Abstract
We report the association of eosinophilic esophagitis (EoE) and hypertrophic cardiomyopathy (HCM) and provide genetic data indicating a linkage between EoE and HCM. The letter begins with an index patient diagnosed with both EoE and HCM and found to have a known genetic mutation for HCM. We then identify an odds ratio of nearly 8 for having both EoE and HCM following review of an electronic medical record with >1,000,000 individuals. Finally, via a candidate gene approach we identify significant association of HCM gene variants in a cohort of EoE patients versus control subjects. Collectively, we have identified a putative interaction between EoE and HCM with clinical and pathogenic implications.
To the Editor
Eosinophilic esophagitis (EoE) usually presents with upper gastrointestinal symptoms, although chest pain can be the primary symptom. We report the association of EoE and hypertrophic cardiomyopathy (HCM) in three patients and genetic data indicating a linkage between EoE and HCM.
The index patient was a 26-year-old Caucasian male who presented with dysphagia and chest pain and was diagnosed with EoE according to consensus criteria.1 After both swallowed and oral glucocorticoid therapy, his dysphagia improved, but he continued to experience chest pain that was increasingly accompanied by palpitations and syncopal episodes post-prandially or with exertion. After an extensive cardiac work-up, he was diagnosed with HCM according to consensus criteria2 (Supplementary Table 1) and heterozygosity for a known, disease-causing missense mutation (E542Q) in the cardiac myosin-binding protein C3 (MYBPC3) gene.3
An electronic chart review of 1,281,475 patient records at Cincinnati Children's Hospital Medical Center (CCHMC) identified 2,100 and 241 possible cases of EoE and HCM, respectively, using ICD-9 codes. Of these, two cases other than the index case met diagnostic criteria for both EoE and HCM. Case #2 was a male with osteogenesis imperfect a type I (OI1), and case #3 was a male with 1p36 deletion syndrome whose deletion was 10.88 megabases and included five actomyosin cytoskeleton–associated genes (ARHGEF16, ACTRT2, PLEKHG5, AJAP1, CTNNBIP1) (Supplementary Table 1). It is important to note that OI1 is a mild form of OI and is not associated with heart or GI symptoms therefore the presence of OI1 is likely a secondary finding. But, it is interesting to note that OI1 is a connective tissue disease associated with collagen 1 deficiency and EoE has recently been associated with other connective tissue diseases.4 Additionally, though patients with 1p36 deletion syndrome are known to have cardiomyopathy there are no reports of EoE associated with this disease. Thus, even with regards to these patients, unexpectedly HCM and EoE are co-occurring. Also, as EoE is not a Mendelian disorder and likely requires multiple genetic hits for disease presentation, it is not surprising to find patients with multiple genetic hits. Using these data the odds ratio is 7.69 (CI 2.46-24.03; p < 0.001) suggesting that the co-occurrence of EoE and HCM is not likely due to chance. It is important to note that our hospital based data exhibits enrichment for EoE (population prevalence 1:2000, our data 1:600) as well as for HCM (population prevalence 1:200,000, our data 1:5000), likely because our center is a tertiary referral center for both EoE and HCM. Although there is enrichment in both conditions, we do not expect that the co-occurrence of both conditions will be affected by this bias, as patients seeking care for EoE are not routinely evaluated by cardiologists and patients seeking care for HCM are not being screened for EoE. Another way to consider what the odds ratio signifies is to examine the expected co-occurrence on the basis of our center's population. From our center's prevalence of EoE (1:600) and HCM (1:5000), random co-occurrence would be expected to be 1:3,000,000. Yet, the observed co-occurrence rate for our center's population was 1:400,000 (or 3 in 1, 281, 475).
Next, we performed a candidate-gene association study assessing the frequency of MYBPC3 genetic variants in EoE vs. non-EoE control cohorts (Supplementary Table 2).5 EoE was significantly associated with 24 single-nucleotide polymorphisms (SNPs) in the linkage disequilibrium block containing MYBPC3, including 6 SNPs in close proximity to the MYBPC3 gene on chromosome 11; this association was significant after permutation analysis (Figure 1 and Supplementary Table 3). One SNP, rs3729986, is a missense mutation (V158M), but is a variant that occurs in 11% of the general population.3 Evaluating the 24 known, HCM-causing genes6, we found 62 EoE-associated genetic variants within 5 kilobases of 17 of the 24 cardiomyopathy genes (P < 0.05) (Supplementary Table 4). EoE-associated variants (P < 0.05) were significantly enriched for variants near cardiomyopathy genes (permuted P < 0.015). Interestingly, of these genes, calreticulin 3 (CALR3) is decreased by 17% (p=0.001) in the epithelial biopsies of EoE patients compared with control individuals as assessed by microarray (Supplementary Figure 1)7. This evidence suggests that HCM-associated genes also contribute to EoE susceptibility. Actomyosin proteins are important in mechanotransduction, a process recognized to be involved in chemotaxis of leukocytes, muscle contraction required for esophageal motility, and migration and proliferation of epithelial cells—processes germane in EoE. Collectively, we have identified a putative interaction between EoE and HCM with clinical and pathogenic implications.
Figure 1.
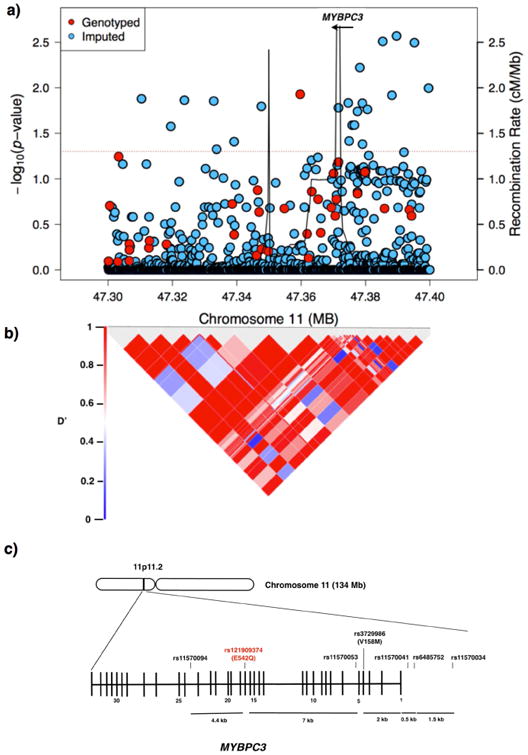
Genetic variants at the MYBPC3 genetic locus. A) Regional association plot in which the red line marks a P value of < 0.05. B) Linkage disequilibrium (LD) heat map of variants in the region showing the LD (D′) between the variants with P values less than 0.05. C) Diagram of the MYPBC3 gene on chromosome (chr) 11. The location of the mutation in the index patient is in red, and single-nucleotide polymorphisms (SNPs) of the candidate gene study are in black. Vertical lines represent exons. Abbreviations: cM/Mb, centimorgans per megabase.
Supplementary Material
Supplementary Figure 1. Relative expression of calreticulin 3 (CALR3) is decreased by 17% (p=0.001) by microarray in the epithelial biopsies of EoE patients (n = 18) compared to normal control biopsies (n = 14). Error bars represent standard error of the means (s.e.m.)
Supplementary Table 1. Summary of patients with hypertrophic cardiomyopathy and eosinophilic esophagitis
Supplementary Table 2. Demographics of eosinophilic esophagitis and control cohorts of candidate-gene association study
Supplementary Table 3. Association between single-nucleotide polymorphisms in the MYBPC3 gene and eosinophilic esophagitisa
Supplementary Table 4. Hypertrophic Cardiomyopathy–Causing Genes having Common Variants Associated with Eosinophilic Esophagitis
Acknowledgments
Funding for this project was provided by the NIAID and NIDDK (U19 AI066738, U19 AI070235, R01 DK076893, R37 A1045898), as well as the Campaign Urging Research For Eosinophilic Diseases (CURED) Foundation, Buckeye Foundation and the Sunshine Foundation through the support of Dave and Denise Bunning. The CoFAR arm of the study was supported by NIH U19 AI066738 by NIAID and NIDDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 2.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy. The Journal of Thoracic and Cardiovascular Surgery. 2011;142:e153–e203. doi: 10.1016/j.jtcvs.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 3.Van Driest SL, Vasile VC, Ommen SR, Will ML, Tajik AJ, Gersh BJ, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–10. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 4.Abonia JP, Wen T, Stucke EM, Grotjan T, Griffith MS, Kemme KA, et al. High prevalence of eosinophilic esophagitis in patients with inherited connective tissue disorders. J Allergy Clin Immunol. 2013;132:378–86. doi: 10.1016/j.jaci.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46:895–900. doi: 10.1038/ng.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian T, Liu Y, Zhou X, Song L. Progress in the molecular genetics of hypertrophic cardiomyopathy: a mini-review. Gerontology. 2013;59:199–205. doi: 10.1159/000346146. [DOI] [PubMed] [Google Scholar]
- 7.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Relative expression of calreticulin 3 (CALR3) is decreased by 17% (p=0.001) by microarray in the epithelial biopsies of EoE patients (n = 18) compared to normal control biopsies (n = 14). Error bars represent standard error of the means (s.e.m.)
Supplementary Table 1. Summary of patients with hypertrophic cardiomyopathy and eosinophilic esophagitis
Supplementary Table 2. Demographics of eosinophilic esophagitis and control cohorts of candidate-gene association study
Supplementary Table 3. Association between single-nucleotide polymorphisms in the MYBPC3 gene and eosinophilic esophagitisa
Supplementary Table 4. Hypertrophic Cardiomyopathy–Causing Genes having Common Variants Associated with Eosinophilic Esophagitis


