional role for many of these DNA methylation changes in the developing human brain.
DNAm changes reflect a shifting neuronal composition
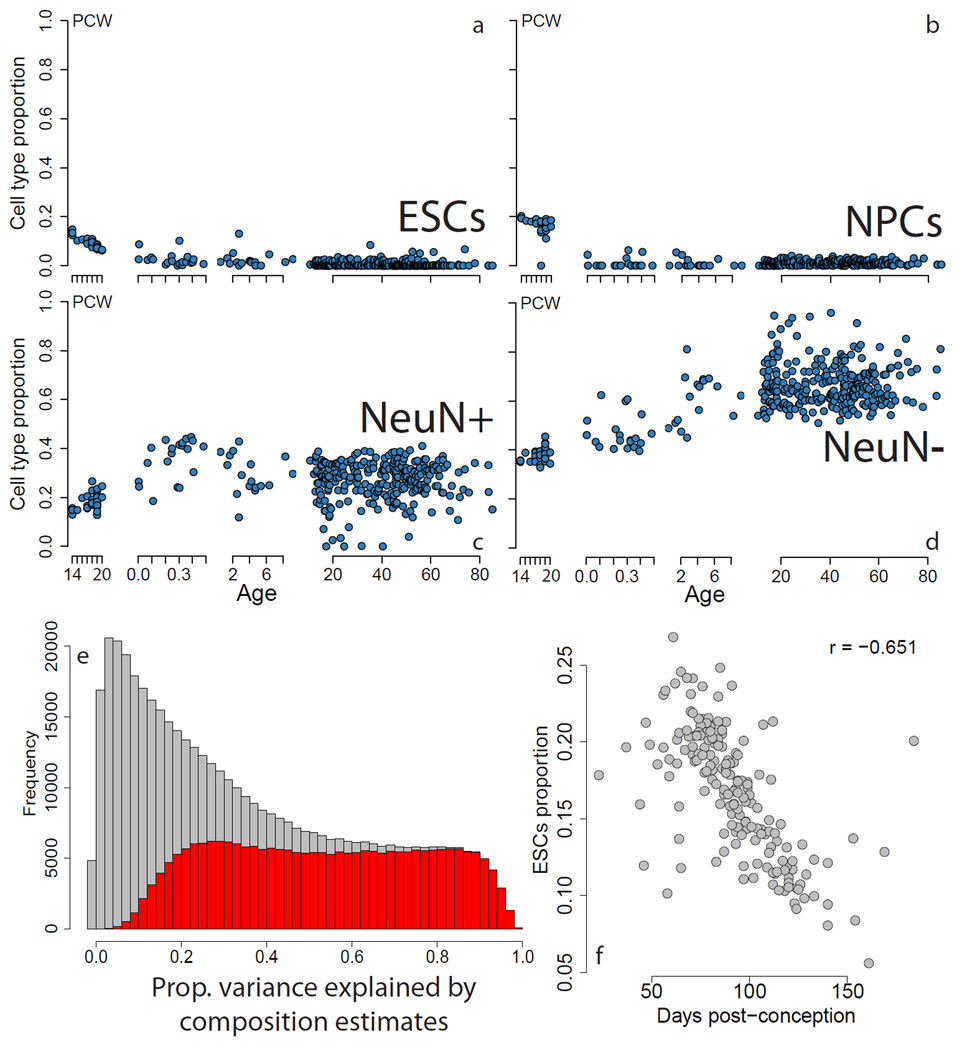
In a small sample of 36 non-psychiatric control subjects (including 32 in this study), we recently uncovered evidence of significant differences in the neuronal composition across the lifespan, including a loss of a progenitor-like epigenetic signature after birth, and a rising non-neuronal signature in postnatal life22. We sought to more fully characterize these composition changes in this much larger sample, and again found strong evidence of age-dependent changes in cell composition based on cell type epigenetic signature analysis (Figure 2). We identified significant linear changes in measures of composition within the second trimester of fetal life, reflecting the large gene expression changes present in this developmentally important time period1, including a decrease in progenitor-like cells based on DNAm signatures found in embryonic (Figure 2A, p= 1.21×10−24) and neural progenitor (Figure 2B, 5.74×10−25) cells, and a rise of mature adult neurons (Figure 2C, p=3.57×10−24) and non-neuronal cells (Figure 2D, p=8.09×10−86). The composition values of the fifth cell type in the dataset (ES-derived dopamine neurons) did not differ between pre- and post-natal samples (p=0.99). These composition profiles explained much of the variability in DNAm levels at individual CpGs, particularly at those differentially methylated from prenatal to postnatal life (Figure 2E). Also, by utilizing the DNAm-derived composition profiles in the expression data, we found significant, but lesser, association with gene expression levels (Supplementary Figure 5). We hypothesize these weakened associations with composition in gene expression data may result from additional layers of unexplored epigenetic regulation, as well as the fact that DNA and RNA from some of these samples were obtained from different tissue dissections.
Figure 2.
A changing neuronal phenotype across brain development. (A–D) Composition proportions per sample plotted versus age; the first subpanel in each represents age in post-conception weeks, and the remaining 3 subpanels show age in years. (E) Proportion of variance, R2, explained by cell composition at each CpG (grey) where the proportion of CpGs showing significant age stage-related (fetal versus postnatal) changes are shown in red. (F) The estimated proportion of ESCs versus post-conception days from Spiers, et al.23 shows strong association. ESCs: embryonic stem cells, NPCs: neural progenitor cells, PCW: post-conception weeks.
We further see these composition changes across brain development in publicly available datasets. For example, similar changes during fetal brain development are seen in a sample of 179 undissected fetal homogenate tissues23 particularly in the proportion of pluripotent-like cells (p=6.03×10−23, Figure 2F, Supplementary Text 5), as well as the other cell types examined (Supplementary Figure 6). Large effects of cellular composition were also observed in the developing and aging postnatal brain using DNAm data on 17 postnatal subjects/brains across 16 brain regions (including 11 neocortical) from the BrainSpan project24 (Supplementary Figure 7, Supplementary Text 5). These convergent findings in combination suggest that DNAm changes in the developing and aging brain reflects a shifting cellular composition – namely loss of progenitor-like cells at birth followed by a rise of non-neuronal cells in postnatal life – that we have previously shown to manifest in the transcriptome.
DNAm changes are enriched for schizophrenia risk loci
We next tested for significant enrichment between the CpGs that display epigenetic differences associated with the prenatal-postnatal transition and genomic loci associated with schizophrenia risk in the latest Psychiatric Genomics Consortium (PGC) genome-wide association study (GWAS)25. Of the 456,513 probes on the Illumina 450k used in differential methylation analysis, 5,476 were within the 108 genome-wide significant risk loci (specifically, the linkage disequilibrium, LD, blocks defined by the loci). These particular CpGs were more likely to be differentially methylated across the fetal-postnatal transition (2,903/5,476, 53.0%, in the loci compared to 228,512/451,037, 50.7% outside; OR=1.10, χ2 p-value=5.75×10−4, Supplementary Table 7). This association was driven by a majority subset of 3,607 CpGs in the PGC regions that were relatively more highly methylated in fetal compared to post-natal life (1,848/3,607; 51.2%, versus 126,674/272,242; 46.5%, OR = 1.21, p-value=2.03×10−8) – the subset of CpGs more highly methylated in postnatal life were not relatively enriched in the SZ GWAS loci (OR=0.98, p=0.67).
We also considered GWAS-positive loci for Alzheimer’s disease26 (AD, N=49), Parkinson’s disease27 (PD, N=29), and type 2 diabetes28 (T2D, N=40) to determine the specificity of our results. Among the CNS disorders, there was no enrichment overall (p > 0.3), with only perhaps marginal enrichment among CpGs that were more highly methylated in fetal life among AD GWAS loci (OR = 1.23, p=0.03), suggesting some specificity to schizophrenia. Interestingly, while we found significant enrichment among the T2D GWAS loci among all birth-associated CpGs (OR=1.19, p=6.56×10−3), this association was driven by CpGs more highly methylated in postnatal life (OR=1.41, p=5.4×10−4) with no association among the CpGs more highly methylated in prenatal life (OR=1.01, p=0.92). This finding may reflect adult lifestyle influences on epigenetic states that are associated with risk for T2D, including diet, body weight and exercise.
DNAm changes associated with the age of illness onset
As a sensitivity analysis to assess the specificity of the fetal-postnatal transition and also to contrast possible neurodevelopmental mechanisms with epigenetic alterations around the time of schizophrenia illness diagnosis, we performed a differential methylation analysis comparing controls between 10–25 years old (N=73) to those greater than 25 (N=190) to identify changes in DNAm associated with the typical age of onset of schizophrenia (see Methods). Here, there were 24,685 CpGs significant at pbonf < 0.05 between these two age groups at lesser effect sizes than those associated with the prenatal/postnatal transition – only 58 CpGs showed changes in DNAm greater than 0.1, making it difficult to perform comparable DMR analyses (see Methods). There were only 313 of the 24,685 CpGs linked to the adolescent period in the PGC2 loci (5.7%), which was not enriched compared to these age of onset-associated CpGs outside these loci (5.4%, OR = 1.06, p=0.32). However, we note that stratifying by directionality resulted in significant enrichment among schizophrenia GWAS loci here among a minority of CpGs (N=144) more highly methylated in adolescence than later adulthood (OR=1.42, p=3.38×10−5). Interestingly, these CpGs also tended to be more highly methylated in fetal than postnatal life (80.6%), consistent with the general trend in GWAS positive loci being associated with CpGs relatively hypermethylated in fetal compared with postnatal life. Overall, these data suggest that epigenetic changes around the age of onset of schizophrenia do not in general reflect genetic risk mechanisms, in contrast to the associations contrasting fetal to postnatal life, but they may contain a subset of risk loci relatively hypermethylated in late postnatal epigenetic regulation.
Extensive genetic regulation of local DNAm levels
Given the statistical enrichment between locations of early developmentally regulated CpGs and genetic risk for SZ, we attempted to directly determine the association between risk genotype among the genome-wide significant loci of SZ and DNAm levels. We genotyped and imputed these samples to the 1000 Genomes reference panel, and retained 7,426,085 common SNPs (MAF > 5%, see Methods) for methylation quantitative trait loci (meQTL) analysis to understand how common genetic variation influences DNAm levels. Here we reintroduced probes on the sex chromosomes (as genotypes across the genome should not correlate with sex) as well as probes with annotated SNPs at the target CpG site (as these could represent true biological signal), while still removing probes with annotated SNPs at the single base extension site (which represent technical signal if associated with genotype), leaving 477,636 CpGs for analysis. We conducted a meQTL analysis in the adult control samples (age > 13, N=258) allowing for up to 20kb between SNPs and CpGs, controlling for factors related to ancestry and global epigenetic variation (see Methods), and identified 4,107,214 significant SNP-CpG methylation associations (meQTLs) at FDR < 1% (corresponding to p < 8.6×10−4, Online Table 2, Online Table 3, Supplementary Text 6). We note that many of these meQTLs identified in the adult samples appear largely consistent in directionality (κ = 0.741) and magnitude (ρ = 0.707) with meQTLs in the smaller set of fetal samples (Supplementary Figure 8). While many may not be cell type-specific, as the composition of the fetal brains differs dramatically from the adult samples (see Figure 2), a subset may impart fetal-specific effects not present in adult subjects29.
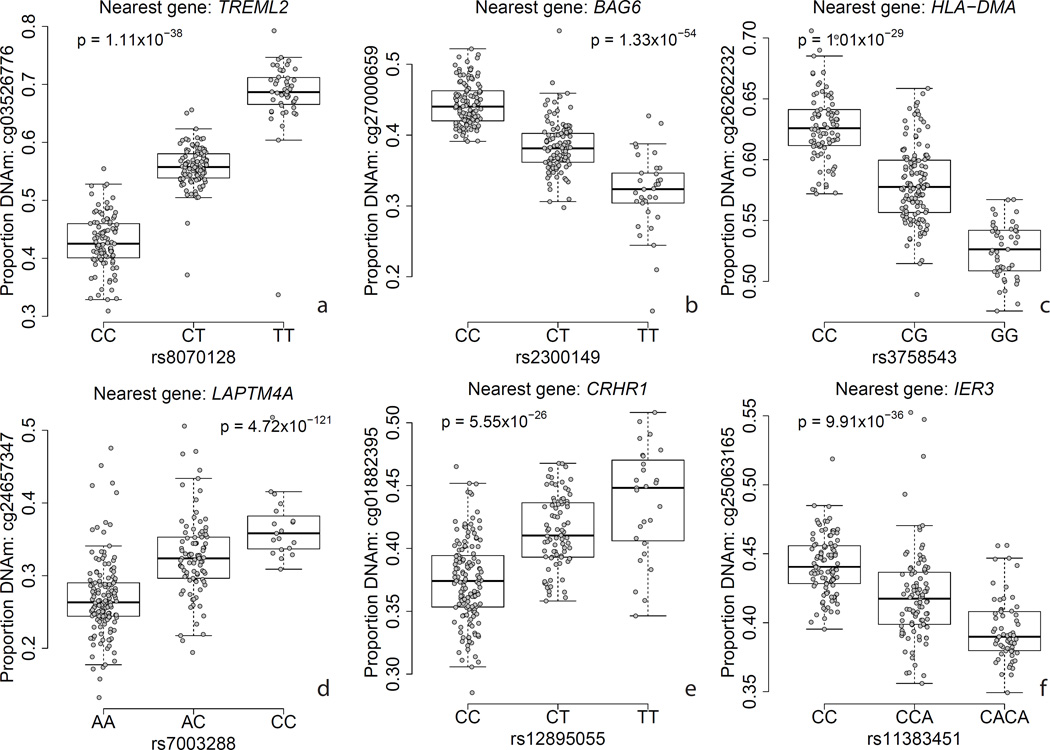
Epigenetic marks like DNAm have been hypothesized to underlie risk for common disease, and potentially mediating genetic risk identified from large genome-wide association studies (GWAS)30. We first examined all genetic variation previously identified in diverse GWAS using the NHGRI-EBI GWAS Catalog, which, at time of access, contains 28,870 genome-wide significant associations across 1,290 disease traits (with 15,930 unique SNPs by rs number)31. We found that 4,208 GWAS-associated SNPs for any trait was a significant DLPFC meQTL in our data (26.5% of all catalog SNPs, and 31.7% of those tested for meQTLs (Supplementary Table 8) – this enrichment of DLPFC meQTLs within GWAS-associated SNPs was highly significant (odds ratio = 1.74, χ2 p < 10−100). Interestingly, the GWAS-associated SNPs were clinically associated with 877 different disorders and/or traits affecting many tissues in the body, further suggesting that many meQTLs are not cell type-specific. We highlight examples of highly significant meQTLs where the clinical risk SNP disrupts a reference CpG dinucleotide in Figure 3 – these epigenetic effects could represent one possible mechanism by which these variants manifest risk30 (Supplementary Text 7).
Figure 3.
Examples of methylation quantitative trait loci (meQTLs) for six GWAS-associated variants with nearby DNA methylation levels. Y-axis: DNA methylation level at a particular probe, X-axis: genotype at a particular SNP, p-value corresponds to the effect of genotype on DNAm level, adjusting for ancestry and epigenetic principal components.
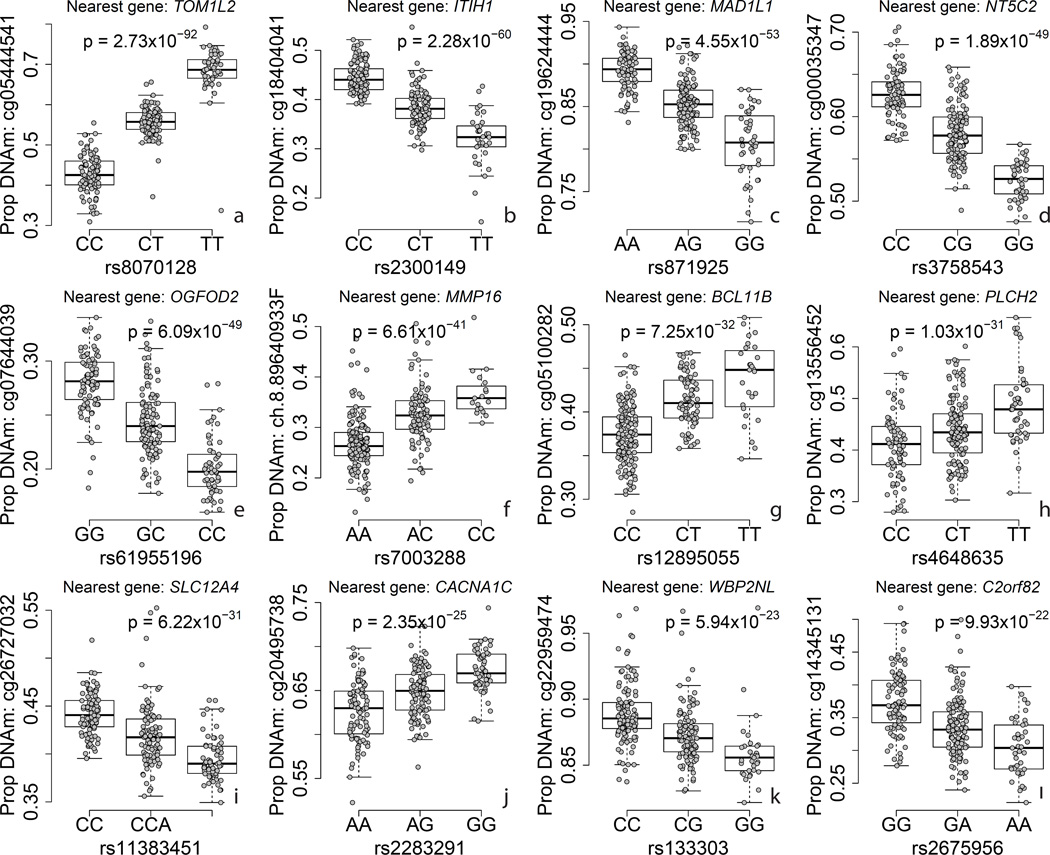
As many of the risk variants for schizophrenia in the NHGRI GWAS Catalog only reached genome-wide suggestive but not significant evidence for association, we last sought to determine the proportion of PGC2 risk genotypes that associated with nearby DNAm levels, which would suggest a possible mechanism of risk. Using all marginally (at p < 1×10−4, N=1302/2107 in our data, see Methods) and genome-wide (at p<5×10−8, 104/111 in our data, see Methods) significant “index” SNPs and their highly correlated proxies (R2 > 0.6) within the PGC2 52 study discovery dataset, we identified that 579/1302 (44.4%) of all marginally significant loci, and 62/104 (59.6%) genome-wide significant loci interrogated, had a risk or proxy SNP that was also a meQTL in our cortex DNAm data (Supplementary Table 9). We hypothesize that these epigenetic signals highlight the particular risk gene in a locus with wide linkage disequilibrium, and we highlight 12 loci that feature meQTL p-values < 1.0×10−20 in Figure 4. These CpGs associated with risk variants map to TOM1L2, ITIH1, MAD1L1, NT5C2, OGFOD2, MMP16, BCL11B, PLCH2, SLC12A4, CACNA1C, WBP2NL, and C2orf82. DNAm levels proximal to risk variants for schizophrenia may therefore influence or possibly mediate the effect of genotype on clinical risk for a large proportion of genome-wide significant loci.
Figure 4.
Examples of methylation quantitative trait loci (meQTLs) for twelve GWAS-positive loci for schizophrenia. Y-axis: DNA methylation level at a particular probe, X-axis: genotype at a particular SNP, p-value corresponds to the effect of genotype on DNAm level, adjusting for ancestry and epigenetic principal components.
DNAm changes and the diagnosis of schizophrenia
We lastly performed a differential methylation analysis for schizophrenia diagnosis at the single CpG level by comparing 191 adult patients with schizophrenia to the 240 (of 335) non-psychiatric controls all with ages greater than 16 (Supplementary Table 1). The schizophrenia patients had relatively typical age of onset (mean = 22.5 years, SD = 7.0 years) and the majority were on anti-psychotic medications at the time of death (64.0%), assessed via chart review and/or toxicology on brain tissue (see Methods). There were similar proportions of race, sex, and causes of death (other than suicide) in the control and schizophrenia group but the patients were more likely to be older and smoke, have a lower tissue pH and longer postmortem interval (PMI). We further note these patients had a relatively young average age at death compared to other post-mortem samples32, potentially reducing the cumulative impact of chronicity, smoking and antipsychotics on DNAm levels. In these adult samples, we did not identify significant differences in composition of any estimated cell types between cases and controls (Supplementary Figure 9A), though cellular composition explained a large component of variability in the data (Supplementary Figure 9B).
We modeled DNAm levels as a function of diagnosis, and adjusted for age, race, and the first four PCs of the negative control probes on the microarrays (see Methods). These negative control PCs were strongly associated with processing plate and microarray slide (Supplementary Figure 10) – only the 4th negative control PC was marginally associated with diagnosis (p=0.003). After initial differential methylation analyses using all 431 samples, we noticed that one of the three processing plates containing both patients with schizophrenia and controls drove much of the differential methylation signal (Supplementary Figure 11), and therefore removed the samples from this plate and reran the analysis on 244 subjects (108 patients with schizophrenia and 136 controls), identifying 2,104 probes/CpGs significant at a Bonferroni-adjusted p < 0.05 (Supplementary Table 10). Almost all of these involved hypomethylation of individual CpGs in patients compared to controls (N=2,043, 97. 1%) with much smaller differences in methylation levels than those that found in the prenatal/postnatal contrast analysis (mean absolute proportion change: 0.013, IQR: 0.010–0.01.8). This pattern of diagnosis (i.e. state) associated hypomethylated CpGs contrasts with the association of risk associated GWAS loci (i.e. trait) with principally relatively hypermethylated CpGs during the fetal and adolescent timeframe. Nevertheless, these differentially methylated CpGs were in or near (within 10kb) genes significantly enriched for embryo development (p=1.9×10−9), cell fate commitment (p=2.68×10−7), and nervous system differentiation (p=1.9×10−7) by Gene Ontology analysis33 (Supplementary Table 11). One of the top differentially methylated CpGs lies in the promoter of HAT1, a histone acetyltransferase primarily targeting lysines on the H4 nucleosome34, perhaps manifesting as differences in histone acetylation in conjunction with DNAm changes35. These hypomethylated CpGs were enriched for being in TSS chromatin states (70.6% compared to 21.7% for the entire array), and depleted for enhancer, repressor and quiescent chromatin states (Supplementary Table 7), suggesting that they may be influencing gene expression. The lack of enrichment of these diagnosis-associated CpGs with adult associated biology suggest that these CpGs reflect environmental influences on early developmental events that increase risk independent of genetic risk influences.
A recent paper identified 4 significant CpGs differentially methylated in a smaller sample of brains of schizophrenia patients (N=20) and controls (N=23) across a discovery and replication cohort36 but we found only one of these CpGs that was directionally consistent and marginally significant in our much larger dataset (near GSDMD (cg26173173) at p=0.02). We failed to replicate the other three CpGs - cg24803255 and cg08171022 were also higher and lower in cases compared to controls, respectively, but neither were significant (p=0.32 and p=0.97, respectively), and cg00903099 was marginally significant (p=0.02) but higher in cases compared to controls in our data rather than the reported hypomethylation. Conversely, only one of our CpGs was marginally significant (at p < 0.05) and directionally consistent in both their discovery and replication datasets when treating each separately, potentially highlighting uncertainties in case control analyses of DNAm, including heterogeneity in the clinical disorder, differing epiphenomena related to diagnosis and illness state, and differences in the ascertained tissue for postmortem human brain studies.
Despite concern about epiphenomena (Supplementary Figure 12, Supplementary Text 8), we did find slight but significant enrichment of our 2,104 diagnosis-associated CpGs within the PGC loci – 40/2,104 CpGs (1.9%) compared to only 1.3% of the rest of CpGs on the array (OR=1.6, p=0.004) but none of these 40 were meQTLs to any SNPs, including the risk-associated SNPs identified in the PGC. Overall, among these 2,104 CpGs showing DNAm level differences between patients and controls, only 97 were genome-wide significant meQTLs, a six fold decrease in enrichment than expected by chance (OR=0.165, p=2.32×10−86, see Methods) even though there was strong global correlation among meQTLs identified in adult controls and in the patients with schizophrenia (Supplementary Figure 13).
Lastly, our data draw parallels to an earlier report of enrichment of DNAm changes during fetal life among schizophrenia diagnosis-related CpGs36. Even in our smaller, but regionally-focused, fetal sample, 1986/2104 of the CpGs associated with diagnosis were significantly differentially methylated between fetal and postnatal life (at pbonf < 0.05, OR = 16.5, enrichment p-value < 10−100). In contrast, these CpGs were strongly depleted (OR = 0.26, p = 1.88×10−15) for those CpGs showing significant differences comparing adolescent to adult controls, reflecting age-related changes occurring near the age of onset of schizophrenia – only 31/2104 CpGs associated with age-related changes around schizophrenia onset as well as diagnosis. These contrasts suggest that the diagnosis-associated differentiated CpGs are not related to epigenetic events germane to illness onset, but appear to reflect lifelong epigenetic states established early in development. This is further supported by the observation that these CpGs largely hypomethylated for diagnosis (compared to adult controls) were relatively highly methylated in fetal life (Supplementary Figure 14, ρ= −0.63, p < 10−20), and appeared to further diverge from fetal levels compared to the adult non-psychiatric controls. Thus, the schizophrenia associations at these CpGs strongly reflect DNAm changes related to early developmental events supporting a neurodevelopmental component not only to genetic risk but also to environmental risk of this debilitating disorder.
Discussion
We identified changes in DNA methylation associated with genetic sequence and developmental stage in one of the largest studies of postmortem human brain tissue to date. The most extensive changes in the epigenome are found at local, regional, and long-range spatial resolutions in comparing prenatal and postnatal specimens that we suggest likely represent in part shifts in neuronal composition across the lifespan, and correspond to strong changes in gene expression profiles. Interestingly, these developmentally associated changes in DNAm were significantly enriched for genomic regions that confer clinical risk for schizophrenia. Many risk variants across the catalog of GWAS positive loci in studies of common medical disorders themselves associate with nearby DNAm levels, termed meQTLs, suggesting potential mechanisms by which genetic risk propagates in the population. Lastly, we show that several thousand individual CpGs demonstrate small but statistically significant difference in DNAm levels comparing adult patients with schizophrenia with controls that did not appear confounded by cellular composition or smoking. The differences found between patients and controls appear to represent epigenetic marks that principally associate with early neurodevelopment and not with events that herald the onset of the disorder or that characterize adult brain biology. Overall, the data suggest that both the genetic and environmental risk components of schizophrenia involve early developmental influences.
The widespread methylome changes that occur across brain development - ranging from early fetal life23, the transition into postnatal life shown here, and through adulthood24 – appear to track first the loss of immature neurons prior to birth followed by the rise of non-neuronal cell types in postnatal life through adulthood (see Figure 2). While the quantitative estimates of cell composition employed here utilized a series of cell types that combined epigenetic data from adult human tissue and derived cellular systems, the proportion of pluripotent-like cells are quite consistent across two independent datasets and brain regions, with ~15% of the cells manifesting this signature by 14 post-conception weeks (Figures 2A,F), and may relate to recent classifications of replicating versus quiescent fetal neurons using single cell analysis37.
Deviations from these essential developmental trajectories during critical windows of development from conception to young adulthood may interfere with the carefully coordinated temporal and spatial dynamics of gene expression through a combination of genetic and epigenetic factors3,5,38,39 that may contribute to risk for schizophrenia and other neurodevelopmental disorders. Indeed, the CpGs that track this changing neuronal phenotype, e.g. those that differ in DNAm levels comparing pre- and post-natal samples, were enriched by genomic location of regions that confer genetic risk for schizophrenia25 as have changes in transcriptome across brain development22,40,41. However the mechanisms by which DNAm changes that track shifting neuronal phenotypes alter risk for schizophrenia appears largely unknown, and will likely require more cell type-specific assays to focus on individual cell populations across brain development to reduce the strong composition effects observed in homogenate brain tissue. This is likely to be a complex conundrum, as composition measures in homogenate tissue did not differ between cases and control samples and controlling for composition did not alter the CpG differences between patients and controls.
Conversely, homogenate brain tissue appears to represent a powerful tool for better understanding how genotypes identified in large population-based GWAS may manifest risk for neurodevelopmental and other brain disorders. Indeed, many meQTLs identified in the DLPFC during adult life appear consistent in fetal life – and reach genome-wide significance in larger fetal samples29 - despite very different cellular compositions, suggesting that many of these variations serve conserved regulatory roles in multiple cell types. Furthermore, DNAm levels may be a more proximal read out of genetic variation than gene expression levels42 – here we identified that 62/104 (59.6%) genome-wide significant genetic loci for schizophrenia risk associated with local DNAm levels, compared to a report suggesting that only 18/108 (16.7%) GWAS-positive loci are eQTLs across the human brain (see Supplementary Table 4, Worksheet 2 in Schizophrenia Working Group of the Psychiatric Genomics25).
The small effect size but significant differences in DNAm levels at individual CpGs between patients with schizophrenia compared to controls highlights the intrinsic tradeoff between statistical gains in increasing sample size versus the clinical and likely by extension molecular heterogeneity of the clinically ill state. The genetic heterogeneity of schizophrenia is reflected in small odds ratios (<1.1) for individual genomic loci that reach genome-wide significance only because of very large samples. These odds ratios look similar in magnitude to our results for differentially methylated CpGs, suggesting likely epigenetic heterogeneity as well. Unlike genetic sequence, which is largely determined at conception, these epigenetic signals are malleable across the lifespan, and likely the many epiphenomena that differentiate patients from controls may leave their marks on the epigenome, perhaps differently in different subpopulations of patients. These epiphenomena include the influence of medical therapy, chronic illness, nutrition, body weight, alcohol and cannabis use, etc. Untangling which epigenetic marks better relate to the causes versus the consequences of illness will be difficult. Indeed, only a fraction of the illness associated CpGs, 4.6%, showed association to nearby genetic variants in the meQTL analysis, further suggesting these findings may be more related to the epiphenomena of the illness state than to the genetic causes of the disorder. Lastly, while these diagnosis-associated CpGs were not confounded by cell composition, it is possible that they have larger effect sizes in individual cellular populations, and new consortia like the psychENCODE project43 can better identify the cellular specificity of, and potentially magnify, these effects.
Further on this point, while we observed significant enrichment of the PGC loci in CpGs differing between patients and controls, this was a marginal enrichment, small in comparison to the enrichment with loci showing epigenetic alterations from prenatal to postnatal life. These results suggest that the majority of DNAm differences associated with the illness state are likely unrelated to genetic mechanisms of causation and implicate environmental factors. In this context and also germane to the issue of state related epiphenomena, it is worth highlighting that the case control differences mapped to genes implicated in early developmental processes, even if not linked with genetic risk variation. Thus, the epigenetic associations with schizophrenia, both in terms of illness state and genetic risk, implicate factors, both genetic and environmental, that track with early development and not adult life. Consistent with this conclusion is the additional observation that epigenetic changes associated with adolescence and early adulthood, the typical time of onset of schizophrenia, did not show enrichment of either genetic risk loci or illness state associated CpG alterations. This observation has potentially sobering implications for attributing a causative role of environmental influences that appear to coincide with the onset of the clinical disorder.
We also explored the relationship of genetic risk loci associated with Alzheimer’s disease, Parkinson’s disease and T2D. None of these disorders showed the enrichment of CpGs hypermethylated during fetal life associated with risk loci for schizophrenia, though the GWAS catalog of risk associated variants is smaller in these cases. Interesting, there was a small enrichment with CpGs showing significant hypermethylation in postnatal life and risk loci for T2D. While the interpretation of this finding is highly speculative, it might reflect adult lifestyle influences on risk for T2D.
In conclusion, the epigenetic landscape represented by DNA methylation in the human brain shows dramatic variation across development. Genetic loci implicated in risk for schizophrenia and other CNS disorders are enriched for loci expressing these shifting epigenetic states, particularly those that change from the transition from prenatal to postnatal life. While these observations do not identify specific molecular mechanisms of the clinical associations, they suggest that there is an important epigenetic intermediate between sequence of risk and cell biology of risk.
Online Methods
Study samples
Brain specimens were donated through the Offices of the Chief Medical Examiners of the District of Columbia and of the Commonwealth of Virginia, Northern District to the NIMH Brain Tissue Collection at the National Institutes of Health in Bethesda, MD, according to NIH Institutional Review Board guidelines (Protocol #90-M-0142). Audiotaped informed consent was obtained from legal next-of-kin on every case. Details of the donation process are described elsewhere44,45. Additional specimens, including the 35 second-trimester fetal brain tissue samples, were obtained via a Material Transfer Agreement with the National Institute of Child Health and Human Development Brain and Tissue Bank. All postnatal non-psychiatric control donors (N=300) were free from psychiatric and/or neurologic diagnoses and substance abuse according to DSM-IV. Every control donor had toxicology screening to exclude for acute drug and alcohol intoxication/use at time of death, and all fetal tissue was also screened for possible in utero drug exposure.
Tissue Processing
All specimens were flash-frozen, and screened for macro- and microscopic neuropathological abnormalities, as previously described44. All specimens with significant evidence of neurological disorders, infarcts or other cerebrovascular abnormalities were excluded from study. Brain pH was measured, and postmortem interval (PMI, in hours) was calculated for every sample. Postmortem tissue homogenates of the prefrontal cortex (dorsolateral prefrontal cortex, DLPFC, BA46/9) were obtained from all subjects. Genomic DNA was extracted from 100 mg of pulverized dorsolateral prefrontal cortex (DLPFC) tissue with the phenol-chloroform method. Bisulfite conversion of 600 ng genomic DNA was performed with the EZ DNA methylation kit (Zymo Research).
DNA Methylation Microarray
DNA methylation was assessed using the Illumina HumanMethylation450 (“450k”) microarray, which measures CpG methylation across >485,000 probes covering 99% of RefSeq gene promoters17. Arrays were run following the manufacturer’s protocols. A percentage of the samples were run in duplicate across multiple processing plates to assess technical variability related to DNA extraction and bisulfite conversion. A total of 675 microarrays were scanned on 534 unique subjects – however, note that all analysis utilized only a single microarray from each sample (see below).
Data Processing and Normalization
Red and green channel intensity files were obtained for each sample in the “idat” file format. These files were processed and normalized using the minfi Bioconductor package in R20. Red and green intensities were mapped to the M(eth) and U(nmeth) channels, and the average intensity for these channels were used to check for low quality samples (0 samples were dropped). Intensities from the sex chromosomes were used to predict sex, and we dropped 8 samples that had predicted sex different from its recorded value (indicating potential sample swaps). Then, the M and U channels were subsequently across-sample quantile normalized using an approach developed by Aryee, et al.20. Briefly, this approach forces the distribution of type I and type II to be the same by first quantile normalizing the type II probes across samples and then interpolating a reference distribution to which the type I probes are normalized, stratified by region (e.g. promoter, shore, island, shelf), which has previously been shown to best minimize the variability between replicates20. For all analyses, we retained a single array in the case of duplicates by choosing the sample that had the closest quality profile (via M and U signal intensity) to all other arrays.
Statistical analyses for differential methylation by development
We modeled differential methylation between pre- (N=35) and post-natal (N=300) non-psychiatric controls using linear modeling approaches. After normalization, probes on the sex chromosomes were dropped (which are more difficult to accurately normalize), as were probes annotated with single nucleotide polymorphisms (SNPs) at the target CpG or single base extension (SBE) site according to dbSNP142 with minor allele frequency > 1%, leaving 456,513 autosomal probes for age-associated DNAm analysis. All three approaches – single CpG, DMR, and block – utilized the below linear model:
where pij is the proportion methylation for probe i and subject j, Fetalj in a binary variable indicating if the j’th sample is pre- or post-natal, and negPCj are the negative control principal components estimated from the microarray background probes. Therefore αi represents the mean methylation proportion/level in the postnatal samples, and βi is the difference in the fetal samples. For CpG-level analyses, we fit the above linear model with the limma R/Bioconductor package46 to obtain mean differences, moderated t-statistics and corresponding p-values, which we adjusted by the number of tests (i.e. Bonferroni correction47) to conservatively control for multiple testing (as neighboring CpGs tend to be correlated48, reducing the number of effective tests). While some previous manuscripts remove all probes containing SNPs in any position, we observe that the single CpGs that are significantly associated with birth are in fact depleted of probes containing SNPs (OR = 0.79, p < 10–100), as 17.0% of significant CpGs contain an annotated probe SNP compared to 20.6% of non-significant probes.
The analyses at the longer spatial scales – DMRs and blocks – both utilized the above linear model; DMR analysis generally finds contiguous CpGs where βi ≠ 0 whereas block analysis first collapses the methylation proportions into one level across neighboring probes per sample (e.g. ), fits the above model. Regional analysis to find differentially methylated regions (DMRs) and “block finding” were performed using the minfi R package49 using the bumphunterEngine and blockFinder functions, respectively, each with 1000 linear model bootstrap iterations18 and a cutoff of 0.1 (corresponding to contiguous probes with a minimum 10% directionally consistent change in DNAm associated with birth, e.g. |β̂i| > 0.1), with other parameters in these minfi functions set to their default values.
CGDMRs were obtained from Lister et al16 which were relative to genome build hg18. We used the corresponding hg18-based probe coordinates from the Illumina 450k manifest file to perform the overlaps between the microarray and WGBS data at the CpG and DMR level, and then used the liftOver tool to map from hg19 to hg18 at the block level (since 450k probes are collapsed to probe groups and given an average coordinate)50.
Secondary analyses for age-of-onset differentially methylated CpGs were fit using the same model above within all control samples greater than 10 years of age, comparing those samples younger than and older than 25 years (replacing the Fetal term in the model).
Analyses of chromatin state data
The 18-chromatin state data, derived using hidden Markov models (HMMs), was obtained for sample E073 in the Epigenome Roadmap project51 (http://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html). These data were derived from a mixture of DLPFC tissue from two subjects (75 year old and 81 year old). The chromatin states overlapping DMRs, blocks, and meQTL SNPs (by genomic coordinates, chr:start-end) were obtained, and compared to a background of all considered probe groups, collapsed probe groups, and all considered SNPs, respectively. Overlap was assessed based on the total coverage (in base pairs) of the chromatin states. Fold changes for enrichment and depletion > 2 were reported. While the project generated data on two fetal brain samples, processed chromatin state data was currently unavailable. Additional details on the chromatin states are available at the above website.
Statistical Analyses for Gene Expression Correlation
Raw gene expression two-color microarray intensity data (available at GSE30272) were loess-normalized as previously described1. Probes were re-annotated to the hg19 genome using the Gemma tool52 leaving 31,699 gene expression probes on 249 samples that had both Illumina 450k DNAm and expression data. Differential expression analysis comparing pre- and post-natal expression levels was performed using limma46. We annotated each 450k probe to its nearest gene in the expression data by distance, and computed the Pearson correlation between proportion DNAm and gene expression level, and converted these correlations to Z-scores and corresponding p-values (e.g. ). For the DMR-expression analysis, we matched each DMR to all probes corresponding to the nearest gene, and retained the most correlated DMR-probe correlation. For block-expression analysis, we identified which probes, and their evidence for differential expression, were present in each block using genomic coordinates.
Composition Estimation
We implemented in silico estimation of the relative proportions of five cell types (ESCs, ES-derived NPCs, and derived dopamine neurons from culture53, and adult cortex neuronal and non-neuronal cells from adult tissue54) using epigenome-wide DNAm data using a recently published algorithm55. All data was obtained using the Illumina HumanMethylation450 (“450k”) microarray platform from GEO56. After normalizing the publicly available data together using the preprocessQuantile function in the minfi Bioconductor package20, we picked the cell type-discriminating probes as outlined by Jaffe and Irizarry57 resulting in 405 unique probes that distinguished the five cell types. Projecting the brain samples onto these profiles results in a composition proportion for each cell type and sample (see Figure 2). We computed the percentage of variance explained (R2), comparing a linear model with all five composition profiles to an intercept-only model at each Illumina 450k probe and gene expression probe.
We downloaded already-processed data from BrainSpan (http://download.alleninstitute.org/brainspan/Methylation/) and Spiers, et al.23 from GEO (under accession GSE58885) and obtained composition estimates per sample using the same 405 probes and five cell types as above. Comparisons between composition estimates and age and/or brain region were performed using linear regression. Re-analysis of Spiers, et al.23 for a main effect of age, adjusting for the composition estimates from the 5 cell types, was performed using limma46.
Enrichment for schizophrenia genetic risk
We analyzed the published 108 regions of schizophrenia risk in the latest Psychiatric Genomics Consortium (PGC) genome-wide association study for schizophrenia25. We calculated a X2-statistic to determine whether CpGs within the PGC regions differed in their developmental DNAm effects, specifically comparing differential methylation effect sizes from the pre- versus post-natal analysis by whether each probe (of the 456,513) fell within the PGC regions (by genomic coordinates, eg chr:start-end) or not.
Genotype data processing
DNA for genotyping was obtained from the cerebella of samples in the collection and performed with either the Illumina Human Hap 650v3,1M Duo V3, or Omni 5M BeadArrays as previously described1. We had genotype data on 520/526 samples measured on the Illumina 450k. Genotypes were called separately by genotyping platform using the crlmm software59, and then cleaned separately for imputation (retaining SNPs with MAF > 0.5% and genotyping missing rate < 10%, then checking sex and heterozygosity)60. Genotypes were phased into haplotypes using SHAPEIT261 and imputed in 6MB chunks using Impute262 to the 1000 Genomes Phase 3 variant set for the autosomes and then Phase 1 variant set for chrX (as Phase 3 data is not available yet). Imputed genotypes were merged across the three platforms following imputation, and SNPs with MAF > 5%, HWE p-value > 1×10−6, and missing rate < 10% were retained across the 520 samples. LD-pruning generated an independent set of SNPs to perform genome-wide clustering to obtain multidimensional scaling (MDS) components for quantitative measures of ancestry.
meQTL analysis
We reintroduced the probes on the sex chromosomes and those CpGs that had a variant at the CpG site (as meQTLs to these CpGs would represent biological, and not technical, signal) resulting in 477,636 Illumina 450k probes and 7,426,085 common variants on 520 subjects. Within the adult control subjects, we modeled the additive effect of genotype (number of minor alleles) on DNAm levels, adjusting for the first five MDS components from the genetic data and the first 11 PCs (based on the 100,000 most inter-individual variable probes for computational efficiency) using the MatrixEQTL package63. We allowed for a maximum distance of 20kb between each SNP and CpG analyzed, resulting in 47,675,913 tests, and we adjusted for multiple testing using a false discovery rate (FDR) threshold of 0.01 to call meQTLs significant. Posthoc analyses of the most significant SNP-CpG pair per probe were calculated separately by Caucasians and African Americans, and then within fetal samples.
We downloaded the entire set of GWAS-suggestive and/or significant variants (by rs number) for thousands of traits from the NHGRI GWAS catalog31. We computed the odds ratio and corresponding p-value for enrichment for GWAS-associated variants by determining which variants were meQTLs and GWAS-associated, meQTLs only, GWAS-associated only, and neither, among those 7,426,085 SNPs considered in our dataset. For the PGC2 analysis, we obtained LD-clumped regions from the discovery dataset from the May 2013 freeze in the Ricopili tool (“PGC_SCZ52_may13”) which contains marginally significant regions (down to p-value < 10−4) and LD-proxies to each index SNP. We matched these SNPs to our data by chromosome and position, and determined which had a meQTL in our dataset.
Schizophrenia differential methylation analysis
We modeled differences in diagnosis, controlling for age, sex, race, and the first 4 PCs from the negative control probes ultimately among 108 patients with schizophrenia and 136 controls. Sex was not associated with diagnosis, and therefore not included as an adjustment variable (p=0.16, see Supplementary Table 1). Smoking status was obtained by toxicology, where those positive for smoking tested positive for either nicotine and/or cotinine, and included in the above model to assess confounding by smoking. Similarly, cell composition estimates were included in the above model to assess potential confounding by composition differences between cases and controls. We performed a Chi-squared test (df=1) to determine whether CpGs showing diagnostic effects were over- or under-enriched for meQTLs.
Supplementary Material
Acknowledgements
We are grateful for the vision and generosity of the Lieber and Maltz Families who made this work possible. We thank the families who donated to this research and we thank Andrew Feinberg for helpful input into data analyses. This work was supported by the Lieber Institute for Brain Development. A.E.J. was partially supported by 1R21MH102791
Footnotes
Data Availability: Raw and processed data are available on the Gene Expression Omnibus at Accession Number: GSE74193. R code for data processing and analysis is available from the GitHub repository: https://github.com/andrewejaffe/devMeth450k.
Disclosure Declaration: The authors have declared that no conflicting interests exist.
Author Contributions:
All authors contributed to the writing of the manuscript, plus the following individual contributions:
AEJ: designed the study, performed the data analysis, oversaw the writing of the manuscript
YG: oversaw the data generation
AD: collected phenotype data on all subjects
RT: performed DNA extractions and contributed to the data generation
TMH: collected brain samples and performed tissue dissections to obtain biological materials
DRW: designed the study, contributed to the data analysis and interpretation of the results, and oversaw the writing of the manuscript.
JEK: collected brain samples, provided clinical interpretation of the results
References
- 1.Colantuoni C, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Numata S, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. American journal of human genetics. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2013;38:138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annual review of nutrition. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 5.Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nature medicine. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberger DR, Levitt P. In: Schizophrenia. Weinberger DR, Harrison PJ, editors. Wiley-Blackwell; 2011. [Google Scholar]
- 7.Heijmans BT, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. American journal of human genetics. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichard JF, Schnekenburger M, Puga A. Long term low-dose arsenic exposure induces loss of DNA methylation. Biochemical and biophysical research communications. 2007;352:188–192. doi: 10.1016/j.bbrc.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Susser ES, Lin SP. Schizophrenia after prenatal exposure to the Dutch Hunger Winter of 1944–1945. Archives of general psychiatry. 1992;49:983–988. doi: 10.1001/archpsyc.1992.01820120071010. [DOI] [PubMed] [Google Scholar]
- 11.Relton CL, Davey Smith G. Is epidemiology ready for epigenetics? International journal of epidemiology. 2012;41:5–9. doi: 10.1093/ije/dys006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortessis VK, et al. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Human genetics. 2012;131:1565–1589. doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schilling E, El Chartouni C, Rehli M. Allele-specific DNA methylation in mouse strains is mainly determined by cis-acting sequences. Genome research. 2009;19:2028–2035. doi: 10.1101/gr.095562.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lienert F, et al. Identification of genetic elements that autonomously determine DNA methylation states. Nature genetics. 2011;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 15.Bird A. Putting the DNA back into DNA methylation. Nature genetics. 2011;43:1050–1051. doi: 10.1038/ng.987. [DOI] [PubMed] [Google Scholar]
- 16.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandoval J, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics : official journal of the DNA Methylation Society. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 18.Jaffe AE, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. International journal of epidemiology. 2012;41:200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nature genetics. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timp W, et al. Large hypomethylated blocks as a universal defining epigenetic alteration in human solid tumors. Genome medicine. 2014;6:61. doi: 10.1186/s13073-014-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaffe AE, et al. Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nature neuroscience. 2015;18:154–161. doi: 10.1038/nn.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiers H, et al. Methylomic trajectories across human fetal brain development. Genome research. 2015;25:338–352. doi: 10.1101/gr.180273.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.BrainSpan. Atlas of the Developing Human Brain. 2011 < http://developinghumanbrain.org>. [Google Scholar]
- 25.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature genetics. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nalls MA, et al. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature genetics. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hannon E, et al. Methylation quantitative trait loci (mQTL) in the developing human brain and their enrichment in genomic regions associated with schizophrenia. Nature neuroscience. 2015 [Google Scholar]
- 30.Liu Y, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nature biotechnology. 2013;31:142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols L, et al. The National Institutes of Health Neurobiobank: a federated national network of human brain and tissue repositories. Biological psychiatry. 2014;75:e21–e22. doi: 10.1016/j.biopsych.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 33.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 34.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Current biology : CB. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 35.Gavin DP, Sharma RP. Histone modifications, DNA methylation, and schizophrenia. Neuroscience and biobehavioral reviews. 2010;34:882–888. doi: 10.1016/j.neubiorev.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pidsley R, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome biology. 2014;15:483. doi: 10.1186/s13059-014-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darmanis S, et al. A survey of human brain transcriptome diversity at the single cell level. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdolmaleky HM, et al. Methylomics in psychiatry: Modulation of gene-environment interactions may be through DNA methylation. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2004;127B:51–59. doi: 10.1002/ajmg.b.20142. [DOI] [PubMed] [Google Scholar]
- 39.Mill J, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. American journal of human genetics. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gulsuner S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parikshak NN, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleinman JE, et al. Genetic neuropathology of schizophrenia: new approaches to an old question and new uses for postmortem human brains. Biological psychiatry. 2011;69:140–145. doi: 10.1016/j.biopsych.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The PsychENCODE Consortium. The PsychENCODE Project. Nature neuroscience. 2015 doi: 10.1038/nn.4156. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 44.Lipska BK, et al. Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biological psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 45.Deep-Soboslay A, et al. Reliability of psychiatric diagnosis in postmortem research. Biological psychiatry. 2005;57:96–101. doi: 10.1016/j.biopsych.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 46.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 47.Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310:170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome research. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.minfi: Analyze Illumina's 450k methylation arrays. 2013 [Google Scholar]
- 50.Farrell CM, et al. Current status and new features of the Consensus Coding Sequence database. Nucleic acids research. 2014;42:D865–D872. doi: 10.1093/nar/gkt1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zoubarev A, et al. Gemma: a resource for the reuse, sharing and meta-analysis of expression profiling data. Bioinformatics. 2012;28:2272–2273. doi: 10.1093/bioinformatics/bts430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim M, et al. Dynamic changes in DNA methylation and hydroxymethylation when hES cells undergo differentiation toward a neuronal lineage. Human molecular genetics. 2014;23:657–667. doi: 10.1093/hmg/ddt453. [DOI] [PubMed] [Google Scholar]
- 54.Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics : official journal of the DNA Methylation Society. 2013;8:290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome biology. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.enrichedRanges: Identify enrichment between two sets of genomic ranges v. 0.0.1 (GitHub, 2014) [Google Scholar]
- 59.Scharpf RB, Irizarry RA, Ritchie ME, Carvalho B, Ruczinski I. Using the R Package crlmm for Genotyping and Copy Number Estimation. Journal of statistical software. 2011;40:1–32. [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson CA, et al. Data quality control in genetic case-control association studies. Nature protocols. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams AL, Patterson N, Glessner J, Hakonarson H, Reich D. Phasing of many thousands of genotyped samples. American journal of human genetics. 2012;91:238–251. doi: 10.1016/j.ajhg.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.