Summary
During aging, oxidized, misfolded, and aggregated proteins accumulate in cells, while the capacity to deal with protein damage declines severely. To cope with the toxicity of damaged proteins, cells rely on protein quality control networks, in particular proteins belonging to the family of heat‐shock proteins (HSPs). As safeguards of the cellular proteome, HSPs assist in protein folding and prevent accumulation of damaged, misfolded proteins. Here, we compared the capacity of all Drosophila melanogaster small HSP family members for their ability to assist in refolding stress‐denatured substrates and/or to prevent aggregation of disease‐associated misfolded proteins. We identified CG14207 as a novel and potent small HSP member that exclusively assisted in HSP70‐dependent refolding of stress‐denatured proteins. Furthermore, we report that HSP67BC, which has no role in protein refolding, was the most effective small HSP preventing toxic protein aggregation in an HSP70‐independent manner. Importantly, overexpression of both CG14207 and HSP67BC in Drosophila leads to a mild increase in lifespan, demonstrating that increased levels of functionally diverse small HSPs can promote longevity in vivo.
Keywords: protein homeostasis, longevity, aging, Drosophila melanogaster, small heat‐shock protein, HSPB family
Introduction
The imbalance in overall protein homeostasis is a crucial factor in the development of heritable age‐related neurodegenerative diseases and during normal aging (Dobson, 2003; Vacher et al., 2005; Zhang et al., 2005; Arslan et al., 2006; Haass & Selkoe, 2007; Morimoto, 2008). Achieving and maintaining the correct three‐dimensional protein structure is a continuous struggle within cells. Firstly, folding of proteins toward an active biological state is challenged by the crowded environment within the cell, which may lead to off‐pathway reactions resulting in protein aggregation (Ellis & Minton, 2006; Engel et al., 2008; Homouz et al., 2008). Protein misfolding can further originate from direct protein damage (e.g., oxidation, thermal denaturation), but can also originate from age‐related mutations, molecular misreading (van Leeuwen et al., 2000), splicing errors (Pettigrew & Brown, 2008), or errors in translation (Parker, 1989; Kramer & Farabaugh, 2007). While cells are challenged by an accumulation of oxidized, misfolded, and aggregation‐prone proteins, their capacity to deal with accumulated protein damage declines with aging (Liu et al., 1989; Bulteau et al., 2002; Ferrington et al., 2005; Naidoo et al., 2008).
As molecular chaperones, heat‐shock proteins (HSPs) play a central role in protein homeostasis: They safeguard protein conformation and folding and assist in the assembly and disassembly of protein complexes, and in protein degradation. By their ability to bind non‐native polypeptides, they maintain their substrates in a state competent for subsequent folding or, when folding is not successful, for degradation by the ubiquitin–proteasome system (Urushitani et al., 2004) or through autophagy (Carra et al., 2008). Hereby chaperones can prevent toxic protein aggregation, and as such, they have been implicated as protectors against age‐related protein folding diseases (Rujano & Kampinga, 2008) and as supporters of healthy aging (Hsu et al., 2003; Morrow & Tanguay, 2003; Walker & Lithgow, 2003; Morley & Morimoto, 2004; Morimoto, 2008). Indeed, activation of all stress‐inducible HSPs, either by overexpression of the heat‐shock factor‐1 (HSF‐1) (Morley & Morimoto, 2004; Morimoto, 2008) or via caloric restriction and the accompanying insulin signaling (Hsu et al., 2003), was shown to delay the onset of protein folding diseases and to induce longevity in otherwise healthy animals.
Interestingly, even the sole overexpression of single members of the small HSP family was shown to support longevity both in Caenorhabditis elegans (Walker & Lithgow, 2003) and in Drosophila melanogaster (Aigaki et al., 2002; Morrow & Tanguay, 2003; Morrow et al., 2004; Wang et al., 2004). The small HSPs tested so far (HSP22, HSP23, HSP26, HSP27) share the capacity to facilitate refolding of stress‐denatured substrates in vitro (Morrow et al., 2006), supporting the hypothesis that maintenance of global protein homeostasis is essential for longevity. To elucidate which Drosophila small HSPs might be the most potent in preventing protein misfolding or the toxic aggregation of protein damage upon aging, we cloned all members of the Drosophila small HSP family (excluding the mitochondrial HSP22) and compared their ability to assist in refolding of stress‐denatured substrates and/or in preventing aggregation of disease‐associated misfolded proteins in living cells. We identified CG14207 as novel and potent small HSP member that exclusively assisted in HSP70‐dependent refolding of stress‐denatured proteins. Furthermore, we report that HSP67BC, which has no role in protein refolding, was the most efficient small HSP preventing toxic protein aggregation in an HSP70‐independent manner. Importantly, overexpression of both CG14207 and HSP67BC in Drosophila leads to increased lifespan, implicating that increased levels of these small HSPs can prevent aging in vivo.
Results
Heat inducibility of the Drosophila small HSPs
To identify all Drosophila small HSPs, we first employed a comprehensive in silico approach and identified 11 candidates (see Experimental procedures). Certain members of all major HSP families are known to be induced upon proteotoxic stresses, including heat shock and exposure to heavy metals (Mosser et al., 1988). To analyze which of the 11 D. melanogaster small HSPs are heat‐inducible, we heat‐shocked Drosophila S2 cells at 38 °C for 30 min and analyzed the small HSP mRNA levels by qPCR. As controls, we measured the mRNA levels of HSC70‐4 and HSC70‐5, two constitutively expressed genes not responsive to heat shock (Ashburner & Bonner, 1979; Ish‐Horowicz et al., 1979), and HSP70Aa, a highly heat‐inducible gene (Fig. 1A). The four classical small HSPs (HSP22, HSP23, HSP26, and HSP27) were all highly induced after a heat shock (Fig. 1B), consistent with previous findings (Marin et al., 1996; Michaud et al., 1997). Apart from CG14207 and CG13133, all the other small HSPs were also found to be heat‐inducible (Fig. 1C). Expression of CG13133 was below the detection limit at both control and heat‐shock conditions. The most strongly induced nonclassical members were HSP67BC and CG4461, while HSP67BA, L(2)EFL, and CG7409 were moderately induced (Fig. 1C).
Figure 1.
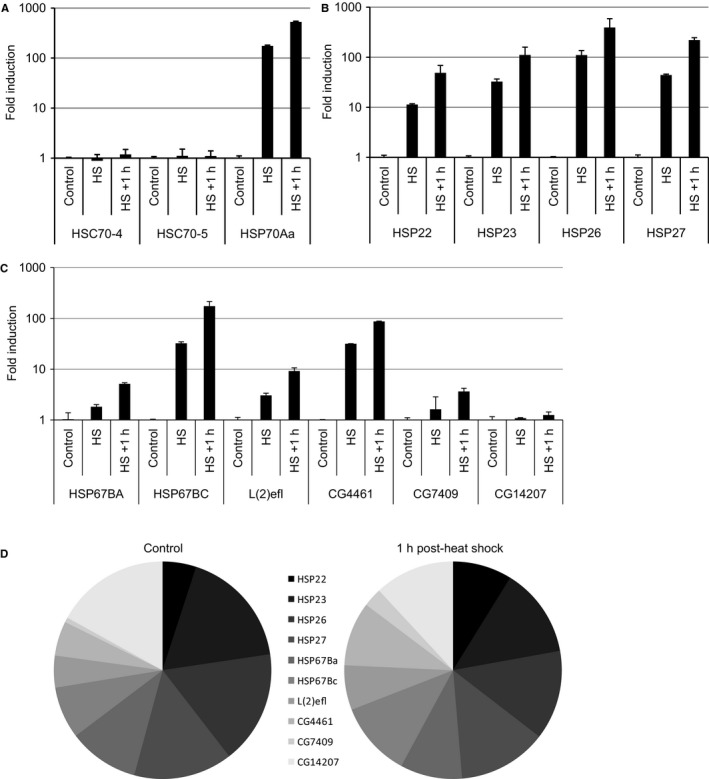
Drosophila melanogaster small heat‐shock proteins (HSP) family and heat inducibility. Transcript levels of HSC70‐4, HSC70‐5, HSP70Aa (A), the classical D. melanogaster small HSP genes (B), and the nonclassical small HSP members (C) were analyzed directly after heat shock (30 min at 38 °C) and 1 h after heat shock using quantitative RT–PCR. Relative mRNA abundance before and after heat shock is depicted in panel D. (CG13133 mRNA was not detected in S2 cells.)
qPCR data were subsequently used to estimate the relative contribution of each member to the total pool of small HSP mRNA before and after heat shock. In S2 cells, the most abundantly constitutively expressed small HSPs are HSP23 (17.6% of the total pool), HSP26 (16.9%), HSP27 (14.7%), and CG14207 (17%) (Fig. 1D). After a heat shock and a recovery period of 1 h, the pool of small HSP mRNA has shifted toward a more homogenous distribution with comparable levels of HSP23, HSP26, HSP27, HSP67BC, HSP67BA, and CG4461 (Fig. 1D).
CG14207 and CG7409 are the most active small HSP members in assisting refolding of heat‐denatured luciferase
It has been shown for several small HSPs that they can maintain substrates in a folding competent form both in vitro and in vivo (Mogk et al., 2003; Cashikar et al., 2005). In vitro, the addition of the HSP70/HSP40 refolding machinery is required for the refolding reaction (Lee et al., 1997). This has been reproduced in living cells using luciferase as a substrate (Nollen et al., 2001; Bryantsev et al., 2007). Here, we tailored the cellular luciferase refolding assay for Drosophila S2 cells and characterized which of the Drosophila small HSPs could enhance luciferase refolding upon overexpression. The mitochondrial HSP22 was excluded from our analyses as our cellular assays were only tailored for the cytosolic and nuclear compartments. We achieved relatively comparable expression levels for all small HSPs (Fig. 2, bottom) although expression of HSP23 and HSP26 was somewhat higher and expression of CG4461 was somewhat lower than that of the other seven sHSPs that showed similar expression levels. Consistent with in vitro data (Morrow et al., 2006), overexpression of the classical small HSPs (HSP23, HSP26, and HSP27) increased luciferase refolding (Fig. 2, top). Although less efficient, overexpression of L(2)EFL also led to improved luciferase refolding, whereas HSP67BA, HSP67BC, CG4461, and CG13133 had no effect. Interestingly, overexpression of CG7409 and the non‐heat‐shock‐inducible CG14207 resulted in the highest level of refolding 1 h after heat shock (Fig. 2, top).
Figure 2.
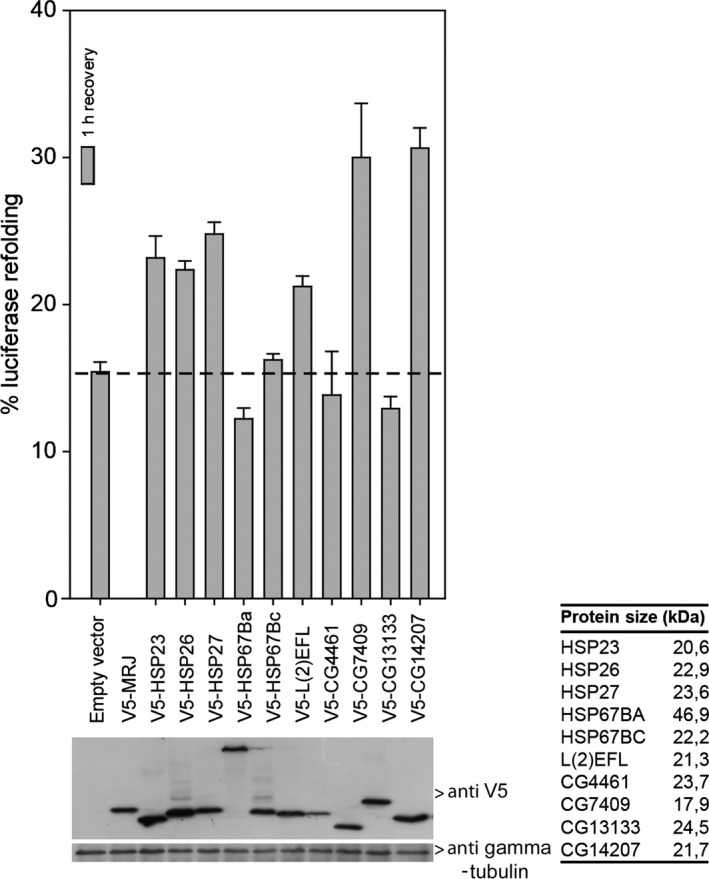
Effect of overexpression of the Drosophila melanogaster small HSP family in S2 cells on refolding heat‐inactivated firefly luciferase. Drosophila melanogaster small HSPs and firefly luciferase were coexpressed in Schneider's S2 cells. Twenty‐four hours after transfection, S2 cells were heated for 30 min at 38 °C reducing luciferase activity to < 5%. Next, cells were re‐incubated for 1 h at 25 °C to allow for (chaperone‐assisted) luciferase refolding. Luciferase activities are plotted relative to the activity in unheated cells (=100%). Data are mean ± SD of three independent experiments. Lower panel shows expression of chaperones using anti‐V5 antibodies.
To analyze whether D. melanogaster small HSPs, like bacterial, plant, and mammalian small HSPs (Lee & Vierling, 2000; Mogk et al., 2003; Bryantsev et al., 2007), also require HSP70 machines for refolding, we first tested which of the D. melanogaster HSP70s could promote refolding of heat‐denatured luciferase. Hereto, we cloned a selection of the D. melanogaster HSP70 family (Table S2, Supporting information) and analyzed their effect on luciferase refolding. Whereas all HSP70 proteins were expressed at equal levels, overexpression of both D. melanogaster HSC70‐2 and HSC70‐4, but not HSP70AA, enhanced luciferase refolding in S2 cells (Fig. 3A). This suggests either that HSP70AA lacks the ability to assist in refolding heat‐denatured luciferase or that specific cofactors that are required for its activity are rate‐limiting under these conditions. Subsequently, we downregulated the individual D. melanogaster HSP70s using dsRNA molecules to assess their role in protein refolding (Table S3, Supporting information). As no available antibodies can distinguish HSP70AA, HSC70‐2, and HSC70‐4, we tested the specificity and effectiveness of the dsRNAs using overexpression of V5‐tagged HSP70AA, HSC70‐2, or HSC70‐4. All dsRNA constructs were specific and significantly downregulated the expression of the targeted V5‐tagged protein (Fig. 3B). Indeed, the luciferase refolding capacity was substantially inhibited upon HSC70‐2 downregulation (Fig. 3C). dsRNA‐mediated knockdown of HSC70‐4 was less effective in downregulating HSC70‐4 and induced no significant effect on refolding (Fig. 3C). In contrast, dsRNA directed against HSP70AA, which did not enhance luciferase refolding upon overexpression, reduced refolding, suggesting that endogenous HSC70AA does play a role in refolding of heat‐denatured luciferase under physiological conditions. Next, we combined overexpression of V5‐HSP27 (Fig. 3D) and V5‐CG14207 (Fig. 3E) with downregulation of HSP70 members to assess the role of HSP70 proteins in sHSP‐mediated refolding. Refolding of luciferase in the presence of both V5‐HSP27 and V5‐CG14207 was considerably reduced by downregulating HSP70AA, HSC70‐2, or HSC70‐4 (Fig. 3D,E). Thus, our results strongly suggest that the refolding capacity of D. melanogaster HSP27 and CG14207 is partially dependent on an intact HSP70 machine.
Figure 3.
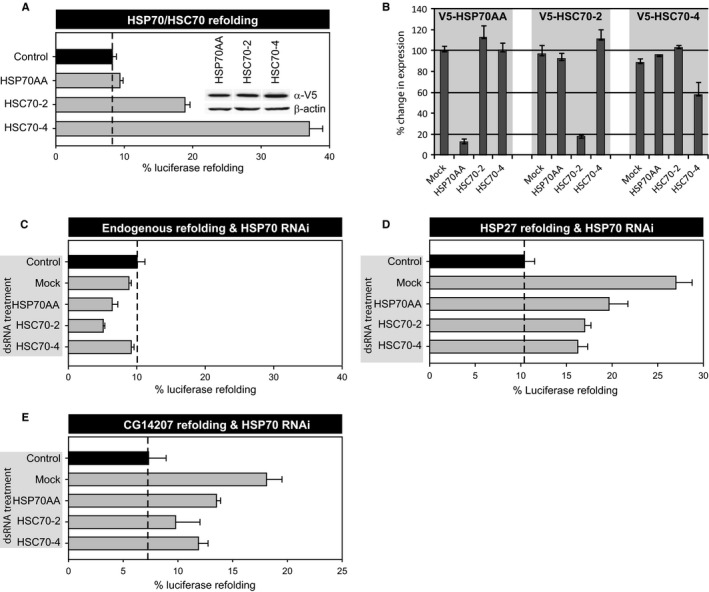
Dependency of HSP27‐ or CG14207‐assisted luciferase refolding on HSP70 (A) Firefly luciferase was coexpressed in Schneider's S2 cells together with eGFP (control) or Drosophila melanogaster HSP70s. Twenty‐four hours after transfection, S2 cells were heated for 30 min at 38 °C reducing luciferase activity to < 5%. Next, cells were re‐incubated for 1 h at 25 °C to allow for (chaperone‐assisted) luciferase refolding. (B–D) Schneider's S2 cells were transfected with dsRNA targeting individual HSP70s, and first RNAi efficiency and specificity were tested. Hereto, the various dsRNAs were coexpressed with V5‐HSP70AA, V5‐HSC70‐2, or V5‐HSC70‐4, and their levels were quantified from Western blots and expressed relative to expression without the dsRNA (B). Next, luciferase refolding was determined in the presence of dsRNA targeting endogenous HSP70AA, HSC70‐2, or HSC70‐4 alone (C) or combined with coexpression of V5‐HSP27 (D) or V5‐CG14207 (E), and luciferase refolding was determined as under panel A. In all panels, luciferase activities are plotted relative to the activity in unheated cells (=100%). Data are mean ± SD of three independent experiments.
HSP67BC is the most potent suppressor of polyglutamine aggregation
Having identified CG14207 as the most potent Drosophila small HSP promoting HSP70‐dependent protein refolding, we next investigated which of the Drosophila small HSPs was the most effective in preventing toxic protein aggregate formation. Several protein folding diseases are characterized by the formation of toxic aggregates such as Alzheimer's disease, Huntington's disease, and amyotrophic lateral sclerosis. Some small HSPs have been reported to suppress the aggregation of such disease‐related proteins (Wilhelmus et al., 2006; Carra et al., 2008), and we have found that this is not always related to their capacity to refold heat‐denatured luciferase (Vos et al., 2010). Therefore, we tested which of the Drosophila small HSPs could suppress aggregation of an EGFP‐tagged huntingtin exon‐1 containing 119 glutamines (EGFP‐HDQ119) in S2 cells. The Dm ortholog of the human DNAJB6, MRJ, previously identified in a screen for suppressors of polyglutamine (polyQ) toxicity (Fayazi et al., 2006; Hageman et al., 2010) was used as a positive control and indeed completely inhibited aggregate formation in S2 cells as demonstrated using filter trap binding (Fig. 4A). Most small HSPs showed only minor or no effects on polyQ aggregate formation. However, overexpression of HSP67BC was very effective in preventing polyQ aggregation (Fig. 4A). Interestingly, this small HSP did not support luciferase refolding in Drosophila S2 cells (Fig. 2) and was identified as the ortholog of the human HSPB8 that in conjunction with BAG3 can reduce polyQ aggregation in mammalian cells (Carra et al., 2010).
Figure 4.
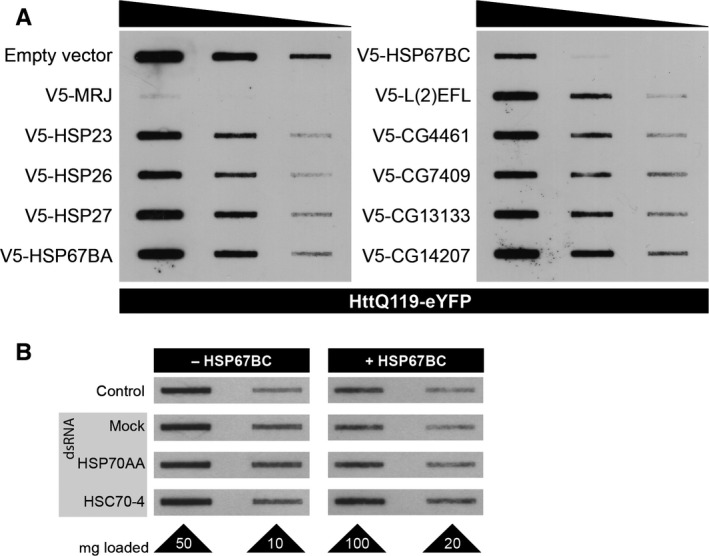
Effect of overexpression of the Drosophila melanogaster small heat‐shock proteins (HSP) in S2 cells on preventing aggregation of polyQ proteins. (A) EGFP‐Htt‐Q119, harboring exon 1 fragment of the huntingtin protein containing 119 glutamine repeats, was coexpressed in Schneider's S2 cells together with an empty vector (control) or the various D. melanogaster small HSPs. MRJ, a well‐known potent inhibitor of polyQ aggregation (Chuang et al., 2002; Fayazi et al., 2006), was included as a positive control. Forty‐eight hours after transfection, S2 cells were lysed and analyzed using the filter trap assay. Serial fivefold dilutions were loaded on cellulose acetate membranes and probed with anti‐GFP antibody. (B) EGFP‐Htt‐Q119 was coexpressed in Schneider's S2 cells without (−) or with (+) HSP67BC together with dsRNA targeting D. melanogaster HSP70Aa or HSC70‐4. PolyQ aggregates were detected using the filter trap assay using two dilutions. Samples coexpressing HSP67BC were loaded at higher concentrations to better visualize trapped aggregation.
Next, we tested whether HSP67BC requires a functional HSP70 machine to prevent polyQ aggregation. RNAi against HSP70AA and HSC70‐4 that resulted in the inhibition of the refolding promoting activity of CG14207 (Fig. 3E) did not lead to an increase in polyQ aggregation (Fig. 4B), indicating that the protective effect of HSP67BC against polyQ aggregation does not require HSP70 activity.
Combined, our findings demonstrate that the Drosophila small HSPs have versatile functions and that HSP70‐independent prevention of protein aggregation (HSP67BC) and HSP70‐dependent stimulation of protein refolding (CG14207) can be separated into two different proteins.
In vivo effects of HSP67BC on polyQ toxicity
To determine whether the observed protective effect of HSP67BC on polyQ aggregation could be extended to an in vivo setting, we employed the ataxin‐3 (SCA3) fly model (Bilen & Bonini, 2007). This fly model expresses the ataxin‐3 gene with 78 CAG repeats under the control of the UAS/gmr‐GAL4 expression system (Brand & Perrimon, 1993), resulting in eye‐specific expression. Cryo‐electron microscopy showed degeneration of the individual hexagonal ommatidia from flies expressing the SCA3‐Q78, which was not observed in wild‐type flies (Fig. S2, Supporting information). This degeneration was also readily visible by light microscopy and was used to score eye degeneration.
Three independent transgenic lines expressing the V5‐tagged HSP67BC (Fig. 5A) showed eye degeneration upon crossing with the SCA3 fly model (Fig. 5A). Downregulation of HSP67BC in two independent RNAi lines (see Experimental procedures and Table S1, Supporting information) led to a small, but significant decrease in endogenous HSP67BC protein levels (Fig. 5C) and aggravated eye degeneration compared to HSP67BC overexpression (Fig. 5D).
Figure 5.
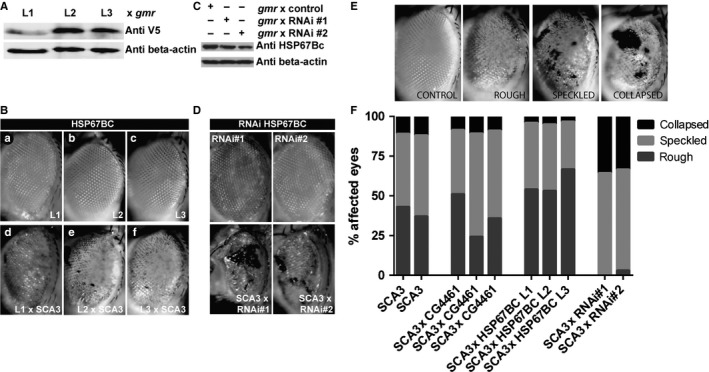
Effect of overexpression of the Drosophila melanogaster small heat‐shock proteins (HSPs) in vivo on reducing polyQ toxicity. (A) Flies transgenic for UAS‐V5‐HSP67BC (L1‐L3) were crossed with the gmr‐GAL4 driver line to verify transgene expression. Protein levels of head tissue are depicted. (B) ataxin‐3 flies (SCA3; gmr‐GAL4 UAS‐SCA3trQ78/+) were crossed with UAS‐V5‐HSP67BC (L1‐L3) transgenic lines (a–c), and the effects on eye degeneration (d–f) were analyzed by light microscopy. (C) Knockdown of endogenous HSP67BC was verified by crossing two UAS‐HSP67BC‐RNAi lines (RNAi#1, RNAi#2) with the gmr‐GAL4‐driver line. Protein levels of head tissue are depicted. (D) The UAS‐HSP67BC‐RNAi lines (RNAi#1, RNAi#2) were crossed with the ataxin‐3 fly line (SCA3; gmr‐GAL4 UAS‐SCA3trQ78/+), and the effects on eye degeneration are depicted. (E) Degenerative eye phenotype was scored by light microscopy into three categories: rough, speckled, and collapsed. For this, the genotype gmr‐GAL4 UAS‐SCA3trQ78/+ was used. Genotype UAS‐SCA3trQ78/+ was used to generate the control image. (F) Quantification of degenerative eye phenotype from the crosses given in B and D as percentage of affected eyes is given. CG4461 overexpression had no effect on SCA3 eye phenotype. HSP67BC overexpression significantly reduced the collapsed eye phenotype (t‐test P < 0.008). HSP67BC knockdown reduced the rough phenotype (t‐test, P < 0.008) and increased speckled and collapsed phenotypes (t‐test, P < 0.03 and P < 0.008, respectively). Total numbers of eyes scored: SCA3, 370; CG4461 overexpression, 110; HSP67BC overexpression, 213; HSP67BC RNAi, 47. Genotypes used for the crosses: SCA3; gmr‐GAL4 UAS‐SCA3trQ78/+, CG4461; UAS‐V5‐CG4461. HSP67BC L1‐L3; UAS‐V5‐HSP67BC, RNAi#1 and RNAi#2; UAS‐HSP67BC‐RNAi (VDRC transformant lines ID 26416 and 26417).
For quantification, eye degeneration was scored into three readily visible degeneration patterns with increasing degenerative phenotypes: rough, speckled, and collapsed (Fig. 5E). As a negative control to further exclude nonspecific effects, we used three lines overexpressing V5‐tagged CG4461, one of the small HSPs that is not active in either refolding luciferase or polyQ aggregation (Figs 2 and 4A). In line with the observation that HSP67BC can suppress the aggregation of polyglutamine containing proteins in cells (Fig. 4A), overexpression of HSP67BC in the SCA3 model significantly reduced the collapsed eye phenotype in all three individual lines tested (Fig. 5F). Downregulation of endogenous HSP67BC in two independent RNAi lines showed both a significant reduction in the rough phenotype and a significant increase in the speckled and collapsed phenotypes, consistent with the anti‐aggregation activity identified in Fig. 4A. The current data are consistent with our previous findings that HSP67BC, the functional ortholog of the human HSPB8, is able to modulate eye degeneration caused by expression of proteins containing expanded polyglutamine repeats (Carra et al., 2010).
Linking chaperone activities to longevity
Finally, we wondered whether the separate activities of HSP67BC and CG14207 on protein stress could have implications during aging. Previous studies have shown that overexpression of HSP22, HSP23, HSP26, and HSP27 can extend lifespan in flies (Aigaki et al., 2002; Morrow et al., 2004; Wang et al., 2004). While for the mitochondrial HSP22 these effects were attributed to improved quality control within mitochondria and likely involve a reduction in oxidative stress (Morrow et al., 2004), the effects of the HSP23, HSP26, and HSP27 can be associated with an improved protein homeostasis in the cytosol and/or nucleus of the cells. Here, we showed that the latter 3 sHSPs have intermediate activities both in assisting refolding of stress‐denatured substrates and in preventing aggregation of disease‐associated misfolded proteins. To uncover whether either one of these activities can also support longevity, we determined the lifespan of male flies overexpressing CG14207 (strong refolder) and HSP67BC (strong anti‐aggregation) using elav‐GAL4 or ey‐GAL4 promoters. Elav‐GAL4 drives expression in the central and peripheral nervous systems (Robinow & White, 1991). The ey‐GAL4 driver, which is traditionally used for eye‐specific expression, also induces expression in the rest of the body (flyatlas http://www.flyatlas.org). To properly evaluate the effects of single gene overexpression on lifespan, we backcrossed all lines over w 1118 males for six generations (Burnett et al., 2011). This led to loss of expression of the transgenes in some lines (Fig. 6A, HSP67Bc‐F1), which served as an additional internal control line for which the lifespan was unaltered compared to the isogenic controls crossed with the driver lines (Fig. 6B,C). Overexpression of HSP67BC (line A7) resulted in a significant, albeit small increase in longevity when expression was driven by either elav‐GAL4 or ey‐GAL4 (Fig. 6B,C). For CG14207, only the line showing the highest expression (Fig. 6A, CG14207‐B7) showed a significant increase in lifespan when expression was driven by elav‐GAL4 or ey‐GAL4. For the other line (CG14207‐B6), the survival curve, however, was unaltered compared to the respective isogenic controls (Fig. 6B,C). Although overexpression of CG14207 and HSP67BC only showed a mild increase in lifespan, our results demonstrate that functionally diverse small HSP members can promote longevity.
Figure 6.
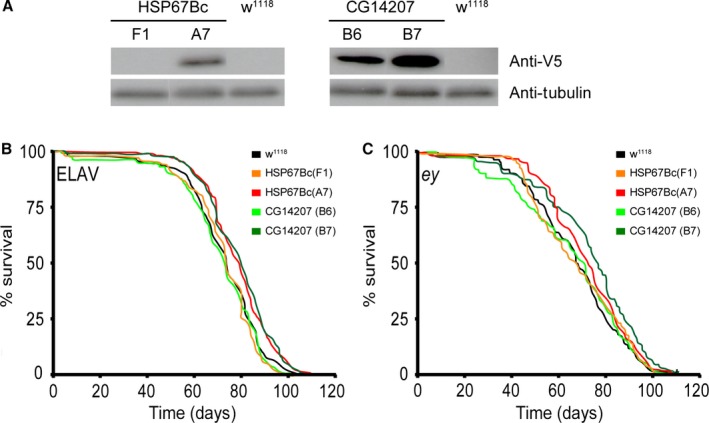
Effect of overexpression of the Drosophila melanogaster small heat‐shock proteins (HSP) in vivo on lifespan. Large cohorts of male flies were analyzed for their lifespan. Overexpression of each sHSP studied (using the ey‐GAL4‐driver) was confirmed by Western blot analysis (A). Isogenic controls (w 1118 lines) or transgene expression lines (UAS‐HSP67Bc(F1), UAS‐HSP67Bc(A7), UAS‐CG14207(B6), and UAS‐CG14207(B7)) were crossed with elav‐GAL4 driver (B) or ey‐GAL4 driver (C) lines, and survival curves of the progeny expressing the transgenes were plotted using Kaplan–Meier and statistically analyzed using log‐rank (Mantel–Cox). As a control, lifespan of the progeny of driver lines crossed with the isogenic controls (w 1118) was determined. Significant differences between survival curves were observed between isogenic control (median survival = 74 days) and HSP67Bc(A7) (median survival = 77.5 days) (P < 0.001), and between the isogenic control (median survival = 74 days) and CG14207(B7) (median survival = 80 days) (P < 0.0001) driven by elav‐GAL4, and significant differences between survival curves were observed between isogenic control (median survival = 68 days) and HSP67Bc(A7) (median survival = 72 days) (P < 0.003), and between the isogenic control (median survival = 68 days) and CG14207(B7) (median survival = 76 days) (P < 0.0001) driven by ey‐GAL4. The total number of flies analyzed was 647 for w 1118, 607 for F1, 522 for A7, 479 for B6, 484 for B7 (panel B), 651 for w1118, 553 for F1, 486 for A7, 484 for B6, and 435 for B7 (panel C).
Discussion
To unravel the functional divergence of small HSPs and their effects on organismal health, we compared the Drosophila small HSPs for heat inducibility their ability to assist in protein refolding and ability to prevent polyQ aggregation. Intriguingly, we identified two small HSPs, which either exclusively supported refolding (CG14207) or prevented aggregation (HSP67BC). CG14207 was the only small HSP that was not heat‐inducible and in addition depended on HSP70 for its refolding activity. HSP67BC, on the other hand, was clearly heat‐inducible and showed the strongest, but HSP70‐independent, activity in preventing polyQ aggregation, while its overexpression did not increase the cellular ability to refold heat‐denatured luciferase. In addition, the overexpression of both single small HSPs resulted in increased lifespan compared to their isogenic controls.
Classical HSP70 activity and anti‐aggregation
Chaperone‐like actions of small HSPs have generally been suggested to depend on HSP70 activity. In protein refolding assays in vitro (Horwitz, 1992; Jakob et al., 1993), aggregation prevention by the ATP‐independent small HSPs has been shown to occur independent of the ATP‐dependent HSP70 machinery, but for efficient refolding, substrate transfer to the HSP70 machine is required (Lee & Vierling, 2000; Mogk et al., 2003). Also, in living cells, refolding assistance by HSPB1 was found to require a functional HSP70 machinery (Bryantsev et al., 2007), and in this study, a similar scenario was found to be true for the best refolding stimulating D. melanogaster small HSP, CG14207. Strikingly, CG14207 was ineffective in preventing polyQ aggregation. Inversely, the best protector against polyQ aggregation, HSP67BC, acted independent of the HSP70 machine and was ineffective in the luciferase refolding assay. We found a similar pattern for members of the human family of small HSP proteins as well, where the efficacy to assist in HSP70‐dependent refolding was almost inversely related to the ability to prevent polyQ aggregation (Vos et al., 2010). One can envision that transfer of nonfoldable substrates to HSP70 would result in a fatuous ATP‐driven substrate binding and release, which may slightly delay aggregation but not prevent it. Indeed, in cells overexpression of HSP70s only marginally affects polyQ aggregation (Chai et al., 1999; Rujano et al., 2007), whereas in vivo HSP70‐mediated rescue of polyQ toxicity only occurs in the presence of semi‐soluble nuclear polyQ aggregates (Warrick et al., 1999; Chan et al., 2000; Cummings et al., 2001). Thus, the different small HSPs with different affinities to substrates and HSP70s may have evolved to serve adequate processing of a broad spectrum of clients. The low HSP70 and high substrate affinity may serve to prevent aggregation of unfoldable substrates and provide a longer time window for ubiquitination and normal proteasomal turnover and/or autophagic processing.
Inducing longevity
Several screens have been performed as an attempt to elucidate the molecular events underlying aging (Zou et al., 2000; Pletcher et al., 2002). This has revealed changed expression of several genes involved in many cellular pathways, including members of HSP families. Instead of looking at physiologically induced aging effects on gene expression, we tried to determine whether individual molecular functions of small HSPs, respectively HSP70‐dependent assistance of (re)folding reactions (CG14207) or HSP70‐independent prevention of polyQ aggregation (HSP67BC), can contribute to an enhanced lifespan in line with the theory that overall protein homeostasis is important for healthy aging (Balch et al., 2008) and that a multitude of molecular chaperones of which the expression is regulated via HSF‐1, a transcriptional regulator of stress‐inducible gene expression, are vital in both the protection against protein folding diseases and aging (Walker et al., 2001; Morley & Morimoto, 2004; Cohen et al., 2006). In line, previous data had already demonstrated that the HSP22, the mitochondrial D. melanogaster small HSP family member, can induce longevity (Morrow et al., 2004). We now show that two other small HSP members that have activities on only one of the two substrates investigated here, one related to acute stress (CG14207) and one to chronic stress (HSP67Bc), also are capable of extending lifespan in Drosophila, demonstrating that increased levels of functionally diverse small HSPs can promote longevity in vivo by different protective activities that yet both contribute to an improved protein homeostasis. Notably, a recent extensive study on proteome remodeling and aggregation in aging C. elegans (Walther et al., 2015) showed that among all chaperones, particularly small HSPs were associated with the increase in protein homeostasis in long‐lived daf‐2 mutant worms, consistent with the view that small HSPs play a pivotal role in the lifespan‐prolonging effect of the insulin/insulin‐like growth factor‐1 signaling pathway.
Experimental procedures
Organisms and growth conditions
For cloning purposes and plasmid isolation, Escherichia coli DH5α (Gibco‐BRL, Gaithersburg, MD, USA) was used and grown at 37 °C in LB medium (1% bacto‐tryptone, 0.5% yeast extract, 0.5% NaCl), supplemented with the appropriate antibiotics when required.
Drosophila Schneider's S2 cells were cultured in Schneider's Drosophila medium (GIBCO, Paisley, UK) supplemented with 10% heat‐inactivated fetal bovine serum (Greiner, Alphen aan den Rijn, the Netherlands), 100 units per mL penicillin, and 100 g mL−1 streptomycin in T25 flasks at 25 °C. For exponential cell growth, cell density was kept between 3 × 105 and 3 × 106 cells mL−1.
Fly stocks (Table S1) were maintained at 22 °C according to standard protocols. GAL4 driver stocks were obtained from the Bloomington Stock Center (Indiana University, USA) (Table S1). The fly stock bearing gmr‐GAL4 UAS‐SCA3Q78 used for the eye‐degeneration screen was generously provided by N. Bonini (University of Pennsylvania, USA) and maintained at 25 °C. Transgenic lines were generated by Genetic Services Inc. (Sudbury, MA, USA) by injection of the pUAS vector, harboring V5‐HSP67BC, V5‐CG4461, or V5‐CG14207, into the w 1118 genetic background. For longevity analyses, HSP‐expressing males were crossed with w 1118 virgins, and next, the female offspring was backcrossed for six generations with male w 1118 to generate isogenic controls. We used the same procedure for the balancer flies. Finally, we crossed the balancers back into the HSP flies. Two independent RNAi lines (transformant ID 26416 (#1) and 26417 (#2)), to downregulate endogenous HSP67BC protein levels, were obtained from the VDRC stock collection (for details, see Table S1).
Molecular techniques, bioinformatics, and plasmid generation
Oligonucleotide primers (Biolegio, Nijmegen, the Netherlands) and plasmids used in this study are listed in Tables S2–S5 (Supporting information). Standard recombinant DNA techniques were carried out essentially as described by Sambrook et al. (1989). Restriction enzymes were used according to the manufacturers' instructions (Invitrogen, Breda, the Netherlands; and New England Biolabs, Ipswich, MA, USA). Vent DNA polymerase (New England Biolabs) was used for preparative polymerase chain reactions. DNA sequencing reactions were carried out by ServiceXS (Leiden, the Netherlands). Blast algorithms (McGinnis & Madden, 2004) were used to screen databases at the National Center for Biotechnology Information (NCBI) (Bethesda, MD, USA).
Drosophila small heat‐shock protein sequences were retrieved from the NCBI database using the D. melanogaster HSP27 sequence as input for a protein BLAST search. Results were analyzed by clustalx (Larkin et al., 2007) and visualized by treeview (Page, 1996). A total of eleven small HSPs were found to be present in the D. melanogaster genome, most of which are located at position 67B on the third chromosome (Fig. S1A, Supporting information). Nearest‐neighbor analysis shows three main groups within the D. melanogaster small HSP family (Fig. S1B). Group A represents the most studied classical small HSPs: HSP22, HSP23, HSP26, and HSP27. Group B consists of L(2)EFL, CG4461, CG7409, and CG14207, while group C contains HSP67BC and CG13133. The Drosophila HSP plasmid library (Fig. 1C) was generated using either cDNA originating from heat‐shocked flies or cDNA from the Gold cDNA library (Bloomington, Indiana University). Primers used for isolation and amplification of individual small HSPs using are listed in Table S2. All PCR products were cloned into the pAc5.1‐V5 plasmid and sequence verified. The pAc5.1‐V5 was generated by annealing two oligonucleotides forming a KpnI overhang at the 5′ end (Table S2). This fragment was ligated into KpnI‐EcoRV‐digested pAc5.1.
The L4440 RNAi feeding vector (Fire Laboratory), containing two opposing T7 sequences, was modified for T/A‐cloning as follows. To remove unneeded nucleotides between both T7 sequences, L4440 was digested with BglII and XhoI. Annealed oligonucleotides (Table S2), designed to provide BglII and XhoI overhangs and two internal XcmI sites, were ligated into digested L4440 leading to L4440‐T/A. Upon digestion with XcmI, this plasmid contains two T‐overhangs, allowing annealing of Taq DNA polymerase‐amplified DNA fragments (A‐overhangs). To generate the dsRNA‐template library, a specific part of the HSP70 and HSC70 genes was amplified using primers listed in Table S3 using Taq polymerase. Subsequently, the A‐tailed PCR products were ligated into the XcmI‐digested L4440‐T/A plasmid.
DNA transfection
Schneider's S2 cells were transfected using either the CaCl2 method or Effectene (Qiagen, Venlo, the Netherlands). Transfection using the CaCl2 was performed as follows. A total of 2.5 μg of plasmid DNA was mixed with 10 μL 2.5 m CaCl2, the volume was adjusted to 100 μL with 0.1 × TE (pH 7.6), and this was added dropwise to 100 μL 2 × HEPES buffer (280 mm NaCl, 1.5 mm Na2HPO4 2H2O, 50 mm HEPES, pH 7.05) while vortexing. Precipitates were allowed to form for 30 min, and then, the solution was added to 1 × 106 cells in a 35‐mm dish. Transfection using Effectene was performed according to the manufacturer's instructions using 0.6 μg of plasmid DNA in combination with 5 μL Effectene.
RNA interference
RNA interference was performed as described previously (Clemens et al., 2000). In short, DNA fragments of variable length coding for specific parts of the target genes (Table S3) were amplified using Taq DNA polymerase. These fragments were cloned into XcmI‐digested L4440‐T/A vector. Using T7‐specific primers, the T7‐flanked target sequence was re‐amplified. Subsequently, dsRNA was generated using the MEGASCRIPT T7 transcription kit (Ambion, Austin, TX, USA). S2 cells were diluted to a final concentration of 1x106 cells/mL in Drosophila serum‐free medium (GIBCO, Paisley, UK). dsRNA was added to the serum‐free medium and incubated for 1 h at 25 °C, followed by the addition of 2 mL complete Schneider's medium. Part of an intron sequence of the human MAZ gene was used as mock dsRNA. One day after dsRNA transfection, cells were transfected with plasmid DNA. Two days later, cells were subjected to either the luciferase refolding assay or the filter trap assay.
Quantitative PCR
Exponentially growing Schneider's S2 cells were resuspended in complete Schneider's Drosophila medium to a final concentration of 1 × 106 cells per mL. A total of 5 mL of cell suspension was heat‐shocked for the indicated temperature and time points using a precision water bath. Total RNA was isolated using the Invisorb Spin Cell RNA mini kit (Westburg, Leusden, the Netherlands). First‐strand cDNA was generated using M‐MLV reverse transcriptase (Invitrogen) using oligo(dT)18 primers (Biolegio, Nijmegen, the Netherlands). Relative changes in transcript level were determined using the iCycler (Bio‐Rad, Hercules, CA, USA) in combination with SYBR Green Supermix (Bio‐Rad, Veenendaal, the Netherlands). Calculations were performed using the comparative CT method according to User Bulletin 2 (Applied Biosystems, Foster City, CA, USA; Livak & Schmittgen, 2001). The PCR efficiencies for all primer pairs (Table S5) were between 85% and 100%. Fold induction was adjusted using RpL32 transcript levels as a standard.
Luciferase refolding assay
The luciferase refolding assay (Michels et al., 1995) was adapted for Schneider's S2 cells as follows. S2 cells were transfected with the pAc5.1‐Luc plasmid, coding for firefly luciferase together with different heat‐shock‐protein‐coding plasmids in a 1:9 ratio. After two days, cells were resuspended in complete Schneider's Drosophila medium containing cycloheximide (2 mg/100 mL). The cell suspension was divided into 400‐μL portions in 1.5‐mL centrifuge tubes. Tubes were placed into custom‐made acrylic glass racks (50 positions), which allowed continuous water flow around the tubes when placed in a water bath. After a heat treatment of 30 min at 38 °C, the tubes were cooled down rapidly by placing them in a 25 °C water bath followed by incubation in a 25 °C incubator. Cell lyses and luciferase measurements were performed as described (Michels et al., 1995). The experiments were performed and measured in triplicate.
Biochemical techniques
SDS‐PAGE and Western blotting were performed by established procedures. Primary antibodies used were monoclonal anti‐EGFP (Clontech, Saint‐Germain‐en‐Laye, France) and monoclonal anti‐V5 (Invitrogen) according to the manufacturers' instruction. The filter trap binding assay was performed as described previously (Carra et al., 2005).
Antibody preparation
The rabbit polyclonal antibody anti‐HSP67BC was raised against the C‐terminal peptide CHKEAGPAASASEPEAK of Drosophila melanogaster HSP67BC coupled to the keyhole limpet hemocyanin.
Lifespan analysis
For lifespan analysis, male flies were selected and maintained at noncrowding conditions at 22 °C. Dead flies were counted three times per week followed by transfer to fresh media. Lifespan curves were visualized in Kaplan–Meier plots and analyzed using a log‐rank (Mantel–Cox) test for significance.
Funding info
This work was supported by Senter (Innovatiegerichte Onderzoeksprogramma genomics Grant IGE03018 to H.H.K.).
Conflict of interest
None declared.
Supporting information
Fig. S1 The Drosophila family of small heat‐shock proteins.
Fig. S2 ataxin‐3 fly model for polyQ‐related eye degeneration.
Table S1 Drosophila lines used in this study.
Table S2 Primers used for molecular cloning.
Table S3 Primers used for the generation of dsRNA and specificities of the dsRNA sequences.
Table S4 Plasmids used in this study.
Table S5 Primers used for qPCR.
References
- Aigaki T, Seong KH, Matsuo T (2002) Longevity determination genes in Drosophila melanogaster . Mech. Ageing Dev. 123, 1531–1541. [DOI] [PubMed] [Google Scholar]
- Arslan MA, Csermely P, Soti C (2006) Protein homeostasis and molecular chaperones in aging. Biogerontology 7, 383–389. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Bonner JJ (1979) The induction of gene activity in Drosophilia by heat shock. Cell 17, 241–254. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW (2008) Adapting proteostasis for disease intervention. Science 319, 916–919. [DOI] [PubMed] [Google Scholar]
- Bilen J, Bonini NM (2007) Genome‐wide screen for modifiers of ataxin‐3 neurodegeneration in Drosophila. PLoS Genet. 3, 1950–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Bryantsev AL, Kurchashova SY, Golyshev SA, Polyakov VY, Wunderink HF, Kanon B, Budagova KR, Kabakov AE, Kampinga HH (2007) Regulation of stress‐induced intracellular sorting and chaperone function of Hsp27 (HspB1) in mammalian cells. Biochem. J. 407, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau AL, Szweda LI, Friguet B (2002) Age‐dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys. 397, 298–304. [DOI] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvári M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, Vazquez‐Manrique RP, Orfila AM, Ackerman D, Au C, Vinti G, Riesen M, Howard K, Neri C, Bedalov A, Kaeberlein M, Soti C, Partridge L, Gems D (2011) Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra S, Sivilotti M, Chavez Zobel AT, Lambert H, Landry J (2005) HspB8, a small heat shock protein mutated in human neuromuscular disorders, has in vivo chaperone activity in cultured cells. Hum. Mol. Genet. 14, 1659–1669. [DOI] [PubMed] [Google Scholar]
- Carra S, Seguin SJ, Landry J (2008) HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy 4, 237–239. [DOI] [PubMed] [Google Scholar]
- Carra S, Boncoraglio A, Kanon B, Brunsting JF, Minoia M, Rana A, Vos MJ, Seidel K, Sibon OC, Kampinga HH (2010) Identification of the Drosophila ortholog of HSPB8: implication of HSPB8 loss of function in protein folding diseases. J. Biol. Chem. 285, 37811–37822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashikar AG, Duennwald M, Lindquist SL (2005) A chaperone pathway in protein disaggregation: HSP26 alters the nature of protein aggregates to facilitate reactivation by HSP104. J. Biol. Chem. 280, 23869–23875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Koppenhafer SL, Bonini NM, Paulson HL (1999) Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J. Neurosci. 19, 10338–10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HY, Warrick JM, Gray‐Board Paulson HL, Bonini NM (2000) Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum. Mol. Genet. 9, 2811–2820. [DOI] [PubMed] [Google Scholar]
- Chuang JZ, Zhou H, Zhu M, Li SH, Li XJ, Sung CH (2002) Characterization of a brain‐enriched chaperone, MRJ, that inhibits Huntingtin aggregation and toxicity independently. J. Biol. Chem. 277, 19831–19838. [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson‐Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE (2000) Use of double‐stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl Acad. Sci. USA 97, 6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A (2006) Opposing activities protect against age‐onset proteotoxicity. Science 313, 1604–1610. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Sun Y, Opal P, Antalffy B, Mestril R, Orr HT, Dillmann WH, Zoghbi HY (2001) Over‐expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum. Mol. Genet. 10, 1511–1518. [DOI] [PubMed] [Google Scholar]
- Dobson CM (2003) Protein folding and misfolding. Nature 426, 884–890. [DOI] [PubMed] [Google Scholar]
- Ellis RJ, Minton AP (2006) Protein aggregation in crowded environments. Biol. Chem. 387, 485–497. [DOI] [PubMed] [Google Scholar]
- Engel R, Westphal AH, Huberts DH, Nabuurs SM, Lindhoud S, Visser AJ, van Mierlo CP (2008) Macromolecular crowding compacts unfolded apoflavodoxin and causes severe aggregation of the off‐pathway intermediate during apoflavodoxin folding. J. Biol. Chem. 283, 27383–27394. [DOI] [PubMed] [Google Scholar]
- Fayazi Z, Ghosh S, Marion S, Bao X, Shero M, Kazemi‐Esfarjani P (2006) A Drosophila ortholog of the human MRJ modulates polyglutamine toxicity and aggregation. Neurobiol. Dis. 24, 226–244. [DOI] [PubMed] [Google Scholar]
- Ferrington DA, Husom AD, Thompson LV (2005) Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 19, 644–646. [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ (2007) Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta‐peptide. Nat. Rev. Mol. Cell Biol. 8, 101–112. [DOI] [PubMed] [Google Scholar]
- Hageman J, Rujano MA, van Waarde MAWH, Kakkar V, Dirks D, Govorukhina N, Oosterveld‐Hut HJM, Lubsen NH, Kampinga HH (2010) A DNAJB chaperone subfamily with HDAC‐dependent activities suppresses toxic protein aggregation. Mol. Cell 37, 355–369. [DOI] [PubMed] [Google Scholar]
- Homouz D, Perham M, Samiotakis A, Cheung MS, Wittung‐Stafshede P (2008) Crowded, cell‐like environment induces shape changes in aspherical protein. Proc. Natl Acad. Sci. USA 105, 11754–11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz J (1992) Alpha‐crystallin can function as a molecular chaperone. Proc. Natl Acad. Sci. USA 89, 10449–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C (2003) Regulation of aging and age‐related disease by DAF‐16 and heat‐shock factor. Science 300, 1142–1145. [DOI] [PubMed] [Google Scholar]
- Ish‐Horowicz D, Pinchin SM, Schedl P, Artavanis‐Tsakonas S, Mirault ME (1979) Genetic and molecular analysis of the 87A7 and 87C1 heat‐inducible loci of D. melanogaster . Cell 18, 1351–1358. [DOI] [PubMed] [Google Scholar]
- Jakob U, Gaestel M, Engel K, Buchner J (1993) Small heat shock proteins are molecular chaperones. J. Biol. Chem. 268, 1517–1520. [PubMed] [Google Scholar]
- Kramer EB, Farabaugh PJ (2007) The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Vierling E (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat‐denatured protein. Plant Physiol. 122, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E (1997) A small heat shock protein stably binds heat‐denatured model substrates and can maintain a substrate in a folding‐competent state. EMBO J. 16, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen FW, Fischer DF, Kamel D, Sluijs JA, Sonnemans MA, Benne R, Swaab DF, Salehi A, Hol EM (2000) Molecular misreading: a new type of transcript mutation expressed during aging. Neurobiol. Aging 21, 879–891. [DOI] [PubMed] [Google Scholar]
- Liu AY, Lin Z, Choi HS, Sorhage F, Li B (1989) Attenuated induction of heat shock gene expression in aging diploid fibroblasts. J. Biol. Chem. 264, 12037–12045. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Marin R, Demers M, Tanguay RM (1996) Cell‐specific heat‐shock induction of Hsp23 in the eye of Drosophila melanogaster . Cell Stress Chaperones. 1, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32, W20–W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S, Marin R, Tanguay RM (1997) Regulation of heat shock gene induction and expression during Drosophila development. Cell. Mol. Life Sci. 53, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels AA, Nguyen VT, Konings AW, Kampinga HH, Bensaude O (1995) Thermostability of a nuclear‐targeted luciferase expressed in mammalian cells. Destabilizing influence of the intranuclear microenvironment. Eur. J. Biochem. 234, 382–389. [DOI] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, Friedrich KL, Schonfeld HJ, Vierling E, Bukau B (2003) Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 278, 31033–31042. [DOI] [PubMed] [Google Scholar]
- Morimoto RI (2008) Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI (2004) Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell 15, 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Tanguay RM (2003) Heat shock proteins and aging in Drosophila melanogaster . Semin. Cell Dev. Biol. 14, 291–299. [DOI] [PubMed] [Google Scholar]
- Morrow G, Samson M, Michaud S, Tanguay RM (2004) Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 18, 598–599. [DOI] [PubMed] [Google Scholar]
- Morrow G, Heikkila JJ, Tanguay RM (2006) Differences in the chaperone‐like activities of the four main small heat shock proteins of Drosophila melanogaster . Cell Stress Chaperones. 11, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DD, Theodorakis NG, Morimoto RI (1988) Coordinate changes in heat shock element‐binding activity and HSP70 gene transcription rates in human cells. Mol. Cell. Biol. 8, 4736–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Master M, Zhu Y, Pack AI (2008) Aging impairs the unfolded protein response to sleep deprivation and leads to proapoptotic signaling. J. Neurosci. 28, 6539–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EA, Kabakov AE, Brunsting JF, Kanon B, Hohfeld J, Kampinga HH (2001) Modulation of in vivo HSP70 chaperone activity by Hip and Bag‐1. J. Biol. Chem. 276, 4677–4682. [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Parker J (1989) Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 53, 273–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettigrew CA, Brown MA (2008) Pre‐mRNA splicing aberrations and cancer. Front Biosci. 13, 1090–1105. [DOI] [PubMed] [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L (2002) Genome‐wide transcript profiles in aging and calorically restricted Drosophila melanogaster . Curr. Biol. 12, 712–723. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K (1991) Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22, 443–461. [DOI] [PubMed] [Google Scholar]
- Rujano MA, Kampinga HH (2008). The HSP70 chaperone machine as guardian of the proteome: implications for protein folding diseases In Heat Shock Proteins in Biology and Medicine (Radons J, Multhoff G, eds). Kerala, India: Research Signpost, pp. 59–85. [Google Scholar]
- Rujano MA, Kampinga HH, Salomons FA (2007) Modulation of polyglutamine inclusion formation by the Hsp70 chaperone machine. Exp. Cell Res. 313, 3568–3578. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989). Molecular Cloning a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Urushitani M, Kurisu J, Tateno M, Hatakeyama S, Nakayama K, Kato S, Takahashi R (2004) CHIP promotes proteasomal degradation of familial ALS‐linked mutant SOD1 by ubiquitinating Hsp/Hsc70. J. Neurochem. 90, 231–244. [DOI] [PubMed] [Google Scholar]
- Vacher C, Garcia‐Oroz L, Rubinsztein DC (2005) Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's disease. Hum. Mol. Genet. 14, 3425–3433. [DOI] [PubMed] [Google Scholar]
- Vos MJ, Zijlstra MP, Kanon B, van Waarde‐Verhagen MAWH, Brunt ERP, Oosterveld‐Hut HMJ, Carra S, Sibon OCM, Kampinga HH (2010) HSPB7 is the most potent polyQ aggregation suppressor within the HSPB family of molecular chaperones. Hum. Mol. Genet. 19, 4677–4693. [DOI] [PubMed] [Google Scholar]
- Walker GA, Lithgow GJ (2003) Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin‐like signals. Aging Cell 2, 131–139. [DOI] [PubMed] [Google Scholar]
- Walker GA, White TM, McColl G, Jenkins NL, Babich S, Candido EP, Johnson TE, Lithgow GJ (2001) Heat shock protein accumulation is upregulated in a long‐lived mutant of Caenorhabditis elegans . J. Gerontol. A Biol. Sci. Med. Sci. 56, B281–B287. [DOI] [PubMed] [Google Scholar]
- Walther DM, Kasturi P, Zheng M, Pinkert S, Vecchi G, Ciryam P, Morimoto RI, Dobson CM, Vendruscolo M, Mann M, Hartl FU (2015) Widespread proteome remodeling and aggregation in aging C. elegans . Cell 161, 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HD, Kazemi‐Esfarjani P, Benzer S (2004) Multiple‐stress analysis for isolation of Drosophila longevity genes. Proc. Natl Acad. Sci. USA 101, 12610–12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick JM, Chan HY, Gray‐Board CY, Paulson HL, Bonini NM (1999) Suppression of polyglutamine‐mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 23, 425–428. [DOI] [PubMed] [Google Scholar]
- Wilhelmus MM, Boelens WC, Otte‐Holler I, Kamps B, de Waal RM, Verbeek MM (2006) Small heat shock proteins inhibit amyloid‐beta protein aggregation and cerebrovascular amyloid‐beta protein toxicity. Brain Res. 1089, 67–78. [DOI] [PubMed] [Google Scholar]
- Zhang X, Smith DL, Meriin AB, Engemann S, Russel DE, Roark M, Washington SL, Maxwell MM, Marsh JL, Thompson LM, Wanker EE, Young AB, Housman DE, Bates GP, Sherman MY, Kazantsev AG (2005) A potent small molecule inhibits polyglutamine aggregation in Huntington's disease neurons and suppresses neurodegeneration in vivo. Proc. Natl Acad. Sci. USA 102, 892–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Meadows S, Sharp L, Jan LY, Jan YN (2000) Genome‐wide study of aging and oxidative stress response in Drosophila melanogaster . Proc. Natl Acad. Sci. USA 97, 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The Drosophila family of small heat‐shock proteins.
Fig. S2 ataxin‐3 fly model for polyQ‐related eye degeneration.
Table S1 Drosophila lines used in this study.
Table S2 Primers used for molecular cloning.
Table S3 Primers used for the generation of dsRNA and specificities of the dsRNA sequences.
Table S4 Plasmids used in this study.
Table S5 Primers used for qPCR.


