Abstract
Reversible protein acetylation is a major regulatory mechanism for controlling protein function. Through genetic manipulations, dietary perturbations, and new proteomic technologies, the diverse functions of protein acetylation are coming into focus. Protein acetylation in mitochondria has taken center stage, revealing that 63% of mitochondrially localized proteins contain lysine acetylation sites. Here we summarize the field, and discuss salient topics that cover spurious versus targeted acetylation, the role of SIRT3 deacetylation, nonenzymatic acetylation, and molecular models for regulatory acetylations that display high and low stoichiometry.
Keywords: metabolic regulation, acetylation, mitochondria, acylation, nonenzymatic, stoichiometry
From Histone Regulation to Mitochondrial Acetylation
Mitochondria function as central mediators of metabolism and energy production [1]. Through the ability to oxidize sugars, fatty acids and amino acids by reducing molecular oxygen and creating a H+ gradient across the inner mitochondrial membrane, mitochondria couple this chemiosmotic gradient to the production of ATP. While it is general knowledge that the majority of cellular ATP is generated by these organelles, mitochondria play central roles in many fundamental cellular processes that include providing precursors for anabolic processes, acting as sentinels of cellular health, and coordinating nuclear–mitochondrial communication. Therefore, revealing previous unknown regulatory networks that operate within mitochondria have broad implications toward our understanding of cellular homeostasis and pathology. Recent research on mitochondria has implicated protein lysine N-ε-acetylation as a major regulatory mechanism for modulating protein function.
The acetylation of histone proteins became the first well established example of biologically functional protein acetylation. Allfrey et al. observed that histones in isolated calf thymus nuclei can be rapidly labeled with radiolabeled acetate and that these acetylated histones were less inhibitory for RNA polymerase [2]. In the late 1990s, the first histone acetyltransferases (HATs) and deacetylases (HDACs) were cloned and linked to the regulation of gene expression on chromatinized templates [3,4]. In this case, acetylation generally correlates with gene expression, acting in part to ‘open up’ chromatin for appropriate transcriptional machinery to access the DNA template. We now know there are many acetyl-CoA dependent histone acetyltransferases and histone deacetylases that function to regulate all DNA-templated processes, which are primarily thought to act through the direct reversible acetylation of histone lysine residues [5–7]. Current evidence supports the idea that certain site-specific acetylation is sufficient to alter nucleosome dynamics and chromatin folding [8,9]. In addition, acetylated-lysines on histones can function as ‘epitopes’ for the recruitment of acetyl-lysine binding domains (e.g., bromodomains) that are contained within large protein complexes such as histone acetyltransferases, methyltransferases, transcriptional co-activators, and ATP-dependent chromatin-remodelers [10].
The acetylation of p53 and α-tubulin were early examples that protein acetylation extends beyond histone proteins [11]. The observation that several deacetylases were localized outside of the nucleus spurred further interest in exploring protein acetylation as a broader phenomenon [12]. Shortly thereafter, the metabolic enzyme acetyl-CoA synthetase was found to be regulated by reversible acetylation in both bacterial and mammalian systems, suggesting that non-histone protein acetylation may be an evolutionarily conserved, general mechanism of metabolic regulation. In this case, acetyl-CoA synthetase activity is controlled by acetylation of a single conserved lysine residue in the active site. Acetylation renders the enzyme inactive, while deacetylation restores full activity [13–15]. Collectively, these results demonstrated the existence of functionally-relevant, non-histone targets, which inspired the use of unbiased discovery methods to identify and characterize other acetylation events. Immunoprecipitation with an anti-acetyllysine antibody followed by liquid chromatography coupled mass spectrometry (LC-MS) was the method of choice and early acetyl-proteomic studies provided lists of acetylated peptides and the corresponding proteins. These catalogs were often dominated by metabolic proteins and particularly enriched with mitochondrial proteins. Such observations suggested that either the method was biased toward highly abundant proteins, or there was something unique to metabolic proteins, especially those resident in the mitochondria. Subsequent proteomic studies have confirmed that the modification is widespread, particularly among metabolic and mitochondrial proteins (Figure 1).
Figure 1. Acetylated Proteins in the Context of Global Metabolism.
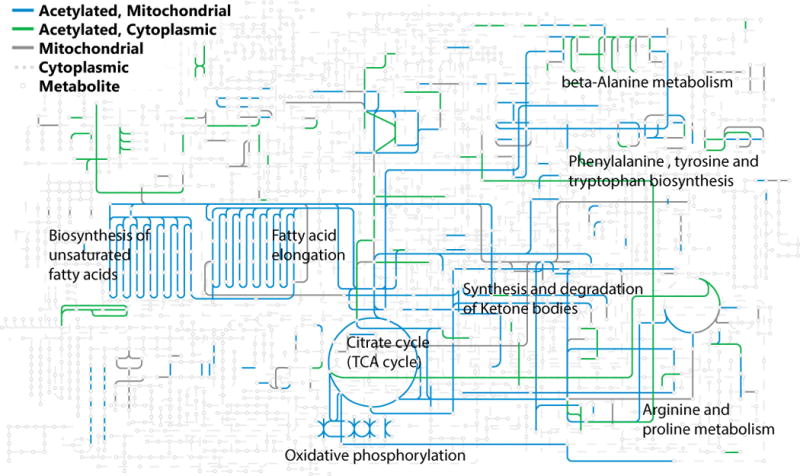
A large proportion of metabolic enzymes are acetylated. Mitochondrial proteins (in blue) and cytoplasmic proteins (in green) that have been found acetylated in M. musculus are overlaid on the KEGG Metabolic Pathways reference pathway. The labeled pathways have had between 8% and 30% of their total lysines acetylated [46,98].
Probing the Extent of the Mitochondrial Acetylome
Growing interest in the protein acetylation field has fueled concerted efforts to characterize the protein acetylome (or acetyl-proteome). By using various nutritional, genetic, and pharmacological model systems, the roster of acetylated lysine sites has expanded rapidly in the past decade (Figure 2). In Mus musculus, the focus of the majority of acetyl-proteomics studies, 13,186 sites across 4,024 proteins have been identified and a full 63% of mitochondrially localized proteins contain acetylation sites. A comparable amount of sites have been found in human cell lines: ~11,000 sites [44–46]. The bulk of this data has been generated in the past four years. In 2011, there were only 552 recorded acetylation sites in Mus musculus [45]. It is not solely the raft of new studies that have added to the compendium. Improvements in antibody reagents, chromatography, and mass-spectrometry technology have increased the proportion of the true acetyl-proteome accessible to each study. For acetylome papers published in 2011, the average number of sites reported was 1,357, and in 2015 the average number of sites per study is 7,784 (Figure 2). It will be interesting to follow these numbers in the upcoming years and assess whether the total will grow substantially as mass-spectrometry technology makes additional gains in sensitivity.
Figure 2. Timeline of Detected Acetylated Peptides per Publication, 2006–2015.
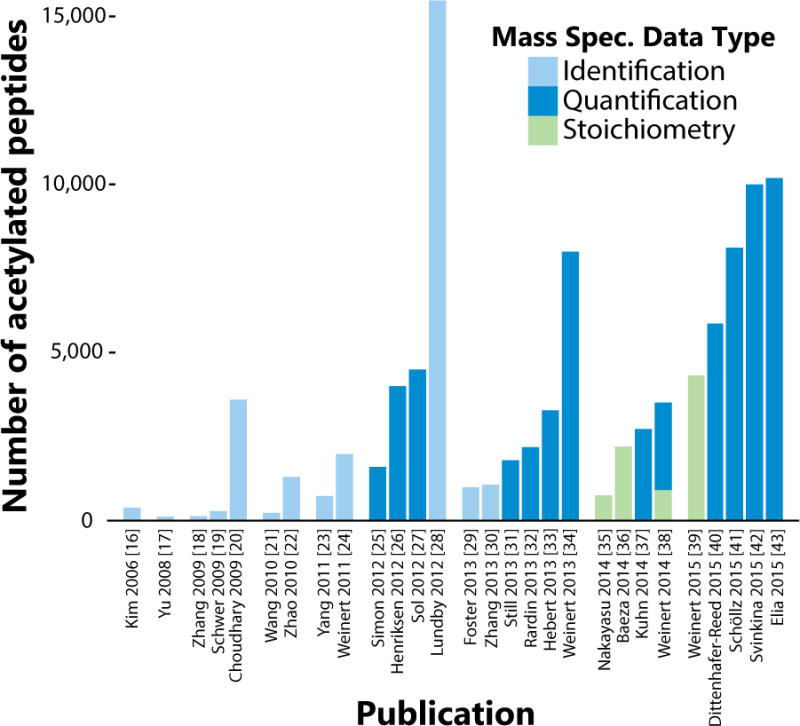
The number of acetylated peptides reported per acetyl-proteome study, grouped by year. Included experiments used liquid chromatography coupled mass spectrometry (LC-MS) to query the acetylation on the proteome level with the data type indicated by fill color. “Identification” studies report a list of peptides, “quantification” studies report the change in relative abundance in an acetyl-peptide between two experimental conditions, and “stoichiometry” studies report the ratio of acetyl-peptide to the amount of acetyl-peptide plus the unmodified corresponding peptide. The papers include experiments performed in the following organisms: M. musculus, H. sapiens, S. enterica, D. melanogaster, S. cerevisiae, R. norvegicus, and E. Coli.
Despite the vast catalog of site-specific mitochondrial acetylation, only a few examples have an established functional consequence. Of the ~700 acetylated mitochondrial proteins, there are 26 proteins that display functional effects when acetylated (Figure 3 and Table 1). This relatively low number is largely due to the technical challenges of acquiring biochemical evidence for altered function. It is even more difficult to determine which biochemical feature is affected: enzyme activity, protein-protein interactions, protein-DNA interactions, stability, localization, allostery, etc. The most rigorous approach to validate a possible regulatory function is to prepare and characterize a fully unacetylated form and a site-specifically acetylated form that is stoichiometrically modified. Genetic incorporation of an acetyl-lysine onto recombinant proteins using an orthogonal acetyllysl-tRNA synthetase–tRNA pair is one such approach. This system produces homogenous recombinant protein containing an acetyl-lysine at defined sites [9,47]. Another commonly used method to probe acetyl-lysine function is using site-directed mutagenesis. A lysine-to-glutamine (K→Q) substitution is often considered an acetyl mimic due to the resemblance of the uncharged functional group. Likewise, a lysine-to-arginine (K→R) substitution preserves the positively charged functional group and is often utilized as an unmodified lysine mimic. However, such mimics are not classical isosteres and accordingly do not always yield the expected results, as illustrated in a recent study on the autoacetylation of a histone acetyltransferase in which both the K→Q and K→R substitutions yielded protein that was ~100-fold less active than the active wild type auto-acetylated species [48]. As an alternative, in vitro acetylation of a target protein can be obtained by using enzymatic or chemical methods [13,49]. The use of a highly specific acetyltransferase can generate a homogenous population of acetylated protein, however the lack of strong sequence specificity of lysine acetyltransferase (KAT) complexes limits this method from widespread use. Reactive acetylating agents, such as acetic anhydride and sulfo-N-hydroxysuccinimide (NHS)-acetate are chemical methods to assess the functional impact of protein acetylation. Both strategies suffer from the difficulty of achieving high stoichiometry and issues of targeting acetylation to only the appropriate lysine residues. These genetic, enzymatic and chemical tools have enabled identification of functionally relevant sites and will continue to be key tools in determining the broader roles of protein acetylation.
Figure 3. Overview of proteins in mitochondrial metabolism with functional consequence caused by reversible acetylation.
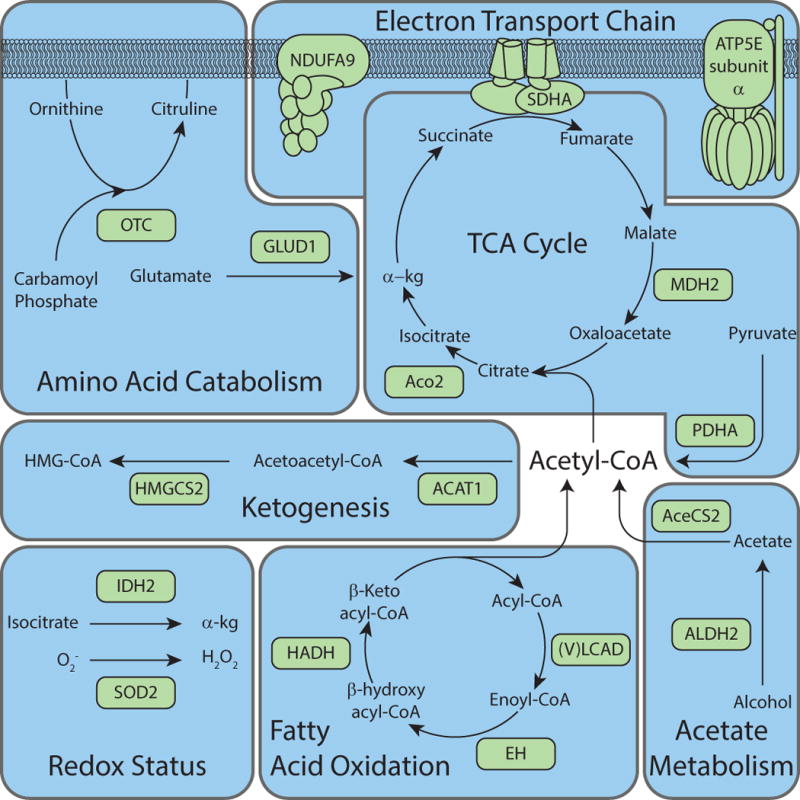
Representative mitochondrial metabolic proteins that contain known regulatory acetylation site(s) are depicted. The pathways represented display only those enzymes or subunits that contain acetylation site(s), which cause a functional consequence on enzymatic activity (See Table 1).
Table 1.
Mitochondrial Proteins with altered function as a consequence of acetylation.
| Gene ID | Gene Name | Organism1 | Tissue | Site | Effect | Validation | Deacetylase | In vitro deacetylase activity with full length protein? | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| ACAT1 | Acetyl-CoA acetyltransferase 1 | Mm | Liver | K260, K265 | Inhibitory | AcK. | Sirt3 | Yes | [31, 40] |
| AccCS2 | Acetyl-CoA synthetase 2 | Mm | K635 | Inhibitory | AcK (PAT) | Sirt3 | Yes | [13] | |
| Hs | K642 | Inhibitory | K→Q | Sirt3 | Yes | [14] | |||
| ACO2 | Aconitase | Mm | Heart | n/a | Stimulatory | Chem AcK | Sirt3 | No | [78] |
| ALDH2 | Aldehyde Dehydrogenase 2 | Hs | n/a | Stimulatory | n/a | Sirt3 | No | [79] | |
| ATP5E | F1F0-ATPase Subunit a | Mm | Liver | n/a | Inhibitory | No | Sirt3 | No | [80] |
| EH/HADH | Enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase | Hs | K165, K171, K346, K584 | Stimulatory | 4K→Q | [22] | |||
| GLUD1 | Glutamate dehydrogenase 1 | Mm | Liver | n/a | Inhibitory | No | Sirt3 | Yes | [50] |
| Hs | n/a | Inhibitory | No | Sirt3 | Yes | [67] | |||
| GOT2 | Glutamate oxaloacetate transaminase | Hs | K159, K185, K404 | Binding | AcK, K→R, K→Q | Sirt3 | No | [81] | |
| HMGCS2 | 3-Hydroxy-3-Methylglutaryl-CoA Synthase 2, Mitochondrial | Mm | Liver | K310, K447, K473 | Inhibitory | K→R | Sirt3 | Yes | [82] |
| IDH2 | Isocitrate Dehydrogenase 2 | Hs | n/a | Inhibitory | No | Sirt3 | Yes | [67] | |
| Mm | Liver, Inner ear, brain | n/a | Inhibitory | No | Sirt3 | Yes | [62] | ||
| Mm | Liver | K413 | Inhibitory | AcK | Sirt3 | Yes | [66] | ||
| LCAD | Long chain acyl-coa dehydrogenase | Hs | Liver | K42 | Inhibitory | K→R | Sirt3 | Yes | [83] |
| Mm | Liver | K318, K322 | Inhibitory | Chemical AcK | Sirt3 | Yes | [49] | ||
| MDH2 | Malate Dehydrogenase 2 | Hs | K185, K301, K307, K314 | Stimulatory | 4K→R | [22] | |||
| Mm | Liver | K239 | Inhibitory | K→Q | Sirt3 | No | [33] | ||
| MRPL10 | Mitochondrial Ribosomal Protein L10 | Mm | Liver | n/a | Inhibitory | No | Sirt3 | Yes | [84] |
| NDUFA9 | NADH Dehydrogenase 1 a Subcomplex 9 | Mm | Liver | n/a | Inhibitory | No | Sirt3 | No | [85] |
| OGG1 | 8-Oxoguanine DNA glycosylase 1 | Hs | n/a | Instability | No | Sirt3 | No | [86] | |
| OPA1 | Optic atrophy 1 | Mm | Heart | K926, K931 | Inhibitory | K→Q, K→R | Sirt3 | Yes | [87] |
| OSCP | Oligomycin sensitivity conferring protein | Hs | n/a | Inhibitory | No | Sirt3 | Yes | [88] | |
| OTC | Ornithine Transcarbamoyltransferase | Mm | Liver | K88 | Inhibitory | No | Sirt3 | Yes | [89] |
| OXCT | Succinyl CoA:3-ketoacid-CoA transferase | Mm | Brain | K451 | Inhibitory | AcK | Sirt3 | Yes | [40] |
| P450scc | P450 Cholesterol side chain cleavage monooxygenase | Hs | K148, K149 | Inhibitory | K→A | Sirt3 | No | [90] | |
| PDHA | Pyruvate Dehydrogenase E1 alpha subunit | Mm | Muscle | K336 | Inhibitory | K→Q, K→R | Sirt3 | No | [91] |
| PPID | Peptidylprolyl cis-trans Isomerase D (Cyclophilin D) | Hs | K145 | Binding | K→Q, K→R | Sirt3 | No | [92] | |
| Hs | n/a | Binding | No | Sirt3 | No | [93] | |||
| SDHA | Succinate Dehydrogenase Complex, Subunit A, Flavoprotein | Mm | Liver | n/a | Inhibitory | No | Sirt3 | No | [94] |
| Mm | Liver | n/a | Inhibitory | No | Sirt3 | Yes | [95] | ||
| SKP2 | F-box protein S-Phase kinase associated protein 2 | Hs | K68, K71 | Mislocalization | K→Q, K→R K→L | Sirt3 | No | [96] | |
| SOD2 | Superoxide Dismutase 2 | Mm | Liver | K122 | Inhibitory | K→R | Sirt3 | Yes | [63] |
| Mm | Liver | K53, K89 | Inhibitory | K→R | Sirt3 | Yes | [65] | ||
| Hs | K68 | Inhibitory | K→Q, K→R | Sirt3 | No | [64] | |||
| VLCAD | Very Long chain acyl-coa dehydrogenase | Hs | Liver | K507 | Inhibitory, Mislocalization | Chemical AcK | Sirt3 | Yes | [97] |
Hs-H. sapiens, Mm-M. musculus
General Trends in Mitochondrial Protein Acetylation
The accumulated evidence suggests that mitochondrial acetylation is widespread and that the acetylation status of many sites is controlled by the enzymatic activity of the NAD+-dependent deacetylase Sirtuin 3 (SIRT3) [32,33,40,50]. Other sirtuin family members that reside in the mitochondria include SIRT4 and SIRT5, however SIRT3 is the only mitochondrial member with robust deacetylation activity [51]. SIRT4 has been shown to possess deacetylation as well as delipoylation and debiotinylation activity, while SIRT5 removes malonyl, succinyl, and glutaryl moieties from lysine residues [52–56]. A definitive mitochondrial acetyltransferase has not been identified, however nonenzymatic acetylation has been discussed as a possible mechanism to explain the extent of protein acylation [57–60]. Baeza et al. demonstrated that the chemical reactivities of lysine residues towards acetyl-CoA (as a function of second order rates) might be sufficient to explain the observed acetylation found in tissues [61].
Although there are some reports that suggest metabolic proteins can be activated upon acetylation, the majority of well documented cases indicate that acetylation of mitochondrial enzymes is an inhibitory mark (Table 1). Overall, the trend suggests that oxidative metabolism is inhibited by higher levels of acetylation among these central metabolic enzymes. Such a regulatory mechanism would serve to sense the overproduction of acetyl-CoA and provide negative feedback to mitochondrial metabolism. Thus, mitochondria utilize targeted acetylation to decrease the flux through metabolic pathways that operate in oxidative mode. Many of these central enzymes are deacetylated by SIRT3, which reverses the inhibitory effect of acetylation, leading to enhanced oxidative metabolism [27,33,62], and the accompanying stimulation of ROS-mitigating systems, such as Isocitrate dehydrogenase (IDH2) and Superoxide Dismutase 2 (SOD2) [62–67]. SIRT3 expression is induced by fasting and chronic caloric restriction [62,68], two conditions that necessitate increased oxidative metabolism of fatty acids and amino acids.
Hebert et al. quantified the fold-change acetylation in SIRT3 wild-type and knockout mice fed either a control or calorie restricted diet [33]. Three classes of acetylation sites emerged from clustering analyses: Class 1 are acetylation sites that were controlled by SIRT3 expression; Class 2, acetylation sites that were primarily affected by a calorie restricted diet; and Class 3, acetylation sites that display minimal or no change under the four conditions [33]. Interestingly, Class 3 sites were enriched in loops with a general acidic stretch of amino acids, while Class 2 sites were enriched in more hydrophobic sequences. Class 1 sites meet the criteria as targets of SIRT3, with basic amino acids as preferred features around the acetylation site [69]. Collectively, these data suggest that Class 3 sites might represent spurious acetylation, whereas Class 1 and 2 are regulated sites with Class 1 sites modulated by SIRT3 and Class 2 sites being partially buried and inaccessible to SIRT3.
Quantitative Acetylomics using Stoichiometry
Relative quantitation-based mass spectrometry has been crucial for understanding acetylation dynamics and mechanisms that modulate mitochondrial metabolism. However these methods do not provide direct information on stoichiometry; that is, the fraction of protein that is modified. Stoichiometric information is necessary to provide a more complete picture for the role of acetylation as a regulatory mechanism. Consider the case where two peptides are quantified in two different biological conditions using a relative quantitation-based method and each peptide displays a 2-fold change. Applying a stoichiometry-based method, the same two peptides are measured as 1 to 2% change and 25 to 50% change. In this scenario, the two ‘2-fold’ changes may have very different effects on the system, given that the level of inhibition (or other regulatory effects) likely scales with the proportion of a protein modified. Whether this scaling is linear or in some way cooperative and nonlinear is an open question. Resolving this question will bring us closer to discerning whether acetyl-peptides in the medium stoichiometry range (5–15%) can have significant biological/regulatory effects.
Two methods have been described for measuring stoichiometric information on a proteomic scale. The first is a direct method that utilizes stable isotope chemical labeling followed by digestion and MS analysis [36]. Comparison of the light (endogenous) and isotopic (chemically labeled) acetyl peptides provides a direct measurement of the stoichiometry. The second method, which is used to estimate stoichiometry, utilizes the quantitative fold-change measurements by correlating relative changes of a modified peptide to a corresponding unmodified peptide in two biological conditions while correcting for overall protein levels [38,39]. This approach circumvents the need to account for varying ionization efficiencies when measuring two different peptides. Both methods are subject to the limitation that they are unable to account for other potential lysine modifications, and so the ratios that are reported are the ratio of the acetyl-peptide to the acetyl-peptide plus the unmodified corresponding peptide.
Initial stoichiometry measurements suggest that the average level of acetylation across all detected lysine-containing peptides is in the 0–5% range, with most acetylated lysines showing levels below 1%. [39]. In Escherichia coli and Saccharomyces cerevisiae, reported estimated acetylation stoichiometry values range from less than 1% – 98% [34,36,38]. Using an indirect approximation, Weinert et al. estimated that 48 SIRT3 regulated sites would exhibit stoichiometry >1% in SIRT3 knockout mice, with one site predicted to be up to 87% acetylated. Together, these observations suggest that most lysine residues detected in MS acetyl-proteomic analyses harbor relatively low acetylation stoichiometry. This raises many questions about the roles played by site-specific acetylation events. One could argue that such low overall stoichiometry means that most acetylation is spurious and of no biological consequence. This argument has merit and is supported by the existence of class 3 acetylation sites that were not altered by SIRT3 expression or dietary restriction. However, such a blanket argument does not account for the existing biochemical and biological data supporting functionally relevant acetylation. Also, the current data is limited to a few biological conditions, and the possibility that cellular stresses could greatly induce high stoichiometry remains to be evaluated. Nevertheless, a logical conclusion is that spurious acetylation co-exists with targeted (functional) acetylation, and the overall steady-state levels are a function of the rate of acetylation, deacetylation and protein turnover, which are subject to metabolic flux, enzyme expression, activity and protein stability.
Regulatory mechanisms that feature low stoichiometry
While it might appear reasonable to discount very low stoichiometry as spurious, and to assign functional meaning to only very high stoichiometry, we must explore the possibility that many regulated acetylation sites with significant but low stoichiometry (e.g. 5–15%) are sufficient to induce clear phenotypes. In the following discussion, we explain potential models where low but significant acetylation can impact function.
Stoichiometry of acylation
The large scale mass spectrometry methods mapping acetyl, malonyl, succinyl, and glutaryl moieties on lysine residues have treated each modification as a unique entity. Each modification alters the charge state from z = +1 (unmodified) to z = 0 (acetyl) or z = −1 (malonyl, succinyl, glutaryl). Additionally, butyrylation and propionylation have also been documented in liver mitochondria [70,71]. Each of these acylations are most likely derived from their Coenzyme A derivatives possibly through a nonenzymatic mechanism [38,59–61,71–73]. Based on these observations, there is potential for other acylations on lysine residues awaiting discovery (Figure 4). Specific acylations may be enriched on metabolic enzymes belonging to the pathway that generates the respective acyl-CoA. For example, 3-hydroxy-3-methylglutarylation and acetoacetylation may be found on enzymes involved in ketogenesis such as acetyl-CoA acetyltransferase 1 (ACAT1), 3-Hydroxy-3-Methylglutaryl-CoA Synthase 2 (HMGCS2) and Hydroxymethylglutaryl-CoA lyase (HMGCL), while isobutarylation (a valine derivative), isovalerylation (a leucine derivative), and α-methylbutarylation (an isoleucine derivative) may be found on enzymes involved in branched chain amino acid catabolism. In a biochemical pathway, sets of different lysine modifications are likely to exist among a protein population, and might have similar regulatory outcomes. Therefore, we need to account for the full distribution of a peptide across all possible modification states (including, but not limited to the above acylations). The current stoichiometry methodology only accounts for the ratio of the unmodified peptide to the corresponding acetylated peptide. Tools to query the full range of possible acylations at the proteome scale are needed.
Figure 4. Described and Potential Lysine Acylations.
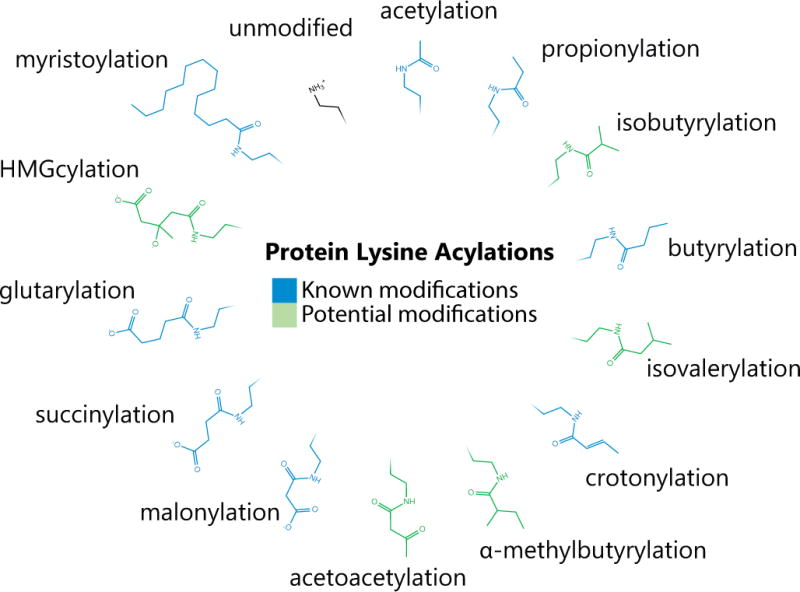
Lysine has been shown to be modified by a number of acyl-groups (in blue), which are often donated from the corresponding acyl-CoA. Acetylation, succinylation [99], propionylation [100], butyrylation [70,101], malonylation [54], glutrarylation [53], myristoylation [102], and crotonylation [103] are among lysine modifications previously discovered. Other acyl-CoAs that are present in the mitochondria are likely to also modify lysines (in green), particularly if the acylation process is primarily nonenzymatically driven.
Shared functional outcome of multi-site acetylation on individual proteins
Some documented cases show that a single acetylation site directly controls enzymatic activity, such as for Acetyl-CoA Synthetase 2 (AceCS2) [13,14]. In other cases, multiple sites affect activity, such as with ACAT1 (sites K260, K265), HMGCS2 (sites K310, K447, K473) and SOD2 (sites K122, K53, K89, K68) (see Table 1). On most proteins, the impact of acetylation is observed consistently across many studies, yet the functional impact of multi-site acetylation has not been experimentally determined. For example, Carbamoyl-phosphate synthase (CPS1) has 59 documented acetylation sites, but no one regulatory acetyl-site in particular is known to affect activity, even though Sirt3−/− mice exhibit CPS1 hyperacetylation and an altered urea cycle, where CPS1 plays a vital role in removing ammonia from the cell [33]. Given that proteins are digested into peptides prior to mass-spectrometry analysis, one cannot discern whether multi-site modifications exist on the same protein molecule or on different protein molecules. There are two mechanisms by which low stoichiometry across multiple sites can have additive effects on activity: 1) acetyl-sites co-occuring on the same molecule could ratchet down (or up) activity in accordance with how many sites are occupied, or 2) acetyl-sites on different molecules with similar functional effects could in each contribute to dampening (or enhancing) activity.
Cumulative effect of low-stoichiometry acetylation across a pathway
While a low level of acetylation on one particular enzyme may have a limited effect on its catalytic capacity, the low-stoichiometry acetylation of multiple enzymes in a metabolic pathway, each causing a small decrease (or increase) in activity, could cumulatively result in physiologically meaningful alteration in metabolite flux. Hebert et al. reports nine multi-enzyme mitochondrial pathways that have perturbed metabolite concentrations in Sirt3−/− mice. Beta oxidation, which was found to have altered metabolites, contains five enzymes targeted by SIRT3 [33]. Similarly, Dittenhafer-Reed et al. demonstrated dysregulated ketone body utilization in the brain of mice lacking SIRT3, a pathway which in brain tissue was found to have 13 predicted SIRT3 targeted acetyl-sites distributed over five proteins involved in ketone body utilization [40]. In light of this, it is quite possible that not one enzyme in particular is culpable, but rather the observed metabolite concentration changes are the additive effects of low stoichiometry modulating several enzymes in the same pathway.
Acetylation and cooperativity in multimeric enzyme complexes
Metabolic enzymes often function as higher-ordered complexes including hetero and homo-oligomerization of polypeptide chains. Glutamate dehydrogenase (GDH) is a homohexamer which displays cooperativity effects upon ligand binding [74,75]. GDH contains an ‘antenna’ domain that protrudes from the structure and was proposed to be an intersubunit communication link during cooperativity [76]. Two lysine sites (K477 and K480 on bovine GDH) are found acetylated at the apex of the antenna and interestingly these sites displayed the highest reactivity towards acetyl-CoA [33,61]. How would acetylation affect the cooperativity of the enzyme complex? What level of stoichiometry would be needed to disrupt protein-protein interactions between multimeric enzymes? Would the acetylated monomer weaken the interactions among the rest of the complex? It is tempting to suggest that acetylation could disrupt complex formation or the communication between monomers in which case having a low stoichiometry would have a non-linear effect.
Acetylation measurements are an aggregate of many levels of biological hierarchies
When acetylation stoichiometry is discussed in a particular biological condition, it is important to note that mass spectrometry measurements collapse many levels of biological hierarchies into one number. In a cell culture study, these data produce one value that represents the average stoichiometry over the whole population of cells in the sample, over the whole population of mitochondria within each cell, and over sub-compartmental, local concentrations of proteoforms, all of which could contain significant heterogeneity. This issue is compounded in studies of bulk tissue, which additionally contain a heterogenous population of cell types and functionality. What appears to be very low stoichiometry in the aggregate could be a combination of highly acetylated populations and populations with no significant modifications, or any intermediate between these extremes. For instance, mitochondria within the same cell or between cells in a population might display very diverse protein acetylation levels, but with the current approaches, these are averaged during analysis. It is intriguing to suggest that mitochondrial dysfunction caused by high levels of protein acetylation might drastically impair functionality organelle-wide and initiate mitophagy. Such a scenario might predict that highly acetylated mitochondria are rapidly turned over which would contribute to low observable stoichiometry under steady state conditions. Such catastrophic events could be induced by supra-physiological levels of mitochondrial acetyl-CoA perhaps combined with loss of SIRT3 function. It would not be unprecedented for acetyl-CoA levels to induce the degradation machinery; cytosolic depletion of acetyl-CoA has recently been shown to stimulate autophagy [77].
A comparison of nuclear and mitochondrial protein (de)acetylation reveals fascinating dichotomies. To date, the general trends suggest that histone and nuclear protein acetylation is enzyme-catalyzed, usually leading to up-regulated processes such as transcriptional activation. In mitochondria, current evidence supports nonenzymatic mechanisms for acetylation, and most acetylated proteins exhibit loss of function, particularly those involved in oxidative metabolism. Both organelles utilize deacetylases to remove these modifications. High levels of acetyl-CoA in the nucleus and cytosol might represent pro-growth or cellular-response states, where acetyl-CoA serves as a precursor for lipid synthesis and as a co-substrate for transcriptional activation. In the mitochondria, increased levels of acetyl-CoA beyond demand is a signal to slow flux through oxidative energy production. By inducing acetyl-CoA dependent protein acetylation, a substrate-level braking system is applied. When energy demands require more oxidative metabolism, induced SIRT3 expression relieves the brake, allowing the cell to push the accelerator for more energy production.
TRENDS BOX.
Improvements in mass-spectrometry-based proteomics have uncovered protein acetylation as a prominent post-translational modification, and the latest methods report fold-change and stoichiometry.
Mitochondria appear to harbor a disproportionately high number of acetylated proteins.
Mitochondrial acetylation is thought to be largely non-enzymatic, mediated by reactive lysine residues and acetyl-CoA.
The NAD+-dependent deacetylase SIRT3 removes mitochondrial acetylation, and loss of SIRT3 function is linked to increased ROS and altered oxidative metabolism.
Protein acetylation in mitochondria typically leads to loss of function in pathways associated with organelle integrity and oxidative metabolism.
Additional endogenous lysine acylations (e.g. succinylation, glutarylation) occur and can be catalytically removed by members of the sirtuin family.
Outstanding questions BOX.
What mechanisms are responsible for mitochondrial protein acetylation?
Is mitochondrial protein acetylation driven by levels and/or flux of acetyl-CoA?
Does mitochondrial protein acetylation occur via non-enzymatic or enzymatic reactions or both?
What are the functional consequences of specific protein acetylation?
Can we distinguish spurious from functionally relevant acetylation?
How does knowledge of site-specific stoichiometry influence our understanding of functionally relevant modifications? For example, can low stoichiometry at multiple sites synergize to modulate catalytic activity or alter protein:protein interactions?
Are there different functional outcomes between diverse acyl-groups on the same lysine residues? For example, is succinylation functionally equivalent to acetylation?
Are there other acylations waiting to be discovered?
Acknowledgments
We would like to thank other members of the laboratory for helpful discussions. This work was supported, in whole or in part, by the National Institutes of Health (NIH) Grant GM065386 (JMD), NIH National Research Service Award T32 GM007215 (JB) and the National Science Foundation Graduate Research Fellowship Program (NSF-GRFP) DGE-1256259 (JB). We apologize to colleagues whose relevant work could not be cited here owing to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allfrey VG, et al. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brownell JE, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 4.Taunton J, et al. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 5.Roth SY, et al. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 6.Guarente L. Sirtuins, Aging, and Medicine. N Engl J Med. 2011;364:2235–2244. doi: 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- 7.Haberland M, et al. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 9.Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piperno G, et al. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 13.Hallows WC, et al. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proceedings of the National Academy of Sciences. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwer B, et al. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proceedings of the National Academy of Sciences. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Starai VJ, et al. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 16.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 17.Yu BJ, et al. The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol. 2008;18:1529–1536. [PubMed] [Google Scholar]
- 18.Zhang J, et al. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwer B, et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L, et al. The fasted/fed mouse metabolic acetylome: N6-acetylation differences suggest acetylation coordinates organ-specific fuel switching. J Proteome Res. 2011;10:4134–4149. doi: 10.1021/pr200313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinert BT, et al. Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci Signal. 2011;4 doi: 10.1126/scisignal.2001902. [DOI] [PubMed] [Google Scholar]
- 25.Simon GM, et al. Quantitative assessment of the impact of the gut microbiota on lysine epsilon-acetylation of host proteins using gnotobiotic mice. Proc Natl Acad Sci U S A. 2012;109:11133–11138. doi: 10.1073/pnas.1208669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henriksen P, et al. Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol Cell Proteomics. 2012;11:1510–1522. doi: 10.1074/mcp.M112.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sol EM, et al. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS One. 2012;7:e50545. doi: 10.1371/journal.pone.0050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundby A, et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012;2:419–431. doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foster DB, et al. The cardiac acetyl-lysine proteome. PLoS One. 2013;8:e67513. doi: 10.1371/journal.pone.0067513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang QF, et al. Reversibly acetylated lysine residues play important roles in the enzymatic activity of Escherichia coli N-hydroxyarylamine O-acetyltransferase. FEBS J. 2013;280:1966–1979. doi: 10.1111/febs.12216. [DOI] [PubMed] [Google Scholar]
- 31.Still AJ, et al. Quantification of Mitochondrial Acetylation Dynamics Highlights Prominent Sites of Metabolic Regulation. J Biol Chem. 2013;288:26209–26219. doi: 10.1074/jbc.M113.483396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rardin MJ, et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci U S A. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hebert AS, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinert BT, et al. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell. 2013;51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Nakayasu ES, et al. A method to determine lysine acetylation stoichiometries. Int J Proteomics. 2014;2014:730725. doi: 10.1155/2014/730725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baeza J, et al. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem. 2014;289:21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhn ML, et al. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One. 2014;9:e94816. doi: 10.1371/journal.pone.0094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinert BT, et al. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinert BT, et al. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 2015 doi: 10.15252/embj.201591271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dittenhafer-Reed KE, et al. SIRT3 mediates multi-tissue coupling for metabolic fuel switching. Cell Metab. 2015;21:637–646. doi: 10.1016/j.cmet.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schölz C, et al. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol. 2015;33:415–423. doi: 10.1038/nbt.3130. [DOI] [PubMed] [Google Scholar]
- 42.Svinkina T, et al. Deep, Quantitative Coverage of the Lysine Acetylome Using Novel Anti-acetyl-lysine Antibodies and an Optimized Proteomic Workflow. Mol Cell Proteomics. 2015;14:2429–2440. doi: 10.1074/mcp.O114.047555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elia AEH, et al. Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA Damage Response. Mol Cell. 2015;59:867–881. doi: 10.1016/j.molcel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Z, et al. CPLM: a database of protein lysine modifications. Nucleic Acids Res. 2014;42:D531–6. doi: 10.1093/nar/gkt1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z, et al. CPLA 1.0: an integrated database of protein lysine acetylation. Nucleic Acids Res. 2011;39:D1029–34. doi: 10.1093/nar/gkq939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calvo SE, et al. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neumann H, et al. Genetically encoding Nɛ-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 48.Albaugh BN, et al. Autoacetylation of the histone acetyltransferase Rtt109. J Biol Chem. 2011;286:24694–24701. doi: 10.1074/jbc.M111.251579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bharathi SS, et al. Sirtuin 3 (SIRT3) protein regulates long-chain acyl-CoA dehydrogenase by deacetylating conserved lysines near the active site. J Biol Chem. 2013;288:33837–33847. doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lombard DB, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feldman JL, et al. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathias RA, et al. Sirtuin 4 Is a Lipoamidase Regulating Pyruvate Dehydrogenase Complex Activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan M, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng C, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10:M111012658–M111.012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park J, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laurent G, et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell. 2013;50:686–698. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner GR, Payne RM. Widespread and enzyme-independent Nɛ-acetylation and Nɛ-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghanta S, et al. Mitochondrial protein acetylation as a cell-intrinsic, evolutionary driver of fat storage: chemical and metabolic logic of acetyl-lysine modifications. Crit Rev Biochem Mol Biol. 2013;48:561–574. doi: 10.3109/10409238.2013.838204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner GR, Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell. 2014;54:5–16. doi: 10.1016/j.molcel.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paik WK, et al. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim Biophys Acta. 1970;213:513–522. doi: 10.1016/0005-2787(70)90058-4. [DOI] [PubMed] [Google Scholar]
- 61.Baeza J, et al. Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chem Biol. 2015;10:122–128. doi: 10.1021/cb500848p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Someya S, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gius D, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y, et al. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 2011;12:534–541. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verdin E, et al. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 66.Yu W, et al. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287:14078–14086. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schlicker C, et al. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 68.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith BC, et al. SIRT3 substrate specificity determined by peptide arrays and machine learning. ACS Chem Biol. 2011;6:146–157. doi: 10.1021/cb100218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fritz KS, et al. Ethanol metabolism modifies hepatic protein acylation in mice. PLoS One. 2013;8:e75868. doi: 10.1371/journal.pone.0075868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pougovkina O, et al. Aberrant protein acylation is a common observation in inborn errors of acyl-CoA metabolism. J Inherit Metab Dis. 2014;37:709–714. doi: 10.1007/s10545-014-9684-9. [DOI] [PubMed] [Google Scholar]
- 72.Wagner GR, Payne RM. Widespread and Enzyme-independent Nε-Acetylation and Nε-Succinylation of Proteins in the Chemical Conditions of the Mitochondrial Matrix. J Biol Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinert BT, et al. Lysine Succinylation Is a Frequently Occurring Modification in Prokaryotes and Eukaryotes and Extensively Overlaps with Acetylation. CellReports. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 74.Wacker SA, et al. Ligand-induced changes in the conformational stability and flexibility of glutamate dehydrogenase and their role in catalysis and regulation. Protein Sci. 2010;19:1820–1829. doi: 10.1002/pro.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alex S, Bell JE. Dual nucleotide specificity of bovine glutamate dehydrogenase. The role of negative co-operativity. Biochem J. 1980;191:299–304. doi: 10.1042/bj1910299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peterson PE, et al. The structure of bovine glutamate dehydrogenase provides insights into the mechanism of allostery. Structure. 1999;7:769–782. doi: 10.1016/s0969-2126(99)80101-4. [DOI] [PubMed] [Google Scholar]
- 77.Mariño G, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014;53:710–725. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 78.Fernandes J, et al. Lysine Acetylation Activates Mitochondrial Aconitase in the Heart. Biochemistry. 2015 doi: 10.1021/acs.biochem.5b00375. at < http://pubs.acs.org/doi/abs/10.1021/acs.biochem.5b00375>. [DOI] [PMC free article] [PubMed]
- 79.Xue L, et al. Acetylation-dependent regulation of mitochondrial ALDH2 activation by SIRT3 mediates acute ethanol-induced eNOS activation. FEBS Lett. 2012;586:137–142. doi: 10.1016/j.febslet.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 80.Bao J, et al. SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radic Biol Med. 2010;49:1230–1237. doi: 10.1016/j.freeradbiomed.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang H, et al. SIRT3-dependent GOT2 acetylation status affects the malateaspartate NADH shuttle activity and pancreatic tumor growth. EMBO J. 2015;34:1110–1125. doi: 10.15252/embj.201591041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shimazu T, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grueter CA, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tong Q, et al. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem. 2010;285:7417–7429. doi: 10.1074/jbc.M109.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahn BH, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng Y, et al. Interaction of Sirt3 with OGG1 contributes to repair of mitochondrial DNA and protects from apoptotic cell death under oxidative stress. Cell Death Dis. 2013;4:e731. doi: 10.1038/cddis.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Samant SA, et al. SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Mol Cell Biol. 2014;34:807–819. doi: 10.1128/MCB.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu YT, et al. Regulation of mitochondrial FoF1ATPase activity by Sirt3-catalyzed deacetylation and its deficiency in human cells harboring 4977 bp deletion of mitochondrial DNA. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2013;1832:216–227. doi: 10.1016/j.bbadis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 89.Hallows WC, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li D, et al. Resveratrol stimulates cortisol biosynthesis by activating SIRT-dependent deacetylation of P450scc. Endocrinology. 2012;153:3258–3268. doi: 10.1210/en.2011-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jing E, et al. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes. 2013;62:3404–3417. doi: 10.2337/db12-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shulga N, et al. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci. 2010;123:894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Wei L, et al. Oroxylin A induces dissociation of hexokinase II from the mitochondria and inhibits glycolysis by SIRT3-mediated deacetylation of cyclophilin D in breast carcinoma. Cell Death Dis. 2013;4:e601. doi: 10.1038/cddis.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cimen H, et al. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Finley LWS, et al. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS One. 2011;6:e23295. doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Inuzuka H, et al. Acetylation-dependent regulation of Skp2 function. Cell. 2012;150:179–193. doi: 10.1016/j.cell.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Y, et al. SIRT3 and SIRT5 regulate the enzyme activity and cardiolipin binding of very long-chain acyl-CoA dehydrogenase. PLoS One. 2015;10:e0122297. doi: 10.1371/journal.pone.0122297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamada T, et al. iPath2.0: interactive pathway explorer. Nucleic Acids Res. 2011;39:W412–5. doi: 10.1093/nar/gkr313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Z, et al. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garrity J, et al. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J Biol Chem. 2007;282:30239–30245. doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- 101.Chen Y, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang H, et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]


