Abstract
Iron is necessary for life, but can also cause cell death. Accordingly, cells evolved a robust, tightly regulated suite of genes for maintaining iron homeostasis. Previous mechanistic studies on iron homeostasis have granted insight into the role of iron in human health and disease. We highlight new regulators of iron metabolism, including iron-trafficking proteins [solute carrier family 39, SLC39, also known as ZRT/IRT-like protein, ZIP; and poly-(rC)-binding protein, PCBP] and a cargo receptor (NCOA4) that is crucial for release of ferritin-bound iron. We also discuss emerging roles of iron in apoptosis and a novel iron-dependent cell death pathway termed ‘ferroptosis’, the dysregulation of iron metabolism in human pathologies, and the use of iron chelators in cancer therapy.
Iron Homeostasis Is a Complex, Highly Regulated Process
Iron is required in a variety of important biological processes including oxygen transport (as heme in hemoglobin), DNA biosynthesis (as a cofactor of ribonucleotide reductase), and ATP generation (as a cofactor for many proteins in the citric acid cycle and electron transport chain); therefore, cells must maintain a sufficient amount of iron. However, iron is redox-active and can generate reactive oxygen species (ROS), leading to oxidative stress and initiation of signaling pathways crucial for cell survival and cell death [1]. To maintain adequate and safe amounts of iron, cells require the coordination of a wide variety of genes, which tightly control both intracellular (reviewed in [2,3]) and systemic (reviewed in [4]) iron metabolism. Extensive research by many groups has revealed key mechanisms in iron homeostasis (Box 1), as well as links between aberrations in iron homeostasis and human disease. The study of iron metabolism continues to be a dynamic field, with many breakthroughs and novel insights in the past several years. In this review we discuss recent advances in the function and regulation of key iron metabolism genes, including: ferritin (FTH1 and FTL), a protein complex that safely concentrates intracellular iron in a mineralized, redox-inactive form for later use; transferrin (TF), an iron-binding serum protein; transferrin receptor 1 (TfR1, TFRC), a plasma membrane protein that allows cellular uptake of iron-loaded transferrin; divalent metal transporter 1 (DMT1, SLC11A2), a metal transporter that is important for TfR1-mediated iron uptake and dietary iron absorption; ferroportin (Fpn, SLC40A1), the only known cellular iron efflux pump; and hepcidin (HAMP), a circulating peptide hormone that regulates serum iron levels by causing ferroportin degradation. We also examine newly identified regulators in iron metabolism, including new membrane iron transporters and cytosolic iron trafficking proteins. In addition, we review new roles for iron in cell death pathways, as well as the importance of aberrant iron metabolism in human diseases such as cancer.
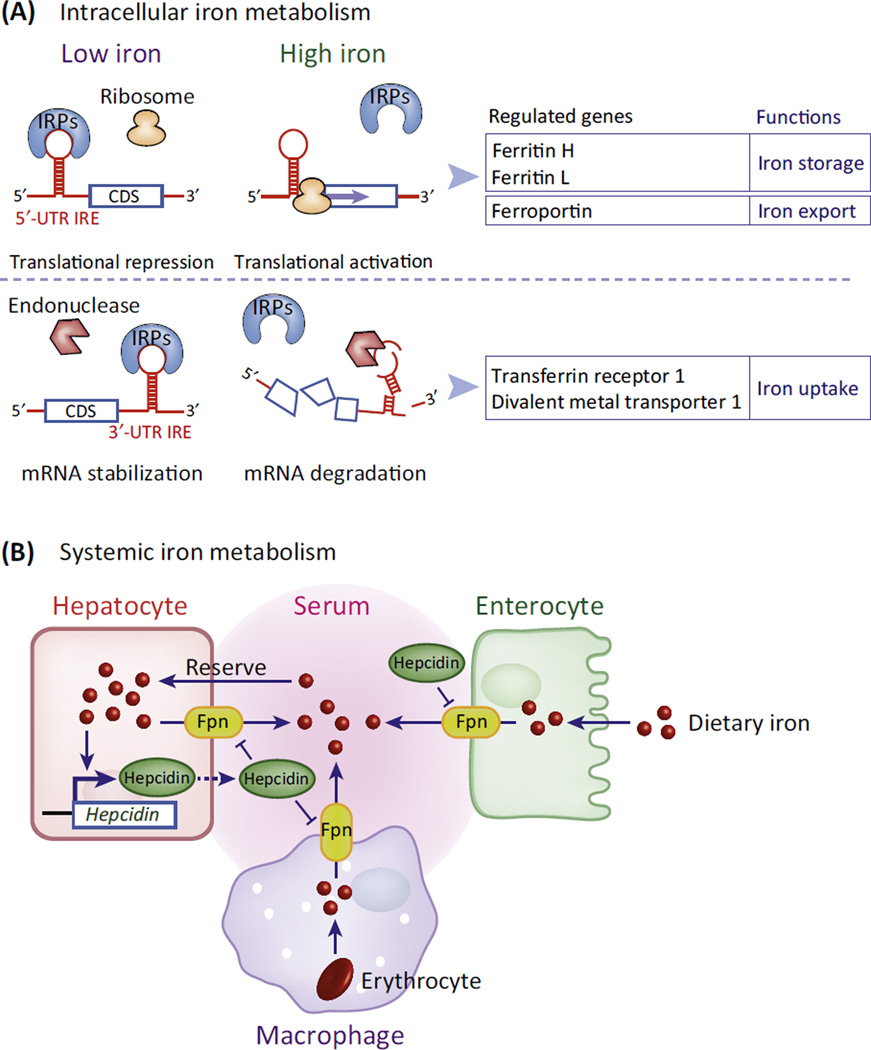
Box 1. Control of Mammalian Iron Metabolism Occurs through Two Distinct, but Connected, Regulatory Systems.
Intracellular iron metabolism is primarily controlled through coordinated post-transcriptional regulation of various iron metabolism genes (reviewed in [2,3]). Many mRNAs involved in iron metabolism contain iron-responsive elements (IREs): stem-loop structures located in 5′ - or 3′ -untranslated regions (UTRs) flanking the coding sequence (CDS) [76]. IREs bind two functionally similar iron regulatory proteins, IRP1 and IRP2. Depending upon whether the IRE is located within the 5′-UTR or in the 3′-UTR, the IRE–IRP interaction has opposite effects on target gene expression. In low iron conditions, 5′-UTR IREs are translationally repressed as a result of IRP blocking ribosome recruitment, whereas 3′-UTR IREs mediate enhanced mRNA stability, ultimately increasing protein levels (Figure IA, Low iron). High iron removes the IRPs, facilitating translational activation of 5′-UTR IRE mRNAs or degradation of 3′-UTR IRE mRNAs via endonuclease attack (Figure IA, High iron). 5′-UTR IREs are usually found in genes that lower the amount of cellular labile iron (i.e., iron that is unbound and redox active) such as ferritin and ferroportin, whereas 3′-UTR IREs are found in genes that facilitate iron uptake such as transferrin receptor 1 and divalent metal transporter 1 (Figure IA) Classically, it was believed that iron affected the IRE–IRP interaction entirely through effects on the IRPs. However, the ability of Fe2+ (and other metal ions) to directly bind to the IRE stem-loop was recently observed [77]. Fe2+ binding to 5′-UTR IREs induces a conformational change in the stem-loop, which decreases its affinity for IRPs [78] and increases its affinity for a ribosome recruitment factor eIF4F [79], showing that the IRE structure itself contributes to post-transcriptional control of gene expression.
In mammals, systemic iron homeostasis is controlled by the hepatocyte-secreted hormone hepcidin (reviewed in [4]). Hepcidin circulates in the serum and binds to the cellular iron exporter ferroportin (Fpn), stimulating Fpn degradation and leading to cellular retention of iron (Figure IB). Hepcidin expression is directly correlated with both cellular and serum iron statuses and is controlled through a complex iron-sensing signaling pathway (reviewed in [4]). Elevated serum hepcidin downregulates Fpn in duodenal enterocytes (which are responsible for dietary iron absorption), macrophages (which contain large amounts of iron from erythrocyte recycling), and hepatocytes (which act as an iron reservoir and export iron as needed). This leads to an overall reduction in serum iron (Figure IB). Some overlap exists between the IRE–IRP system and the hepcidin–Fpn axis (reviewed in [3]) because Fpn is regulated by IRPs though its 5′-UTR IRE.
Figure I. Mechanisms of Intracellular and Systemic Iron Homeostasis.
(A) Intracellular iron homeostasis is predominantly controlled by the post-transcriptional control of iron metabolism genes via the iron responsive element–iron regulatory protein (IRE–IRP) system. The iron-sensitive IRE–IRP interaction regulates the translation rate or mRNA stability of mRNAs depending upon the location of the IRE in the 5′- or 3′-untranslated region (UTR). (B) Systemic iron homeostasis is regulated by the circulating peptide hormone hepcidin, which binds to ferroportin (Fpn) on the plasma membrane and induces Fpn internalization and degradation on various cell types.
New Proteins Are Involved in Iron Trafficking and Utilization
In recent years several new proteins have been identified as key players in intracellular iron trafficking and utilization. This section discusses novel iron transporters, iron chaperones, and ferritin-shuttling proteins.
SLC39/ZIP Family Transporters Are a New Class of Iron-Trafficking Proteins
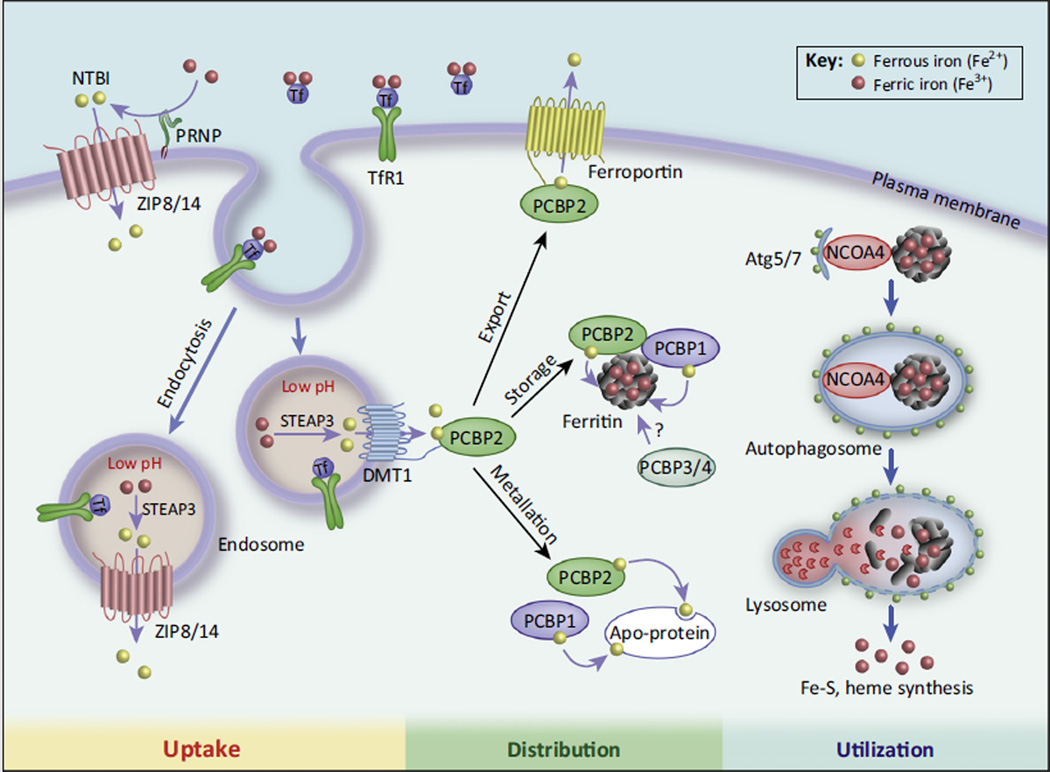
The solute carrier family 39 (SLC39), also known as the ZRT/IRT-like protein (ZIP) family, contains 14 members and has classically been understood to comprise transmembrane zinc transporters that pump extracellular Zn2+ into the cell [5]. However, recent studies have uncovered a role for two ZIP family proteins in iron transport [6]. ZIP14 was found to mediate the uptake of non-transferrin-bound iron (NTBI) [7] by directly transporting NTBI across the cell membrane [8] (Figure 1, left, ‘Uptake’). ZIP14-mediated NTBI transport is enhanced by the Fe3+ to Fe2+ ferrireductase activity of the prion protein PRNP [9]. ZIP14 is also capable of transporting transferrin-bound iron from the endosome to the cytoplasm, similarly to DMT1 [10], although subsequent studies have revealed that this is likely not the major function of ZIP14 [11]. ZIP8, the closest relative to ZIP14 [6], was also found to transport NTBI [12] (Figure 1, left, ‘Uptake’). Interestingly, both ZIP8 and ZIP14 show maximal iron transport above pH 7, near physiological pH [8,12], whereas DMT1 is most efficient at pH 5.5 [13], which corresponds to the pH of acidified endosome. The widely disparate pH activities suggest that ZIP8/14 and DMT1 may have different biological roles. Indeed, a recent report utilizing primary rat hippocampal neurons showed that ZIP8 is the major NTBI transporter whereas DMT1 was principally responsible for transferrin-bound iron transport from the endosome [11].
Figure 1. New Mechanisms Regulating Intracellular Iron Metabolism.
The ZRT/IRT-like protein (ZIP) family transporters, ZIP8 and ZIP14, were recently identified as crucial for transporting non-transferrin bound iron (NTBI) after reduction of NTBI by prion protein (PRNP). In the acidic endosome, Fe3+ is released from transferrin (Tf) and free Fe3+ is reduced to Fe2+ by six-transmembrane epithelial antigen of prostate 3 (STEAP3) and transported to the cytoplasm by divalent metal transporter 1 (DMT1) and ZIP8/14. Poly-(rC)-binding protein 1 (PCBP1) and PCBP2 are cytosolic iron chaperones that deliver Fe2+ to apo-proteins (metallation), such as hypoxia-inducible factor (HIF) prolyl hydroxylases), ferroportin (iron export), and ferritin (oxidation to Fe3+ and storage). Nuclear receptor coactivator 4 (NCOA4)-mediated autophagy of iron-loaded ferritin releases iron for utilization in cellular processes (see text).
ZIP8 and ZIP14 appear to have nonredundant functions because they have distinct tissue expression profiles and knockout mouse models show different phenotypes. Knockout of ZIP8 in mice causes failures in hematopoiesis and organogenesis of the spleen, liver, kidney, and lung, with associated perinatal lethality [14]. Zip8−/− mice exhibited profound anemia and have significantly lowered amounts of iron and zinc in various tissues, with the hematopoietic defect seemingly being caused by the combination of iron and zinc deficiency. By contrast, Zip14−/− mice are viable and have adequate iron stores, but exhibit dwarfism owing to aberrant bone growth. The dwarfism was found to primarily be the result of altered zinc, rather than iron, metabolism, indicating that zinc transport is more important than iron transport by ZIP14 for normal growth [15]. However, a more recent study uncovered a crucial role for ZIP14 in iron transport by knocking out ZIP14 in two mouse models for hereditary hemochromatosis, a family of iron overload diseases [16]. Hemochromatosis results in a progressive accumulation of iron primarily in the liver, resulting in hepatic fibrosis and potentially hepatocellular carcinoma. Knocking out ZIP14 in Hfe−/− (hereditary hemochromatosis protein) or Hfe2−/− (hemojuvelin) mice [modeling type 1 and type 2 (juvenile) hemochromatosis, respectively] prevented iron accumulation in the liver. ZIP14 knockout also prevented iron overload in Hfe2−/− mouse pancreas, but not heart [16], which could be attributable to the high pancreatic but low cardiac expression of ZIP14 [17]. Uncovering the importance of ZIP14 in iron loading provides a new target in the treatment of iron overload diseases. Namely, inhibition of ZIP14 could potentially block hepatic and pancreatic iron overloading, and prevent subsequent organ damage.
The expression of ZIP8 and ZIP14 is regulated differently than other iron transporter genes such as TfR1 and DMT1, both of which are controlled post-transcriptionally by changes of mRNA stability through the 3′ untranslated region (3′ -UTR) iron-responsive element (IRE)–iron regulatory protein (IRP) interaction (Box 1) [2,3]. By contrast, ZIP8 and ZIP14 mRNA stability is unaffected by iron, and ZIP protein levels are positively correlated with iron status, in that iron loading increases ZIP8/14 protein amount whereas iron deficiency decreases protein amount [12,18].
In summary, ZIP8 and ZIP14 are newly recognized iron transporters with important roles in the uptake of NTBI and in iron overload diseases. Further investigation will be necessary to determine the importance of ZIP iron transport in other contexts, such as disease states with disordered iron homeostasis (anemia, cancer, etc.).
PCBP Family Proteins Are Important Iron Chaperones for Cytosolic Iron Distribution
PCBPs are a family of four proteins that were originally identified for their high affinity binding to tracts of polycytosine and have been studied as regulators of gene expression (reviewed in [19]). Each member of the PCBP family contains three highly conserved K homology (KH) domains and can form complexes with other family members. Recently, PCBP1 and PCBP2 were identified as cytosolic iron chaperones that are responsible for delivering iron to ferritin, the protein that is primarily responsible for storing intracellular iron in a non-toxic, mineralized state [20,21] (Figure 1, center, ‘Distribution’). PCBP3 and PCBP4 are expressed at much lower levels, with a limited tissue distribution and an unclear biological role, but there is some evidence that they can also act as iron chaperones and that PCBP3 can bind to ferritin [21].
To date, the majority of the research on the iron chaperone function of the PCBP family has focused on PCBP1 and PCBP2. Both PCBP1 and PCBP2 are necessary to form a stable ternary complex with ferritin, and ferritin–PCBP interactions were dependent upon iron-loaded PCBP1/2 [21]. In addition to ferritin, PCBP1 and PCBP2 can also deliver iron to proteins that require non-heme iron as a cofactor (‘metallation’, Figure 1, center, ‘Distribution’), such as hypoxia-inducible factor (HIF) prolyl and asparginyl hydoxylases [22], and deoxyhypusine hydroxylase [23]. PCBP2 is loaded with iron through binding to the iron transporter DMT1. Iron transfer to PCBP2 is mediated through interaction of the second KH domain of PCBP2 with the N-terminal cytoplasmic domain of DMT1 [24]. After transfer of Fe2+, PCBP2 subsequently dissociates from DMT1 (Figure 1, center, ‘Distribution’). By contrast, PCPB1 does not interact with DMT1, and the mechanism of apo-PCBP1 iron acquisition is still undetermined. A comprehensive screen of both iron-loaded and apo-PCBP1 and -2 binding to other iron metabolism genes (ZIP transporters, etc.) is crucial for understanding overall cytoplasmic iron shuttling and may shed light on PCBP1 iron loading.
Many questions about the roles and mechanisms of the PCBP proteins in iron homeostasis remain unanswered. For instance, it was noted that PCBP2 can also bind to the iron exporter ferroportin (Fpn) [24] (Figure 1, center, ‘Distribution’), but that association remains to be characterized in depth, including what effects, if any, PCBP2 has on Fpn iron export. In addition, the question of what roles, if any, PCBP3 and PCBP4 have in intracellular iron homeostasis remains to be answered.
Lysosomal Ferritin Degradation Involves both Autophagy and Non-Autophagic Pathways
Ferritin stores ferric iron in a mineralized, non-toxic state. However, ferritin-bound iron cannot be utilized by the cell. Consequently, iron must be released from ferritin to be biologically useful, usually through lysosomal degradation of ferritin [25–28]. Ferritin protein turnover is a constant process in cells. While the rate of ferritin degradation appears to be the same in iron-deficient and iron-replete conditions, the delivery mechanism of ferritin to lysosomes seems to be different: autophagy is responsible for delivering ferritin to the lysosome during iron deficiency, whereas a non-autophagic pathway dominates during iron sufficiency [26].
Recently, nuclear receptor coactivator 4 (NCOA4) was identified as the protein responsible for mediating ferritin autophagy [29,30]. NCOA4 was originally identified as a coactivator of several nuclear receptors but was later found to primarily be localized in the cytoplasm and have roles in various biological processes independent of its coactivator function (reviewed in [31]). Two groups found that NCOA4 is greatly enriched in the autophagosome, and identified ferritin as an NCOA4 binding partner [29,30] (Figure 1, right, ‘Utilization’). Knockdown of NCOA4 in various cell types prevented ferritin degradation in iron-deficient conditions [30], suggesting the importance of NCOA4 in ferritin autophagy. By contrast, autophagy-deficient mouse embryonic fibroblasts (MEF) produced by knockout of the key autophagy mediators Atg5 or Atg7 degraded ferritin in the lysosome only in iron-replete conditions and not during iron deficiency [26]. Collectively, lysosomal ferritin degradation appears to involve autophagy in iron deficiency but a non-autophagic pathway dominates during iron sufficiency. NCOA4 knockdown also resulted in an increase in IRP2 and TfR1 [30], likely from a decrease in labile iron owing to ferritin protein accumulation. Furthermore, Ncoa4−/− mice exhibit profound iron deposits within spleen macrophages, suggesting that NCOA4-mediated ferritin autophagy is crucial for splenic iron homeostasis [29].
Ferritin stores iron in a 24-subunit multimer that is a combination of ferritin heavy (FTH1) and light (FTL) chains. NCOA4 was found to interact with the 24-subunit ferritin lattice rather than ferritin monomers in a FTH1-dependent manner [29], but several important aspects of the interaction remain to be characterized, including identification of the NCOA4 and FTH1 interacting domains. In addition, the ratio of FTH1 and FTL protein expression is plastic in various physiological conditions and tissue types (reviewed in [32,33]), and the ability of NCOA4 to mediate autophagy of different ferritin lattice compositions needs to be tested.
Aberrations of ferritin degradation in cancer are an ongoing area of research. The non-autophagic ferritin degradation pathway in iron-replete cells is deficient in various tumor-derived cells (HeLa, MCF-7, and Hepa1–6) compared to normal cells, allowing cancer cells to maintain a higher level of ferritin protein during times of iron sufficiency/excess and potentially explaining why some cancer-derived cells are resistant to iron toxicity [26]. Reactivation of non-autophagic ferritin degradation may increase iron toxicity in cancer, and requires further investigation to determine if this is a viable therapeutic strategy.
Modulating ferritin autophagy has provided some interesting avenues for cancer treatment. The anti-malaria drug artesunate was found to accumulate within lysosomes of cancer cells and enhance lysosomal activity, accelerating autophagic degradation of ferritin and increasing lysosomal iron stores [34]. Artesunate reacts with the increased lysosomal iron to generate ROS, leading to apoptosis in cervical and liver cancer cells [34,35]. As expected, treatment with an iron chelator or knockdown of NCOA4 could prevent artesunate-induced apoptosis [34], indicating that lysosomal iron derived from the ferritin–NCOA4 autophagic pathway is necessary for artesunate-mediated apoptosis.
Iron Is an Important Mediator of Cell Death
Iron is deeply linked to cell death; traditionally, iron is thought to contribute to cell death pathways through ROS production (reviewed in [36]). Recently, an ROS-independent role for iron in modulating apoptosis (programmed cell death) was defined. In addition, an entirely new mode of iron-dependent cell death, termed ‘ferroptosis’, was described.
Iron Inhibits Alternative Splicing of Fas and Facilitates Production of a Pro-Apoptotic Isoform
The cell death receptor Fas exists in two isoforms that are generated by alternative splicing. Fas is a proapoptotic receptor with a transmembrane domain encoded by exon 6. Alternative splicing of Fas pre-mRNA results in exclusion of exon 6 and generates a soluble, anti-apoptotic Fas isoform [37,38]. A recent genome-wide screen identified iron as a crucial suppressor of serine/arginine-rich splicing factor 7 (SRSF7, also known as 9G8), an RNA splicing regulator and important mediator of Fas exon 6 exclusion. Iron inhibits SRSF7 RNA-binding activity, without affecting overall SRSF7 expression or localization, by competing with zinc in the RNA-binding zinc-knuckle domain of SRSF7 [39,40]. Through this mechanism, iron inhibited SRSF7 binding to Fas pre-mRNA and subsequent exon 6 exclusion, leading to an increase in the expression of the pro-apoptotic, exon 6-containing Fas isoform [39]. Further studies will be necessary to confirm the suppression mechanism of SRSF7 splicing activity as well as the regulation of other alternative splicing events by iron.
With the recent identification of ZIP8 and ZIP14 as iron transporters (see above and Figure 1), there exists a potential relationship between ZIP transporters and Fas-induced apoptosis. Given that ZIP14 is crucial for the development of hepatic iron overload in hereditary hemochromatosis [16], ZIP14-mediated iron transport may be a contributing factor to liver cell death in hemochromatosis patients by enhancing expression of the pro-apoptotic Fas isoform. A thorough characterization of any relationship between ZIP transporters and Fas-induced apoptosis might yield another link between cellular iron homeostasis and cell death.
Ferroptosis Is a Novel Form of Iron-Dependent Cell Death
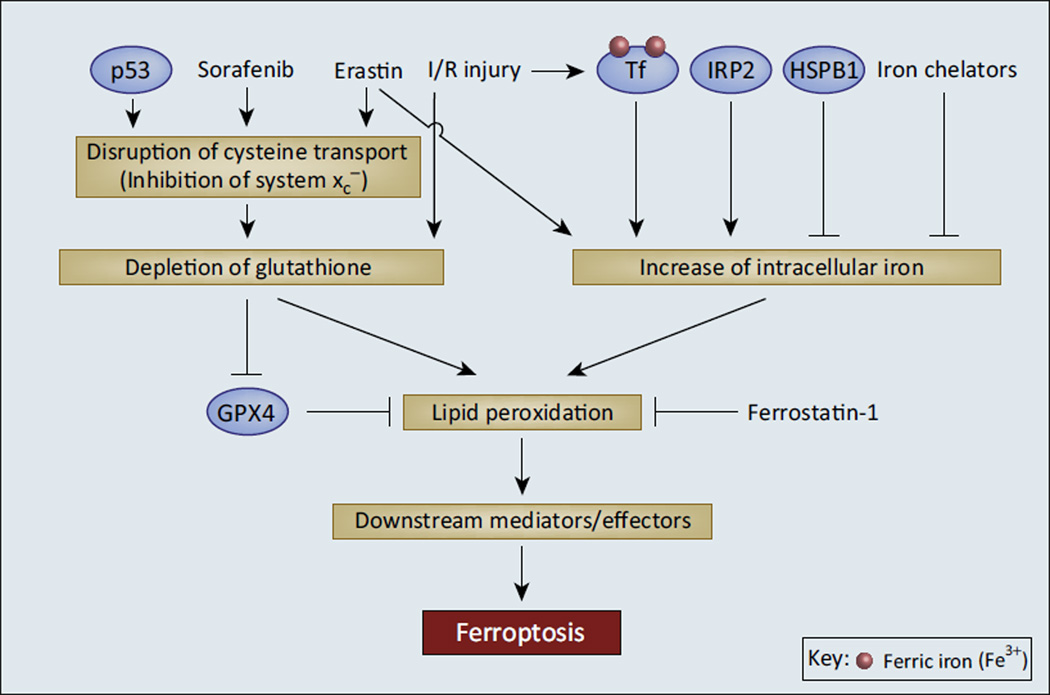
Ferroptosis, an iron-dependent cell death pathway that is non-apoptotic, non-necroptotic, and non-autophagic, was first described in 2012 [41]. Ferroptosis is characterized by lipid peroxidation that is generated at least partially by ROS from heme-containing NADPH oxidase (NOX) family enzymes [41]. Ferroptosis can be inhibited by iron chelation as well as by several novel small molecules (e.g., ferrostatin-1 [41] and liproxstatin-1 [42]) but not by inhibitors of apoptosis (zVAD) or necroptosis (necrostatin-1) [41]. Although the exact role of iron in ferroptosis is not fully understood, there is considerable evidence that iron is a necessary component. IRP2 was identified as an essential gene for the induction of ferroptosis in HT-1080 cells (Figure 2) [41], likely because of its role in iron accumulation and regulation of iron metabolism genes through the IRE–IRP system (Box 1). Recently, a study identified a role for the serum iron-carrier protein transferrin in ferroptosis induction [43]. This study noted that, whereas MEFs grown in serum- and amino-acid-free media showed only modest apoptosis, replenishing the media with serum caused very extensive cell death that was neither apoptotic nor necroptotic. Further investigation determined that two serum components, transferrin and glutamine, were necessary to induce the cell death phenotype [43]. Iron chelation, immunodepletion of transferrin from the serum, and RNAi knockdown of TfR1 all independently prevented cell death. In addition, recombinant iron-loaded (holo-) but not iron-free (apo-) transferrin enhanced cell death [43]. Erastin, a synthetic small molecule that induces ferroptosis [41], required both transferrin and glutamine to effectively kill cells, while ferrostatin 1 (ferroptosis inhibitor) blocked cell death induced by amino acid starvation, suggesting that this form of cell death is indeed ferroptosis (Figure 2) [43].
Figure 2. Intracellular Iron and Antioxidant Depletion Are Crucial Mediators of Ferroptosis.
Ferroptosis appears to occur as a consequence of two cellular events: disruption of the cellular antioxidant pool and an increase of intracellular iron, leading to a buildup of toxic lipid peroxides. Chemical compounds such as erastin and sorafenib as well as p53 signaling disrupt cysteine transport, leading to depletion of cellular antioxidant glutathione (GSH). GSH depletion also occurs during ischemia/reperfusion (I/R) injury, wherein oxidative stress overwhelms the cellular antioxidant capacity. Loss of GSH inhibits glutathione peroxidase 4 (GPX4), a key lipid reduction enzyme. Increase in intracellular iron by erastin treatment, iron regulatory protein 2 (IRP2) activity, and iron-loaded transferrin after I/R injury also contribute to the accumulation of lipid peroxides and death by ferroptosis. Inhibitors of ferroptosis function by detoxifying lipid peroxides (ferrostatin 1) or decreasing intracellular iron [iron chelators, heat shock protein B1 (HSBP1)]. The pathway from lipid peroxides to ferroptotic cell death is still not determined; future studies should show if lipid peroxides are the terminal effectors of ferroptosis or if additional ferroptosis mediator/effector molecules are downstream.
Modulating nutrient availability to cells in culture can be seen as a model for ischemia/reperfusion (I/R) injury, a process that often causes oxidative organ injury and cell death. Indeed, ferroptosis was observed in the kidney [44] and heart [43] after I/R (Figure 2). The identification of ferroptosis as a method of cell death after reperfusion grants mechanistic insight into the role of iron and the efficacy of iron chelation therapy in I/R injury, and generates new therapeutic strategies through the use of synthetic ferroptosis inhibitors. Indeed, treatment with ferroptosis inhibitors can reduce post-reperfusion tissue damage in mouse liver [42] and kidneys [44].
Signaling pathways that affect iron metabolism have also been shown to modulate ferroptosis. Heat shock proteins (HSPs) are inducible by heat shock factor (HSF) transcription factors under various types of stress. HSPB1 (also known as HSP-27) was shown to decrease intracellular iron concentrations through inhibition of TfR1 recycling [45,46]. Knockdown of HSPB1 or HSF1 sensitized cancer cells in culture and xenograft models to erastin-induced ferroptosis through accumulation of intracellular iron and an associated increase of lipid peroxidation, whereas overexpression of HSPB1 inhibited ferroptosis [47]. Given that HSPB1 levels are often elevated in tumors [48], inducing ferroptosis by blocking the HSF1–HSPB1 axis is a tempting therapeutic target that warrants further study.
Several other proteins have been identified as crucial regulators and mediators of ferroptosis, including the tumor suppressor p53 [49], the lipid peroxide reduction enzyme glutathione peroxidase 4 (GPX4) [42,50,51], and the glutamate-cystine antiporter (also known as system xc−, and is crucial for glutathione biosynthesis) [52] (Figure 2). Erastin induces ferroptosis through inhibition of system xc−. Sorafenib, which has traditionally been used as a cancer therapy due to its broad inhibition of kinases, has also been shown to inhibit system xc− and induce ferroptosis in various tumors [52,53]. No clear mechanistic link between iron metabolism and the ferroptotic activity of these proteins has been established.
Much work has been done characterizing ferroptosis since its initial report in 2012, shedding light on biological events leading to ferroptosis and their associated regulators (Figure 2). In addition, several important distinctions have been made between ferroptosis and apoptosis (Table 1). However, many important questions about ferroptosis remain to be answered, including defining the precise role of iron in ferroptotic induction (e.g., is it merely an ROS generator or is it a cofactor for a protein important for ferroptosis?), the identification of effector molecules and proteins downstream of lipid peroxidation, a greater understanding of the biochemical and molecular features of ferroptosis that distinguish it from other forms of cell death, and the characterization of ferroptosis in various cell death contexts and disease states.
Table 1.
Biochemical and Morphological Comparison Between Ferroptosis and Apoptosis
| Characteristic | Ferroptosis | Apoptosis | Refs |
|---|---|---|---|
| Iron-dependent? | Yes | No | [41,43] |
| Caspase activation (cleavage)? | No | Yes | [41–44] |
| Blocked by caspase-inhibitors (e.g., zVAD)? | No | Yes | [41–44] |
| Lipid peroxides required as a signaling/effector molecule? |
Yes | No | [41,42,51] |
| Inhibited by lipid peroxide reducing agent (e.g., ferrostatin-1)? |
Yes | No | [41,42,44] |
| Defining morphological feature(s) | Small mitochondria with increased membrane density |
Plasma membrane blebbing, chromosome condensation/margination |
[41] |
| Causes inflammation and recruitment of immune cells |
Yes | No | [80] |
Dysregulated Iron Metabolism Supports Cancer Growth and Survival
Iron has long been associated with cell proliferation and tumorigenesis, and cancer cells are known to stockpile intracellular iron through dysregulation of iron metabolism, in part by upregulating genes involved in iron uptake (reviewed in [54]). Sixteen iron regulatory genes were found to be predictive of outcome in breast cancer, and patients that had tumors with low iron import and/or high iron export (i.e., lower intracellular iron) experienced significantly more-favorable prognoses [55]. We discuss here recent findings for the role of the iron exporter ferroportin (Fpn) down-regulation in promoting cancer growth, as well as the use of iron chelators in cancer therapy.
Inhibition of Fpn-Mediated Iron Efflux Promotes Cancer Growth
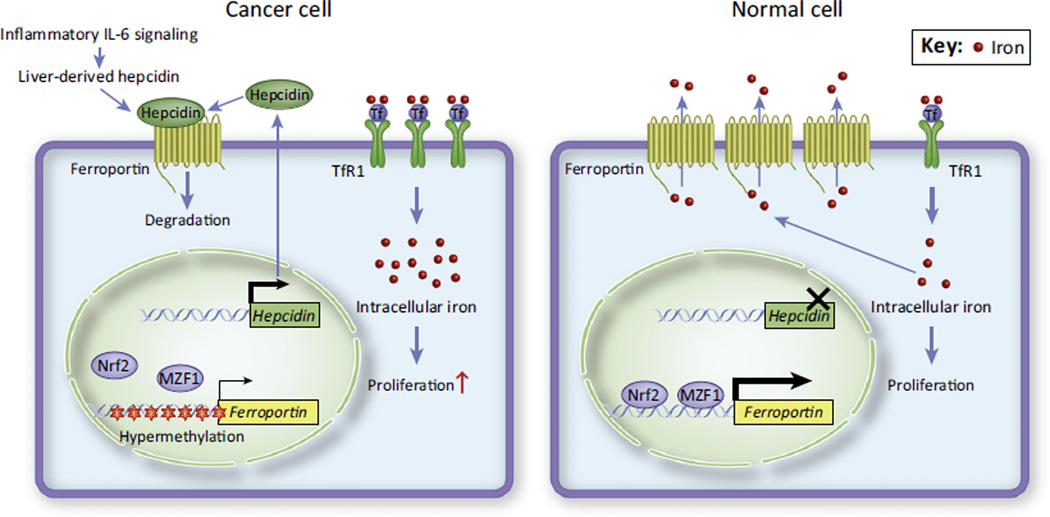
Fpn is the only known vertebrate iron efflux pump, and is regulated post-transcriptionally by the IRE–IRP system and hepcidin (Box 1). Fpn is downregulated in malignant breast epithelium [56,57], leading to an overall increase in tumor iron stores and significantly faster cell proliferation (Figure 3). Breast cancer cells express extremely elevated amounts of hepcidin compared to nonmalignant cells [56], and this could presumably act in an autocrine or paracrine fashion to downregulate Fpn in breast tumors. In addition, breast cancer patients often have higher liver-derived circulating hepcidin (Box 1) as a result of inflammatory interleukin-6 (IL-6) signaling (Figure 3), which contributes to tumor Fpn downregulation and increased cancer growth [57]. Furthermore, the FPN promoter is hypermethylated in breast cancer, which inhibits transcriptional activation of the FPN gene by nuclear factor (erythroid-derived 2)-like-2 (Nrf2) and myeloid zinc finger 1 (MZF1) transcription factors [58] (Figure 3). Fpn suppresses cancer growth through limiting the availability of iron, therefore downregulation of Fpn by hepcidin upregulation or inhibition of FPN transcription promotes tumor progression. Indeed, a cohort of 276 estrogen-receptor-positive breast cancer patients showed that patients with high Fpn-low hepcidin tumors had a 10 year metastasis-free survival rate of 89%, whereas patients with low Fpn/high hepcidin tumors had a 10 year metastasis-free survival rate of only 65% despite similar therapeutic regimens [56]. Consequently, Fpn and hepcidin may be effective prognostic indicators for breast cancer and might be a useful guide for therapeutic strategies; for example, patients with low Fpn/high hepcidin tumors (that is, tumors with high intracellular iron) may be good candidates for iron chelation therapy (see below). Altered Fpn expression has been found not only in breast cancer but also in prostate cancer [59] and myelomas [60]. Further investigation is necessary to determine the importance of the Fpn–hepcidin axis in other tumor types.
Figure 3. The Ferroportin–Hepcidin Axis Is Altered in Cancers.
Normal breast and prostate cells (right) contain relatively low intracellular iron as a result of high ferroportin, low hepcidin, and low TfR1 expression. By contrast, breast and prostate cancer cells (left) often have elevated TfR1 as well as low ferroportin expression because of hypermethylation of the ferroportin promoter which inhibits Fpn transcription factors (Nrf2, MZF1). In addition, post-translational repression of Fpn by tumor- and liver-derived hepcidin further blocks iron efflux. Consequently, these cancer cells can aberrantly accumulate intracellular iron, leading to increased cell proliferation and an aggressive cancer phenotype.
Iron Chelators Are Promising Anticancer Agents
Malignant cells rely on dysregulated iron metabolism for increased proliferation and survival; therefore, reducing the amount of iron available to tumors is an obvious therapeutic strategy. In addition to approaches such as targeting overexpressed TfR1 in various cancer cells [61–63], iron chelators, generally used in the treatment of iron overload diseases, have been considered in cancer treatment since the 1980s [64,65]. However, poor pharmacological properties reduced their potential clinical impact as chemotherapeutics. Newer-generation chelators show more promising pharmacokinetics and pharmacodynamics [66], leading to a resurgence in the study of iron chelation as an effective antineoplastic.
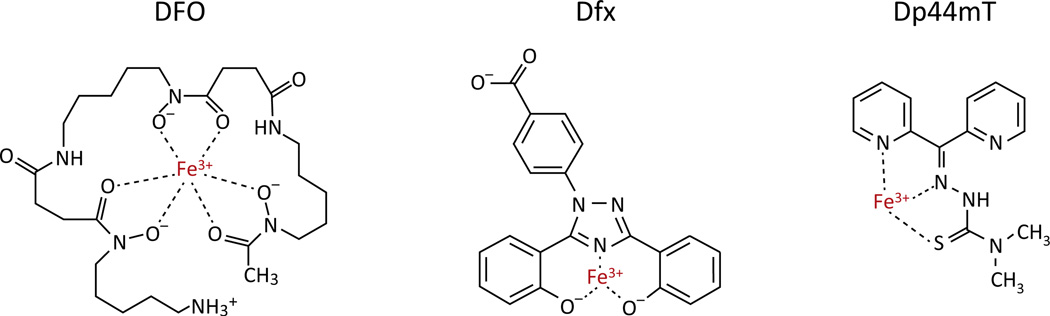
Iron chelators deplete intracellular iron and may paradoxically contribute to ROS generation depending on the number of chemical bonds used to chelate an iron atom (Box 2). Deferasirox (Dfx), a tridentate chelator that allows iron redox cycling, is an orally available iron chelator that has promise as an antineoplastic. Dfx has been shown to inhibit the growth of various types of malignant cells in cell culture [66,67] and in xenograft models [68,69]. There is also a case report of Dfx contributing to complete remission in a patient with recurrent acute myelogenous leukemia [70]. A relatively new class of tridentate chelators, the thiosemicarbazones, has been investigated in recent years. One member, Dp44mT, can chelate iron or copper, and either metal can participate in ROS generation [71,72] through redox cycling involving thioredoxin reductase 1 and glutathione reductase [73].
Box 2. Two Classes of Iron Chelators Sequester Iron as Redox-Active or -Inactive Species.
Iron can coordinate with up to six ligands. Hexadentate iron chelators such as the bacteria-derived siderophore DFO (deferoxamine) form six bonds with a central iron atom, filling the coordination sphere of iron and rendering it inert (Figure I). This class of chelators acts by tightly sequestering iron and preventing its use in cellular processes (converting to‘non-labile’ iron), leading to overall depletion of intracellular iron levels and slowdown of cellular metabolism. Tridentate chelators, such as the synthetic chelators Dfx (deferasirox) and Dp44mT (Figure I), also prevent cellular utilization by binding to iron, but leave several ligand-coordination sites available on the iron atom. After reduction of chelated Fe3+ to Fe2+ in the lysosome, tridentate chelated iron can generate ROS. As a result, this class of chelator shows antiproliferative effects through two mechanisms: it removes iron from the biologically available pool and facilitates the generation of damaging ROS by redox cycling.
Figure I. Hexadentate and Tridentate Chelators Sequester Iron with Different Chemistries.
Deferoxamine (DFO) forms six bonds with a central iron atom, rendering the Fe3+ inert. Tridentate chelators, such as Dfx and Dp44mT, only form three bonds with iron, allowing iron reduction and Fenton chemistry with subsequent reactive oxygen species (ROS) generation.
Beyond inducing cell death through ROS generation, iron chelation can exhibit anticancer effects through multiple mechanisms (Figure 4). These include preventing deoxyribonucleotide synthesis through blocking the activity of ribonucleotide reductase, inducing cell cycle arrest by downregulating cyclins and cyclin-dependent kinases, and upregulating tumor suppressor genes such as TP53 and PTEN (reviewed in [66]), activating mitophagy [74], and perturbing various signaling pathways including kinase cascades, signal transducer and activator of transcription 3 (STAT3), transforming growth factor-β (TGF-β), and Wnt (reviewed in [66]). The myriad targets of iron chelation indicate that, although cell death after iron overload primarily seems to be a consequence of ROS generation [75], cell death from iron chelation is a complex, multimodal mechanism (Figure 4).
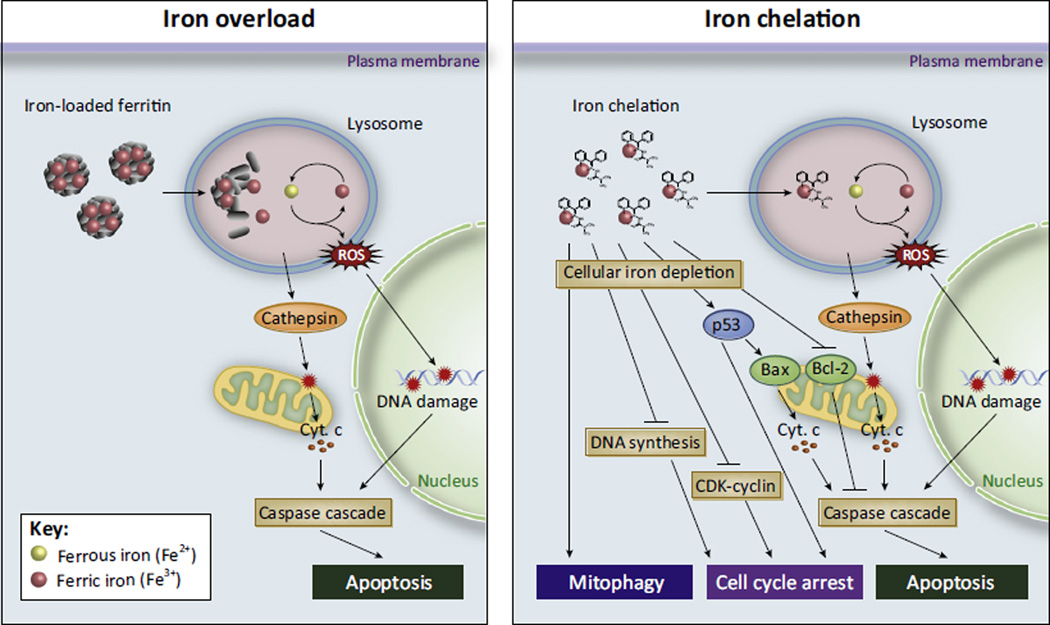
Figure 4. Both Iron Excess and Deficiency Are Deleterious to Cells.
Cell death by iron overload (left) is thought to be the result of reactive oxygen species (ROS) generation, particularly in the lysosome. Iron-overloaded cells accumulate iron inside lysosomes as a consequence of the degradation of iron-loaded ferritin. Inside the lysosome, iron can cycle between Fe3+ and Fe2+, generating ROS and leading to lysosomal membrane permeabilization and DNA damage. Cathepsin and other lysosomal proteases induce release of mitochondrial cytochrome c (Cyt. c) and other pro-apoptotic proteins, leading to downstream caspase activation and apoptosis. Iron chelation (right) has multiple antiproliferative/pro-death effects on the cell. Iron chelators can induce mitophagy, cell cycle arrest, and apoptosis through various other mechanisms, including blocking DNA biosynthesis through inhibition of ribonucleotide reductase, downregulation of cyclins and cyclin-dependent kinases (CDKs), induction of the tumor suppressor p53, downregulation of the apoptosis inhibitor Bcl-2, and upregulation of apoptosis activator Bax.
The Future of Iron Biology
The field of human iron biology is dynamic and expanding. Great strides in understanding iron metabolism have been made in the past several years, including the identification of new essential proteins in human iron metabolism and novel roles for iron in both normal cellular processes and in pathologies such as cancer. However, despite all the recent advances, many unanswered questions remain and the extension of new mechanistic knowledge to human health outcomes remains a crucial goal (see Outstanding Questions). With the recent improvements in high-throughput techniques and the growth of the ‘-omics’ fields (transcriptomics, proteomics, etc.), the next several years should yield even more exciting insights into how our cells regulate, and are regulated by, iron.
Outstanding Questions.
What are the additional mechanisms of crosstalk between intracellular and systemic iron homeostasis?
How do ZIP8 and ZIP14 contribute to iron dysregulation in various disease states (iron overload, cancer, etc.)? What are the roles of other ZIP family members in iron trafficking?
What are the degradation mechanisms of ZIP8 and ZIP14 proteins?
How does PCBP1 acquire iron? What proteins interact commonly or specifically with PCBP1 and PCBP2? What is the importance of PCBP3/4?
What is the mechanism of nonautophagic ferritin degradation in iron-replete cells? How is this pathway dysregulated in cancer?
How is NCOA4-mediated autophagy affected by different ferritin-lattice compositions?
What are the other targets of the splicing factor SRSF7? How does iron affect the splicing of these mRNAs?
What is the precise role of iron in ferroptosis? Do additional iron metabolism genes contribute to ferroptosis induction or inhibition?
What proteins/molecules exist downstream of lipid peroxidation in ferroptosis? How are they affected by iron?
How can pharmaceutical compounds be utilized to induce or inhibit ferroptosis? How does this affect human health outcomes such as cancer or reperfusion injury?
What is the importance of the ferroportin/hepcidin axis in tumor types other than breast and prostate cancers? Does the current generation of iron chelators have clinical relevance as anticancer agents? Can their effectiveness in model systems be extended to human patients?
Trends.
Dysregulation of iron metabolism contributes to various human pathologies, including iron overload diseases and cancer.
Several new proteins have been identified as crucial iron traffickers and chaperones with important connections to human health.
Ferroptosis is a unique cell death pathway that is iron-dependent, non-apoptotic, non-necroptotic, and non-autophagic.
Dysregulation of the ferroportin/hepcidin regulatory axis contributes to tumor progression and is predictive of patient outcomes.
New iron chelators utilize more specific mechanisms to elicit anticancer activity.
Acknowledgments
A.R.B. was supported by National Institutes of Health (NIH) Training Grant T32ES007046 from the National Institute of Environmental Health Sciences, and this work was supported by NIH Research Grants RO1GM088392 and RO1GM095550 from the National Institute of General Medical Sciences to Y.T.
References
- 1.Ray PD, et al. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacKenzie EL, et al. Intracellular iron transport and storage: from molecular mechanisms to health implications. Antioxid. Redox Signal. 2008;10:997–1030. doi: 10.1089/ars.2007.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hentze MW, et al. Two to tango: regulation of mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Ganz T, Nemeth E. Hepcidin and iron homeostasis. Biochim. Biophys. Acta. 2012;1823:1434–1443. doi: 10.1016/j.bbamcr.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeong J, Eide DJ. The SLC39 family of zinc transporters. Mol. Aspects Med. 2013;34:612–619. doi: 10.1016/j.mam.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenkitkasemwong S, et al. Physiologic implications of metal-ion transport by ZIP14 and ZIP8. Biometals. 2012;25:643–655. doi: 10.1007/s10534-012-9526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liuzzi JP, et al. Zip14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13612–13617. doi: 10.1073/pnas.0606424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinilla-Tenas JJ, et al. Zip14 is a complex broad-scope metal-ion transporter whose functional properties support roles in the cellular uptake of zinc and nontransferrin-bound iron. Am J. Physiol. Cell Physiol. 2011;301:C862–C871. doi: 10.1152/ajpcell.00479.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripathi AK, et al. Prion protein functions as a ferrire-ductase partner for ZIP14 and DMT1. Free Radic. Biol. Med. 2015;84:322–330. doi: 10.1016/j.freeradbiomed.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Netal. ZRT/IRT-like protein 14 (ZIP14) promotes the cellular assimilation of iron from transferrin. J. Biol. Chem. 2010;285:32141–32150. doi: 10.1074/jbc.M110.143248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji C, Kosman DJ. Molecular mechanisms of non-transferrin-bound and transferring-bound iron uptake in primary hippocampal neurons. J. Neurochem. 2015;133:668–683. doi: 10.1111/jnc.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang CY, et al. ZIP8 is an iron and zinc transporter whose cell-surface expression is up-regulated by cellular iron loading. J. Biol. Chem. 2012;287:34032–34043. doi: 10.1074/jbc.M112.367284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunshin H, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 14.Galvez-Peralta M, et al. ZIP8 zinc transporter: indispensable role for both multiple-organ organogenesis and hematopoiesis in utero. PLoS ONE. 2012;7:e36055. doi: 10.1371/journal.pone.0036055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hojyo S, et al. The zinc transporter SLC39A14/ZIP14 controls G-protein coupled receptor-mediated signaling required for systemic growth. PLoS ONE. 2011;6:e18059. doi: 10.1371/journal.pone.0018059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkitkasemwong S, et al. SLC39A14 is required for the development of hepatocellular iron overload in murine models of hereditary hemochromatosis. Cel Metab. 2015;22:138–150. doi: 10.1016/j.cmet.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam H, et al. ZIP14 and DMT1 in the liver, pancreas, and heart are differentially regulated by iron deficiency and overload: implications for tissue iron uptake in iron-related disorders. Haematologica. 2013;98:1049–1057. doi: 10.3324/haematol.2012.072314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao N, et al. An iron-regulated and glycosylation-dependent proteasomal degradation pathway for the plasma membrane metal transporter ZIP14. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9175–9180. doi: 10.1073/pnas.1405355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhury A, et al. Heterogeneous nuclear ribonucleo-proteins (hnRNPs) in cellular processes: focus on hnRNP E1's multifunctional regulatory roles. RNA. 2010;16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H, et al. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2008;320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leidgens S, et al. Each member of the poly-RC)-binding protein 1 (PCBP) family exhibits iron chaperone activity toward ferritin. J. Biol. Chem. 2013;288:17791–17802. doi: 10.1074/jbc.M113.460253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nandal A, et al. Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metab. 2011;14:647–657. doi: 10.1016/j.cmet.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frey AG, et al. Iron chaperones PCBP1 and PCBP2 mediate the metallation of the dinuclear iron enzyme deoxyhypusine hydroxylase. Proc. Natl. Acad. Sci. U.S.A. 2014;111:8031–8036. doi: 10.1073/pnas.1402732111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanatori I, et al. Chaperone protein involved in transmembrane transport of iron. Biochem. J. 2014;462:25–37. doi: 10.1042/BJ20140225. [DOI] [PubMed] [Google Scholar]
- 25.Kidane TZ, et al. Release of iron from ferritin requires lysosomal activity. Am. J. Physiol. Cel Physiol. 2006;291:C445–C455. doi: 10.1152/ajpcell.00505.2005. [DOI] [PubMed] [Google Scholar]
- 26.Asano T, et al. Distinct mechanisms of ferritin delivery to lysosomes in iron-depleted and iron-replete cells. Mol. Cel. Biol. 2011;31:2040–2052. doi: 10.1128/MCB.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishi-Itakura C, et al. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cel Sci. 2014;127:4089–4102. doi: 10.1242/jcs.156034. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Lysosomal proteolysis is the primary degradation pathway for cytosolic ferritin and cytosolic ferritin degradation is necessary for iron exit. Antioxid. RedoxSignal. 2010;13:999–1009. doi: 10.1089/ars.2010.3129. [DOI] [PubMed] [Google Scholar]
- 29.Dowdle WE, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 2014;16:1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 30.Mancias JD, et al. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kollara A, Brown TJ. Expression and function of nuclear receptor co-activator 4: evidence of a potential role independent of co-activator activity. Cel. Mol. Life Sci. 2012;69:3895–3909. doi: 10.1007/s00018-012-1000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torti FM, Torti SV. Regulation of ferritin genes and protein. Blood. 2002;99:3505–3516. doi: 10.1182/blood.v99.10.3505. [DOI] [PubMed] [Google Scholar]
- 33.Arosio P, et al. Ferritins: a family of molecules for iron storage, antioxidation and more. Biochim. Biophys. Acta. 2009;1790:589–599. doi: 10.1016/j.bbagen.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Yang ND, et al. Artesunate induces cell death in human cancer cells via enhancing lysosomal function and lysosomal degradation of ferritin. J. Biol. Chem. 2014;289:33425–33441. doi: 10.1074/jbc.M114.564567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamacher-Brady A, et al. Artesunate activates mitochondrial apoptosis in breast cancer cells via iron-catalyzed lysosomal reactive oxygen species production. J. Biol. Chem. 2011;286:6587–6601. doi: 10.1074/jbc.M110.210047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon SJ, Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 2014;10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 37.Cheng J, et al. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994;263:1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- 38.Cascino I, et al. Three functional soluble forms of the human apoptosis-inducing Fas molecule are produced by alternative splicing. J. Immun. 1995;154:2706–2713. [PubMed] [Google Scholar]
- 39.Tejedor JR, et al. Genome-wide identification of Fas/CD95 alternative splicing regulators reveals links with iron homeostasis. Mol. Cell. 2015;57:23–38. doi: 10.1016/j.molcel.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 40.Cavaloc Y, et al. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixon SJ, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cel. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedmann Angeli JP, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cel Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao M, et al. Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linkermann A, et al. Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 2014;111:16836–16841. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arrigo AP, et al. Hsp27 consolidates intracellular redox homeostasis by upholding glutathione in its reduced form and by decreasing iron intracellular levels. Antioxid. Redox Signal. 2005;7:414–422. doi: 10.1089/ars.2005.7.414. [DOI] [PubMed] [Google Scholar]
- 46.Chen H, et al. Heat shock protein 27 downregulates the transferrin receptor 1-mediated iron uptake. Int. J. Biochem. Cell Biol. 2006;38:1402–1416. doi: 10.1016/j.biocel.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Sun X, et al. HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene. 2015;34:5617–5625. doi: 10.1038/onc.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oesterreich S, et al. The small heat shock protein hsp27 is correlated with growth and drug resistance in human breast cancer cell lines. Cancer Res. 1993;53:4443–4448. [PubMed] [Google Scholar]
- 49.Jiang L, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang WS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsushita M, et al. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J. Exp. Med. 2015;212:555–568. doi: 10.1084/jem.20140857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dixon SJ, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3:e02523. doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Louandre C, et al. Iron-dependent cell death of hepatocellular carcinoma cells exposed to sorafenib. Int. J. Cancer. 2013;133:1732–1742. doi: 10.1002/ijc.28159. [DOI] [PubMed] [Google Scholar]
- 54.Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat. Rev. Cancer. 2013;13:342–355. doi: 10.1038/nrc3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller LD, et al. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 2011;71:6728–6737. doi: 10.1158/0008-5472.CAN-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinnix ZK, et al. Ferroportin and iron regulation in breast cancer progression and prognosis. Sci. Transl. Med. 2010;2:43ra56. doi: 10.1126/scisignal.3001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang S, et al. Disordered hepcidin-ferroportin signaling promotes breast cancer growth. Cel Signal. 2014;26:2539–2550. doi: 10.1016/j.cellsig.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y, et al. Disordered signaling governing ferroportin transcription favors breast cancer growth. Cell Signal. 2015;27:168–176. doi: 10.1016/j.cellsig.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Tesfay L, et al. Hepcidin regulation in prostate and its disruption in prostate cancer. Cancer Res. 2015;75:2254–2263. doi: 10.1158/0008-5472.CAN-14-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu Z, et al. Decreased ferroportin promotes myeloma cell growth and osteoclast differentiation. Cancer Res. 2015;75:2211–2221. doi: 10.1158/0008-5472.CAN-14-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daniels TR, et al. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta. 2012;1820:291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horonchik L, Wessling-Resnick M. The small-molecule iron transport inhibitor ferristatin/NSC306711 promotes degradation of the transferrin receptor. Chem. Biol. 2008;15:647–653. doi: 10.1016/j.chembiol.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Byrne SL, et al. Ferristatin II promotes degradation of transferrin receptor-1 in vitro and in vivo. PLoS ONE. 2013;8:e70199. doi: 10.1371/journal.pone.0070199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blatt J, Stitely S. Antineuroblastoma activity of desferoxamine in human cell lines. Cancer Res. 1987;47:1749–1750. [PubMed] [Google Scholar]
- 65.Reddel RR, et al. Cell cycle effects of iron depletion on T-47D human breast cancer cells. Exp. Cell Res. 1985;161:277–284. doi: 10.1016/0014-4827(85)90085-0. [DOI] [PubMed] [Google Scholar]
- 66.Lui GY, et al. Targeting cancer by binding iron: dissecting cellular signaling pathways. Oncotarget. 2015;6:18748–18779. doi: 10.18632/oncotarget.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohyashiki JH, et al. The oral iron chelator deferasirox represses signaling through the mTOR in myeloid leukemia cells by enhancing expression of REDD1. Cancer sci. 2009;100:970–977. doi: 10.1111/j.1349-7006.2009.01131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lui GY, et al. The iron chelator, deferasirox, as a novel strategy for cancer treatment: oral activity against human lung tumor xenografts and molecular mechanism of action. Mol. Pharmacol. 2013;83:179–190. doi: 10.1124/mol.112.081893. [DOI] [PubMed] [Google Scholar]
- 69.Ford SJ, et al. Deferasirox (ICL670A) effectively inhibits oesophageal cancer growth in vitro and in vivo. Br. J. Pharmacol. 2013;168:1316–1328. doi: 10.1111/bph.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fukushima T, et al. Iron chelation therapy with deferasirox induced complete remission in a patient with chemotherapy-resistant acute monocytic leukemia. Anticancer Res. 2011;31:1741–1744. [PubMed] [Google Scholar]
- 71.Richardson DR, et al. Dipyridyl thiosemicarbazone chelators with potent and selective antitumor activity form iron complexes with redox activity. J. Med. Chem. 2006;49:6510–6521. doi: 10.1021/jm0606342. [DOI] [PubMed] [Google Scholar]
- 72.Lovejoy DB, et al. Antitumor activity of metal-chelating compound Dp44mT is mediated by formation of a redox-active copper complex that accumulates in lysosomes. Cancer Res. 2011;71:5871–5880. doi: 10.1158/0008-5472.CAN-11-1218. [DOI] [PubMed] [Google Scholar]
- 73.Myers JM, et al. Redox activation of Fe(III)-thiosemicarbazones and Fe(III)-bleomycin bythioredoxin reductase: specificity of enzymatic redox centers and analysis of reactive species formation by ESR spin trapping. Free Radic. Biol. Med. 2013;60:183–194. doi: 10.1016/j.freeradbiomed.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Allen GF, et al. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Terman A, Kurz T. Lysosomal iron, iron chelation, and cell death. Antioxid. Redox Signal. 2013;18:888–898. doi: 10.1089/ars.2012.4885. [DOI] [PubMed] [Google Scholar]
- 76.Kuhn LC. Iron regulatory proteins and their role in controlling iron metabolism. Metalomics: Int. Bio. Sci. 2015;7:232–243. doi: 10.1039/c4mt00164h. [DOI] [PubMed] [Google Scholar]
- 77.Khan MA, et al. Direct Fe2+ sensing by iron-responsive messenger RNA:repressor complexes weakens binding. J. Biol. Chem. 2009;284:30122–30128. doi: 10.1074/jbc.M109.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma J, et al. Fe2+ binds iron responsive element-RNA, selectively changing protein-binding affinities and regulating mRNA repression and activation. Proc. Natl. Acad. Sci. U.S.A. 2012;109:8417–8422. doi: 10.1073/pnas.1120045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khan MA, et al. Rapid kinetics of iron responsive element (IRE) RNA/iron regulatory protein 1 and IRE-RNA/eIF4F complexes respond differently to metal ions. Nucleic Acids Res. 2014;42:6567–6577. doi: 10.1093/nar/gku248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Linkermann A, et al. Regulated cell death and inflammation: an auto-amplification loop causes organ failure. Nat. Rev. Immunol. 2014;14:759–767. doi: 10.1038/nri3743. [DOI] [PubMed] [Google Scholar]