Abstract
MicroRNAs (miRNAs) are a key component of the noncoding RNA family. The underlying mechanisms involved in the interplay between the tumor microenvironment and cancer cells involve highly dynamic factors such as hypoxia and cell types such as cancer-associated fibroblasts and macrophages. Although miRNA levels are known to be altered in cancer cells, recent evidence suggests a critical role for the tumor microenvironment in regulating miRNA biogenesis, methylation, and transcriptional changes. Here, we discuss the complex pro-tumorigenic symbiotic role between tumor cells, the tumor microenvironment, and miRNA deregulation.
Keywords: MicroRNAs, Hypoxia, DICER, Drosha, miRNA biogenesis, tumor progression
Introduction
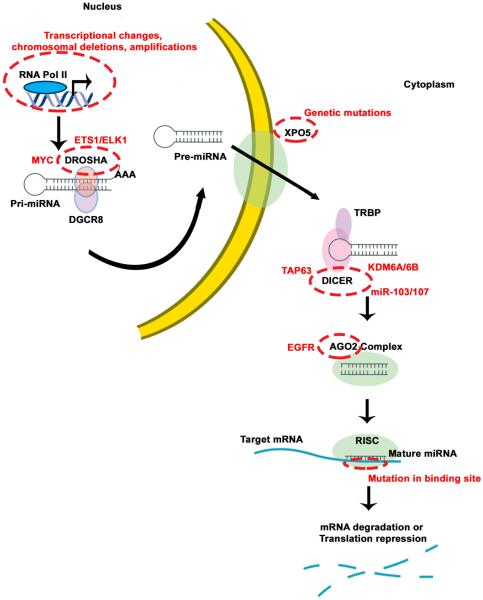
MicroRNAs (miRNAs), a key component of the noncoding RNA family, are involved in multiple cellular functions (1). Since the discovery of these short RNA molecules in Caenorhabdidtis elegans, miRNAs have been recognized to play multifaceted roles in controlling cellular functions by repressing target genes (1–6). MiRNA genes in humans and many other organisms are located in varying genomic contexts, which include intergenic and intragenic noncoding RNA regions in introns or sometimes within an exon of the gene. Mature miRNA biogenesis starts with RNA polymerase II processing of long non-protein coding RNA primary transcripts, called precursor miRNAs (7). These transcripts are further processed by DROSHA and its binding partners, such as DGCR8, leading to primary miRNAs (pri-miRNAs). After these pri-miRNAs are translocated into the cytoplasm via exportin 5, they bind to DICER and RNA-induced silencing complex (RISC), which includes argonaute proteins. In conjunction with RISC, a guide strand helps to navigate the mature miRNAs to the target mRNA, consequently resulting in downregulation of target genes (7) (Figure 1).
Figure 1. Summary of canonical miRNA biogenesis pathway.
MicroRNA genes are transcribed from intergenic or intragenic regions of noncoding RNA transcripts mediated by RNA polymerase II, called primary miRNAs (pri-miRNA). These long pri-miRNAs are processed by the DROSHA-DGCR8 complex to form precursor miRNAs (pre-miRNAs) of approximately 60 nucleotides in length. EXPO5 mediates the export of these pre-miRNAs to the cytoplasm for further processing by DICER. DICER is a ribonuclease, which cleaves pre-miRNAs to form mature miRNAs of approximately 20 nucleotides in length. One of the strands of mature miRNA (guide strand) gets incorporated into RISC (RNAi induced silencing complex) involving DICER and AGO2 enzymes to target mRNAs to cause degradation or translational suppression of gene. The canonical miRNA biogenesis pathway is significantly perturbed in cancer by several proteins at various stages, as highlighted. At the gene level, transcripts are altered in cancer by transcription factors such as MYC or by epigenetic modifications. DROSHA mediated miRNA processing is suppressed in cancer by hypoxia, involving ETS1/ELK1 transcriptional repression of the DROHSA gene. Several studies have highlighted DICER downregulation in cancer mediated by several factors such as TAP63, hypoxia-mediated epigenetic changes, and miR-103/107. EGFR has been reported to bind to AGO2, resulting in phosphorylated AGO2 with decreased association to RISC. TRBP is TAR RNA binding protein.
Although miRNA biogenesis is a tightly regulated process, deregulation of miRNAs caused by alterations in the biogenesis pathway proteins, including DROSHA, DICER, and AGO2, has been recognized to occur in cancer cells (8–10). In addition to autonomous cancer cell gene changes, the tumor microenvironment can directly influence miRNA levels. These alterations can occur as a result of either biogenesis defects under the influence of hypoxia (11–15) or miRNA transcriptional changes (16–18). Despite biogenesis defects and global downregulation in miRNAs (8, 9, 14, 15, 19–21), many oncogenic miRNAs are significantly increased in cancer (16, 22–27). Mechanisms by which expression of oncogenic miRNAs is increased in cancer are diverse and individual miRNA dependent (e.g., increased transcription of specific miRNAs). Here, we summarize recent advances in understanding the complex interplay between miRNA deregulation and the tumor microenvironment.
Part I: Cancer cells and deregulation of miRNAs
Key downregulated enzymes in the miRNA biogenesis pathway in cancer
More than 6 years ago, downregulation of DROSHA and DICER, 2 key enzymes involved in miRNA biogenesis, was reported in many cancers, including ovarian, lung, and breast cancers (8, 9). Such changes are functionally relevant because cells with deficient biogenesis exhibit defects in miRNA processing (9). Since then, several other studies have demonstrated the importance of downregulated DROSHA and DICER expression in an array of cancer types (19–21, 28, 29); this finding is often associated with poor patient survival.
Possible mechanisms for DROSHA regulation include transcriptional activation via MYC (28) or downregulation via ADARB1 (19) proteins. DROSHA was found to be transcriptionally increased by MYC (28), leading to increased miRNA processing in A549 lung cancer cells. However, other independent groups using patient sample analysis of DROSHA expression have shown that DROSHA downregulation in lung cancer was correlated with poor survival (8, 30). These observations suggest intratumoral heterogeneity in cancer. Downregulation of DROSHA expression by ADARB1 in chronic lymphocytic leukemia can lead to decreased miR-15/16 expression and increased oncogenic signaling (19). For DICER, direct binding of Tap63 transcription factor to the DICER promoter has been demonstrated, and DICER downregulation owing to loss of Tap63 in cancer has been observed (31). In that study, loss of DICER led to decreased miR-130b and increased cancer cell invasive potential. Mutant p53 has also been shown to result in DICER downregulation in a p63-independent manner (32), suggesting that DICER downregulation in cancer contains multiple layers of complexity. This is further illustrated by the observation that some miRNAs target the DICER 3' untranslated region. Two independent studies have shown that miR-103/107 (33) and Let-7 (34) can target DICER, and loss of these miRNAs is related to increased tumor growth. MiR-200 was one of the main miRNAs downregulated by low DICER as a result of miR-103/107 direct targeting, and this led to increased cancer metastasis (33).
In addition to DROSHA and DICER, other enzymes in the miRNA biogenesis pathway, such as TARBP2 and AGO2, have also been reported to be downregulated in cancer. In sporadic and hereditary carcinomas, mutations leading to a truncated form of TARBP2 protein can impair DICER function (35). Downregulation of TARBP2 protein expression in cancer stem cells was shown to be important for pro-metastasis signaling (36). EGFR-dependent AGO2 phosphorylation impairs AGO2 binding to DICER, resulting in decreased miRNA biogenesis (12).
Although downregulation of these key enzymes involved in biogenesis is important for cancer progression, additional alterations in miRNAs (unrelated to biogenesis enzymes) have also been reported. For example, DNA damage induces ATM kinase-mediated phosphorylation of KH-type splicing regulatory protein, which leads to increased processing of a select set of miRNAs (37). This observation is important because cancer cells contain several upregulated miRNAs despite the decrease in DROSHA or DICER enzymes, suggesting that alternative mechanisms process some of the miRNAs involved in oncogenic signaling. Likewise, Hippo protein sequesters DDX17 and leads to decreased miRNA production (38). A genetic defect in XPO5 (39) that prevents precursor miRNAs from being exported to the cytoplasm for processing by DICER has also been reported. In this study, a genetic mutation in XPO5 resulted in entrapment of precursor miRNAs in the nucleus. Also, genomic studies showed a tumor-promoting role for mutant XPO5, via increased expression of oncogenes such as EZH2, MYC, and KRAS due to loss of the corresponding targeting miRNAs.
Key miRNAs downregulated in cancer and implications
Some of the main miRNAs downregulated in cancer are those in the miR-200 family. These miRNAs are involved in many diverse functions, such as induction of epithelial-to-mesenchymal transition (EMT) via downregulation of E-cadherin and consequent increases in ZEB proteins (40–42). MiR-200 targets ETS1, and loss of miR-200–mediated repression of ETS1 under hypoxic conditions leads to angiogenic responses in cancer cells (43). We have demonstrated that miR-200 influences angiogenesis indirectly via downregulation of CXCL1 and IL-8, which are major cytokines in the tumor microenvironment (44). Overall, miR-200 acts as a master regulator of several cancer cell signaling pathways, and targeting this miRNA could be an important strategy for cancer treatment.
Members of the Let-7 family can also regulate cancer stem cells by targeting HRAS and HMGA2 (27, 45). Additional roles of Let-7 relate to cell proliferation and regulation of several cell cycle regulators (46). In silico and tumor sample analyses have shown that a master regulatory network of miRNAs is involved in the mesenchymal phenotype of cancer cells (47). Some of the key miRNAs identified were miR-506, miR-200, and miR-25 (47). These networks can lead to increased tumor metastasis. Specifically, miR-506 was shown to target SNAI2, an EMT-promoting protein, and overexpression of miR-506 in cancer cells resulted in decreased tumor metastasis. MiR-10b is another important miRNA induced by TWIST1, which targets HOXD10, leading to increased RHOC protein levels and, subsequently, increased metastasis (18).
Oncogenic miRNAs in tumor progression
Oncogenic miRNAs targeting key tumor suppressor genes have been reported. Considering the significant downregulation in miRNAs due to defective miRNA biogenesis, it is of great interest to understand how these oncogenic miRNAs are increased in cancer. Two of the major oncogenic miRNAs reported are the miR-17-92 cluster (48, 49) and miR-21 (50–52). Noncoding RNA C13orf25 encodes the miR-17-92 cluster and is known to be upregulated in several cancers (48). Amplification of the 13q31-32 locus is attributed to increased expression of this noncoding RNA region, resulting in increased expression of miRNAs in the cluster. Two of the miRNAs in this cluster, miR-17 and miR-20a, target E2F1, a cell cycle regulator involved in cell division and apoptosis (49). In breast cancer, miR-21 showed a significant increase in expression and was correlated with poor patient survival. PDCD4, a protein with a role in promotion of cellular apoptosis was the prime target of miR-21 in breast and colon cancers, leading to increased tumor growth (50, 51). One of the main questions concerning oncogenic miRNAs is how does the expression of a select set of oncogenic miRNAs remain at an elevated level despite the decreases in miRNA biogenesis in cancer cells. One answer could be selective processing of miRNAs by binding to RNA binding proteins such as KSRP (53). Some miRNAs (e.g. miR-21) bind to KSRP and the entire pre-miRNA–KSRP complex gets loaded into RISC at higher affinity, leading to increased processing (53). Recently, hypoxia was found to result in downregulation of miRNAs in cancer cells via decreased DROSHA and DICER (11, 14, 15). Hypoxia in the tumor is a dynamic process, and it is possible that miRNAs which are oncogenic in nature are processed in the normoxia phase of the tumor and diffused into other areas to regulate gene expression and promote tumor growth. Alternatively, another potential explanation for increased oncogenic miRNAs in cancer is via transcriptional increase at precursor levels. Considering that miRNA biogenesis is decreased in cancer, but not completely lost, it is possible that the high input of precursor miRNAs to the remaining miRNA processing enzymes would result in expression of these miRNAs. However, these two theories will require further investigation.
Collectively, studies suggest that a complex interplay of miRNAs and their corresponding targets in cancer lead to augmented cancer growth or metastasis. The ability of miRNAs to target multiple genes provides an opportunity to interrupt this oncogenic network; this concept is discussed further in Part III.
PART II: Tumor microenvironmental factors influence tumor progression through miRNA deregulation
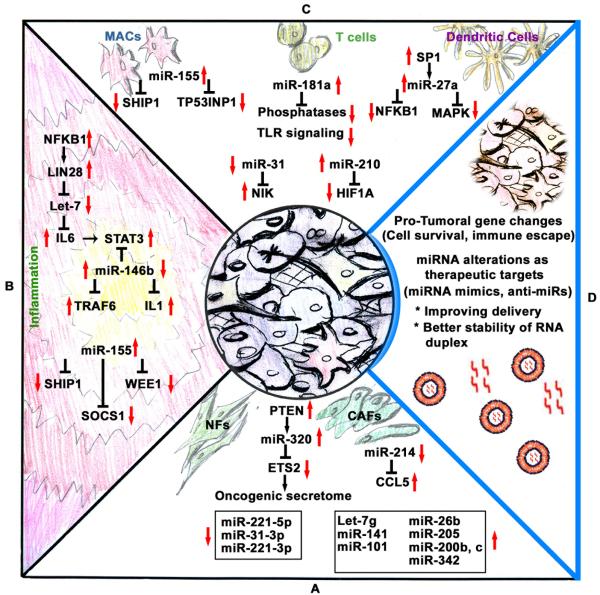
Tumor growth and metastasis are highly dependent on interactions between the tumor and the associated microenvironment. For every step in tumor growth and metastasis, intricate molecular interactions occur among tumor microenvironmental cells, such as fibroblasts and immune-related cells. Additional factors associated with cancer cells, such as hypoxia, as well as tumor-derived factors such as cytokines also influence the tumor microenvironment. Conversely, proteins secreted from cells in the tumor microenvironment can influence cancer cells. Several miRNAs have been shown to play critical roles in the interactions between the tumor and the tumor microenvironment (Figure 2). In the following sections, we summarize and discuss the potential of these findings to inform a better understanding of processes involved in cancer growth and metastasis and opportunities for innovative clinical interventions.
Figure 2. Illustration of cancer cells and tumor microenvironment-deregulated microRNA target networks leading to tumor growth and progression.
Panel A, MiRNAs play a very important role in the transformation of normal fibroblasts (NFs) to cancer-associated fibroblasts (CAFs). For example, miR-320 targets ETS2 and controls oncogenic secretome secretion. This oncogenic secretome converts NFs to CAFs in the tumor microenvironment, leading to increased tumor growth via inflammation. Panel B, Inflammation in the tumor microenvironment results in alterations in several key miRNAs, such as Let-7 and miR-155, which target a multitude of mRNAs that are involved in pro-inflammatory signaling. Panel C, Macrophages (MACs), T cells, and dendritic cells, all of which are important immune cells found in the tumor microenvironment, deregulate miRNAs that promote tumor growth. Panel D, Key challenges in developing miRNA therapeutics include developing novel tumor targeting nanoparticle delivery systems and better stable miRNA mimics or anti-miRs.
MiRNAs and cancer-associated fibroblasts (CAFs)
CAFs are known to influence tumor growth by modulating inflammation or direct cell-to-cell communication. In addition, fibroblasts provide a stromal framework to cancer cells during early growth and development, leading to malignant transformation. These extensive roles have been reviewed elsewhere (54, 55). One of the early studies involving the role of miRNAs in the transformation of normal fibroblasts to CAFs focused on Pten-regulated miR-320 (56) (Figure 2, Panel A). Using a Pten knockout mouse model, the authors found that loss of Pten in stromal fibroblasts resulted in activation of an oncogenic secretome (56). Downregulation of miR-320 and upregulation of one of its direct targets, ETS2, with loss of PTEN is a key event in oncogenic secretome signaling, leading to increased angiogenesis and tumor formation (56). MiR-320 was found to regulate CAF-secreted proteins, including MMP9, MMP2, LOXL2, and EMILIN2, which are known to enhance tumor metastasis by programming the tumor microenvironment via degradation of extracellular matrices.
A miRNA array-based analysis comparing normal fibroblasts with CAFs identified a miRNA signature of CAFs: 3 miRNAs were upregulated (miR-221-5p, miR-31-3p, and miR-221-3p) and 8 miRNAs were downregulated (miR-205, miR-200b, miR-200c, miR-141, miR-101, miR-342-3p, Let-7g, and miR-26b) in CAFs compared with normal fibroblasts (57) (Figure 2, Panel A). Many of these miRNAs are either functionally oncogenic (upregulated miRNAs) or tumor suppressors (downregulated miRNAs). However, the role played by these miRNAs in fibroblasts is not well understood. One could speculate that these miRNAs alter the chemokines secreted by fibroblasts (e.g., miR-320) or could alter fibroblast phenotypes to change tumor stromal compartments to facilitate migration and invasion. In CAFs isolated from ovarian cancer samples, 2 other miRNAs, miR-31 and miR-214, were downregulated and miR-155 was upregulated (58). Expression of miR-155 in normal fibroblasts resulted in the conversion of the fibroblasts to a CAF-like phenotype (58). In addition, the authors identified CCL5, an important chemokine in the tumor microenvironment, as a target of miR-214, which is downregulated in CAFs (58). These data support the idea that miRNAs in fibroblasts could alter the tumor microenvironment by changing proteins such as chemokines to have a pro-growth phenotype.
Tumor-related inflammation, immune cells, and miRNAs
Inflammation plays a pivotal role in the development and progression of cancer through modulation of immune cells, cytokines, and angiogenesis (59). Considering the role of miRNAs in modulating genes related to inflammation, such as those regulating cytokines (60), it is not surprising that miRNAs can influence tumor inflammation, leading to pro-growth features. For example, Let-7 is reported to be involved in an epigenetic switch, leading to tumor transformation (61) (Figure 2, Panel B). Increased transcriptional activity of LIN28 leads to reduced Let-7 and de-repression of the IL-6 cascade involving STAT3, leading to transformation of normal cells into cancer cells owing to significantly increased inflammation (61). Interestingly, this functions as a positive loop because IL-6 can activate NF-κB. IL-6 signaling-mediated STAT3 activation is not limited to Let-7 miRNA regulation alone. In a breast cancer model, increased STAT3 signaling was observed and loss of miR-146b owing to methylation in the promoter region was reported (26). Members of the miR-146 family are reported to be elevated in a NF-κB–dependent manner, regulating innate immune responses (62). These complex signaling networks are highlighted in Figure 2, Panel B. MiR-146b targets the tumor necrosis factor receptor–associated factor 6 and IL-1 receptor–associated kinase 1 genes, which are involved in Toll-like receptor cytokine signaling (62). Inhibition of this signaling could be an important step for cancer cells to interfere with the immune response in severe inflammatory settings during cancer initiation and development.
Another important miRNA involved in the modulation of immune responses and inflammation is miR-155. Multiple studies have reported that miR-155 promotes growth in several types of cancer, including breast and lung cancers. Oncogenic miR-155 downregulates SHIP1, an important modulator of immune responses, which is involved in activation of AKT signaling during the cellular response to lipopolysaccharide (63) (Figure 2, Panel B). The role of miR-155 in targeting WEE1, an important cell cycle regulator involved in DNA damage response during inflammation and during cancer development, has also been reported (64). A tight link between miR-155 levels and DNA damage leading to increased mutation rates under inflammatory conditions has been suggested (64). MiR-155 deficiency leads to accumulation of Socs-1, causing defective cytokine signaling through Stat5 (65). Using mouse models, Dudda et al. demonstrated that enforced Socs-1 silencing augmented tumor destruction (65).
In addition to miRNAs deregulating key cytokines and inflammatory responses, leading to modulation of immune responses, a direct role of miRNAs in immune cells such as T cells and B cells has been reported. MiR-181a expression in mature T cells increases the sensitivity of T cells to antigens, and inhibition of miR-181a results in impaired selection of antigens (66) (Figure 2, Panel C). Downregulation of multiple phosphatases by miR-181a leads to a reduction of T cell receptor signaling (66). This is highly relevant to cancer development, considering that high CD8+ T cell influx is observed during inflammation and cancer development.
In adult T cell leukemia, constitutively active NF-κB signaling is reported to have a causative role in cancer development (67). MiR-31 is lost in adult T cell leukemia and negatively regulates the NF-κB pathway by directly targeting NF-κB–inducing kinase, leading to apoptosis resistance (67) (Figure 2, Panel C). In addition, hypoxia-upregulated miR-210 (68) acts in a feedback loop to regulate HIF1-α, a key regulator of the transcription of genes related to TH17 polarization (69). Thus, miR-210 may act as an important regulator under disease conditions involving hypoxia to modulate immune responses to cancer antigens, although further investigation is needed to clearly define the multiple roles of miR-210.
Another detailed study of the role of miRNAs in B cell–related lymphoma development focused on the role of miR-21 as an oncogenic miRNA (23). Overexpression of miR-21 led to a pre–B cell malignant lymphoid-like phenotype, and when miR-21 was inactivated, the tumors regressed owing to apoptosis (23). Increased expression of miR-21 in CAFs was found to be induced by SMAD7/TGF-beta signaling (70), which resulted in increased inflammation. MiRNAs can activate Toll-like receptor signaling by acting as ligands. Consistent with this function, miR-21 and miR-29a have been found to be secreted from cancer cells and bind to murine Tlr7 and human TLR8, suggesting a role for these miRNAs as ligands for protein molecules (71). This leads to an activated inflammatory response in the tumor microenvironment, contributing to aggressive tumor behavior.
Tumor-associated macrophages are a key component of the tumor microenvironment and are known to promote an inflammatory network to modulate immune responses. MiR-155 and miR-342-5p have emerged as important regulators of inflammatory responses (25). MiR-342-5p directly targets AKT1 and increases levels of pro-inflammatory mediators such as NOS2 and IL-6 in macrophages via upregulation of miR-155. Although these findings were related to atherosclerosis, they are highly relevant to cancer and inflammation in the tumor microenvironment because inflammation can drive malignant transformation. In a study of primary murine macrophages, O'Connell et al. found that after the macrophages' exposure to inflammation stimulants, miR-155 levels were significantly increased via Toll-like receptor ligands through myeloid differentiation factor 88 or TRIF-dependent pathways (72). Later, the same group identified inositol phosphatase SHIP1 as a primary target of miR-155. Comparing Ship1 levels between LPS-treated wild-type and miR-155−/− primary macrophages, the authors demonstrated that Ship1 is repressed by physiologically regulated miR-155 (63) (Figure 2, Panel C). MiR-511 also modulates genetic programming of tumor-associated macrophages. Restoration of miR-511 led to a decreased pro-tumoral gene signature in tumor-associated macrophages, as well as reduced tumor growth (73).
In addition to macrophages, dendritic cells can influence tumor growth. For example, dendritic cell signaling via SP1 transcription factor–mediated increased expression of miR-27a can lead to altered NF-κ B and MAPK activity (74) (Figure 2, Panel C). As a result of the hampered cytokine signaling, increased levels of miR-27a led to decreased dendritic cell–mediated differentiation of Th1 and Th17 cells and increased tumor growth in vitro and in vivo (74).
Role of hypoxia in miRNA biogenesis
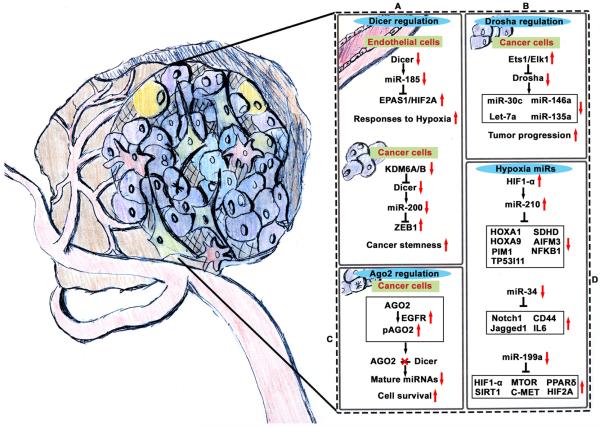
Hypoxia is common in the tumor microenvironment and can influence tumor progression by altering cancer and host cell interactions and molecular signaling. During tumor growth and metastasis, cancer cells encounter significant amounts of hypoxia owing to improperly developed and tortuous blood vessels. Key contributions of hypoxia to cancer progression, with an emphasis on protein signaling and clinical implications, are highlighted elsewhere (75, 76). In human endothelial cells, DICER-dependent miR-185 was found to be decreased under chronic hypoxic conditions, and this resulted in increased HIF2-α (Figure 3, Panel A); however, biological endpoints have yet to be defined in this setting (11). Interestingly, suppression of angiogenesis after complete loss of Dicer has been reported (77). In tumors from Dicer−/− mice, a significant increase in hypoxia was found to be caused by reduced angiogenesis resulting from de-repression of a HIF1-inhibiting factor, Fih1 (77).
Figure 3. Tumoral hypoxia functions as a master regulator of microRNAs.
Panel A, Hypoxia leads to decreased DICER expression in a HIF-dependent manner in endothelial cells and via methylation of DICER in cancer cells. Panel B, DROSHA is downregulated under hypoxic conditions by 2 transcription factors, ETS1 and ELK1, which bind to the DROSHA promoter region. This binding results in downregulation of DROSHA expression through promoter methylation. Panel C, AGO2, an important enzyme component of the RNA-induced silencing complex, is functionally downregulated via phosphorylation by EGFR under hypoxic conditions in cancer cells. The downregulation of these 3 key biogenesis components under hypoxic conditions results in various gene changes important for cancer cell survival and tumor metastasis. Panel D, Several miRNAs are regulated by hypoxia through mechanisms unrelated to biogenesis. For example, miR-210 is upregulated by the HIF1-α transcription factor and is involved in several hypoxia cancer cell signaling pathways. Also, miR-34 and miR-199a are significantly downregulated under hypoxic conditions, leading to altered prometastatic signaling.
In patients with breast cancer, tumor hypoxia is associated with reduced DICER expression (14). The underlying mechanism of hypoxia was found to be related to inhibition of oxygen-dependent H3K27me3 demethylase KDM6A/B, which resulted in increased DICER promoter methylation, leading to downregulation of DICER under hypoxic conditions (15) (Figure 3, Panel A). Functionally, this leads to decreased processing of the miR-200 family, resulting in EMT and associated stem cell phenotypes (14). In a parallel study, a significant reduction in miRNA biogenesis was found to occur as a result of decreased DROSHA and DICER in ovarian and breast cancers. ETS1 and ELK1 mediate DROSHA promoter methylation under hypoxic conditions, resulting in decreased expression of DROSHA (14) (Figure 3, Panel B). This decrease in DROSHA (via ETS1/ELK1) and DICER (via KDM6A/B) results in a global decrease in mature miRNAs. Cells under hypoxic conditions showed consistent upregulation of the pro-metastatic genes RHOB1, TAGLN, SRTAD1, TXNIP, JAG1, CTGF, and JUN, owing to downregulation of corresponding miRNAs Let-7a, miR-135a, miR-146a, and miR-30c (14) (Figure 3, Panel B).
Another important protein in the miRNA biogenesis pathway is AGO2, which is part of the RNA-induced silencing complex. In cancer cells, under hypoxic conditions, EGFR phosphorylates AGO2 at Tyr 393, resulting in decreased AGO2 function (12) (Figure 3, Panel C). AGO2 phosphorylation was found to result in decreased DICER-AGO2 interaction, leading to decreased miRNA maturation and function (12). However, another study reported that AGO2 protein levels were increased owing to post-translational changes in hydroxylation under hypoxic conditions (78). Collectively, hypoxia plays a multi-faceted role in deregulating miRNAs, leading to tumor progression (Figure 3).
Functional implications of hypoxia-deregulated miRNAs
By comparing breast cancer cell lines cultured under normoxic and hypoxic conditions, Kulshreshtha et al. identified a miRNA signature of hypoxia (68). One of the miRNAs in this group was miR-210, a transcriptional target of HIF1-α (16) (Figure 3, Panel D). AGO2 immunoprecipitation and RNA sequencing analysis has identified more than 50 potential gene targets of miR-210, and these targets are involved in the response to hypoxia, which improves cell survival. In orthotopic mouse models of head and neck and pancreatic cancers, loss of miR-210 resulted in decreased tumor initiation or growth (16). The role of miR-210 in mitochondrial dysfunction under hypoxic conditions has also been reported. MiR-210 was identified as one of the highly upregulated miRNAs in samples from patients with advanced-stage lung cancer (79).
Microarray-based mRNA pathway analyses have suggested that cell lines with increased miR-210 have increased apoptosis. However, target analysis showed that miR-210 targets SDHD, leading to stabilization of HIF1-α and cell survival under hypoxic conditions (79). Another study showed that miR-210 plays a cytoprotective role by targeting apoptosis-inducing factor mitochondrion-associated 3 (AIFM3), known to induce cell death (24) (Figure 3, Panel D). Negative regulation of NF-κ B in murine macrophages by miR-210, resulting in decreased cytokines, suggests that the role of miR-210 is not limited to cancer cell signaling (80). Increased miR-210 levels in the placenta result in decreased IL6/STAT signaling (81). MiR-210 is also involved in TH17 differentiation. HIF1A is reported to be a target of miR-210 in T cells, and under hypoxic conditions, deletion of miR-210 promoted TH17 differentiation (69). TH17 differentiation could lead to either pro- or antitumor effects; thus, the role of miR-210 in TH17 differentiation under hypoxic conditions is an important question to be answered.
MiR-34 is downregulated under hypoxic conditions and influences cancer cells and the tumor microenvironment (57). NOTCH1 and JAG1 are targets of miR-34a, and transfection of cells with miR-34a was found to result in reversal of EMT (57). In prostate cancer, miR-34 was found to be involved in cancer stem cell signaling by directly targeting CD44 (82). In colorectal cancer, downregulation of miR-34 resulted in increased IL6 signaling, leading to EMT and cancer metastasis (83). Altogether, these data suggest that miR-34 is part of the mechanism that leads to a hypoxia-induced increase in cancer metastasis (Figure 3, Panel D).
Another miRNA proven to play an important role in cell response to hypoxia is miR-199a. Targeting of MTOR and c-MET by miR-199a resulted in increased sensitivity to doxorubicin (84) (Figure 3, Panel D). Targeting of PPARδ by miR-199a in the setting of cardiac hypoxia resulted in a metabolic shift toward glycolysis. Mice treated with antagomir-199a displayed improved cardiac function and restored mitochondrial fatty acid oxidation (85). Although that study was not conducted with a cancer mouse model, its findings demonstrate the importance of miR-199a in modulating hypoxia metabolism. Recently, the role of miR-199 in the regulation of HIF-1α and HIF-2α signaling in ovarian cancer has been reported (86) (Figure 3, Panel D). Decreased miR-199a expression under hypoxic conditions resulted in increased HIF levels. Exogenous expression of miR-199a decreased HIF levels, cell migration, and ovarian cancer metastasis (86).
Part III: Therapeutic targeting of miRNA deregulation
MiRNAs have a unique advantage for targeted therapy because single miRNAs can target multiple genes. As highlighted in earlier reviews, miRNA or siRNA delivery to tumors is an attractive, yet challenging opportunity for improving therapy for cancer (87–90). Some of the major challenges and current advances are highlighted below.
Finding the right target
One of the early miRNA therapeutic strategies that showed significant impact on tumor growth was the delivery of miR-34a and Let-7 in lung cancer models (91). In this study, effective delivery of miRNA mimics miR-34a and Let-7 was demonstrated in orthotopic models of non-small cell lung cancer. Encouraging results from preclinical studies involving miR-34a in several types of cancers have increased efforts to move miR-34a delivery as a therapeutic strategy into clinical trials (92). Delivery of miR-200 in ovarian, lung, breast, and renal cancer preclinical models significantly reduced tumor metastasis and angiogenesis and induced vascular normalization by targeting IL-8 and CXCL1 (44). Combining miRNA with siRNA is another attractive approach that may allow a “boosting” effect for targeting oncogenic pathways. Combined systemic delivery of miR-520d-3p with EPHA2 siRNA resulted in robust antitumor effects (93). In this study, miR-520d was shown to target EPHA2 and EPHB2, and combining the miR-520d replacement with siRNA-mediated depletion of EPHA2 resulted in synergistic effects on reducing tumor growth. These and other studies demonstrate the use of miRNA mimics to replenish the lost miRNAs as a viable option for cancer therapy.
Designing a delivery system for miRNAs
One of the major challenges in developing miRNA therapeutics is the high vulnerability of RNA molecules to nucleases. Hence, design of novel nanoparticle platforms is needed to allow intracellular delivery with minimal toxicity while providing protection to RNA molecules from nucleases. Several lipid-based carriers (91, 94) have been developed and tested in preclinical models, and some are in clinical trials. For example, MRX34, a lipid-based nanoparticle–miR-34 system, is currently in phase I clinical testing and has shown great promise (92). Another approach is use of neutral liposomal particle 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (94). This delivery platform has already been used for miR-200 (44), miR-520d (93), and miR-506 (47) in several cancer types in preclinical models. Use of DOPC for delivery of siRNA against EPHA2 is currently in phase I clinical trials. Completely unrelated to cancer, use of miRNAs as therapy has been employed against hepatitis C virus infections (95). Inhibition of miR-122 by locked nucleic acid–based inhibitor resulted in a significant reduction in hepatitis C virus RNA in patients, as reported in a phase 2a clinical trial (NCT01200420). Apart from these clinical delivery platforms, several novel siRNA or miRNA delivery systems are under development. One such effort includes spherical nucleic acid nanoparticle conjugates with a gold nanoparticle core. Using this system, siRNA against EGFR was successfully delivered to skin and showed significant reduction in EGFR levels (96). In glioblastoma models, delivery of siRNAs against Bcl2L12 resulted in a significant reduction in tumor growth, and these nanoparticles were able to cross the blood-brain barrier, providing a potentially significant advance in treating brain cancers (97). In colon cancer xenograft mouse model, delivery of miR-145 and miR-33a using Polyethylenimine (PEI) particles resulted in significant reduction in tumor growth (98). Use of adenovirus in delivering miRNAs is another approach. Using this approach, investigators delivered miR-26a to liver tumors; the study showed a significant reduction in tumor growth with restored miR-26a expression (99). If the results from the above approaches continue to show success and enter into the clinic, use of noncoding RNAs as therapeutics could emerge as a key technology for treatment of cancer and other diseases (Figure 2, Panel D).
Conclusion
In summary, we have highlighted recent advances in the understanding of tumor microenvironmental interactions mediated by miRNAs. As highlighted in Figures 2 and 3, several miRNAs target important cancer cell regulatory molecules and are involved in a complex network of signaling between cancer cells and the tumor microenvironment. In addition to their involvement in direct cell-to-cell signaling, several miRNAs are secreted through microvesicles or exosomes and affect cancer cell growth and metastasis. All of these microenvironmental changes are suggestive of a complex signaling network between tumor cells and stromal components and conditions such as hypoxia, CAFs, and endothelial cells (Figures 2 and 3). Some of the current challenges in RNAi and miRNA therapeutics involve selecting the right target and optimizing the delivery systems. Advances in RNAi and miRNA therapeutics have enabled us to target miRNA alterations in a highly specific and robust manner in preclinical models. Nevertheless, studies of miRNA-mediated interactions, specifically those focused on understanding the origin of miRNA alterations, are needed to improve targeted therapy.
Statement of significance.
MicroRNAs play a central role in cell signaling and homeostasis. In this article, we provide insights into the regulatory mechanisms involved in the deregulation of miRNAs in cancer cells and the tumor microenvironment and discuss therapeutic intervention strategies to overcome this deregulation.
Acknowledgments
The authors apologize to their many colleagues whose work could not be cited owing to space constraints. We would like to thank Dr. Rebecca Previs for helping with illustration work. The authors thank Ms. Erica Goodoff (Department of Scientific Publications, The University of Texas MD Anderson Cancer Center) for the English editing of this manuscript.
Funding support: Work in this manuscript is supported, in part, by the National Institutes of Health (CA016672, CA109298, UH3TR000943, P50 CA083639, P50 CA098258, CA182905, P50 CA100632, P50 CA097007, P50 CA127001, U54 CA151668 and U24 CA143835), Cancer Prevention and Research Institute of Texas (RP110595), Knowledge GAP MD Anderson grant, Ovarian Cancer Research Fund, Inc. (Program Project Development Grant), Department of Defense (OC073399, W81XWH-10-1-0158), Marcus Foundation, Red and Charline McCombs Institute for the Early Detection and Treatment of Cancer, RGK Foundation, the Duncan Family Institute for Cancer Prevention and Risk Assessment, the Gilder Foundation, Blanton-Davis Ovarian Cancer Research Program, the Betty Anne Asche Murray Distinguished Professorship (A.K.S.), and Alan M. Gewirtz Leukemia & Lymphoma Society Scholar award (G.A.C). R.R. is supported in part by the Russell and Diana Hawkins Family Foundation Discovery Fellowship.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Esteller M. Non-coding RNAs in human disease. Nature reviews Genetics. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Spizzo R, Nicoloso MS, Croce CM, Calin GA. SnapShot: MicroRNAs in Cancer. Cell. 2009;137:586–e1. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 5.Boehm M, Slack FJ. MicroRNA control of lifespan and metabolism. Cell cycle (Georgetown, Tex. 2006;5:837–40. doi: 10.4161/cc.5.8.2688. [DOI] [PubMed] [Google Scholar]
- 6.Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Current Opinion in Genetics & Development. 2005;15:200–5. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer science. 2005;96:111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. The New England journal of medicine. 2008;359:2641–50. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annual review of medicine. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 11.Ho JJ, Metcalf JL, Yan MS, Turgeon PJ, Wang JJ, Chalsev M, et al. Functional importance of Dicer protein in the adaptive cellular response to hypoxia. The Journal of biological chemistry. 2012;287:29003–20. doi: 10.1074/jbc.M112.373365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–7. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Lai TC, Jan YH, Lin FM, Wang WC, Xiao H, et al. Hypoxia-responsive miRNAs target argonaute 1 to promote angiogenesis. The Journal of clinical investigation. 2013;123:1057–67. doi: 10.1172/JCI65344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rupaimoole R, Wu SY, Pradeep S, Ivan C, Pecot CV, Gharpure KM, et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat Commun. 2014;5 doi: 10.1038/ncomms6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Beucken T, Koch E, Chu K, Rupaimoole R, Prickaerts P, Adriaens M, et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:5203. doi: 10.1038/ncomms6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Molecular cell. 2009;35:856–67. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews Genetics. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 18.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 19.Allegra D, Bilan V, Garding A, Dohner H, Stilgenbauer S, Kuchenbauer F, et al. Defective DROSHA processing contributes to downregulation of MiR-15/-16 in chronic lymphocytic leukemia. Leukemia. 2014;28:98–107. doi: 10.1038/leu.2013.246. [DOI] [PubMed] [Google Scholar]
- 20.Dedes KJ, Natrajan R, Lambros MB, Geyer FC, Lopez-Garcia MA, Savage K, et al. Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. European journal of cancer. 2011;47:138–50. doi: 10.1016/j.ejca.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Torres A, Torres K, Paszkowski T, Jodlowska-Jedrych B, Radomanski T, Ksiazek A, et al. Major regulators of microRNAs biogenesis Dicer and Drosha are down-regulated in endometrial cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2011;32:769–76. doi: 10.1007/s13277-011-0179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–65. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 24.Mutharasan RK, Nagpal V, Ichikawa Y, Ardehali H. microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. American journal of physiology Heart and circulatory physiology. 2011;301:H1519–30. doi: 10.1152/ajpheart.01080.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Y, Nazari-Jahantigh M, Chan L, Zhu M, Heyll K, Corbalan-Campos J, et al. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation. 2013;127:1609–19. doi: 10.1161/CIRCULATIONAHA.112.000736. [DOI] [PubMed] [Google Scholar]
- 26.Xiang M, Birkbak NJ, Vafaizadeh V, Walker SR, Yeh JE, Liu S, et al. STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-kappaB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Science signaling. 2014;7:ra11. doi: 10.1126/scisignal.2004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Zhao X, Gao P, Wu M. c-Myc modulates microRNA processing via the transcriptional regulation of Drosha. Scientific reports. 2013;3:1942. doi: 10.1038/srep01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X, Liao Q, Chen P, Li X, Xiong W, Ma J, et al. The microRNA-processing enzymes: Drosha and Dicer can predict prognosis of nasopharyngeal carcinoma. Journal of cancer research and clinical oncology. 2012;138:49–56. doi: 10.1007/s00432-011-1058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaz-Garcia CV, Agudo-Lopez A, Perez C, Lopez-Martin JA, Rodriguez-Peralto JL, de Castro J, et al. DICER1, DROSHA and miRNAs in patients with non-small cell lung cancer: implications for outcomes and histologic classification. Carcinogenesis. 2013;34:1031–8. doi: 10.1093/carcin/bgt022. [DOI] [PubMed] [Google Scholar]
- 31.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–90. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller PA, Trinidad AG, Caswell PT, Norman JC, Vousden KH. Mutant p53 regulates Dicer through p63-dependent and -independent mechanisms to promote an invasive phenotype. The Journal of biological chemistry. 2014;289:122–32. doi: 10.1074/jbc.M113.502138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 34.Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–7. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 35.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nature genetics. 2009;41:365–70. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.De Vito C, Riggi N, Cornaz S, Suva ML, Baumer K, Provero P, et al. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer cell. 2012;21:807–21. doi: 10.1016/j.ccr.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Wan G, Berger FG, He X, Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Molecular cell. 2011;41:371–83. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer cell. 2010;18:303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 41.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. The Journal of biological chemistry. 2008;283:14910–4. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & development. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. The Journal of biological chemistry. 2011;286:2047–56. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, et al. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung SY, Liao CH, Wu HP, Hsiao WC, Wu IH, Jinpu, et al. Loss of let-7 microRNA upregulates IL-6 in bone marrow-derived mesenchymal stem cells triggering a reactive stromal response to prostate cancer. PloS one. 2013;8:e71637. doi: 10.1371/journal.pone.0071637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer research. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 47.Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer cell. 2013;23:186–99. doi: 10.1016/j.ccr.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2009;28:140–5. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 51.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. The Journal of biological chemistry. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 52.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. Rna. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schauer IG, Sood AK, Mok S, Liu J. Cancer-associated fibroblasts and their putative role in potentiating the initiation and development of epithelial ovarian cancer. Neoplasia. 2011;13:393–405. doi: 10.1593/neo.101720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nature reviews Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 56.Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, et al. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320 Nature cell biology. 2012;14:159–67. doi: 10.1038/ncb2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du R, Sun W, Xia L, Zhao A, Yu Y, Zhao L, et al. Hypoxia-induced down-regulation of microRNA-34a promotes EMT by targeting the Notch signaling pathway in tubular epithelial cells. PloS one. 2012;7:e30771. doi: 10.1371/journal.pone.0030771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitra AK, Zillhardt M, Hua Y, Tiwari P, Murmann AE, Peter ME, et al. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer discovery. 2012;2:1100–8. doi: 10.1158/2159-8290.CD-12-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tili E, Michaille JJ, Croce CM. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunological reviews. 2013;253:167–84. doi: 10.1111/imr.12050. [DOI] [PubMed] [Google Scholar]
- 61.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7113–8. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tili E, Michaille JJ, Wernicke D, Alder H, Costinean S, Volinia S, et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4908–13. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dudda JC, Salaun B, Ji Y, Palmer DC, Monnot GC, Merck E, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38:742–53. doi: 10.1016/j.immuni.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–61. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Yamagishi M, Nakano K, Miyake A, Yamochi T, Kagami Y, Tsutsumi A, et al. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-kappaB pathway in adult T cell leukemia and other cancers. Cancer cell. 2012;21:121–35. doi: 10.1016/j.ccr.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 68.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, et al. A microRNA signature of hypoxia. Molecular and cellular biology. 2007;27:1859–67. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H, Flach H, Onizawa M, Wei L, McManus MT, Weiss A. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nature immunology. 2014;15:393–401. doi: 10.1038/ni.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Q, Zhang D, Wang Y, Sun P, Hou X, Larner J, et al. MiR-21/Smad 7 signaling determines TGF-beta1-induced CAF formation. Scientific reports. 2013;3:2038. doi: 10.1038/srep02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2110–6. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Squadrito ML, Pucci F, Magri L, Moi D, Gilfillan GD, Ranghetti A, et al. miR-511-3 p modulates genetic programs of tumor-associated macrophages. Cell reports. 2012;1:141–54. doi: 10.1016/j.celrep.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 74.Min S, Li L, Zhang M, Zhang Y, Liang X, Xie Y, et al. TGF-beta-associated miR-27 a inhibits dendritic cell-mediated differentiation of Th1 and Th17 cells by TAB3, p38 MAPK, MAP2K4 and MAP2K7. Genes and immunity. 2012;13:621–31. doi: 10.1038/gene.2012.45. [DOI] [PubMed] [Google Scholar]
- 75.Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer metastasis reviews. 2007;26:225–39. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 76.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–72. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen S, Xue Y, Wu X, Le C, Bhutkar A, Bell EL, et al. Global microRNA depletion suppresses tumor angiogenesis. Genes & development. 2014;28:1054–67. doi: 10.1101/gad.239681.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu C, So J. Davis-Dusenbery BN, Qi HH, Bloch DB, Shi Y, et al. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Molecular and cellular biology. 2011;31:4760–74. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell death and differentiation. 2011;18:465–78. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qi J, Qiao Y, Wang P, Li S, Zhao W, Gao C. microRNA-210 negatively regulates LPS-induced production of proinflammatory cytokines by targeting NF-kappaB1 in murine macrophages. FEBS letters. 2012;586:1201–7. doi: 10.1016/j.febslet.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 81.Kopriva SE, Chiasson VL, Mitchell BM, Chatterjee P. TLR3-induced placental miR-210 down-regulates the STAT6/interleukin-4 pathway. PloS one. 2013;8:e67760. doi: 10.1371/journal.pone.0067760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature medicine. 2011;17:211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rokavec M, Oner MG, Li H, Jackstadt R, Jiang L, Lodygin D, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. The Journal of clinical investigation. 2014;124:1853–67. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer research. 2010;70:5184–93. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 85.el Azzouzi H, Leptidis S, Dirkx E, Hoeks J, van Bree B, Brand K, et al. The hypoxia-inducible microRNA cluster miR-199a approximately 214 targets myocardial PPARdelta and impairs mitochondrial fatty acid oxidation. Cell metabolism. 2013;18:341–54. doi: 10.1016/j.cmet.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 86.Joshi HP, Subramanian IV, Schnettler EK, Ghosh G, Rupaimoole R, Evans C, et al. Dynamin 2 along with microRNA-199a reciprocally regulate hypoxia-inducible factors and ovarian cancer metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5331–6. doi: 10.1073/pnas.1317242111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheng CJ, Saltzman WM, Slack FJ. Canonical and non-canonical barriers facing antimiR cancer therapeutics. Current medicinal chemistry. 2013;20:3582–93. doi: 10.2174/0929867311320290004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rupaimoole R, Han HD, Lopez-Berestein G, Sood AK. MicroRNA therapeutics: principles, expectations, and challenges. Chinese journal of cancer. 2011;30:368–70. doi: 10.5732/cjc.011.10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nature reviews Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nature reviews Drug discovery. 2013;12:847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Molecular therapy: the journal of the American Society of Gene Therapy. 2011;19:1116–22. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Frontiers in genetics. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nishimura M, Jung EJ, Shah MY, Lu C, Spizzo R, Shimizu M, et al. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer discovery. 2013;3:1302–15. doi: 10.1158/2159-8290.CD-13-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Landen CN, Jr., Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer research. 2005;65:6910–8. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 95.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. The New England journal of medicine. 2013;368:1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 96.Zheng D, Giljohann DA, Chen DL, Massich MD, Wang XQ, Iordanov H, et al. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11975–80. doi: 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ibrahim AF, Weirauch U, Thomas M, Grunweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer research. 2011;71:5214–24. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 99.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]