Abstract
Multiple myeloma (MM) is a hematological malignancy of clonal plasma cells in the bone marrow (BM). The microenvironment plays a key role in MM cell survival and drug resistance through release of soluble factors, expression of adhesion molecules and release of extracellular vesicles (EV). The aim of this manuscript is to use proteomic profiling of EVs as a tool to identify circulating tumor associated markers in MM patients. First, we characterized the EV protein content obtained from different MM cell lines. Then, we established differences in protein abundance among EVs isolated from MM patient serum and BM and the serum of healthy donors. These data show the Major Histocompatibility Complex Class I is highly enriched in EVs of MM cell lines and MM patient's serum. Next, we show that CD44 is highly expressed in the EVs isolated from the corticosteroid resistant MM cell line, MM.1R. Furthermore, CD44 was found to be differentially expressed in EVs isolated from newly diagnosed MM patients. Finally through ELISA analysis, we establish the potential of serum CD44 as a predictive biomarker of overall survival. These results support the analysis of EVs as an easily accessible source for MM biomarkers.
Graphical Abstract
INTRODUCTION
Multiple myeloma (MM) is a hematological malignancy characterized by clonal plasma cells (PCs) in the bone marrow (BM) and accounts for approximately 20,000 diagnoses and 10,000 deaths annually in the US [1,2]. MM cells are dependent on the BM microenvironment (e.g. BM stromal cells, macrophages etc) and create a network with surrounding cells [3-6]. These cells play a pivotal role in the regulation of MM cell survival and drug resistance by direct interactions through adhesion molecules causing cell adhesion mediated drug resistance (CAM-DR) or soluble factors including supportive cytokines (e.g. IL-6, IL-8, and VEGF) or exosomes (or extracellular vesicles; EVs) [7,8]. EVs are membrane-covered cell fragments of variable size (30-1,000 nm), which contain specific protein and RNA cargo [9-12]. BM stromal cells and MM cells can mutually exchange information through soluble factors including cytokines, surface molecules and EVs. Additionally, EV also have been recently reported to induce survival and drug resistance in human MM cells in vitro, but the potential use of their RNA and protein cargo as source of biomarkers for disease initiation and drug resistance have been only partially explored [4,8].
Recently, we established the use a global label-free quantification method to determine the relative amount of proteins identified from EVs obtained from MM cell lines [13]. By protein array analysis, Roccaro et al. showed that MM bone marrow myeloid stromal cell (MM BM-MSC) derived EV contain increased levels of oncogenes and protein kinases compared to normal BM-MSC vesicles, suggesting that EV associated protein abundance can play a key role in the patho-biology of MM [4]. Although the protein array used from the authors have highlighted the presence of specific set of proteins in the EV of MM BM-MSC, our global systematic proteomic analysis of EV isolated from MM cells and from the peripheral blood of MM patients, allow the identification of differential EV protein content without prior knowledge.
Here we are reporting a systematic proteomic analysis of EVs derived from MM cell lines, blood from MM patients, and BM from MM patients. To characterize a specific repertoire of EV-proteins from MM cells, we first measured the global EV protein content of several MM cell lines and the content of circulating EV in MM primary patient samples and compared the results to healthy donors. We identify the Major Histocompatibility Complex Class I (MCHI) and its binding protein β2-Microglobulin (β2-MG) enriched in the EV isolated from all MM cell lines and from MM patients. Because MHCI and β2-MG molecules as antigen presenting complex bind antigens (peptides) generated mainly from degradation of cytosolic proteins by the proteosome, it is not surprising that MM cells, which for nature exhibit an increase proteosome activity, are releasing EV enriched in those two molecules [14].
Previously published proteomic data has shown, that in other forms of cancer, EVs isolated from drug resistant cell lines display a different protein content compared to their corresponding parental cells [15-17]. We analyzed the differences in the EV protein content between corticosteroid resistant (MM.1R) and sensitive (MM.1S) cell lines. We identified single-chain transmembrane glycoprotein CD44 as the most differentially expressed protein in the EV of MM.1R [18-20]. Our data are consistent with the previously published role of CD44 to mediate resistance to dexamethasone and Lenalidomide in MM cells and with the fact that CD44 has been identified on EV derived from a number of cancer cell types [21-27]. Additionally, we identified CD44 as a key molecule in EVs of MM patients and confirmed its role as a prognostic marker.
MATERIALS & METHODS
Antibodies
Antibodies and dilutions used were as follows: β2-Microglobulin (1:10,000), Lamin B1 (1:1000) (Abcam, Cambridge, MA), CD44 (1:1000, Santa Cruz Biotechnology, Santa Cruz, CA) and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Cell Signaling Technology, Boston, MA). Anti-mouse & anti-rabbit IgG-HRP (GE Healthcare, Piscataway, NJ). CD44-FITC (1:20, BD Biosciences, Brea, CA). Unless otherwise noted secondary antibody dilutions were 1:5000.
Cell Line Tissue Culture
MM.1R and RPMI-8226 cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured as recommended by ATCC. Briefly, Cell lines were maintained at 4 × 105 cells/mL by incubation at 37 °C with 5 % CO2 in complete growth medium (RPMI-1640 with fetal bovine serum (10 %, FBS), glutamine (2 mM, GlutaMAX), penicillin-G (50 U/mL) and streptomycin (50 μg/mL, Life Technologies, Grand Island, NY)). For vesicle isolation, 1-2 × 108 cells were serum starved for 48 h prior to vesicle enrichment.
Cell Line Derived Extracellular Vesicle Isolation
MM derived extracellular vesicles were isolated as originally described by Théry et al. [28]. Serum starved cells and media were differentially centrifuged at 300 × g for 10 min at 4 °C and 2000 × g for 20 min at 4 °C. Supernatant was centrifuged at 10,000 × g for 30 min at 4 °C and ultracentrifuged at 100,000 × g for 70 min at 4 °C with vacuum. Ex vivo samples were uniformly processed, fresh. Cells were spun at 300 × g for 10 min at 4 °C and 4,500 × g for 20 min at 4 °C. Supernatant was centrifuged at 10,000 × g for 30 min at 4 °C and ultracentrifuged at 100,000 × g for 70 min at 4 °C with vacuum. The resulting pellets were resuspended in 1 mL of PBS and pooled. Pooled preparations were again ultracentrifuged at 100,000 × g as described previously. Vesicle containing pellets were frozen on dry ice and stored at −80 °C.
Flow Cytometry
Serum starved MM.1R and RPMI-8226 cell lines were analyzed for apoptosis and cell death by Annexin V and propidium iodide flow cytometry. Approximately 1 × 105 serum starved cells were washed with PBS and placed into Annexin V-FITC and Propidium Iodide staining solution (BD Biosciences, San Jose, CA) for 15 min in the dark at room temperature. Cells were washed and immediately analyzed on a BD LSRII (BD Biosciences, San Jose, CA). RPMI-8226 cells were gated to eliminate confounding cellular debris (Supplemental Data 1).
CD44-FITC staining was conducted on serum-starved cells using a Beckman Coulter FC500 (Brea, CA) flow cytometer. 8 × 105 starved cells were stained at 1:20 in PBS for 30 minutes on ice in the dark. Cells were washed and immediately analyzed. All computational analyses were conducted using the FlowJo Software (ver. 10.0.7r2, Tree Star Inc., Ashland, OR).
Cryo-Transmission Electron Microscopy (cryo-TEM)
MM1.R and RPMI-8226 derived extracellular vesicles were prepared for cryo-TEM as described previously [13]. Briefly, 4 μL of vesicle suspensions were applied to glow discharged lacey carbon coated copper grids (400 mesh, Pacific Grid-Tech, San Francisco, CA) in a controlled environment (22 °C and 95 % relative humidity) using an automated vitrification device (FEI Vitrobot Mark IV, FEI, Hillsboro, OR) and flash-frozen in liquid ethane. Vesicles were visualized in a FEI Tecnai G2 F20 ST transmission electron microscope (TEM, FEI, Hillsboro, OR) operated at 200 kV under low dose radiation. Images were captured on a Gatan Ultrascan CCD camera (38,000× magnification).
Nanoparticle Tracking Analysis
Cell lines were seeded at 3.0 × 108 in serum free media and grown overnight as described above. Supernatant was collected and sequentially centrifuged at 3,220 × g for 10 minutes then ultracentrifuged at 10,000 × g for 30 minutes. Peripheral blood from healthy donors and patients previously diagnosed with monoclonal gammopathy of undetermined significance, smoldering and active MM were collected in EDTA Vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ) and sequentially centrifuged with supernatant transfers after 1,000 × g for 10 minutes, 4500 × g for 15 minutes, and 10,000 × g for 30 minutes [29,30]. Remaining supernatant was homogenized then diluted in PBS, as necessary. Size distribution analysis was carried out on a Nanosight NS300 (Malvern Instruments Ltd., Malvern, UK) where two separate vesicle preparations for each cell line and single MM patient and healthy donor preparations were analyzed five times each. Batch capture was conducted on an sCMOS camera with variable shutter length and frame rate, 1000 shutter setting and 400 camera gain. Computational analysis was carried out on the Nanoparticle Tracking Analysis Software (Malvern Instruments Ltd., Malvern, UK) version 2.3 build 0033.
Cell Line Vesicle Sample Preparation & Liquid Chromatography Mass Spectrometry (LC-MS/MS)
Extracellular vesicles and cellular lysates were prepared for LC-MS/MS analysis as described previously [13,31]. In triplicate, 1 × 105 serum starved cells or vesicle isolations were dissolved in ammonium bicarbonate (50 mM, Sigma Aldrich, St. Louis, MO) with Rapigest SF surfactant (0.5 %, Waters, Milford, MA) and sequencing grade modified trypsin (800 ng, Promega, Madison, WI). Suspensions were incubated overnight (>16 h) at 37 °C. 98 % formic acid (Acros Organics, Geel, Belguim) was added to approximately 30 % (v/v) and samples were incubated at 37 °C for 30 min. Rapigest and insoluble material was removed by centrifugation at 21,000 × g (3×). Peptides were dried and resuspended in 20 μL of loading buffer (2 % acetonitrile with 0.1 % formic acid (aq)). Peptide concentrations were estimated by the absorbance at 280 nm (NanoDrop ND-1000, NanoDrop, Wilmington, DE).
Extracellular vesicle and cellular lysate peptides (1-2 μg) were loaded onto a Dionex Ultimate 3000 capillary/nano HPLC (Dionex, Sunnyvale, CA) for RP-HPLC separation. Samples were directly introduced into a ThermoFisher LTQ Orbitrap XL mass spectrometer (ThermoFisher, Waltham, MA) for top-5 data dependent mass analysis. The Orbitrap XL was equipped with a captive spray micro/nanospray ionization source for ion generation (Michrom Bioresources Inc, Auburn, CA). Peptide separations were carried out on a 0.2 mm × 150 mm C18 column (5 μm, 300 Å, Michrom Bioresources Inc., Auburn, CA) at a flow rate of 2 μL/min. Mobile phases: A) HPLC water (J.T. Baker, Center Valley, PA):0.1 % (v/v) formic acid and B) acetonitrile (EMD Millipore, Billerica, MA):0.1 % (v/v) formic acid. The RP-HPLC separation was conducted at 2 % mobile phase B with a linear increase to 5 % B at 12 min, 15 % at 40 min, 30 % at 170 min, 55 % at 240 min, 85 % at 265 min and 90% at 270 min. A column wash was conducted at 90 % B for five minutes with a system equilibration to 2 % B over 24 min. The LTQ Orbitrap XL was operated in positive ion mode with the following dynamic exclusion settings: repeat count=3, repeat duration=30.00, exclusion list size=500, exclusion duration=350 s, exclusion mass width of ±1.50 m/z. MS/MS data was searched against the UniprotKB complete H. sapiens proteome (20,237 entries as of 18Sep12) using the MassMatrix search engine (v 2.4.2) [32-35]. Search parameters were as follows: three missed cleavages, MS1 ion tolerance of ± 10 ppm and a MSn ion tolerance of ± 0.8 Da. Keratin identifications (cytoskeletal, epidermal and cuticle) were considered contaminant proteins and removed from the analysis (listed in Supplemental Data 2-5). False discovery (FDR) estimations were made using target database reversed sequences. Protein IDs and spectral count data was parsed and combined using an in-house python application [36]. Retention of positive protein identifications were based upon a 5 % FDR and 2 unique peptide matches or 2 max decoys.
Acquisition and Preparation of Primary Serum & Bone Marrow Aspirate Derived Vesicles
Newly diagnosed transplant-ineligible MM patients enrolled on a Gruppo Italiano Malattie EMatologiche dell'Adulto (GIMEMA) phase III clinical trial (NCT#01063179) with adequate material available for analysis, were used to study serum levels of CD44 [37]. On a total of 511 patient enrolled in the trial, 233 had a serum sample at diagnosis that was suitable for our analysis. Clinical data at diagnosis, including ISS (International Staging System) stage and FISH profile, and overall survival were available. The international staging system is defined as: MGUS (serum monocolonal protein <3g/dl, clonal bone marrow plasma cells <10% and absence of end organ damage), SMM (serum monocolonal protein ≥3g/dl, clonal bone marrow plasma cells ≥10% and absence of end organ damage) and active multiple myeloma (presences of serum or urinary monoclonal protein, clonal bone marrow plasma cells ≥10% and evidence of end organ damage) [29]. Fluorescent in Situ Hybridization (FISH) analyses were performed using purified CD138+ cells obtained from bone marrow at diagnosis [38]. Patients carrying del17p13, t(4;14), or t(14;16) were classified as high cytogenetic risk and those with no chromosomal abnormality, t(11;14) or del(13q14) were considered standard risk [39]. 13 age-matched non-cancer subjects were used as a control having had no history of past or present neoplastic disease and no monoclonal gammopathy of undetermined significance (MGUS). All MM patients and healthy subjects included in the analyses were enrolled on IRB approved protocols and provided informed consent. For MM patients and healthy subjects, serum samples were stored at −80 °C.
The non-cancer, MGUS, smoldering MM (sMM) and active MM samples of plasma used to extract extracellular vesicles were derived from The Buckeye Surveillance, Contact and Research MM Registry (IRB #2010C0126). The plasma extracted from cancer free patients’ significant others consented for peripheral blood sample banking, allowing for an age-matched, were also used. EVs were isolated from serum by differential centrifugation/filtration from a total of 10 mL of blood (serum collection tube) obtained from 40 individual MM patients and 15 individual healthy donors, as previously published [13].
Serum and Bone Marrow Vesicle Sample Preparation & LC-MS/MS
Healthy donor serum and MM serum/bone marrow derived vesicles were prepared for LC-MS/MS analysis as described above for cell line derived vesicles. 1-2 μg of vesicle peptides were separated on a Thermo Scientific Easy-nLC II (Thermo Scientific, Waltham, MA) nano-HPLC system coupled to a hybrid Orbitrap Elite ETD (Thermo Scientific, Waltham, MA) mass spectrometer for mass analysis as described by Yi et al. [40]. In-line desalting was accomplished using a reversed-phase trap column (100 μm × 20 mm, Magic C18 AQ, 5 μm, 200 Å, Michrom Bioresources, Auburn, CA) followed by peptide separation on a reversed-phase column (75 μm × 250 mm, Magic C18 AQ, 5 μm 100 Å, Michrom Bioresources, Auburn, CA) directly mounted on the electrospray ion source. A 90-minute gradient from 7 % to 35 % acetonitrile in 0.1 % formic acid was conducted at a flow rate of 300 nL/minute. The heated capillary temperature was set to 300 °C and a spray voltage of 2,250 V was applied to the electrospray tip. The Orbitrap Elite instrument was operated in top 20 data-dependent mode, switching automatically between MS survey scans in the Orbitrap (AGC target value 1,000,000, resolution 240,000, and injection time 250 milliseconds) with MS/MS spectra acquisition in the linear ion trap (AGC target value of 10,000 and injection time 100 milliseconds). Collision-induced dissociation (CID) used normalized collision energy of 35 %. Selected ions were dynamically excluded for 30 seconds with a list size of 500 and exclusion mass width of ±10 ppm. MS/MS data was searched as described above for cell line vesicles. Keratin identifications are provided in Supplemental Data 8-11.
Label-Free Relative Quantitation
Label-free relative quantitation was performed using the approach originally described by Liu et al. and Colinge et al. [41,42]. Spectral counts do not include modified, or semi-tryptic peptides and shared peptides from multiple protein isoforms contribute equally to each isoform. The MS/MS data for each sample along with biological and/or technical replicates was combined using an in-house developed application into parsimonious protein lists. Spectral count quantitation was performed on the top protein matches with distinct peptide sequences (two or more) in at least a single sample and a protein score above the decoy threshold. The decoy threshold was set by the protein score for the third decoy ID or the decoy score that exceeds the false discovery rate of 5 %. Quantitation of spectral counts was limited to those proteins with at least one observation in each replicate with a minimum of 5 total spectral counts. The spectral count data and FDRs estimations are provided in Supplemental Data 2-5 & 8-11. Raw MS/MS spectral counts were adjusted to center, normalize and cluster across the arrays and genes using the average linkage function of the Cluster 3.0 software (C Clustering Library 1.50). Visualization of clustered spectral counts was conducted using the Java Tree View (ver. 1.1.6r2) software.
Computational Annotations and Bioinformatics
2-way and 4-way Venn diagrams of protein identifications were generated using the BioVenn (http://www.cmbi.ru.nl/cdd/biovenn/) and Venny web applications (http://bioinfogp.cnb.csic.es/tools/venny/) [43]. Gene ontological annotations were determined by the use of the PANTHER Classification System (http://www.pantherdb.org/) [44,45].
Immunoblotting
Extracellular vesicle preparations and cell pellets were lysed using 50 mM Tris at pH 7.5, 150 mM NaCl, 10 % Glycerol, 1.0 % NP-40, 0.1 % SDS supplemented with protease and phosphatase inhibitors. Protein concentrations were estimated by Bradford assay and equivalent quantities of the lysates were run on 4-15 % Tris-HCl SDS-PAGE TGX gels (Bio-Rad, Richmond, CA). Proteins were transferred to nitrocellulose and blotted for CD44, β2-MG, and GAPDH using either the ECL Western Blotting Detection Reagents (GE Healthcare, Piscataway, NJ) or SuperSignal West Femto Kit (Pierce Biotechnology, Rockford, IL).
Enzyme-linked immunosorbent assay (ELISA)
The ELISA performed for CD44 content in primary serum was conducted as described by the manufacturer (Abcam, Cambridge, MA). Briefly, primary serum was diluted 1:40 in Standard Diluent Buffer. In duplicate onto a 96-well plate, 100 μL of each sample, each standard and 1× control solution was added to the appropriate well and incubated for 1 h. All incubations were conducted at room temperature unless otherwise noted. The plate was washed, biotinylated anti-CD44 was added to each well and solutions were incubated for 30 min (repeated). Added 100 μL 1× Streptavidin-HRP and allowed to stand for 30 min. In the dark, 100 μL of Chromogen TMB substrate was added to each well and incubated for 15 min. 100 μL of Stop Reagent was added and absorbance at 450 nm was read on a spectrophotometer. Calculations for CD44 content were made based upon the standard curve and sample dilution factors.
Statistical Analysis
Overall survival was defined as the time from study enrollment to the time of death. Patients who did not die during the study follow up period were censored at the end of study follow up or the date of last follow up. Associations between baseline disease characteristics and overall survival were estimated through Cox proportional hazards models (Supplemental Data 19) and Kaplan and Meier. The assumption of proportional hazards was assessed through tests of the Schoenfeld residuals with no violations observed. All reported p-values are two-sided and analyses were performed using the statistical program Stata (version 13, StataCorp, College Station, TX).
RESULTS
MHC Class I and Beta2-Microglobulin are Commonly Enriched in MM Cell Lines Derived EV
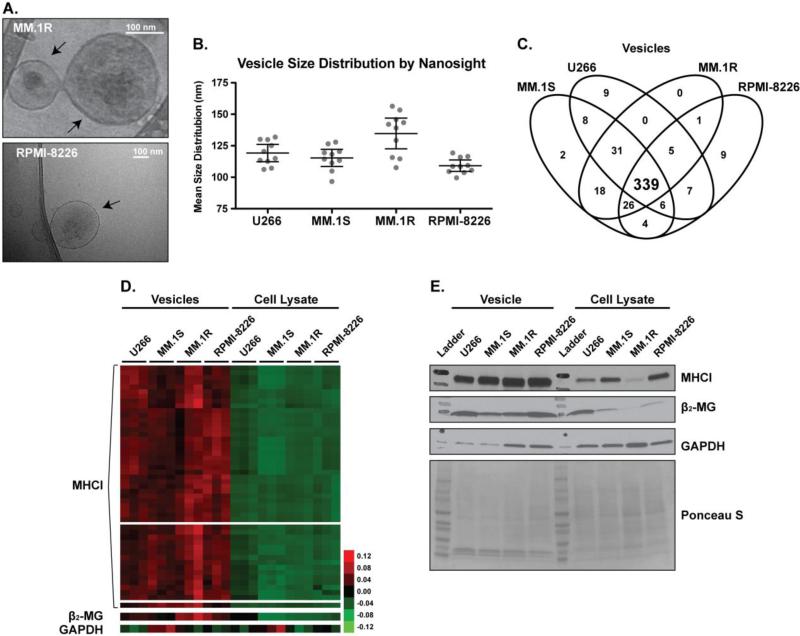
To further corroborate our previously published data in which we have shown MM cell lines produce EV with similar physical characteristics, we extended our proteomic analysis to the EV population derived from an additional MM cell line, RPMI-8226. Additionally, we analyzed vesicles from the dexamethasone (corticosteroid) resistant, MM.1R, cells. These cells are derived from the parental, MM.1S, cell line, that we previously analyzed and published [13]. Annexin V/Propidium Iodide (Annexin/PI) staining was performed in the MM cells used as source of EV. Less than 2 % apoptosis was observed, supporting the idea that EV are not the result of programmed cell death (Supplemental Figure 1). Cryo-Transmission Electron Microscopy (Cryo-TEM) and Nanoparticle Tracking Analsysis (NTA) show that the EV diameters are approximately 75 and 175 nm in all MM cell lines we analyzed (Figures 1A & 1B) [13].
Figure 1. MM cell line EVs are enriched for MHCI and β2-MG.
A) Cryo-TEM images of vesicles derived from MM.1R and RPMI-8226 cells. B) Mean size distributions for vesicles derived from U266, MM.1S, MM.1R and RPMI-8226 cell lines by Nanosight analysis (bars indicate 95 % CI). C) 4-way Venn diagram of vesicle protein identifications from MM cell lines. D) Expanded cluster map of extracellular vesicle and cellular lysate spectral counts for MHCI, β2-MG and GAPDH from MM cell lines E) Immunoblot of extracellular vesicle and cellular lysates for MHCI, β2-MG and GAPDH from MM cell lines. Ponceau S is provided for loading comparison. Collectively, these data show MM cell line derived vesicles are enriched in MHCI and β2-MG.
To further assess whether the EV isolated from these two cell lines could also be enriched in specific secreted proteins, a shotgun proteomics approach was applied to the cellular and derived EV fraction for protein identification [13]. Comparison of the identified MM EV proteins from the current study (RPMI-8226 and MM.1R) and our previous study (MM.1S and U266) shows a large number of overlapping proteins (339) between the four data sets (Figure 1C & Supplemental Data 2-5). Of the 497 total proteins identified, 77 were uniquely found, when compared to global lysate protein IDs, in the EVs of the 4 MM cell lines. Gene ontological analysis of the 77 proteins show a diverse range of molecular functions, biological processes, cellular components and protein classes (Supplemental 6). Interestingly, these data show that the MHCI and its associated binding protein, β2-MG, were among the most abundant communally enriched proteins across the 4 MM cell line derived EV analyzed (Figure 1D, 1E & Supplemental Data 7). In addition, β2-MG expression in the EV fraction was correlated with EV-MHCI protein levels (Figures 1D & 1E). These results support the hypothesis that an active antigen-presenting complex is secreted through EV, as previously reported in other cellular systems [46]. The observed enrichments could be associated with the enhanced proteosomic activity and consequent increase of processing antigens of myeloma cells compared to other forms of cancer [14].
MHCI and B2-MG Are Enriched in EV Isolated from the Serum of Newly Diagnosed MM Patients
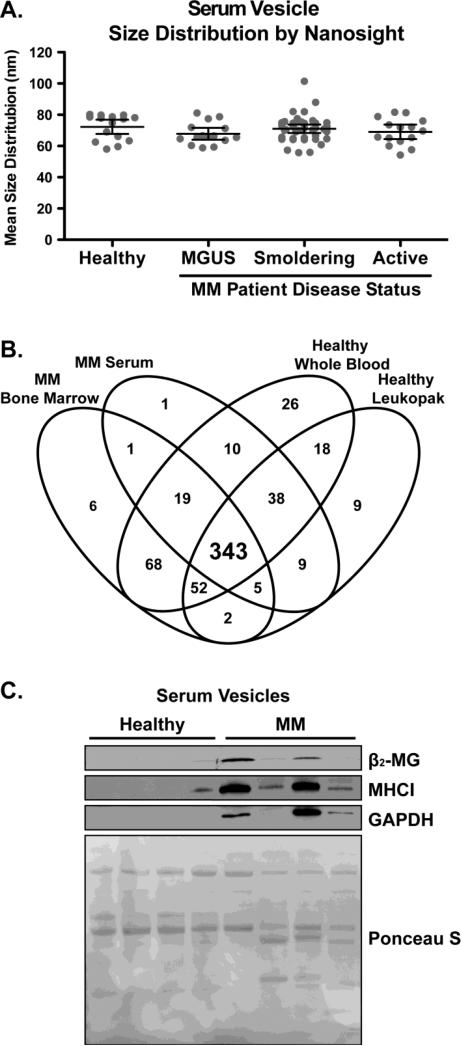
The clinical translation of in vitro models is of utmost importance. Hence, we assessed whether as in the case of cell lines, we could enrich a uniform EV population from the serum of different donors. NTA analysis was performed to evaluate the size distributions of the patient derived serum EVs of symptomatic MM (active MM), asymptomatic MM termed smoldering MM (sMM), premalignant MM termed Monoclonal Gammopathy of Undetermined Significance (MGUS) and from healthy donors. As shown in Figure 2A, the patient derived EV are between 50-100 nm in diameter with no statistically significant difference in numbers and size distributions between any of the groups by one-way ANOVA (p=0.4088). The NTA analysis provides results that are consistent with previous reports for serum derived EV diameters [47]. LC-MS/MS analysis, which was previously used for EV derived MM cell lines, was conducted on the EVs isolated from two newly diagnosed MM patients by collecting serum and bone marrow aspirates. The protein identifications and spectral count data from each sample type are provided in Supplemental Data 8-11. A comparison of the protein identifications by LC-MS/MS show many ubiquitously identified proteins (343) in each sample type (Figure 2B). These results are similar to those observed in the cell line EV data (Figure 1C). Clustering analysis confirmed enrichment of MHCI in the serum and in the BM of MM patients compared to healthy donors (Supplemental Data 11). Additionally, β2-MG was identified in these samples, but spectral counts were below the filter limits for the clustering analysis as described in the methods section. Immunoblot analysis validates enrichment of MHCI and β2-MG in the EV isolated from newly diagnosed MM patients compared to healthy donors with β2-MG expression correlated with EV-MHCI protein levels (Figure 2C).
Figure 2. MM Patient EVs are enriched for MHCI and β2-MG.
A) Mean serum vesicle size distributions from healthy donors (n=3) and MM patients of different stages (MGUS n=3, Smoldering n=8, Active n=3, error bars indicate 95 % CI). B) Venn diagram of overlapping protein identifications from MM serum, MM bone marrow aspirates and healthy donor peripheral blood samples (whole blood & leukopaks). C) Immunoblot for β2-MG, MHCI and GAPDH from serum of representative random healthy donors (lanes 1-4) and MM patients (lanes 5-8). Ponceau S is provided for loading comparison. Collectively, these results illustrate an enrichment of MHCI and β2-MG in the MM ex vivo EV samples when compared to healthy donor EVs.
CD44 is Enriched in the EVs Isolated from Dexamethasone Resistant MM Cells
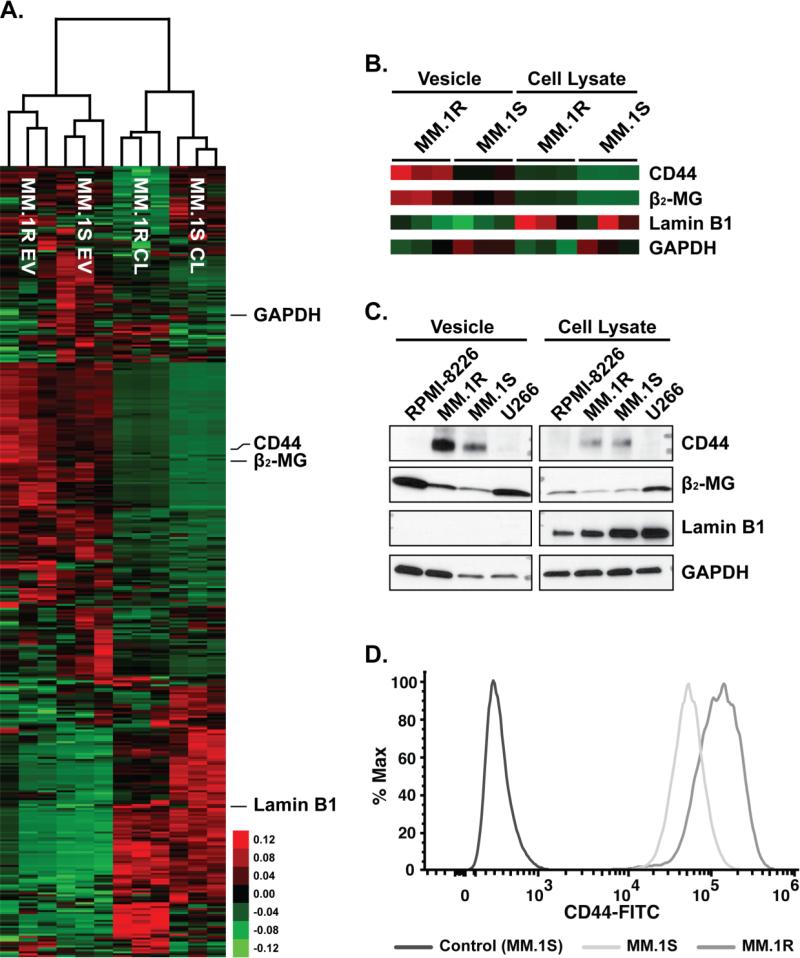
To assess whether MM derived EVs could harbor proteins associated with mechanisms of drug resistance, the EV and cellular protein contents from the dexamethasone resistant MM cells, MM.1R, were compared with those obtained from the sensitive parental cell line, MM.1S. By LC-MS/MS, we observed that the EV fraction obtained from MM.1R cells were significantly enriched in the main receptor of hyaluronic acid, CD44. Interestingly, CD44 expression has been extensively associated with mechanisms of drug resistance, not only in MM, but also in other forms of cancer [26,27,48,49]. CD44 protein enrichment was observed in the MM.1R derived EV, but no differences were found in the cellular protein levels of CD44 between sensitive (MM.1S) and resistant cells (MM.1R, Figure 3A & 3B). Immunoblot analysis of the EVs isolated from the MM cell lines confirms the enrichment of CD44 in the MM.1R EVs compared to other MM cell line's EVs, including the MM.1S cell derived EVs. The enrichment of EVCD44 from the MM.1R cells, compared to MM.1S, correlates to an increase of CD44 on the cell surface of MM.1R compared to MM.1S cells (Figure 3D). Because GAPDH seems to be differentially present in EV fraction, Lamin B1 was used to assess the purity of our EV preparation compared to the cellular fraction. Furthermore, β2-MG staining was used to show enrichment of the EV preparation (Figure 3C). These data support the hypothesis that membrane CD44 could be responsible of mechanisms of drug resistance, as also previously suggested by Vincent et al [50].
Figure 3. Corticosteroid resistant cell lines harbor EVs with proteins associated drug resistance.
A) A cluster map of 403 extracellular vesicle and cellular lysate protein spectral counts from the resistant (MM.1R) and parent (MM.1S) cell lines with at least one observation in each replicate of greater than five spectral counts. B) Expanded cluster maps of CD44, β2-MG and GAPDH from the MM.1R and MM.1S data. C) Immunoblot for CD44, β2-MG, Lamin B1 and GAPDH from extracellular vesicle and cellular lysates. D) Flow cytometry for surface expression of CD44 from the MM cell lines. Collectively, these data show an enrichment of CD44 in the corticosteroid resistant cell line MM.1R EVs compared the parent MM.1S cell line.
Identification of Extracellular Vesicles as the Primary Location of Serum CD44 in MM Patients
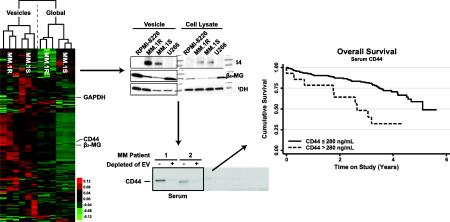
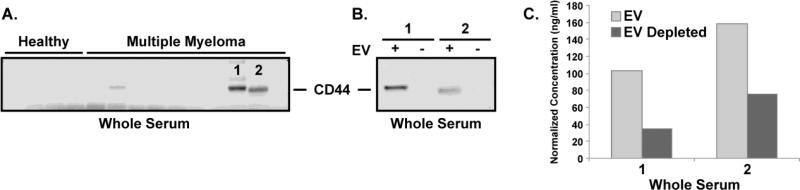
The glycoprotein CD44 has been shown to play a key role in various cellular functions in normal cells, such as signaling, cell adhesion and microenvironmental interactions [51]. However in hematological malignancies, CD44 has been implicated in increased invasiveness, cancer cell trafficking and resistance to apoptosis [51]. In our study, CD44 was identified not only in the EV of MM resistant cells but also in primary patients with variable abundance (Figure 3 & Supplemental Data 12). Through immunoblotting, we show CD44 is contained in the EVs isolated, not only from MM cells, but also from MM patient serum (32 newly diagnosed MM patients and 13 healthy individuals). Figure 4A shows a representative blot for CD44, from whole serum, with additional blots provided in Supplemental Data 13. These data illustrate the variable expression of CD44 in several MM patients’ serum when compared to healthy individuals. To confirm the localization of CD44 to the EV of the MM patient serum, the serum of two patients showing high CD44 levels, in labeled 1 & 2 in Figure 4A, was depleted of EV by ultracentrifugation. Figure 4B shows the immunoblot for CD44 from the samples not depleted (+) and depleted (−) of EV. These results demonstrate a reduced CD44 content when serum is depleted of EVs, suggesting the CD44 in the patent's serum is mainly localized to the EV. The depletion of CD44 from the serum, by removing the EV, was further validated by an ELISA analysis (Figure 4C). Similarly to Figure 4B, the ELISA results, of those samples labeled 1 & 2 in Figure 4A not depleted and depleted of EV, show a reduction in CD44 content in the serum depleted of EV. Collectively, these results show the CD44 content of MM patient serum is primarily localized to the circulating EV fraction.
Figure 4. Identification of the extracellular vesicles as the primary location of serum CD44 in MM patients.
A) Immunoblot for CD44 from whole serum of healthy individuals and MM patients. B) Immunoblot for CD44 from whole serum (+) and vesicle depleted serum (−) from those samples labeled 1 & 2 in Figure 4A. C) ELISA for CD44 from whole serum and vesicle depleted serum from those samples labeled 1 & 2 in Figure 4A. Collectively, these results show CD44 is primarily located in the serum vesicles of MM patients.
Serum CD44 Levels in Newly Diagnosed MM Patients is Associated with Overall Survival
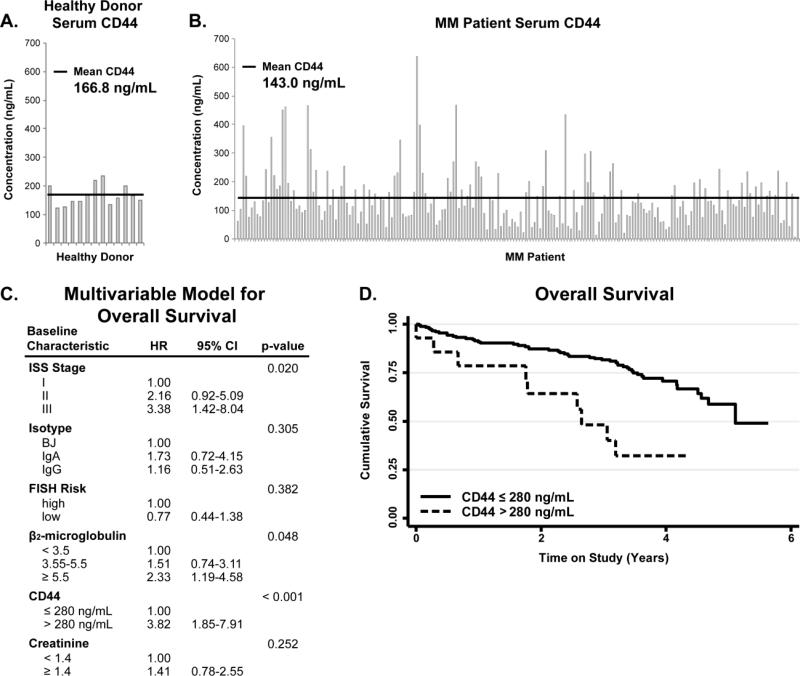
Our results indicated increased expression levels of CD44 in the EVs isolated from corticosteroid resistant MM cells, possibly suggesting a role in drug resistance. EV associated CD44 is differentially expressed in the serum of MM patients and increased expression of specific CD44 isoforms on the surface of MM cells by immunohistochemistry has been correlated with poor prognosis and decreased overall survival [52]. Hence, we hypothesize that serum CD44 could be used as a prognostic marker in MM patients, which are normally treated in combinatorial therapy as standard of care. ELISA assays were conducted for CD44 from the serum collected from 13 healthy individuals and 233 newly diagnosed MM patients prospectively enrolled in a Phase 3 clinical trial including corticosteroid treatment. Patients received Velcade–Melphalan–Prednisone (VMP) or Velcade–Melphalan–Prednisone–Thalidomide followed by maintenance with Velcade–Thalidomide (VMPT–VT) through the parent clinical trial (NCT#01063179). CD44 values for the healthy individuals range from 235.69-122.32 ng/mL while the MM patients have a much wider range, 6.43-637.51 ng/mL (Figure 5A & 5B, Supplemental Data 14). The statistical analysis of the ELISA data shows no statistical difference between the means of MM patients (143.0 ng/mL) and healthy donor (166.8 ng/mL) CD44 levels based on a student's t-test (p=0.5070). However, these data show that the CD44 levels in the MM patient serum are highly variable and maybe useful for stratifying MM patients at diagnosis. In fact, one approach to improve risk prognostication in patients with MM is to use new technologies to stratify patients based on distinct outcomes [53]. Outcome prediction for most patients is based on the International Staging System (ISS) and the presence or absence of specific fluorescent in-situ hybridization abnormalities. Additional biomarkers are necessary to improve precision. Our data show that CD44, which has been involved in mechanisms of Myeloma drug resistance and therapeutic outcome, is present in soluble form in the serum of MM patients and it can be released from MM cells through EV [27,54]. Therefore, we decided to evaluate its possible utility as non-invasive biomarker. To address this question, we measured serum CD44 levels in a large cohort of newly diagnosed MM patients who were uniformly treated and followed, correlating serum CD44 levels with clinical outcome to test their prognostic impact in a multivariate model [37].
Figure 5. Identification of serum CD44 as a predictor of overall survival in MM patients.
A) ELISA for CD44 from 13 non-cancer donor age matched serum. B) ELISA for CD44 from MM patient serum in 233 newly diagnosed MM patients. C) Multivariable model for overall survival, HR: hazard ratio, 95% CI: 95% confidence interval. D) Kaplan Meier survival curve for those patients with serum CD44 concentrations greater than 280 ng/mL. Collectively, these data show serum CD44 levels predict overall survival in multiple myeloma patients.
Of the 233 newly diagnosed patients analyzed for CD44, 87 died during the course of follow up with a five-year overall survival of 56.6 % (95% CI: 44.8%-66.8%). In age-adjusted models, a strong increased risk of death was associated with increased ISS stage, elevated β2-MG and increased serum CD44 (Figure 5C). When modeled together, following multiple imputation of missing ISS stage, serum CD44 and ISS stage remain significantly associated with overall survival (Figure 5C). The risk of death increased significantly when CD44 was greater than the maximum value in normal controls of 280 ng/mL after adjusting for age and stage (Figure 5D, adjusted hazard ratio: 3.00, 95% CI: 1.36-6.45). Collectively, these data establish serum CD44 as a potential biomarker of overall survival in multiple myeloma.
DISCUSSION
EVs are becoming a research focus due to their roles in cancer cell biology such as immune evasion, therapeutic resistance, proliferation and metastases (recently reviewed in [55,56]). EV from MM BM-MSCs contain miRNA that are taken up by MM cells and influence tumor growth and dissemination, and it is not known if this is bi-directional and if MM cells derived EV are also responsible to affect the cancer microenvironment towards their survival advantage [4]. While numerous studies of vesicle characterization and biology have been conducted in many cancer models, the role of EV in MM remains relatively unstudied. Roccaro et al. recently established implications for the importance of EV released by BMSC in MM biology through characterization of the protein content of the MM BM-MSC derived vesicles by protein array [4]. They found that when compared to normal BM-MSC, the MM derived BMMSC EV had higher expression of oncogenes and protein kinases [4]. They establish a functional relevance of EV to the MM microenvironment, however these results focus only on the BM-MSC derived vesicles and the protein spectrum analyzed was limited.
Here we found that EVs isolated from MM cells are enriched in MHC-1 antigen presenting complex and its binding protein β2-MG, this observation is compatible with the enhanced proteasome activity of MM cells compared to other cancers and the ability of functional MHC-1 to bind and present peptides, generated from protein degradation by the proteasome [14]. A proportional enrichment was also observed in the peripheral blood of MM patients for MHCI when compared to healthy donors by LC-MS/MS and MHCI and β2-MG immunoblot analysis. It is well known that the serum level of β2-MG is an important prognostic factor in MM. However, vesicle β2-MG represents only a small percentage (data not shown) of the total serum β2-MG, suggesting that two separate β2-microglobulin populations coexist in the serum of patients. However, further studies are required to investigate their different biological functions.
CD44 has been identified on extracellular vesicles derived from a number of cell types and locations, including B cells, bladder and colorectal cancer cells, saliva and urine [21-25]. Specifically in MM, CD44 causes adhesion of MM cells to BMSC and induction of IL-6, the primary cytokine for MM cell survival, secretion by the BMSC [51]. Additionally, specific alternatively spliced variants of CD44, such as CD44v9, have been studied on MM cells linking CD44 expression to advancing disease status and shorter overall survival [52].
Our experiments show that CD44 is particularly enriched in the EV fraction of corticosteroid resistant MM.1R cells and is differentially expressed in the EV fraction of MM patients. Our serum depletion experiments show that CD44 is primarily localized to the peripheral EV in MM patients. Furthermore, ELISA assays for CD44 content from the serum of 254 newly diagnosed MM patients enrolled in a Phase 3 randomized trial show highly variable CD44 levels [13]. Additionally, those patients with >280 ng/mL serum CD44 showed a reduced overall survival time. These results suggest the potential use of CD44 as a prognostic biomarker. Because previously published data have specifically associated CD44 as survival protein for MM cells responsible in increasing MM cell adhesion to BMSC and in inducing IL-6 secretion by the BMSC [51], it is not surprising that increased level of soluble CD44 can be associated with decrease survival of MM patients. The presence of CD44 in EV of other cancers highlights the key role of this protein as hallmark of cancerous EV. Moreover, its differential expression between healthy subjects and MM patients suggests it could be a selective target for cancer vesicles.
Supplementary Material
Highlights.
Characterized the vesicular protein content from MM cell lines and MM patients.
MHC Class I is enriched in vesicles of MM cell lines and patients.
CD44 is highly enriched in corticosteroid resistant MM cell lines.
Serum CD44 is a predictive biomarker of overall survival in MM.
SIGNIFICANCE.
Extracellular vesicles are becoming a research focus due to their roles in cancer cell biology such as immune evasion, therapeutic resistance, proliferation and metastases. While numerous studies of vesicle characterization and biology have been conducted in many cancer models, the role of EV in MM remains relatively unstudied. Here we found that EVs isolated from MM cells are enriched in MHC-1 antigen presenting complex and its binding protein β2-MG, this observation is compatible with the enhanced proteasome activity of MM cells compared to other cancers and the ability of functional MHC-1 to bind and present peptides, generated from protein degradation by the proteasome. Additionally, our experiments show that CD44 is particularly enriched in the EV fraction of corticosteroid resistant MM.1R cells and is differentially expressed in the EV fraction of MM patients. This is of high significance due to the established role of CD44 in adhesion of MM cells to BMSC and induction of IL-6, the primary cytokine for MM cell survival, secretion by the BMSC. Furthermore, ELISA assays for CD44 content from the serum of 254 newly diagnosed MM patients enrolled in a Phase 3 randomized trial show highly variable CD44 levels and those patients with >280 ng/mL serum CD44 showing a reduced overall survival time. These results suggest the potential use of CD44 as a prognostic biomarker in MM.
ACKNOWLEDGEMENTS
This work was supported by the Ohio State University Pelotonia Fellowship Program (A.R.), by Ohio State University Pelotonia Idea grant (F.P., D.B.), in part from NIH grants (R01 CA107106, P01 CA124570 and RC2 AG036559), NSF (EEC-0425626 and EEC-019790) and funded in part by Multiple Myeloma Opportunities for Research & Education (MMORE) and in part from Sidney Kimmel Foundation Scholar Award (F.P.). The cryo-TEM data were obtained at the TEM facility at the Liquid Crystal Institute, Kent State University, supported by the Ohio Research Scholars Program Research Cluster on Surfaces in Advanced Materials. The authors thank Dr. Min Gao for technical support provided for the TEM experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
Authors have no conflict of interest to declare.
REFERENCES
- 1.American Cancer Society Cancer Facts & Figures. 2011:1–68. < http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/docu ments/document/acspc-031941.pdf>.
- 2.Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2:175–187. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- 3.Reagan MR, Ghobrial IM. Multiple Myeloma-Mesenchymal Stem Cells: Characterization, Origin, and Tumor-Promoting Effects. Clin Cancer Res. 2012;18:342–349. doi: 10.1158/1078-0432.CCR-11-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roccaro AM, Sacco A, Maiso P, Azab AK, Tai Y-T, Reagan M, et al. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest. 2013;123:1542–1555. doi: 10.1172/JCI66517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vacca A, Ribatti D, Roccaro AM, Frigeri A, Dammacco F. Bone marrow angiogenesis in patients with active multiple myeloma. Semin Oncol. 2001;28:543–550. doi: 10.1016/s0093-7754(01)90022-3. [DOI] [PubMed] [Google Scholar]
- 6.Mahindra A, Hideshima T, Anderson KC. Multiple myeloma: biology of the disease. Blood Rev. 2010;24:S5–S11. doi: 10.1016/S0268-960X(10)70003-5. [DOI] [PubMed] [Google Scholar]
- 7.Damiano JS, Cress AE, Hazlehurst LA, Shtil AA, Dalton WS. Cell adhesion mediated drug resistance (CAM-DR): role of integrins and resistance to apoptosis in human myeloma cell lines. Blood. 1999;93:1658–1667. [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Hendrix A, Hernot S, Lemaire M, De Bruyne E, Van Valckenborgh E, et al. Bone marrow stromal cell-derived exosomes as communicators in drug resistance in multiple myeloma cells. Blood. 2014;124:555–566. doi: 10.1182/blood-2014-03-562439. [DOI] [PubMed] [Google Scholar]
- 9.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. BBA-Gen Subjects. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113:3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- 11.Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 12.Wubbolts R, Leckie RS, Veenhuizen PTM, Schwarzmann G, Möbius W, Hoernschemeyer J, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 13.Harshman SW, Canella A, Ciarlariello PD, Rocci A, Agarwal K, Smith EM, et al. Characterization of Multiple Myeloma Vesicles by Label-Free Relative Quantitation. Proteomics. 2013;13:3013–3029. doi: 10.1002/pmic.201300142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards CM, Lwin ST, Fowler JA, Oyajobi BO, Zhuang J, Bates AL, et al. Myeloma cells exhibit an increase in proteasome activity and an enhanced response to proteasome inhibition in the bone marrow microenvironment in vivo. Am J Haematol. 2009;84:268–272. doi: 10.1002/ajh.21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pokharel D, Padula MP, Lu JF, Tacchi JL, Luk F, Djordjevic SP, et al. Proteome analysis of multidrug-resistant, breast cancer-derived microparticles. J Extracell Vesicles. 2014;3:24384. doi: 10.3402/jev.v3.24384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi D-Y, You S, Jung JH, Lee JC, Rho JK, Lee KY, et al. Extracellular vesicles shed from gefitinib-resistant nonsmall cell lung cancer regulate the tumor microenvironment. Proteomics. 2014;14:1845–1856. doi: 10.1002/pmic.201400008. [DOI] [PubMed] [Google Scholar]
- 17.Ma X, Chen Z, Hua D, He D, Wang L, Zhang P, et al. Essential role for TrpC5-containing extracellular vesicles in breast cancer with chemotherapeutic resistance. Proc Natl Acad Sci USA. 2014;111:6389–6394. doi: 10.1073/pnas.1400272111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahrens T, Assmann V, Fieber C, Termeer CC, Herrlich P, Hofmann M, et al. CD44 is the principal mediator of hyaluronic-acid-induced melanoma cell proliferation. J Invest Dermatol. 2001;116:93–101. doi: 10.1046/j.1523-1747.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- 19.Ahrens T, Sleeman JP, Schempp CM, Howells N, Hofmann M, Ponta H, et al. Soluble CD44 inhibits melanoma tumor growth by blocking cell surface CD44 binding to hyaluronic acid. Oncogene. 2001;20:3399–3408. doi: 10.1038/sj.onc.1204435. [DOI] [PubMed] [Google Scholar]
- 20.Naor D, Sionov RV, Ish-Shalom D. CD44: structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 21.Buschow SI, van Balkom BWM, Aalberts M, Heck AJR, Wauben M, Stoorvogel W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol Cell Biol. 2010;88:851–856. doi: 10.1038/icb.2010.64. [DOI] [PubMed] [Google Scholar]
- 22.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi D-S, Park JO, Jang SC, Yoon YJ, Jung JW, Choi D-Y, et al. Proteomic analysis of microvesicles derived from human colorectal cancer ascites. Proteomics. 2011;11:2745–2751. doi: 10.1002/pmic.201100022. [DOI] [PubMed] [Google Scholar]
- 24.Mathivanan S, Lim JWE, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE, et al. Proteomic Analysis of Human Parotid Gland Exosomes by Multidimensional Protein Identification Technology (MudPIT) J Proteome Res. 2009;8:1304–1314. doi: 10.1021/pr800658c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohwada C, Nakaseko C, Koizumi M, Takeuchi M, Ozawa S, Naito M, et al. CD44 and hyaluronan engagement promotes dexamethasone resistance in human myeloma cells. Eur J Haematol. 2008;80:245–250. doi: 10.1111/j.1600-0609.2007.01014.x. [DOI] [PubMed] [Google Scholar]
- 27.Bjorklund CC, Baladandayuthapani V, Lin HY, Jones RJ, Kuiatse I, Wang H, et al. Evidence of a role for CD44 and cell adhesion in mediating resistance to lenalidomide in multiple myeloma: therapeutic implications. Leukemia. 2014;28:373–383. doi: 10.1038/leu.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3. Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 29.International Myeloma Working Group Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 2003;121:749–757. [PubMed] [Google Scholar]
- 30.Rajkumar SV. Multiple myeloma: 2012 update on diagnosis, risk-stratification, and management. Am J Haematol. 2012;87:78–88. doi: 10.1002/ajh.22237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro JP, Biswas S, Merchant AS, Satoskar A, Taslim C, Lin S, et al. A quantitative proteomic workflow for characterization of frozen clinical biopsies: laser capture microdissection coupled with label-free mass spectrometry. J Proteomics. 2012;77:433–440. doi: 10.1016/j.jprot.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu H, Yang L, Freitas MA. A robust linear regression based algorithm for automated evaluation of peptide identifications from shotgun proteomics by use of reversed-phase liquid chromatography retention time. BMC Bioinformatics. 2008;9:347. doi: 10.1186/1471-2105-9-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, Zhang L, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J Proteome Res. 2008;7:138–144. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu H, Freitas MA. Monte carlo simulation-based algorithms for analysis of shotgun proteomic data. J Proteome Res. 2008;7:2605–2615. doi: 10.1021/pr800002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palumbo A. Bortezomib-Melphalan-Prednisone-Thalidomide Followed by Maintenance With Bortezomib-Thalidomide Compared With Bortezomib-Melphalan-Prednisone for Initial Treatment of Multiple Myeloma: A Randomized Controlled Trial. J Clin Oncol. 2010;28:5101–5109. doi: 10.1200/JCO.2010.29.8216. [DOI] [PubMed] [Google Scholar]
- 38.Ross FM. The T(14;20) Is a Poor Prognostic Factor in Myeloma but Is Associated with Long-Term Stable Disease in Monoclonal Gammopathies of Undetermined Significance. Haematologica. 2010;95:1221–1225. doi: 10.3324/haematol.2009.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munshi NC. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood. 2011:4694–4700. doi: 10.1182/blood-2010-10-300970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi EC, Lee H, Aebersold R, Goodlett DR. A microcapillary trap cartridge-microcapillary high-performance liquid chromatography electrospray ionization emitter device capable of peptide tandem mass spectrometry at the attomole level on an ion trap mass spectrometer with automated routine operation. Rapid Commun Mass Spectrom. 2003;17:2093–2098. doi: 10.1002/rcm.1150. [DOI] [PubMed] [Google Scholar]
- 41.Liu H, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 42.Colinge J, Chiappe D, Lagache S, Moniatte M, Bougueleret L. Differential proteomics via probabilistic peptide identification scores. Anal Chem. 2005;77:596–606. doi: 10.1021/ac0488513. [DOI] [PubMed] [Google Scholar]
- 43.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–10. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vraetz T, Ittel TH, van Mackelenbergh MG, Heinrich PC, Sieberth HG, Graeve L. Regulation of β2-microglobulin expression in different human cell lines by proinflammatory cytokines. Nephrol Dial Transplant. 1999;14:2137–2143. doi: 10.1093/ndt/14.9.2137. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159–1167. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 48.Yang X, Iyer AK, Singh A, Milane L, Choy E, Hornicek FJ, et al. Cluster of Differentiation 44 Targeted Hyaluronic Acid Based Nanoparticles for MDR1 siRNA Delivery to Overcome Drug Resistance in Ovarian Cancer. Pharm Res. 2015;32:2097–2109. doi: 10.1007/s11095-014-1602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoon C, Park DJ, Schmidt B, Thomas NJ, Lee H-J, Kim TS, et al. CD44 expression denotes a subpopulation of gastric cancer cells in which Hedgehog signaling promotes chemotherapy resistance. Clin Cancer Res. 2014;20:3974–3988. doi: 10.1158/1078-0432.CCR-14-0011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Vincent T, Mechti N. IL-6 regulates CD44 cell surface expression on human myeloma cells. Leukemia. 2004;18:967–975. doi: 10.1038/sj.leu.2403333. [DOI] [PubMed] [Google Scholar]
- 51.Hertweck MK, Erdfelder F, Kreuzer K-A. CD44 in hematological neoplasias. Ann Haematol. 2011;90:493–508. doi: 10.1007/s00277-011-1161-z. [DOI] [PubMed] [Google Scholar]
- 52.Stauder R, Van Driel M, Schwarzler C, Thaler J, Lokhorst HM, Kreuser ED, et al. Different CD44 splicing patterns define prognostic subgroups in multiple myeloma. Blood. 1996;88:3101–3108. [PubMed] [Google Scholar]
- 53.Kumar SK, Uno H, Jacobus SJ, Van Wier SA, Ahmann GJ, Henderson KJ, et al. Impact of gene expression profiling-based risk stratification in patients with myeloma receiving initial therapy with lenalidomide and dexamethasone. Blood. 2011;118:4359–4362. doi: 10.1182/blood-2011-03-342089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinstock M, Aljawai Y, Morgan EA, Laubach J, Gannon M, Roccaro AM, et al. Incidence and clinical features of extramedullary multiple myeloma in patients who underwent stem cell transplantation. Br J Haematol. 2015;169:851–858. doi: 10.1111/bjh.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metast Rev. 2013;32:623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med. 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.