Summary
All organisms must replicate their genetic information accurately to ensure its faithful transmission. DNA polymerase errors provide an important source of genetic variation that can drive evolution. Understanding the origins of genetic variation will inform our understanding of evolution and the development of genetic diseases. A number of factors have been proposed to influence mutagenesis [1–10]. Here, we used mutation accumulation lines, whole-genome sequencing and whole-transcriptome analysis to study the locations and rate at which mutations arise in bacteria with as little selection bias as possible [11, 12]. Our analysis of greater than 7,000 replication errors in over 180 sequenced lines that underwent a total of more than 370,000 generations has provided new insights into how DNA polymerase errors sculpt genetic variation and drive evolution. Homopolymer run enrichment outside of genes causes insertions and deletions in these regions. Genes encoded in the lagging strand are transcribed such that RNA polymerase and DNA polymerase collide head-on. Head-on genes have been proposed to mutate at a higher rate than genes transcribed codirectionally with DNA polymerase progression due to conflicts between transcription and DNA replication [6, 10]. We did not detect associations between the number of base pair substitutions in genes and their orientation or expression. Strikingly, any higher mutation rate for head-on genes can be explained by differing sequence composition between the leading and lagging strands and the error bias for DNA polymerase in specific sequence contexts. Therefore, we find local sequence context is the major determinant of mutagenesis in bacteria.
Results and Discussion
DNA is the storage medium for genetic information throughout cellular life. Errors made by DNA polymerase during normal DNA replication have the potential to introduce genetic variation and drive evolution. Many studies have imposed a selection or stress prior to analysis of mutational reporters [1, 6, 10], which limits observation to a small number of possible mutations in a restricted sequence context. Recent work using mutational reporters suggests that genes encoded on the lagging strand, i.e., transcribed such that RNA polymerase will collide head-on with DNA polymerase, have higher mutation rates and evolve more quickly than genes transcribed codirectionally [6, 10]. A translesion DNA polymerase, PolY1, has been suggested to mediate this effect [10], but the contribution of the replicative DNA polymerase is unclear. Local sequence context has a strong effect on DNA polymerase error rate in vivo [9], and this cannot be fully appreciated when using mutational reporters. Therefore, a combination of genome-wide and transcriptome-wide approaches is necessary to determine the combined effects of sequence composition, gene expression and gene orientation on mutation rate. In prior bacterial mutation accumulation (MA) line work, the effect of transcription on mutation was either not determined or the mutations analyzed were not likely to be caused by DNA polymerase errors [2, 4, 9, 13]. Integrating our MA line analysis with RNA-seq data we were able to determine to what extent sequence context, gene expression and gene orientation impact DNA polymerase accuracy. Our study shows that head-on genes have a slightly higher mutation rate than codirectional genes, and this difference can be explained entirely by the differing sequence composition of head-on genes and the error bias of DNA polymerase within specific sequence contexts.
In order to determine factors impacting mutation occurrence in vivo, we observed mutations caused by DNA polymerase errors during DNA replication throughout the genome of the model bacterium Bacillus subtilis using MA lines [11, 12]. MA lines make use of repeated bottlenecks to reduce selection against highly deleterious mutations. We inactivated DNA mismatch repair (MMR) and determined where and in what context base pair substitutions (BPSs) and insertions and deletions (indels) were produced by DNA polymerase.
Our results are briefly summarized in Table S1 and a full list of the variants is presented in Table S2. Loss of mutSL gave rise to a 60-fold increase in overall mutation rate (Table S1), which is similar to what we observed using rifampin resistance as a measure of mutation occurrence (Figure S1). In the absence of mutSL an average of 15.5 generations pass between mutations, yielding a DNA polymerase error rate of one error resulting in a mutation per ≈ 59,000,000 nucleotides replicated. In wild type B. subtilis DNA replication is accurate, with an average of 909 generations between mutations (Table S1). This is similar to E. coli, which undergoes ≈ 1000 generations between mutations [4]. WalJ is a 5′ → 3′ exonuclease involved in MMR in Bacillus species [14] (Table S1 and Figure S1). Because ablation of mutL endonuclease activity and deletion of walJ and mutSL all yield defects in MMR with identical mutation spectra (Figure S1D), for the remainder of this study, we binned data from the mutL[E468K], ΔwalJ and ΔmutSL and lines such that 6952 mutations were present in the combined total of 118 MMR-lines and 295 mutations were present in the wild type MMR+ lines.
To assess the possibility of selection bias in our study we determined the ratio of nonsynonymous to synonymous base pair substitutions. We found the nonsynonymous to synonymous ratio was 3.0 and 2.1 in MMR+ and MMR- lines, respectively (Figure S2A). Monte Carlo simulation of expected ratios of nonsynonymous to synonymous substitutions revealed that there was a slight enrichment of nonsynonymous substitutions in our data compared to expected distributions (Figure S2A). This is in close agreement with prior MA line work in B. subtilis and indicates that selection bias was minimized throughout the MA line procedure [9].
Globally, indels appeared to be evenly distributed throughout the genome (Figure S2B). However, upon closer examination of the relationship between indel and gene locations, we determined that indels were enriched outside of coding sequences (CDSs) (Figure 1A). Long homopolymer runs are also enriched intergenically (Figure S3A). In agreement with other studies, indel mutation rate in our dataset increases exponentially with increasing homopolymer run length [4] (Figure S3B). This yields a very high rate of indel occurrence in homopolymer runs longer than five nucleotides. Given that indels are enriched between CDSs and indel rate is very high in homopolymers, we tested whether intergenic enrichment of indels was caused by concomitant enrichment of homopolymer runs.
Figure 1. Indels in homopolymer runs are enriched outside of coding regions.
All starts and ends of CDSs were aligned at relative position zero. Indels were counted in 50 bp bins, offset by 25. Negative distances indicate the indel was 5′ to a CDS start site or 3′ to a CDS end site. The lines in each plot represent the locally weighted polynomial regression (loess) fit to the data. (A) The number of indels found in each bin without correcting for homopolymer run bias. (B) Uncorrected indel counts separated by the homopolymer run length in which the indel was produced. (C) Expected indel count in each bin after applying a correction for homopolymer run bias (see Supplemental Experimental Procedures). See also Figure S3.
Most indels observed immediately outside CDSs were in homopolymer runs ranging in length from six to eight (Figure 1B). Because the reference genome contains only 28 homopolymer runs of length nine and two homopolymers of length ten, few indels were observed in homopolymers of these lengths. Correcting for the bias in homopolymer run distribution, we found that enrichment of indels outside of CDSs can be explained by enrichment of homopolymer runs in these regions (Figure 1). We propose that CDSs containing long homopolymer runs have been selected against due to the high propensity of long homopolymers to obtain indels, which will cause frameshifts and loss of CDS function.
Grouping transitions into their four possible categories, we found that complementary transitions, i.e. C → T and G → A, or T → C and A → G, accumulated symmetrically between the two replichores regardless of whether MMR was intact (Figure 2B and C). This distribution cannot be explained by bias in the distribution of nucleotides between the replichores, as our calculation of mutation rate in Figure 2C was normalized to the number of each nucleotide found in the reference sequence of each replichore. A similar complementary symmetry was observed in undomesticated B. subtilis, Mesoplasma florum, E. coli and S. cerevisiae [4, 8, 9]. We attribute the complementary symmetry of transition accumulation between the replichores to differences in the fidelity of leading and lagging strand replication [1].
Figure 2. Transitions display complementary symmetry between replichores.
(A) Schematic representation of the B. subtilis chromosome. DNA replication initiates at oriC, which is at position zero in the reference genome, and proceeds bidirectionally toward terC, which is at 1.97 Mb in the reference genome. The left and right replichores of the chromosome are in green and black, respectively. (B) Cumulative distributions of the indicated types of transitions along the genome. The origin of replication is indicated by the vertical dashed red line and the terminus is at both ends. (C) A barplot displaying the mutation rate for the indicated types of transitions binned by replichore. The mutation rate is normalized to the number of each base in each replichore as described in Supplemental Experimental Procedures. Error bars represent 95% confidence intervals determined by bootstrapping. All comparisons between left and right replichores are statistically significant with the exception of T → C transitions in MMR+ lines. MMR intact refers to wild type data and MMR deficient refers to the pooled data for ΔmutSL, ΔwalJ, and mutL[E468K].
It has recently been suggested that head-on oriented genes have an increased mutation rate due to collisions between RNA and DNA polymerases [6, 10]. We therefore tested whether expression of CDSs and their orientation relative to replication impacted CDS mutation rate by performing RNA-seq and integrating transcript abundance with MA line data for BPSs. We performed multiple linear regression using the number of BPSs in each CDS as the dependent variable against CDS orientation, length, steady-state transcript abundance (RPKM), the interaction between orientation and length, and the interaction between orientation and RPKM (see Equation S5 and Supplementary Methods). We found that orientation of a CDS was a significant predictor of the number of BPSs it accumulated only if outliers were included in the regression (Figure 3). Performing a separate regression using only those genes within three standard deviations of the mean length or RPKM (11 head-on and 90 codirectional of the total 4163 CDSs were determined to be outliers) yielded gene length as the only significant predictor of the number of BPSs in a CDS (Figure 3 and Table S3). We conclude that in the absence of strong selective pressure for specific BPSs and when virtually all sequence contexts can be interrogated, gene expression and orientation has no detectable effect on mutation rate. It is possible, however, that more generations and observed mutations would enable us to detect an effect of gene orientation. Therefore, we performed Monte Carlo simulation of millions of generations to determine if the mutation rates of head-on and codirectional genes are impacted by local sequence context.
Figure 3. Regression of base pair substitution count against coding sequence length, expression and orientation.
(A) A graphical representation of linear regression analysis. Blue triangles indicate head-on CDSs and red circles represent codirectional CDSs. Lines indicate the linear fit to the data, and the shaded region around each line indicates the 95% confidence interval for the fit. The plot on the left includes all CDSs and the plot on the right excludes CDSs greater or less than three standard deviations from either the mean length or expression (RPKM). See Table S4 for a summary of each CDS including which were determined to be outliers. (B) A table listing the variables determined to be significantly associated with the average number of BPSs found in CDSs either with or without outliers. See Table S3 for detailed results and Equation S5 for the regression model.
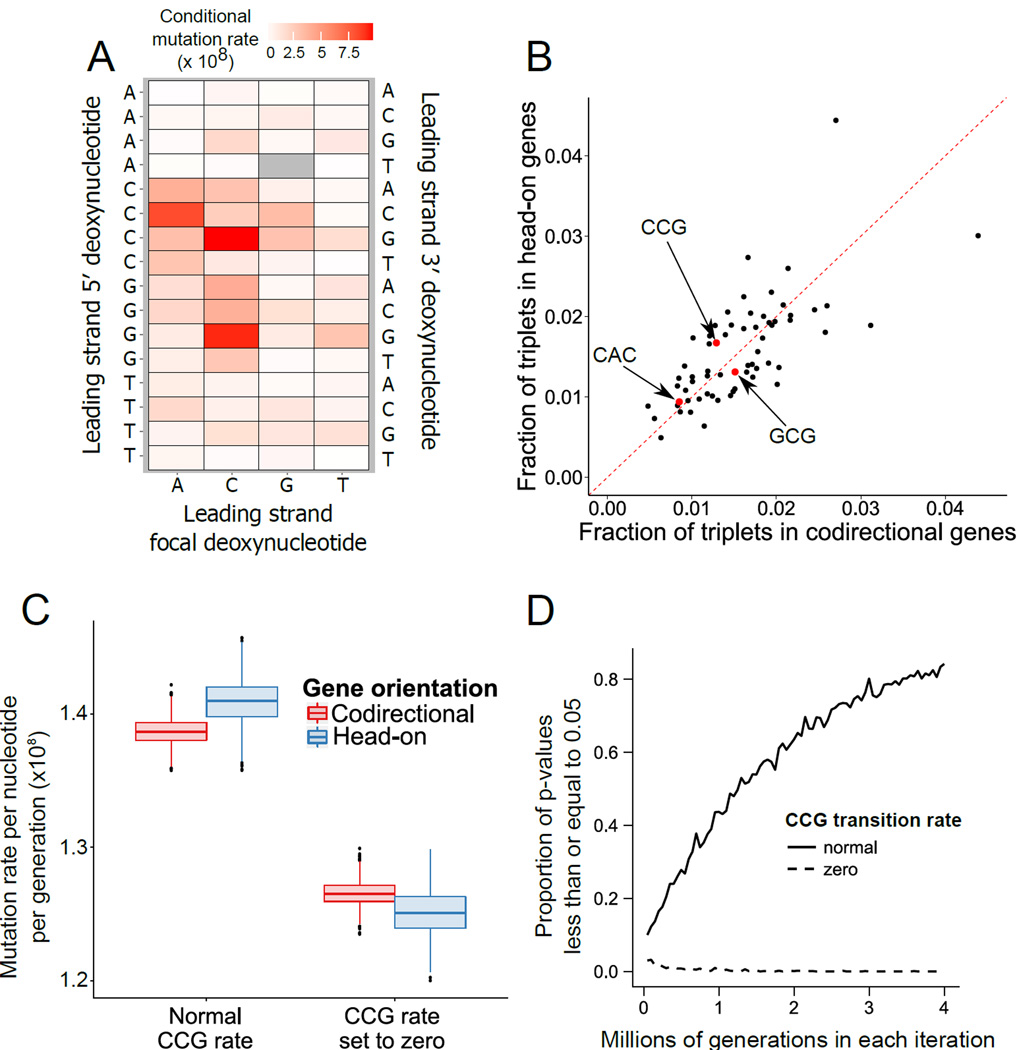
Sequence context has been shown to affect DNA polymerase error rate in vitro and mutation rate in vivo in a variety of experimental systems [8, 9, 15–18]. We therefore asked how sequence context affects DNA polymerase error rate in our experiment. We binned all transitions in our MMR- data (5798 transitions) into one of the 64 possible triplet sequence contexts, depending on the reference base that underwent transition (focal base) and the bases 5′ and 3′ of the transition as present in the leading strand. Similar to prior work, the triplets with the highest transition rate were 5′-CCG-3′, 5′-GCG-3′, and 5′-CAC-3′ [9]. The effect of neighboring sequence context on DNA polymerase error rate is very strong, as the transition rate of 5′-CCG-3′ was 403-fold greater than that of the triplet with the lowest transition rate, 5′-AGT-3′ after normalizing to the number of each triplet present in the leading strand of the genome (Figure 4A). The effect of neighboring leading strand sequence context in MMR- lines was symmetrical between the replichores (Figure S3C), and a similar trend was observable in MMR+ lines, although we were unable to analyze the two replichores independently due to loss of resolution from having only 198 transitions in the MMR+ data (Figure S3D). Transversions were rare, with 113 in MMR-lines and 56 in MMR+ lines. We were therefore unable to test effects of context on transversion occurrence. We conclude that transition rate is highly influenced by neighboring nucleotide context. We next tested whether context-dependent transition rate and sequence composition could result in different transition rates for head-on versus codirectional CDSs.
Figure 4. Increased mutation rate in head-on genes due to sequence composition.
(A) The transition rate for the focal base in each of the 64 possible triplet nucleotide sequences in MMR-lines is shown. Rates are normalized to the number of times each triplet is present in the leading strand. (B) The leading strand triplet composition of head-on CDSs is plotted versus that of codirectional CDSs. “Fraction of triplets” in the axis labels refers to the number of a given triplet divided by the total number of all triplets present in the leading strand of either head-on or codirectional CDSs. The triplets with the highest transition rates are plotted as larger red dots and indicated by arrows. The red dashed line indicates a slope of one so that differences between head-on and codirectional genes may be easily noticed. (C) Monte Carlo simulations were performed to generate transitions using the MMR-context-dependent transition rates shown in panel A. The boxplots on the left indicate the distribution of transition rates for codirectional (red) and head-on (blue) CDSs using the MMR-context-dependent transition rates. Boxplots on the right indicate the distribution of transition rates with the 5′-CCG-3′ triplet rate set artificially to zero. Each pair of boxplots represents the distribution of mutation rates resulting from 1,000 independent simulations of 500,000 generations. (D) The simulation performed in C was carried out at a range of generations per iteration. For each number of generations, 1,000 iterations was performed and hypothesis testing was carried out to test whether head-on genes had a mutation rate greater than that of codirectional genes. The proportion of those 1,000 p-values less than or equal to 0.05 is plotted in the y-axis against the number of generations per iteration. See also Figure S3.
Triplet nucleotide composition in the leading strand of head-on and codirectional genes differs substantially, and the triplet with the highest transition rate, 5′-CCG-3′, accounts for a greater proportion of the triplets in head-on CDSs than in codirectional CDSs (Figure 4B). Given the overabundance of 5′-CCG-3′ in head-on CDSs, we tested whether head-on CDSs had a higher transition rate. Although our multiple linear regression analysis in Figure 3 did not detect a difference in the BPS rate of head-on and codirectional genes, we surmised that our MA lines may not have undergone sufficient generations to detect a true underlying difference. We therefore carried out 1,000 independent iterations of simulating 500,000 generations. We began by introducing transitions according to the context-dependent transition rates in MMR- lines. This method reproduced the experimental MMR- transition rate, with a simulated genome-wide transition rate of 1.37×10−8 per generation per nucleotide compared to the experimental rate of 1.38×10−8 per generation per nucleotide. Such remarkable concurrence between the experimental transition rate and the transition rate we expect based only on the effects of sequence context reveals that local sequence context is the most important factor impacting transition rate due to replicative DNA polymerase errors.
Our simulations revealed that head-on genes do, indeed, have a higher transition rate than codirectional genes (Figure 4C). To test to what extent 5′-CCG-3′ in the leading strand caused head-on genes to obtain transitions at a higher rate, we performed another 1,000 independent iterations of 500,000 generations with 5′-CCG-3′ transition rate set artificially to zero. The resulting simulated mutation rate was overall lower, further displaying the importance of context-dependent transition rate in determining mutation rate. The distributions of simulated transition rates in head-on and codirectional genes also revealed a reversal of the former trend, with head-on genes having a lower transition rate when 5′-CCG-3′ transition rate was set to zero (Figure 4C), showing that 5′-CCG-3′ abundance in the leading strand of head-on genes is partly responsible for their higher mutation rate. To assess the power of our experiment to detect the difference in transition rate between head-on and codirectional genes we carried out the simulation over a range of generations. We tested the hypothesis that head-on genes have a higher transition rate than codirectional genes after each of the 1,000 iterations for each number of generations. The proportion of p-values ≤ 0.05 is an indicator of the power of the experiment at a given number of generations. As expected, the power of our experiment to detect a greater transition rate in head-on genes increased with increasing number of generations only when 5′-CCG-3′ transition rate was normal (Figure 4D), suggesting that the difference observed after 1,000 iterations of 500,000 generations in Figure 4C reflects a true underlying difference in the transition rate. Using separate Monte Carlo simulations we determined that triplet nucleotide composition of the leading strand also changed more quickly in head-on genes in a manner that is partially dependent on overabundance of 5′-CCG-3′ in head-on CDSs (Figure S3E). Therefore, differing sequence composition of head-on and codirectional CDSs drives a higher rate of change in the sequence of head-on CDSs.
Mutations drive evolution and genetic disease. What is the source of mutagenesis? There is no single answer to this question, as many potential sources of mutations exist under varying circumstances. Errors made during DNA replication under relatively stress-free growth conditions are one source of mutagenesis in bacteria, and evolution occurs due to natural selection acting on genomes which are dynamic due to many potential sources of mutagenesis. Recent models propose that genes encoded on the lagging strand evolve more quickly due to a higher mutation rate caused by replication/transcription conflicts in a manner resembling stress-induced mutagenesis [6, 10]. This may be an accurate depiction of the nature of mutagenesis when specific conditions are met. However, observing mutations that accumulate genome-wide in the near absence of exogenous stress, we find that a different model is able to describe the evolution of head-on genes.
Our work shows that local sequence context is the major determinant of mutagenesis. Genes in which RNA and DNA polymerases collide head-on have a higher mutation rate than codirectional genes. Under the relatively stress-free conditions of our MA line procedure, this can be explained by an overabundance of a triplet nucleotide sequence with an extremely high transition rate, 5′-CCG-3′, in the leading strand of head-on genes.
Experimental Procedures
Genome sequencing and RNA-seq alignments have been deposited to the SRA under accession number SRP067020. The list of strains used in this study is presented in Table S5. Detailed experimental procedures can be found in the Supplemental Information.
Supplementary Material
Highlights.
Intergenic regions are enriched for indels
Local sequence context strongly impacts DNA replication accuracy in vivo
Sequence context in head-on genes can lead to higher mutation rate
Acknowledgments
We wish to acknowledge Heather Schroeder for help with statistical analyses. We thank Peter Burby and Drs. Jue Wang and Lindsay Matthews for comments and feedback on the manuscript. G.A.S. was supported in part by NSF REU award MCB1050948. J.W.S. and W.G.H. were each supported in part by the NIH National Research Service Award T32 GM007544 from the National Institute of General Medical Sciences. Additionally, this work was supported by NIH grant R01 GM107312 to L.A.S..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
Conceptualization, J.W.S. and L.A.S.; Methodology, J.W.S. and L.A.S.; Software, J.W.S.; Formal Analysis, J.W.S.; Investigation, J.W.S., W.G.H. and G.A.S.; Visualization, J.W.S.; Writing – Original Draft, J.W.S.; Writing – Review & Editing, J.W.S. and L.A.S.; Supervision, J.W.S. and L.A.S.; Funding Acquisition, J.W.S., W.G.H., G.A.S. and L.A.S..
The authors have no conflict of interest to declare.
References
- 1.Fijalkowska IJ, Jonczyk P, Tkaczyk MM, Bialoskorska M, Schaaper RM. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:10020–10025. doi: 10.1073/pnas.95.17.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lind PA, Andersson DI. Whole-genome mutational biases in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17878–17883. doi: 10.1073/pnas.0804445105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma X, Rogacheva MV, Nishant KT, Zanders S, Bustamante CD, Alani E. Mutation hot spots in yeast caused by long-range clustering of homopolymeric sequences. Cell Rep. 2012;1:36–42. doi: 10.1016/j.celrep.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaibley VM, Zawistowski M, Wegmann D, Ehm MG, Nelson MR, St Jean PL, Abecasis GR, Novembre J, Zollner S, Li JZ. The influence of genomic context on mutation patterns in the human genome inferred from rare variants. Genome Res. 2013;23:1974–1984. doi: 10.1101/gr.154971.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. Accelerated gene evolution through replication-transcription conflicts. Nature. 2013;495:512–515. doi: 10.1038/nature11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster PL, Hanson AJ, Lee H, Popodi EM, Tang H. On the mutational topology of the bacterial genome. G3 (Bethesda) 2013;3:399–407. doi: 10.1534/g3.112.005355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lujan SA, Clausen AR, Clark AB, MacAlpine HK, MacAlpine DM, Malc EP, Mieczkowski PA, Burkholder AB, Fargo DC, Gordenin DA, et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;24:1751–1764. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung W, Ackerman MS, Gout JF, Miller SF, Williams E, Foster PL, Lynch M. Asymmetric Context-Dependent Mutation Patterns Revealed through Mutation-Accumulation Experiments. Mol Biol Evol. 2015;32:1672–1683. doi: 10.1093/molbev/msv055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Million-Weaver S, Samadpour AN, Moreno-Habel DA, Nugent P, Brittnacher MJ, Weiss E, Hayden HS, Miller SI, Liachko I, Merrikh H. An underlying mechanism for the increased mutagenesis of lagging-strand genes in Bacillus subtili. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1096–E1105. doi: 10.1073/pnas.1416651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrick JE, Lenski RE. Genome dynamics during experimental evolution. Nat Rev Genet. 2013;14:827–839. doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halligan DL, Keightley PD. Spontaneous Mutation Accumulation Studies in Evolutionary Genetics. Annu Rev Ecol Evol S. 2009;40:151–172. [Google Scholar]
- 13.Foster PL, Lee H, Popodi E, Townes JP, Tang H. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E5990–E5999. doi: 10.1073/pnas.1512136112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Yung M, Li L, Hoch JA, Ryan CM, Kar UK, Souda P, Whitelegge JP, Miller JH. Evidence that YycJ is a novel 5'-3' double-stranded DNA exonuclease acting in Bacillus anthracis mismatch repair. DNA repair. 2013;12:334–346. doi: 10.1016/j.dnarep.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Kunkel TA, Schaaper RM, Beckman RA, Loeb LA. On the fidelity of DNA replication. Effect of the next nucleotide on proofreading. The Journal of biological chemistry. 1981;256:9883–9889. [PubMed] [Google Scholar]
- 16.Petruska J, Goodman MF. Influence of neighboring bases on DNA polymerase insertion and proofreading fidelity. The Journal of biological chemistry. 1985;260:7533–7539. [PubMed] [Google Scholar]
- 17.Sinha NK. Specificity and efficiency of editing of mismatches involved in the formation of base-substitution mutations by the 3'----5' exonuclease activity of phage T4 DNA polymerase. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:915–919. doi: 10.1073/pnas.84.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu YO, Siegal ML, Hall DW, Petrov DA. Precise estimates of mutation rate and spectrum in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2310–E2318. doi: 10.1073/pnas.1323011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.