Abstract
Exosomes are membranous vesicles that are released by a variety of cells into the extracellular microenvironment and are implicated in intercellular communication. As exosomes contain RNA, proteins and lipids, there is a significant interest in characterizing the molecular cargo of exosomes. Here, we describe ExoCarta (http://www.exocarta.org), a manually curated web-based compendium of exosomal proteins, RNAs and lipids. Since its inception, the database has been highly accessed (>54,000 visitors from 135 countries). The current version of ExoCarta hosts 41,860 proteins, >7,540 RNA and 1,116 lipid molecules from more than 286 exosomal studies annotated with ISEV minimal experimental requirements for definition of extracellular vesicles. Besides, ExoCarta features dynamic protein-protein interaction networks and biological pathways of exosomal proteins. Users can download most often identified exosomal proteins based on the number of studies. The downloaded files can further be imported directly into FunRich (http://www.funrich.org) tool for additional functional enrichment and interaction network analysis.
Keywords: Exosomes, FunRich, Extracellular vesicles, Protein-protein interactions
Graphical abstract
Introduction
Exosomes are small (30-150 nm) membranous vesicles secreted by a variety of cells into the extracellular microenvironment [1, 2]. During physiological and pathological conditions, inward budding of late endosomal membranes results in the accumulation of intraluminal vesicles (ILVs) within the multivesicular bodies (MVBs) [3, 4]. Fusion of the MVBs with the plasma membrane results in the release of ILVs into the extracellular microenvironment and from here on referred to as exosomes [5]. During their biogenesis, exosomes are packaged with proteins, RNA and lipids that is reflective of the host cell [6]. As exosomes are secreted by many cell types, conserved in most organisms and transport signaling molecules between cells, they have been implicated in intercellular communication [3, 7, 8]. More importantly, exosomes are detected in bodily fluids and are known to be highly stable reservoirs of disease biomarkers [9-11] prompting numerous studies aimed at cataloging the exosomal proteins, RNAs and lipids using integrated high-throughput OMICS techniques [12]. Here, we describe ExoCarta, an online database that catalogues exosome specific data pertaining to proteins, RNAs and lipids. ExoCarta (http://www.exocarta.org) was initially described in 2009 [13] and since its launch, two more databases (Vesiclepedia [12] and EVpedia [14]) have been described. Whilst, both Vesiclepedia and EVpedia catalog data from multiple types of extracellular vesicles, ExoCarta is a primary resource of exosomal cargo [15] and contains annotations on the isolation and characterization methods. The new version of ExoCarta database has more than 2-fold data compared to the last release in 2012 and have new additional features including the annotation of International Society of Extracellular Vesicles standards and dynamic protein-protein interaction networks. Since its launch, ExoCarta has been visited by >54,000 users from more than 135 countries.
Database design and structure
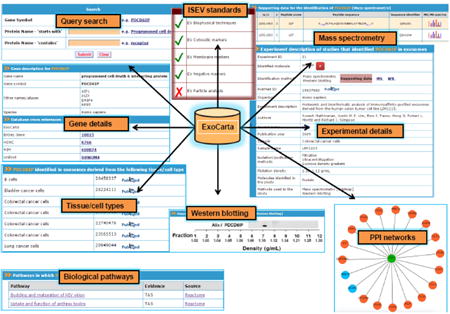
ExoCarta is developed using Zope (http://www.zope.org), a leading open source web application server written in object-oriented programming language Python. MySQL is used as a backend data storage system. The Data-Driven Document JavaScript (D3JS) module coupled with the JSON (JavaScript Object Notation) module for data exchange (http://d3js.org/) is used in developing dynamic protein-protein interaction networks. ExoCarta has a user friendly web-based graphical query system (Fig. 1) which can be used to search proteins and RNA detected in exosomes isolated from more than 10 different species. As the exosomal studies catalogued in ExoCarta are manually curated from the literature, the annotations include precise details on the tissue/cell types from which exosomes were isolated and the methods employed to isolate exosomes. Furthermore, using browse option, the users can find exosomal datasets based on content types (RNAs, proteins and lipids), species, tissue/cell types and gene symbols. To facilitate further analyses, ExoCarta allows for free download of all the exosomal datasets.
Figure 1. Snapshot of ExoCarta structure and features.
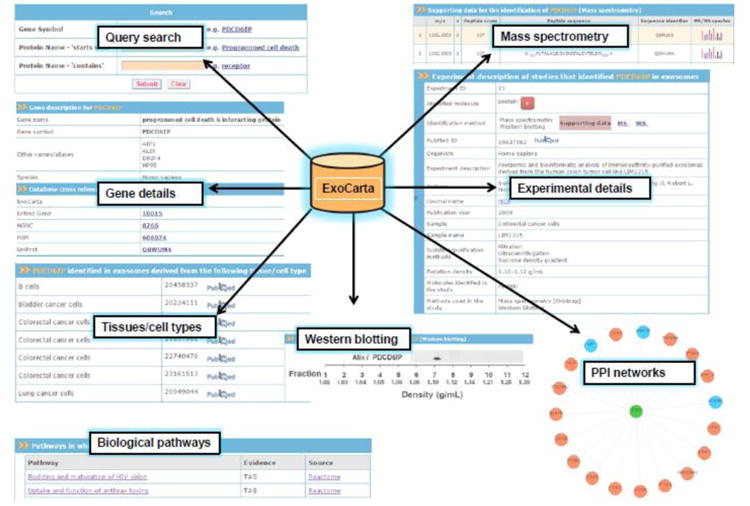
ExoCarta database query search results and different types of data including mass spectrometry, Western blotting, biological pathways and protein-protein interaction (PPI) networks pertaining to exosomal proteins are depicted. In addition, exosomes characterization in accordance to ISEV standards is also provided.
Annotation and database features
ExoCarta datasets
In the last 3 years alone, the studies on exosomes have significantly increased (Fig. 2A) giving rise to multidimensional data [1, 16]. Hence, these publicly available datasets have been manually curated by expert biologists and uploaded into the ExoCarta database. In addition, many researchers have contributed data to ExoCarta aiding in community annotation. Currently, ExoCarta hosts 41,860 protein, >7,540 RNA and 1,116 lipid entries cataloged from 10 different species.
Figure 2. Statistics of exosomal studies in last 3 years and list of top 20 most often identified exosomal proteins.
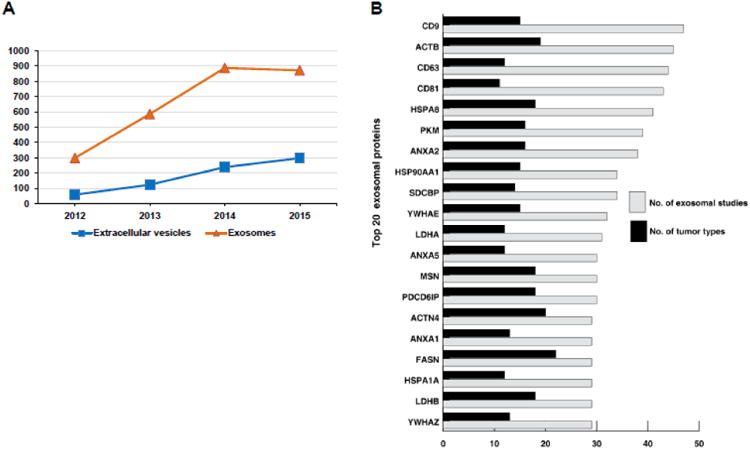
A) Statistics of exosomal studies in the last 3 years A search was performed in PubMed on 15 September 2015 to identify articles using the term exosomes and extracellular vesicles. The list was manually validated to remove non-extracellular vesicles studies. The analysis clearly shows a surge in the publication of articles in exosomes and extracellular vesicles in the last 3 years. Other terms such as microvesicles, prostasomes and ectosomes were not used in the PubMed search. B) Top 20 most often identified exosomal proteins and its involvement in number of cancer types. This figure illustrates top 20 most often identified exosomal proteins based on the number of exosomal studies. The grey bar depicts number of exosomal studies and black bar depicts different cancer types in which they are known to be mutated.
Database features
ExoCarta as a reservoir of candidate exosomal markers and mutant oncogenic proteins
Over the past decade, exosomes have gained significant interest due to their pleiotropic effects that influences the physiology of the neighboring cells. Multiple studies on exosomes from different tissue/cell types or cell lines under normal and pathophysiological conditions has alluded to the presence of exosomal enriched proteins [1, 17, 18]. To further facilitate these investigations, ExoCarta encompasses proteins detected in exosomes and hence could be exploited to identify proteins enriched in exosomes. Furthermore, proteins that are targeted for secretion via exosomes in a specific cell/tissue type can be identified using ExoCarta. For instance, mutant proteins that are present exclusively in diseased cells are known to be secreted via exosomes [11, 18]. It has been proposed that mutations in more than 1% of human genes are associated with tumorigenesis [19]. Currently, ExoCarta contains more than 20% of the proteins that are known to be mutated as per COSMIC database [20]. Among the top 20 often identified exosomal proteins, many of the proteins including CD9, ACTB, CD63, CD81, HSPA8, PKM2, ANXA2, HSP90A1, SDCBP, YWHAE, LDHA, MSN, PDCP6IP, ANXA5, FASN, ACTN4, LDHB, ANXA1, HSPA1A and YWHA are known to be mutated in multiple cancer types (Fig. 2B). Interestingly, majority of these proteins are known to be involved in exosome biogenesis, sorting and secretion. However, the precise role of these underlying mutations in exosomal biogenesis remains unexplored.
Implementation of standards for minimal requirements of exosomal studies
One of the key updated features in ExoCarta includes the implementation of annotations in accordance to the standards proposed by International Society of Extracellular Vesicles (ISEV) as the minimal requirements for exosomal studies [1]. Expert curators annotated five different features from every article in ExoCarta such as the biophysical methods (e.g., transmission electron microscopy) to characterize exosomes, the detection of cytosolic exosomal enriched proteins (e.g., Alix, TSG101), detection of membrane proteins (e.g., CD9, CD63), non-exosomal proteins (e.g., gp96) and particle analysis (e.g., nanoparticle tracking analysis). Each annotated study in ExoCarta is now also presented with yes or no for all the five criteria listed above further allowing users to quickly assess the characterization of exosomes.
Dynamic protein-protein interaction networks of exosomal proteins
A non-redundant protein-protein interaction network for all the exosomal proteins catalogued in ExoCarta was generated using HPRD [21] and BioGRID [22] databases. Using D3js and JSON modules, a highly dynamic protein-protein interaction network graphics is depicted in ExoCarta for all the exosomal proteins. Within these networks, the interacting partners already identified in exosomes are colored differently to distinguish from the rest of the nodes. Besides, users can search for particular interacting partners within the network. In addition to the functional implication of the interaction network, the interacting proteins can be utilized to understand exosomal target cell uptake. For instance, tetraspanins TSPAN8 and CD9 are implicated in regulating exosomal uptake by the target cell and hence the interacting partners of CD9 (Fig. 1) might provide clues to decipher the regulation of target cell uptake [23].
Gene Ontology and biological pathways of exosomal proteins
To further understand the role of exosomal proteins in different biological pathways and underlying functions, the exosomal proteins catalogued in ExoCarta are linked to respective pathways and Gene Ontology terms which are compiled from Reactome [24] and Gene Ontology [25] databases, respectively.
Download features and community annotation
ExoCarta datasets including proteins, RNAs and lipids can be freely downloaded at http://www.exocarta.org/download in tab delimited text file formats. Besides, users can also download the list of most often identified (100) exosomal proteins catalogued in ExoCarta. The downloaded datasets can be imported into FunRich [26], a free standalone gene set enrichment tool, in order to perform enrichment analysis. To further minimize cumbersome manual annotations and to maintain the database up-to-date, the exosomal research community can submit their datasets with minimum requirements directly to ExoCarta (http://www.exocarta.org/data_submission). The users are given credits in the ExoCarta webpage (http://www.exocarta.org/credits) for their contributions and have the freedom of making the exosomal data private until published.
Conclusions
ExoCarta is a manually curated, web-based and freely accessible database which hosts exosome specific proteins, RNAs and lipid contents from 10 different organisms. Since its launch, ExoCarta has been accessed by >54,000 visitors from 135 countries. We believe, ExoCarta would serve as a discovery resource tool and aid researchers in understanding the molecular mechanisms of exosomal biogenesis, sorting and secretion under normal and diseases conditions.
Research Highlights.
ExoCarta is a web-based compendium of exosomal proteins, RNAs and lipids
ExoCarta database is manually curated by expert biologist and also allows community annotation.
ExoCarta host protein-protein interaction networks and biological pathways of exosomal proteins
Nearly 25% of the human proteins are known to be secreted via exosomes
Acknowledgments
SM is supported by the Australian Research Council Discovery project grant (DP130100535), Australian Research Council DECRA (DE150101777) and U54-DA036134 supported by the NIH Common Fund through the Office of Strategic Coordination/Office of the NIH Director. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- COSMIC
Catalogue of Somatic Mutations in Cancer
- ILV
Intraluminal Vesicles
- MVB
Multivesicular bodies
- D3JS
Data-Driven Document Java Script
- JSON
Java Script Object Notation
- HPRD
Human Protein Reference Database
- BioGRID
Biological General Repository for Interaction Datasets
- FunRich
Functional enrichment
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. Journal of extracellular vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. Journal of proteomics. 2010;73:1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of cell biology. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gangoda L, Boukouris S, Liem M, Kalra H, Mathivanan S. Extracellular vesicles including exosomes are mediators of signal transduction: are they protective or pathogenic? Proteomics. 2015;15:260–71. doi: 10.1002/pmic.201400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nature reviews Immunology. 2009;9:581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 6.Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Molecular & cellular proteomics : MCP. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cossetti C, Iraci N, Mercer TR, Leonardi T, Alpi E, Drago D, et al. Extracellular vesicles from neural stem cells transfer IFN-gamma via Ifngr1 to activate Stat1 signaling in target cells. Molecular cell. 2014;56:193–204. doi: 10.1016/j.molcel.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samuel M, Bleackley M, Anderson M, Mathivanan S. Extracellular vesicles including exosomes in cross kingdom regulation: a viewpoint from plant-fungal interactions. Frontiers in Plant Science. 2015;6 doi: 10.3389/fpls.2015.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. Journal of extracellular vesicles. 2014:3. doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13:3354–64. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 11.Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clinical applications. 2015;9:358–67. doi: 10.1002/prca.201400114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS biology. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 14.Kim DK, Lee J, Kim SR, Choi DS, Yoon YJ, Kim JH, et al. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics. 2015;31:933–9. doi: 10.1093/bioinformatics/btu741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic acids research. 2012;40:D1241–4. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. Journal of extracellular vesicles. 2012:1. doi: 10.3402/jev.v1i0.18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minciacchi VR, You S, Spinelli C, Morley S, Zandian M, Aspuria PJ, et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6:11327–41. doi: 10.18632/oncotarget.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keerthikumar S, Gangoda L, Liem M, Fonseka P, Atukorala I, Ozcitti C, et al. Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes. Oncotarget. 2015;6:15375–96. doi: 10.18632/oncotarget.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nature reviews Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic acids research. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, et al. Human Protein Reference Database--2009 update. Nucleic acids research. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, et al. The BioGRID interaction database: 2015 update. Nucleic acids research. 2015;43:D470–8. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rana S, Yue S, Stadel D, Zoller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. The international journal of biochemistry & cell biology. 2012;44:1574–84. doi: 10.1016/j.biocel.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 24.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic acids research. 2014;42:D472–7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gene Ontology C. The Gene Ontology in 2010: extensions and refinements. Nucleic acids research. 2010;38:D331–5. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]


