Highlight
Determination of mesophyll and bundle sheath cell-specific transcriptomes for the monocot NAD-ME C4 plant switchgrass reveals both evolutionary divergence and common elements in C4 establishment.
Key words: C4 photosynthesis, carbon fixation, cell-specific transcriptomics, comparative transcriptomics, switchgrass.
Abstract
Almost all C4 plants require the co-ordination of the adjacent and fully differentiated cell types, mesophyll (M) and bundle sheath (BS). The C4 photosynthetic pathway operates through two distinct subtypes based on how malate is decarboxylated in BS cells; through NAD-malic enzyme (NAD-ME) or NADP-malic enzyme (NADP-ME). The diverse or unique cell-specific molecular features of M and BS cells from separate C4 subtypes of independent lineages remain to be determined. We here provide an M/BS cell type-specific transcriptome data set from the monocot NAD-ME subtype switchgrass (Panicum virgatum). A comparative transcriptomics approach was then applied to compare the M/BS mRNA profiles of switchgrass, monocot NADP-ME subtype C4 plants maize and Setaria viridis, and dicot NAD-ME subtype Cleome gynandra. We evaluated the convergence in the transcript abundance of core components in C4 photosynthesis and transcription factors to establish Kranz anatomy, as well as gene distribution of biological functions, in these four independent C4 lineages. We also estimated the divergence between NAD-ME and NADP-ME subtypes of C4 photosynthesis in the two cell types within C4 species, including differences in genes encoding decarboxylating enzymes, aminotransferases, and metabolite transporters, and differences in the cell-specific functional enrichment of RNA regulation and protein biogenesis/homeostasis. We suggest that C4 plants of independent lineages in both monocots and dicots underwent convergent evolution to establish C4 photosynthesis, while distinct C4 subtypes also underwent divergent processes for the optimization of M and BS cell co-ordination. The comprehensive data sets in our study provide a basis for further research on evolution of C4 species.
Introduction
C4 species are among the world’s most important food, feed, and bioenergy crops, including maize (Zea mays), sugarcane (Saccharum officinarum), sorghum (Sorghum bicolor), and switchgrass (Panicum virgatum) (Brutnell et al., 2010; John et al., 2014). C4 plants have evolved the C4 cycle pathway, which creates a CO2 pump that concentrates CO2 around the carboxylating enzyme Rubisco (Christin et al., 2009). In most C4 species, this is achieved through integrating the two CO2 assimilation pathways spatially into two discrete cell types, namely mesophyll (M) and bundle sheath (BS) cells (Sheen, 1999; John et al., 2014). C4 plants can achieve high photosynthetic efficiency and consequently decrease photorespiration. These characteristics enable C4 plants to thrive in tropical and subtropical environments that induce high rates of photorespiration in C3 plants (Brutnell et al., 2010).
In C4 plants, CO2 fixation is a two-step process. Atmospheric CO2 is initially fixed by phosphoenolpyruvate carboxylase (PEPC) in the cytosol of M cells. The resulting four-carbon dicarboxylic acid oxaloacetate (OAA) is converted to malate or aspartate. These C4 acids are then transferred into the inner compartment of the BS, where they are decarboxylated to release CO2 to the Calvin cycle (Hatch, 1987; Furbank and Taylor, 1995; Furbank et al., 2004). The mechanism of decarboxylation in BS cells has been traditionally divided into three different C4 types; NAD-malic enzyme (NAD-ME), NADP-malic enzyme (NADP-ME), and phosphoenolpyruvate carboxykinase (PEPCK) (Hatch, 1987), with some degree of flexibility or co-existence (Wang et al., 2014) (Fig. 1). However, only NAD-ME and NADP-ME subtypes are considered to be exclusive subtypes from specific lineages; both contain the PEPCK cycle as the supplementary photosynthetic pathway (Wang et al., 2014; Liu and Osborne, 2015).
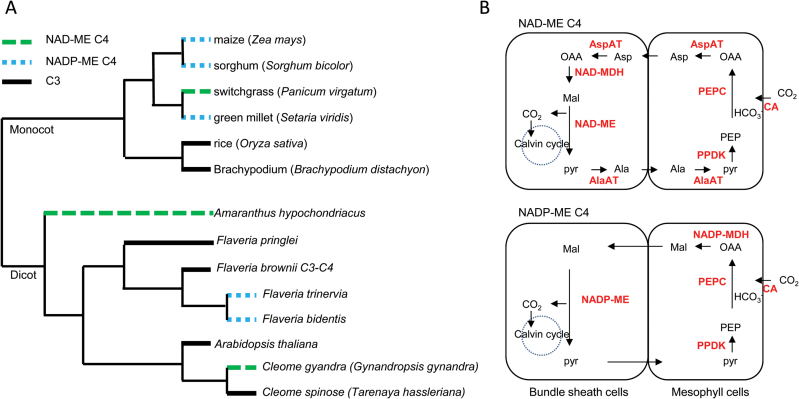
Fig. 1.
Phylogenetic tree of selected C4 and C3 species and carbon fixation process of C4 leaves. (A) Phylogenetic analysis of C4 model species with rice, Arabidopsis, and Brachypodium C3 model species, drawn based on Bennetzen et al. (2012). (B) The major biochemical cycles in NAD-ME and NADP-ME subtypes of C4 photosynthesis. AlaAT, aspartate aminotransferase; AspAT, aspartate aminotransferase; CA, carbonic anhydrase; PEPC, phosphoenolpyruvate carboxylase; PEPCK, phosphoenolpyruvate carboxykinase; NAD-ME, NAD-dependent malic enzyme; NAD-MDH, NAD-dependent malate dehydrogenase; NADP-ME, NADP-dependent malic enzyme; NADP-MDH, NADP-dependent malate dehydrogenase; PPDK, pyruvate/orthophosphate dikinase; Ala, alanine; Asp, aspartate; Mal, malate; Pyr, pyruvate; OAA, oxaloacetic acid; PEP, phosphoenolpyruvate. (This figure is available in colour at JXB online.)
C4 photosynthesis is thought to have first arisen ~30 million years ago and is found in >66 independent lineages of monocotyledons and dicotyledons (Aubry et al., 2014; Burgess and Hibberd, 2015) (Fig. 1). Most of the C4 species occur in the grasses (~4600) and sedges (~1600) of monocots, while only 1600 C4 species are found in the dicots (Gowik and Westhoff, 2011). The mechanism of establishment of C4 photosynthesis is still unclear. Genes involved in C4 photosynthesis recruited existing C3 chloroplastic ancestors and did not evolve de novo (Gowik and Westhoff, 2011; Maier et al., 2011). C4 photosynthesis may have emerged through gene duplication, promoter insertion, alteration of leaf structure, and establishment of the photorespiratory CO2 pump (Gowik and Westhoff, 2011; Maier et al., 2011).
A distinct set of transcripts are preferentially expressed in the fully specialized M or BS cells to enable their co-operation in carbon fixation (Majeran et al., 2005). Comparison of transcriptomes for M and BS cell-specific gene expression in different lineages of C4 plants will provide insight into the commonality and differentiation of regulatory mechanisms for the compartmentalization of C4 photosynthesis in its independent evolutionary origins (Burgess and Hibberd, 2015). Only limited lineages have so far been assessed for M and BS cell-specific profiles; two for the monocot NADP-ME subtype and one for the dicot NAD-ME subtype. Chang et al. (2012) and Li et al. (2010) generated M and BS transcriptome profiles from mature leaves of maize, a monocot NADP-ME subtype. Pairwise comparisons of M and BS transcriptomes have been conducted for maize versus Setaria viridis (monocot NADP-ME subtype), and maize versus Cleome gynandra (dicot NAD-ME subtype) (Li et al., 2010; Chang et al., 2012; Aubry et al., 2014; John et al., 2014). However, the transcript expression profiles of M and BS cells in a monocot NAD-ME subtype C4 plant have yet to be determined, resulting in the absence of a global comparison of cell-specific transcriptomes in the two distinct subtypes of monocot and dicot C4 plants.
Switchgrass (Panicum virgatum L.) is being targeted as a source of biomass for biofuel production (McLaughlin and Kszos, 2005; Bouton, 2007), and is here selected as the representative of the monocotyledonous NAD-ME-type C4 plant (Christin et al., 2009). We first defined the transcriptomic profiles of M and BS cells from switchgrass by manually isolating M and BS cells from mature leaves. We further applied comparative transcriptome analysis to dissect the evolutionary convergence and divergence of monocot and dicot C4 plants: switchgrass, maize, Setaria viridis, and Cleome gynandra. Our study provides an overview of the functional differentiation and co-ordination of M and BS cells in the C4 photosynthesis pathway, and provides insights into the possible evolutionary pathway of differentiation in C4 photosynthesis.
Materials and methods
Plant material, growth conditions, and enzyme assays
Switchgrass plants (cultivar Alamo-AP13) were grown in the greenhouse at 28 °C and 16/8h day/night conditions, using supplemental lighting from halide lamps (250mol photons m−2 s−1). For age gradient experiments, leaves were harvested from the first fully expanded leaves to the fourth leaves of three-node stage plants. For leaf developmental gradient experiments, leaves were harvested as the top, middle, and base sections from the second or third leaf of three-node stage plants. Samples were immediately frozen in liquid nitrogen and extracted as described (Sommer et al., 2012). Enzyme activity measurements of NAD-ME, NADP-ME, PEPCK, aspartate aminotransferase (AspAT), and alanine aminotransferase (AlaAT) were conducted using coupled spectrophotometric assays as described previously (Sommer et al., 2012). All assays were performed at 28 °C. The concentration of total soluble protein was determined by the Bradford method (Bradford, 1976) using the Bio-Rad protein assay reagent at 595nm.
Fixation, cryosectioning, and cell-specific micro-dissection
The middle sections from the second or third leaf of three-node stage plants were fixed and subsequently used for micro-dissection. A Leica CM 1850 cryostat (Leica Microsystems Inc., Bannockburn, IL, USA) was used for obtaining cross-sections of switchgrass leaves, which were processed by fixation and cryoprotection as described previously (Srivastava et al., 2010). We found that the optimum longitudinal section thickness was 10 μm. For obtaining M and BS cells, leaf sections were manually dissected with a scalpel blade under a fluorescence stereo microscope. Around 1000 cells were collected into each tube and stored at −80 °C, and every 5–10 tubes were used together for RNA isolation.
RNA extraction and analysis
Total RNA was extracted from manually harvested M and BS cells using an RNeasy Micro Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. RNA quality and quantity were estimated using an RNA 6000 Pico chip on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). RNA was then amplified using an Arcturus® RiboAmp® HS PLUS 2-round kit (Life Technologies, CA, USA) following the manufacturer’s protocol (Li et al., 2010). At least 1ng of starting total RNA was used for amplification of each replicate. The quality of amplified RNA was checked with the Bioanalyzer 2100, using the RNA 6000 nanochip.
Construction of cDNA libraries and sequencing procedure
As previously described (Rao et al., 2014), 1 μg of total RNA for each sample was used to construct an RNA sequencing (RNA-seq) library using TruSeq RNA Sample Prep Kits v2 (Illumina Inc., San Diego, CA, USA), according to the manufacturer’s instructions, at the Genomics Core Facility at the Noble Foundation (Ardmore, OK, USA). Each library was indexed. Six libraries with different indexes were pooled together in one lane for 100bp paired-end sequencing. The Hiseq2000 run was conducted at the Genomics Core Facility of the Oklahoma Medical Research Foundation (Oklahoma City, OK, USA).
Determination of transcript abundance
All paired-end Illumina reads were trimmed using in-house PERL scripts with two filters, quality scores of ≤20 from the end of each read, and poly(A) (forward sequencing) or poly(T) (reverse sequencing) tails >15bp in length. Reads <50bp in length after trimming were discarded along with their mates. The trimmed reads were mapped to the Switchgrass genome Panicum virgatum v1.1 (http://phytozome.jgi.doe.gov/) using Bowtie 2 version 2.0.0 (Langmead and Salzberg, 2012) and TopHat version 2.0.10 (Kim et al., 2013) in conjunction with SAMtools version 0.1 (Li et al., 2009) with default parameters. Based on Panicum virgatum v1.1 annotation, normalized gene expression levels were calculated in FPKM (fragments per kilobase of exon model per million mapped fragments) (Trapnell et al., 2010) using Cufflinks version 2.1.1 (default settings, set to 100 mean inner distance for paired-end reads; Trapnell et al., 2013) and in TPM (transcripts per million) (Wagner et al., 2012) using RSEM version 1.2.23 (default settings; Li and Dewey, 2011). For differential expression testing, alignments of reads from Tophat v2.0.10 were counted to genes using HT-SEQ with union mode (Anders et al., 2015), then analyzed using DESeq2 (Love et al., 2014). A Benjamini–Hochberg corrected P-value of <0.05 was set to identify differentially expressed genes. Raw data are provided in Supplementary Table S1 at JXB online.
Data annotation
Panicum virgatum v1.1 gene annotation was downloaded from the Phytozome v10.3 website (http://phytozome.jgi.doe.gov/). Classification for cell type-enriched genes was based on MapMan mappings of their Arabidopsis homologs (Thimm et al., 2004). Significant functional enrichment was determined using Fisher’s exact test with Benjamini–Hochberg multiple testing correction [false discovery rate (FDR) ≤0.1]. Identification of transcription factors (TFs) in Panicum virgatum v1.1 genes was based on their Arabidopsis and rice homologs and the annotation from the Plant Transcription Factor Database v3.0 (Jin et al., 2014). Identification of homologous genes among switchgrass, maize, and S. viridis was performed using alignments of Panicum virgatum v1.1, Setaria italica v2.1, and Zea mays v5b.60 protein sequences obtained from Phytozome v10.3 using blast+ tools. The syntenic gene set of maize, sorghum, and rice was obtained from Schnable et al. (2012) and that of switchgrass, S. viridis, and maize was generated for pairwise comparisons using SynMap (Lyons et al., 2008) with default parameters (score ≥90).
Real-time quantitative reverse transcription–PCR
Total RNA was isolated from the second and third leaf, leaf sheath, and stem of three-node stage switchgrass plants. cDNA synthesis, primer design, and real-time quantitative reverse transcription–PCRs (qRT–PCRs) were performed as previously described (Rao et al., 2014). qRT–PCR was performed in triplicate for each sample, and three biological replicates were evaluated for each gene tested. Data were collected and analyzed using PikoReal Software (Thermo Scientific). PCR efficiency was estimated using PikoReal software, and transcript levels were determined by relative quantification using the actin gene as a reference (Gimeno et al., 2014).
In situ hybridization
For in situ hybridization, samples of middle sections from the second or third leaf of three-node stage switchgrass plants were harvested. Tissue fixation, dehydration, and paraffin embedding were performed according to Long’s protocol (http://www.its.caltech.edu/~plantlab/protocols/insitu.pdf). Pre-hybridization, hybridization, and washing were conducted on the robotic GenePaint™ system (Tecan Inc., Durham, NC, USA) following the manufacturer’s instructions (Zhou et al., 2011).
Construction of phylogenetic trees
For phylogenetic reconstruction, multiple protein sequences were aligned using the ClustalW algorithm (Larkin et al., 2007). Neighbor–Joining (NJ) phylogenetic trees were generated by the MEGA 6.0 program (Tamura et al., 2013), using 1000 replicates of bootstrap analysis. Protein sequences were obtained from a previous report (Maier et al., 2011).
Accession numbers
The data sets supporting the results of this article are available in the NCBI Sequence Read Archive (SRA) repository, NCBI SRA accession no. SRX1160366.
Results and Discussion
Activity of C4 enzymes in the switchgrass leaf
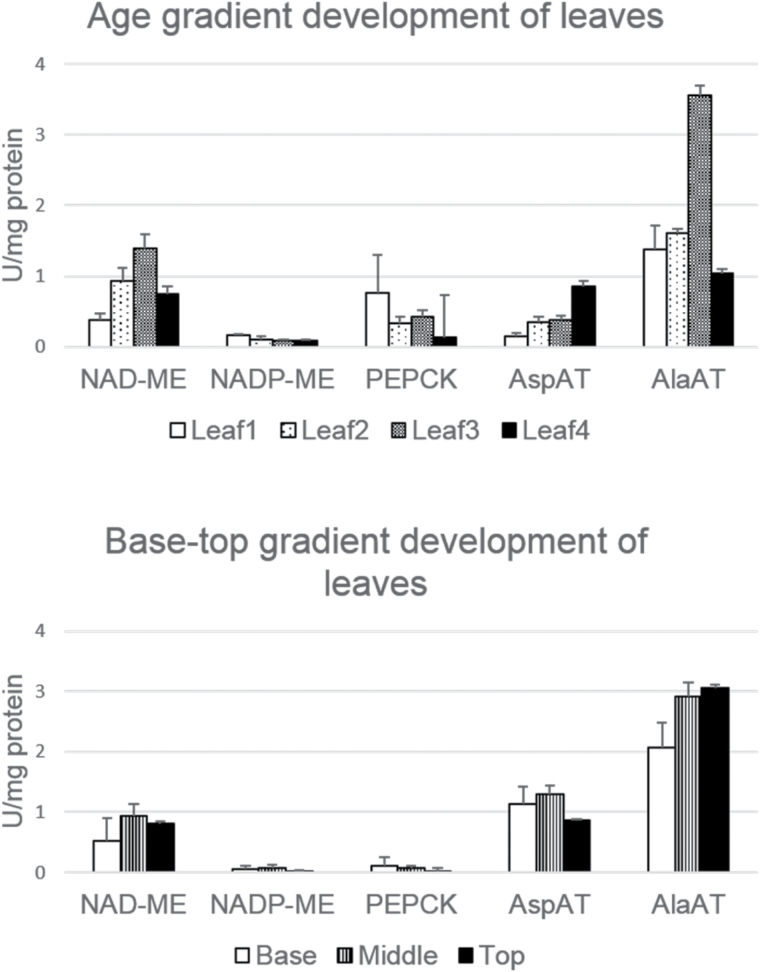
To determine the differences in CO2 fixation enzymes along a developmental gradient in switchgrass leaves as a basis for selection of tissues for transcriptomic analysis, we measured the activities of enzymes involved in C4 photosynthetic pathways in crude leaf extracts. For the age gradient experiment, we selected from the first to the fourth leaf blades as young, mature, and old samples from plants with three nodes, in the elongation stage of the plant (Moore et al., 1991). The three decarboxylating enzymes NAD-ME, NADP-ME, and PEPCK were assayed, as well as AspAT and AlaAT (Fig. 2; Supplementary Table S2). This analysis confirmed that switchgrass performs C4 photosynthesis of the NAD-ME type with a minimal PEPCK decarboxylating pathway (Warner et al., 1987). Compared with young and old samples, the mature leaves (the second and third leaves) showed significantly higher activity of NAD-ME, AspAT, and AlaAT. A base to top gradient experiment was further conducted on the second and the third leaves to assess area-related differences in C4 enzyme activities of mature leaves. The classical NAD-ME subtype enzymes NAD-ME and AlaAT showed lower activities in the base section of leaves compared with the top or middle sections. This is consistent with the results of transcriptomic and C4 enzyme activity assays in maize leaves, since cells in the leaf base are younger with minimal photosynthetic activity (Li et al., 2010; Pick et al., 2011). Considering the C4 enzyme activity and the size of the leaf cells, the middle sections in the mature zones of the second and third leaves were selected for subsequent experiments.
Fig. 2.
Enzyme activities in age and base–top gradient development of switchgrass leaves. One unit (U) of enzyme activity is defined as the amount of enzyme required to produce 1.0 µmol of product in 1min. Means and standard errors from three independent biological replicates are plotted. The significant differences by the Student’s t-test between pairwise samples are provided in Supplementary Table S2 at JXB online.
Isolation of bundle sheath and mesophyll cells
Cross-sections from selected parts of the switchgrass leaf showed the typical Kranz anatomy structure of NAD-ME subtype C4 plants; a layer of small M cells surrounding the layer of large BS cells with the inner mestome sheath and vascular tissue (Supplementary Fig. S1) (Edwards and Walker, 1983; Edwards et al., 2001). As observed in a previous study (Warner et al., 1987), the >10 μm width of BS and M cells makes hand dissection feasible (Supplementary Fig. S1). Hand dissection has been successfully applied for tissue-specific and single-cell analysis in plant research, and can be simply carried out by use of a razor blade (Outlaw and Zhang, 2001). In this study, leaf fragments were cut into longitudinal cryosections with optimal thickness of 10 μm, and BS and M cells were obtained by manual micro-dissection under a fluorescence stereo microscope (Supplementary Fig. S1). RNA isolated from BS and M cells (Supplementary Fig. S2) was of suitable quality for amplification, library construction, and sequencing.
Deep sequencing, read mapping, and identification of expressed genes
Duplicated BS and M cell samples from switchgrass leaves were sequenced on the HiSeq™ Sequencing System to generate paired-end reads. Clean sequence reads for BS and M samples were mapped to the switchgrass reference genome Panicum virgatum v1.1, resulting in 79% of the cleaned reads mapped to the reference genome (Table 1). Considering the high complexity of the tetraploid switchgrass genome (Sharma et al., 2012) and the fact that Panicum virgatum v1.1 represents a partial genome sequence, a moderate ratio of mapped reads is expected.
Table 1.
Statistics of the transcriptome data
| Parameter | Value |
|---|---|
| Read length | 100 bp |
| Read type | Paired |
| Total reads | 144 129 974 |
| Trimmed reads | 111 728 008 |
| Mapped reads | 87 920 267 (79%) |
| Detected genes (mapped read >1) | 46 716 |
| Differentially expressed genes | 9752 |
Normalized transcript levels were calculated using RSEM version 1.2.23 (Li and Dewey, 2011) and Cufflinks version 2.1.1 (Trapnell et al., 2013) in terms of TPM (Wagner et al., 2012) and FPKM (Trapnell et al., 2010). To define ‘differentially expressed genes’, we used the criterion of adjusted P≤0.05 between the two RNA samples. On this basis, 5122 and 4630 genes were considered as differentially expressed in BS and M cells, respectively. Moreover, increasing the criterion of difference in fold value, more genes were enriched in BS cells than in M cells (Table 2). For example, 332 genes in BS but only 199 genes in M showed a 16-fold difference in expression level. This finding is consistent with observations in the other C4 plants maize (Chang et al., 2012) and S. viridis (John et al., 2014), and suggests that BS cells play a more important role in C4 photosynthesis and may also differentially perform other metabolic processes (Chang et al., 2012).
Table 2.
Numbers of expressed genes, differentially expressed genes, and TF genes in BS and M samples
| Parameter | BS cells | M cells |
|---|---|---|
| Differentially expressed genes (adjustws P<0.05) | 5,122 | 4,630 |
| Differentially expressed genes (ratio >2) | 4,343 | 3,886 |
| Differentially expressed genes (ratio >4) | 2,444 | 2,126 |
| Differentially expressed genes (ratio >8) | 923 | 695 |
| Differentially expressed genes (ratio >16) | 332 | 199 |
| Differentially expressed TF genes | 175 | 243 |
Estimation of C4 pathway transcript abundance in the switchgrass leaf
The main difference in the photosynthetic pathway between C3 and C4 plants is that in C4 plants the CO2 concentration and assimilation mechanisms are divided between M and BS cells (Chang et al., 2012). Gene families that encode proteins with key roles in C4 photosynthesis were detected in our M/BS transcriptome data set (Supplementary Table S3). Among them, 25 genes were selected for further analysis due to strong cell-specific expression (Table 3). The fold change between M and BS samples was similar using either the FPKM or TPM value (Supplementary Table S3). Because TPM eliminates statistical biases compared with FPKM (Wagner et al., 2012), we used TPM to present expression levels of C4 genes in the following sections.
Table 3.
Differential expression of C4 photosynthesis-specific genes between M and BS cells
| Gene | Role | Cell type | ID | M (TPM) | BS (TPM) | Log 2 (M/BS) | Adjusted P |
|---|---|---|---|---|---|---|---|
| CA | Core pathway | M | Pavir.J05107 | 10360.08 | 2559.41 | 1.74 | 0.0043498 |
| PEPC | Core pathway | M | Pavir.Da00871 | 404.89 | 132.94 | 1.35 | 0.0158103 |
| PPDK | Core pathway | M | Pavir.J15849 | 10630.50 | 1774.50 | 2.24 | 1.567E-09 |
| ASP-AT(M) | Core pathway | M | Pavir.Eb03232 | 2699.88 | 559.12 | 1.79 | 5.627E-07 |
| ASP-AT(BS) | Core pathway | BS | Pavir.J06248 | 91.64 | 1045.03 | –3.27 | 1.32E-17 |
| ALA-AT | Core pathway | BS/M | Pavir.Ba00407 | 4.82 | 17.78 | –2.09 | 5.823E-08 |
| NADP-MDH | Core pathway | M | Pavir.Fa00047 | 193.63 | 39.84 | 1.71 | 4.326E-07 |
| NADP-ME | Core pathway | BS | Pavir.Eb00308 | 188.46 | 4081.74 | –4.58 | 3.438E-53 |
| NAD-ME | Core pathway | BS | Pavir.Ia01553 | 18.64 | 695.89 | –4.84 | 8.162E-89 |
| NAD-MDH | Core pathway | BS | Pavir.J08443 | 108.92 | 1566.72 | –4.35 | 0 |
| PEPCK | Core pathway | BS | Pavir.Ia03881 | 9.77 | 59.16 | –2.56 | 5.193E-06 |
| RBCS | Calvin cycle | BS | Pavir.Ca02105 | 279.46 | 8643.50 | –5.18 | 0 |
| RCA | Calvin cycle | BS | Pavir.Hb00014 | 23.07 | 627.48 | –4.64 | 8.738E-75 |
| PRK | Calvin cycle | BS | Pavir.Aa01018 | 68.43 | 1903.17 | –4.86 | 1.096E-41 |
| CP12 | Calvin cycle | BS | Pavir.J38955 | 0.85 | 19.27 | –3.91 | 1.398E-11 |
| RPI | Calvin cycle | BS | Pavir.Bb00418 | 130.41 | 2702.07 | –4.52 | 1.1E-171 |
| RPE | Calvin cycle | BS | Pavir.Ia04312 | 133.34 | 3492.42 | –4.72 | 1.788E-31 |
| TKL | Calvin cycle | BS | Pavir.Da02287 | 44.74 | 1504.46 | –5.28 | 1.22E-242 |
| SBP | Calvin cycle | BS | Pavir.J03580 | 6.71 | 287.19 | –4.91 | 2.529E-50 |
| FBP | Calvin cycle | BS | Pavir.J33737 | 7.74 | 81.39 | –3.36 | 5.467E-24 |
| FBA | Calvin cycle | BS | Pavir.J02023 | 56.93 | 1424.97 | –4.53 | 1.08E-244 |
| PGK | Calvin cycle | M | Pavir.J00566 | 5471.47 | 1559.81 | 1.33 | 0.0048193 |
| GADPH(A) | Calvin cycle | BS | Pavir.Ca01865 | 30.68 | 54.80 | –1.07 | 0.0002747 |
| GADPH(B) | Calvin cycle | M | Pavir.J23272 | 3301.02 | 1316.49 | 1.17 | 4.605E-14 |
| TPI | Calvin cycle | M | Pavir.J26504 | 815.95 | 108.90 | 2.62 | 1.611E-33 |
CA, carbonic anhydrase; PEPC, phosphoenolpyruvate carboxylase; PPDK, pyruvate/orthophosphate dikinase; AspAT, aspartate aminotransferase; AlaAT, alanine aminotransferase; NADP-MDH, NADP-dependent malate dehydrogenase; NADP-ME, NADP-dependent malic enzyme; NAD-MDH, NAD-dependent malate dehydrogenase; NAD-ME, NAD-dependent malic enzyme; PEPCK, phosphoenolpyruvate carboxykinase; RBCS, ribulose bisphosphate carboxylase small chain; RCA, Rubisco activase; PRK, phosphoribulokinase; CP12, Calvin cycle protein CP12; RPI, ribose 5-phosphate isomerase; RPE, ribulose-phosphate 3-epimerase; TKL, transketolase; SBP, sedoheptulose-bisphosphatase; FBP, fructose-1,6-bisphosphatase; FBA, fructose-bisphosphate aldolase; PGK, phosphoglycerate kina; GADPH, glyceraldehyde-3-phosphate dehydrogenase; TPI, triosephosphate isomerase.
The C4 marker genes encoding carbonic anhydrase (CA), pyruvate orthophosphate dikinase (PPDK), PEPC, and NAD-dependent malate dehydrogenase (NADP-MDH; Fig. 1B) that are involved in carbon fixation (Chang et al., 2012) were preferentially expressed in M cells, whereas NAD-MEs, NAD-MDH, and PCK that are involved in releasing CO2 from C4 acid (Chang et al., 2012), and nine genes encoding enzymes involved in the Calvin cycle, showed much higher transcript levels in BS cells than in M cells. In addition, phosphoglycerate kinase (PGK), glyceraldehyde 3-phosphate dehydrogenase B subunit (GADPH-B), and triosephosphate isomerase (TPI), that work in M cells to allow balancing of reducing equivalents between the two cell types (John et al., 2014), were preferentially expressed in the M cells.
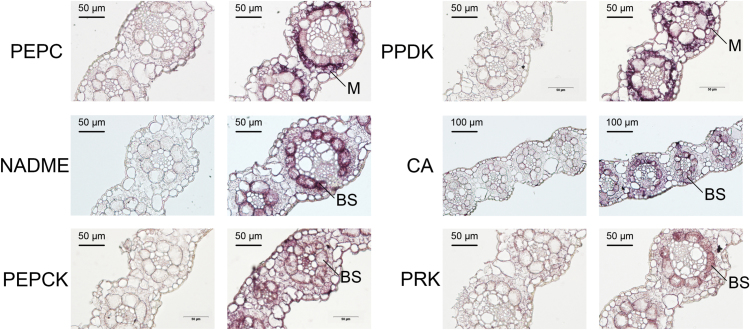
On the basis of previously published microarray analyses (Zhang et al., 2013), C4 marker genes in switchgrass displayed a higher expression level in leaf blade compared with other major tissues and organs such as leaf sheath and stem (Supplementary Fig. S3). The high expression of C4 marker genes in the mature switchgrass leaf was confirmed through qRT–PCR analysis (Supplementary Fig. S3). To validate further the digital expression profile, NAD-ME, CA, PEPCK, phosphoribulokinase (PRK), PEPC, and PPDK were selected for localization analysis by in situ hybridization. Labeled antisense probes were used to hybridize with the target mRNAs in situ, while labeled sense probes were used as negative controls (Srivastava et al., 2010). The sequences of the primers used for qRT–PCR and in situ hybridization are given in Supplementary Table S4. As expected, NAD-ME, CA, PEPCK, and PRK were preferentially expressed in BS cells, whereas PEPC and PPDK transcripts were enriched in M cells (Fig. 3). These results are consistent with those from the transcriptome data set, and indicate that our isolation methods caused only low-level cross-contamination of M and BS cells.
Fig. 3.
In situ hybridization of C4-related gene transcripts in mature leaves. (This figure is available in colour at JXB online.)
Convergence in C4 transcript abundance among monocot and dicot C4 plants
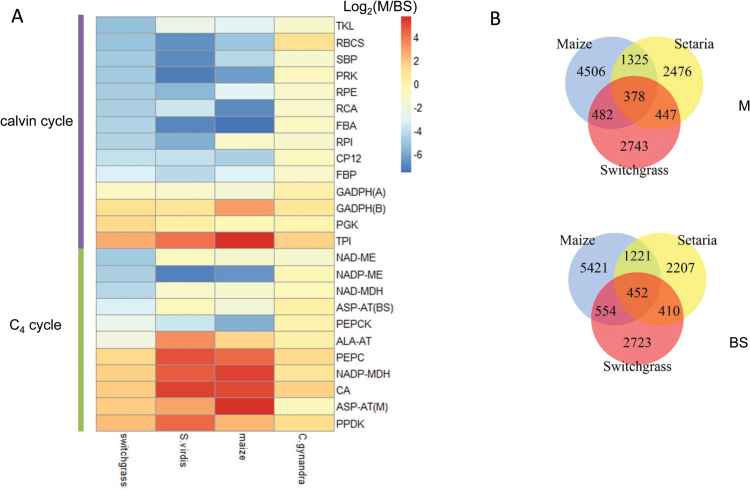
We compared C4 transcript abundance in the M and BS transcriptomes from switchgrass (present work), S. viridis (John et al., 2014), maize (Chang et al., 2012), and C. gynandra (Aubry et al., 2014) (Fig. 4; Supplementary Table S5). Maize, S. viridis, and switchgrass belong to the C4 grasses, and are classified into two distinct subtypes, NADP-ME and NAD-ME (Wang et al., 2014). The dicot plant C. gynandra performs NAD-ME-type C4 photosynthesis (Sommer et al., 2012). The major biochemical processes for C4 carbon fixation in the two C4 photosynthetic subtypes are shown in Fig. 1B. Transcripts involved in the C4 cycle showed very similar compartmentalization between M and BS cells in the grass species; genes encoding enzymes involved in CO2 fixation (CA, PEPC, PPDK, and NADP-MDH) and the Calvin cycle [ribulose bisphosphate carboxylase small chain (RBCS), Rubisco activase (RCA), PRK, ribose 5-phosphate isomerase (RPI), ribulose-phosphate 3-epimerase (RPE), transketolase (TKL), sedoheptulose-bisphosphatase (SBP), fructose-1,6-bisphosphatase (FBP), and fructose-bisphosphate aldolase (FBA)] were enriched in M and BS cells, respectively. Transcripts from the C. gynandra transcriptome showed reduced enrichment in BS cells, possibly due to post-transcriptional regulation (Aubry et al., 2014). The consistency of C4 core gene expression preference in four distinct C4 lineages highlights the evolutionary convergence in the C4 pathway.
Fig. 4.
Global comparison of transcript abundance within M and BS cells of four C4 species. (A) Fold change of expression level of genes involved in C4 metabolism and the Calvin cycle in M and BS cells from switchgrass, S. viridis, maize, and C. gynandra. (B) Venn diagrams show the number of homologous genes differentially expressed in M and BS cells from switchgrass, S. viridis, and maize. (This figure is available in colour at JXB online.)
The GADPH of land plants includes two plastidic tetramer isoforms named A4 and A2B2 (Fermani et al., 2012). Transcripts encoding GADPH-B were clearly enriched in M cells in the four species, consistent with the role of the A2B2 isoform in catalyzing the reducing step of the Calvin cycle in M cells. In contrast, transcripts encoding GADPH-A accumulated in both cell types, with a slightly higher ratio in BS cells. Furthermore, transcripts encoding CP12, a small protein involved in regulation of the Calvin cycle by forming a supercomplex with PRK and GADPH (Groben et al., 2010; Lopez-Calcagno et al., 2014), were exclusively found in BS cells. This suggests that in BS cells, CP12 and A4 GADPH mainly function together for regulation of the activity of PRK, in agreement with the observation of formation of a stable CP12–A4 GADPH complex in the dark (Fermani et al., 2012).
PEPCK, the decarboxylation enzyme that utilizes OAA to release CO2 in the cytosol (Hatch, 1987), is enriched in BS cells at moderate expression levels in all four C4 species. This is consistent with the detection of PEPCK activity in switchgrass, maize, and C. gynandra (Prendergast et al., 1987; Sommer et al., 2012), and suggests that a mixed and flexible decarboxylation system commonly exists in NAD-ME and NADP-ME subtypes, through an additional service of the PEPCK cycle (Furbank, 2011; John et al., 2014).
Divergence in C4 transcript abundance between NAD-ME and NADP-ME subtypes
C4 plants have evolved different means to decarboxylate organic carbon compounds to release CO2 at the site of Rubisco; NADP-ME decarboxylates malate to pyruvate in chloroplasts, whereas NAD-ME decarboxylates malate to pyruvate in mitochondria (Brautigam et al., 2014) (Fig. 1B). Transcripts encoding NADP-ME, rather than NAD-ME or NAD-MDH, show enrichment in BS cells in two different lineages of the NADP-ME subtype (maize and S. viridis), whereas NAD-ME is enriched in BS cells in the monocot switchgrass and the dicot C. gynandra (Fig. 4). In the phylogenetic tree constructed with known NAD-ME sequences, four switchgrass NAD-ME genes detected in our M/BS transcriptome data set are classified into two distinct clusters, and photosynthetic NAD-ME in switchgrass belongs to the NAD-ME 2 group (Supplementary Fig. S4).
Interestingly, transcripts of two switchgrass genes (PvEa00263 and PvEb00308) that are annotated as chloroplastic NADP-ME showed high accumulation in BS cells. The high expression level of NADP-ME genes in switchgrass leaves has been observed in several previous reports (Zhang et al., 2013; Meyer et al., 2014; Palmer et al., 2015) and here was confirmed by qRT–PCR analysis using multiple pairs of primers (Supplementary Fig. S3; Supplementary Table S4). To address the evolutionary origin of switchgrass NADP-MEs, a phylogenetic tree was constructed from protein sequence alignments of NADP-MEs from C4 and C3 plants (Supplementary Fig. S5). The two highly expressed switchgrass NADP-ME genes clustered with the C4 NADP-ME of S. viridis, and share identity to the non-photosynthetic plastidic NADP-ME in maize (Saigo et al., 2004), which presents high intrinsic NADP-ME activity but is expressed constitutively at low levels (Maier et al., 2011). Considering the low NADP-ME activity detected in switchgrass leaves, it is likely that in switchgrass either strong post-transcriptional or translational control leads to degradation or loss of activity of NADP-ME in BS cells, or the two genes encode low activity NADP-ME isoforms. To assess further the possible derivation of C4 functional decarboxylation enzyme from the same ancestral genomic region (Schnable et al., 2012), we identified the syntenic orthologs of NAD-ME and NADP-ME isoforms among C4 grasses and outgroup C3 rice (Supplementary Fig. S6). Interestingly, NAD-ME 2 genes (functional C4 group) were syntenic within C4 grasses (switchgrass, S. viridis, maize, and sorgum), whereas NAD-ME 1 genes were syntenic between C4 grasses and C3 rice. Genes in non-C4 and C4 NADP-ME groups were syntenic in switchgrass, S. viridis, maize, and sorghum. Overall, it is possible that the photosynthetic and non-photosynthetic isoforms found in C4 plants could present at some level within the most recent common ancestor (Washburn et al., 2015) but may or may not have been recruited into C4 photosynthesis depending on selective pressure.
In NAD-ME and PEPCK subtypes, carboxylation and decarboxylation are linked by the transfer of aspartic acid and alanine in M/BS cells, whereas NADP-ME subtypes mainly recruit malate/pyruvate shuttles between the two cell types (Weber and von Caemmerer, 2010). The amino acid aspartate in the chloroplast of M cells is generated by chloroplastic AspAT, then enters the BS cells where it is converted into OAA in the mitochondria by mitochondrial AspAT (Weber and von Caemmerer, 2010) (Fig. 1B). In switchgrass, transcripts encoding chloroplastic and mitochondrial AspAT are abundant in M and BS cells, respectively, which supports a function in maintaining the ammonia balance between M/BS cell types (Weber and von Caemmerer, 2010). In contrast, AspAT shows reduced expression in BS cells in the NADP-ME subtypes maize and S. viridis. Transcripts encoding AlaAT are significantly accumulated in both M and BS cells of C. gynandra, consistent with a dual function in converting from/into alanine in M/BS cells. In contrast, the AlaAT in the NADP-ME subtypes (maize and S. viridis) displayed unequal distribution in M and BS cells. However, the apparent low expression level of the AlaAT gene in switchgrass is possibly artifactual due to the incompleteness of the switchgrass genome. At least 20 expressed sequence tags (ESTs) were predicted to represent genes encoding AlaAT in switchgrass (Wang et al., 2012; Zhang et al., 2013); one EST (AP13CTG00178) was highly expressed in mature leaves with similar expression pattern to other C4 genes (SupplementaryFig. S3), but is not represented in the current genome assembly Panicum virgatum v1.1.
Differences in C4 shuttle/transport between NAD-ME and NADP-ME subtypes
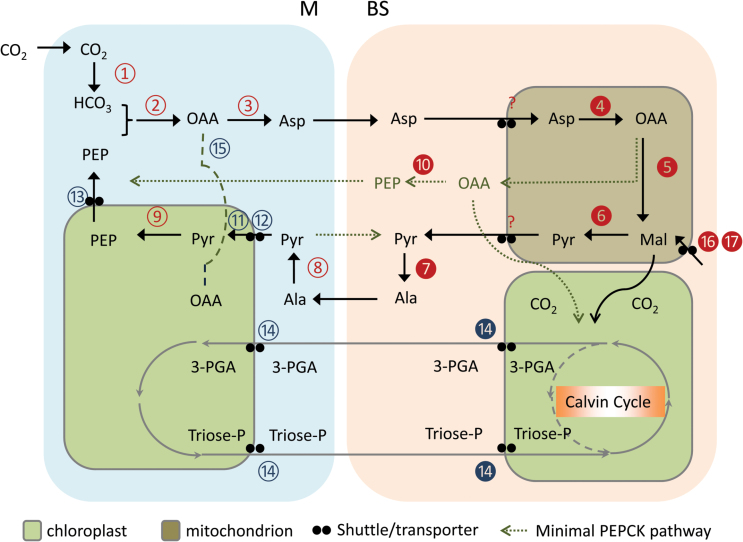
C4 photosynthesis recruits a high rate of metabolic exchange between the two main cell types, which requires multiple transporters in M and BS chloroplast envelopes to facilitate this metabolic collaboration (Majeran and van Wijk, 2009). Based on the present study and previous work (Majeran and van Wijk, 2009; Weber and von Caemmerer, 2010; Brautigam et al., 2014), the detailed processes of C4 photosynthesis in the NAD-ME subtype are summarized in Fig. 5.
Fig. 5.
Model for NAD-ME with minimal PEPCK pathway. 1, carbonic anhydrase (CA); 2, phosphoenolpyruvate carboxylase (PEPC); 3, chloroplastic aspartate aminotransferase (AspAT); 4, mitochondrial aspartate aminotransferase (AspAT); 5, NAD-dependent malate dehydrogenase (NAD-MDH); 6, NAD-dependent malic enzyme (NAD-ME); 7, alanine aminotransferase (AlaAT) in bundle sheathes cells; 8, alanine aminotransferase (AlaAT) in mesophyll cells; 9, pyruvate/orthophosphate dikinase (PPDK); 10, phosphoenolpyruvate carboxykinase (PEPCK); 11, sodium bile acid symporter (BASS) homolog; 12, sodium: hydrogen antiporter (NHD); 13, phosphate/phosphoenolpyruvate translocator (PPT); 14, triose phosphate/phosphate translocator (TPT); 15, dicarboxylate transporter (DiT1); 16, malate phosphate antiporter (DIC); 17, phosphate/protons symporter (PIC). Ala, alanine; Asp, aspartate; Mal, malate; Pyr, pyruvate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; 3-PGA, 3-phosphoglycerate, Triose-P, triose phosphate. (This figure is available in colour at JXB online.)
For both NAD-ME-type and NADP-ME-type C4 plants, pyruvate is used to regenerate the acceptor PEP in M cells and is thereby required to transfer from the cytosol to the chloroplast. However, plants may evolve different strategies to pump pyruvate into these two subtypes (Majeran and van Wijk, 2009). In switchgrass, we observed high enrichment of transcripts corresponding to two genes (Pv.Ea02237 and Pv.Eb01809) encoding the plastidic sodium-dependent pyruvate transporter BASS2 (Furumoto et al., 2011) and moderate enrichment of transcripts corresponding to a plastidic proton:sodium symporter NHD1 gene (Pv.Ba03078) in M cells. The preferential enrichment of BASS2 and NHD1 transcripts in M cells was also found in the NAD-ME subtype C. gynandra (BADSS2, AT2G26900; and NHD1, AT3G19490), but not in maize and S. viridis which, as NADP-ME subtype plants, use the sodium/pyruvate symporter (Chang et al., 2012). This supports the hypothesis that in NAD-ME subtype plants, sodium-dependent transporters take pyruvate into M cell chloroplasts and NHD1 exports sodium from the chloroplast in order to maintain the sodium gradient (Brautigam et al., 2011) (Fig. 5).
In both NAD-ME and NADP-ME subtype plants, PEP generated from pyruvate in M cell chloroplasts is exported into the cytosol by a PEP/phosphate translocator (PPT) (Brautigam et al., 2011). Similarly in maize and S. viridis (Chang et al., 2012), two transcripts (Pv.Fa00799, M TPM=913; and Pv.Fb00666, M TPM=650) encoding PPT were enriched in M cells, whereas the other three (Pv.J33600, Pv.Ea00394, and Pv.J15038) were enriched in BS cells. This suggests that PPT encoded by Pv.Fa00799 and Pv.Fb00666 may specifically export PEP and import triose phosphate in the M chloroplasts. The triose phosphate translocator (TPT) is thought to export triose phosphates and 3-phosphoglycerate (3-PGA) from chloroplasts to the cytosol in M and BS cells; this is necessary to generate half reducing equivalents for the Calvin cycle (Knappe et al., 2003; Weber and von Caemmerer, 2010). In switchgrass, maize, and S. viridis (Chang et al., 2012), genes encoding TPT were expressed at a high level in both M and BS cells. In C. gynandra, PPT (AT5G33320) and TPT (AT5G46110) display high expression levels in both cell types as well. This suggests that C4 plants recruit PPT and TPT to transfer PEP/phosphate and triose phosphate, respectively, in both NAD-ME and NADP-ME subtypes.
For NADP-ME type C4 photosynthesis, DiT1 (a 2-oxoglutarate/malate transporter) exchanges OAA/malate across the M cell chloroplast envelope membrane and DiT2 imports malate into BS chloroplasts (Majeran and van Wijk, 2009; Chang et al., 2012). These processes are not required for NAD-ME-type C4 photosynthesis (Brautigam et al., 2011). Genes encoding DiT1 were preferentially expressed in M cells and genes encoding DiT2 were only weakly expressed in both M and BS cell types in switchgrass (Supplementary Table S6). No increased transcript abundance of DiT1 in C4 (C. gynandra) was detected compared with its closely related C3 species (C. spinosa) (Brautigam et al., 2011). This suggests that DiT1 may not act specifically for C4 metabolite transport in M cells in the NAD-ME subtype because it dually acts as the malate valve and as a 2-oxoglutarate/malate transporter for photorespiratory nitrogen recycling (Kinoshita et al., 2011). For NAD-ME-type C4 photosynthesis, malate is required to be generated from OAA in mitochondria or transported into mitochondria (Fig. 5). We found that transcripts encoding the mitochondrial malate/phosphate carrier DIC (Pavir.Fb01780) and the mitochondrial phosphate transporter PIC2 (Pavir.Aa00590) were significantly enriched in switchgrass BS cells with a high expression level. Similar BS cell-type enrichment and high expression of genes encoding two mitochondrial transporters (DIC, AT2G22500; and PIC2, AT5G14040) (Brautigam et al., 2014; Jia et al., 2015) was also observed in C. gynandra, but DIC expression was not enriched in BS cells of maize and S. vidiris. This suggests a requirement for the combination of the malate phosphate antiporter DIC with the phosphate/proton transporter PIC2 to pump malate into mitochondria in NAD-ME species, but not in NADP-ME species (Fig. 5).
Global comparisons of the M and BS transcriptomes among monocot and dicot C4 plants
To define the extent to which gene expression patterns are conserved or divergent in monocot and dicot C4 plants, the same adjusted P≤0.05 between M and BS samples was applied as the definition of ‘differentially expressed genes’ for switchgrass, S. viridis (John et al., 2014), maize (Chang et al., 2012), and C. gynandra (Aubry et al., 2014). Among S. viridis, maize, and C. gynandra, 5049 and 4631 transcripts, 6691 and 7647 transcripts, and 372 and 338 transcripts were enriched in M or BS cells, respectively. In our switchgrass transcriptome data set, 4630 and 5122 genes were considered as differentially expressed in M or BS cells, respectively (Supplementary Table S6). The higher numbers in switchgrass and maize are probably due to the highly duplicated and complex natures of their genomes (John et al., 2014).
Switchgrass, S. viridis, and maize are close relatives among the C4 grasses (Fig. 1A). To determine if homologous genes underlie C4 photosynthetic development in C4 grasses, homology alignment was applied to differentially expressed genes in switchgrass, S. viridis, and maize (Fig. 5B). A total of 378 and 452 homologous genes are preferentially expressed in M and BS cells of these three C4 grasses. Pairwise comparison showed that S. viridis and maize share the most homologs that are preferentially expressed in M and BS cells, whereas fewer homologs are shared between S. viridis and switchgrass, and between switchgrass and maize. Classification into the same subtype contributes to this higher degree of similarity in M- and BS-enriched genes of maize and S. viridis, though switchgrass and S. viridis have a very close phylogenetic relationship (Fig. 1A) (Bennetzen et al., 2012).
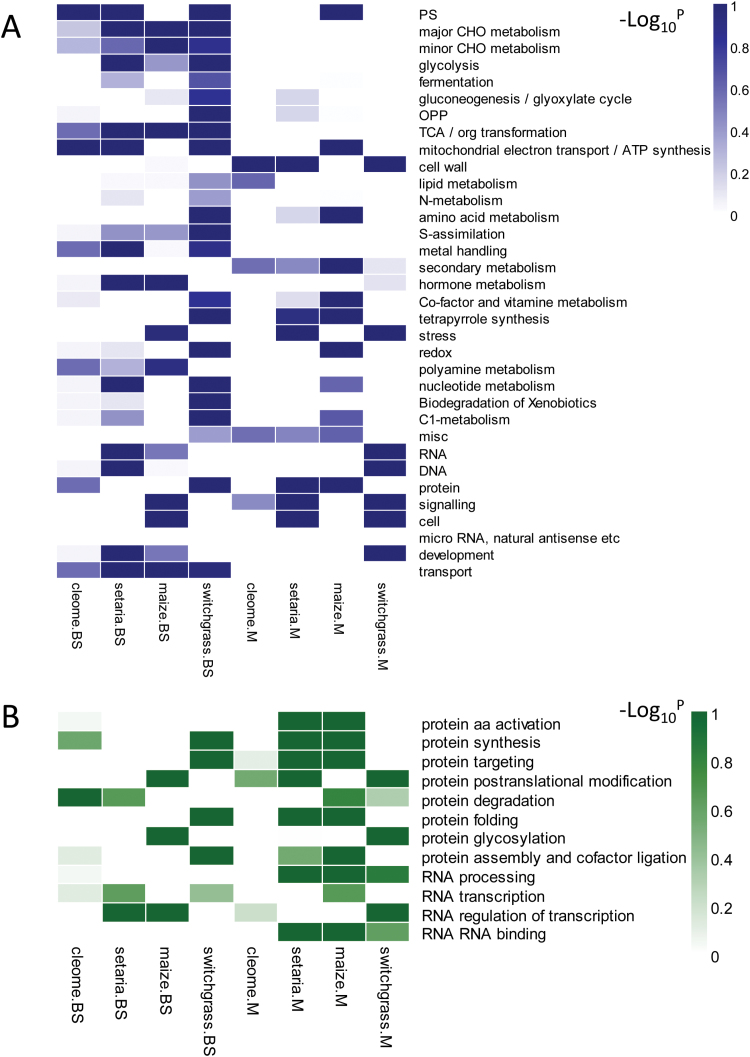
To compare the functional differentiation between M and BS cells in C4 plants, the biological functions of cell-specific enriched genes were visualized using Mapman. Between 74% and 100% of the cell-specific enriched genes in the four C4 species could be assigned into 35 main categories (Fig. 6; Supplementary Table S7). To evaluate the convergence in the distribution of each category in C4 plants, we compared the gene number assigned into categories for pairs of species in M and BS cells, respectively. The Pearson’s correlation coefficient (r 2) ranged from 0.68 to 0.96, indicating a high degree of convergence in distribution of biological processes in the four distinct C4 species (Supplementary Table S7).
Fig. 6.
Functional distribution of BS and M cell type-enriched genes in switchgass, S. viridis, maize, and C. gynandra. Cell type-enriched functional groups in main categories (A) and in subcategories of protein and RNA (B) were identified by Fisher exact test (FDR<0.1 for each group). (This figure is available in colour at JXB online.)
Using the Fisher exact test with FDR ≤0.1, we identified two and one, 12 and five, seven and nine, and 14 and seven main functional groups significantly enriched in M and BS cells of C. gynandra, S. viridis, maize, and switchgrass, respectively (Fig. 6A). Similarity in distribution of genes in certain categories was observed in these four C4 species. For example, considering that primary carbon assimilation is restricted to BS cells, transcripts in the categories of the TCA (tricarboxylic acid) cycle and major CHO (carbohydrate) metabolism were significantly enriched in BS cells of S. viridis, maize, and switchgrass, and moderately enriched in BS cells of C. gynandra. The preferential expression of genes involved in transport in BS cells is consistent with the fact that these cells are surrounded by vascular tissue for release and transport of metabolites to other plant parts (Christin et al., 2009).
However, many differences were detected in functional enrichment between M and BS cells in the four C4 plants. For instance, transcripts encoding maize genes in the photosystem (PS) and mitochondrial electron transport/ATP synthesis categories were more abundant in M cells, whereas in the other three C4 species, these genes were preferentially expressed in BS cells. This is possibly because maize is depleted in PSII in the BS cells to lower oxygen production in these cells (Gowik and Westhoff, 2011).
Further analysis of subcategories within the main categories of protein and RNA revealed more divergence in functional enrichment within the four C4 plant species (Fig. 6B; Supplementary Fig. S7). Interestingly, genes involved in protein synthesis, targeting, and folding displayed preferential expression in BS cells in switchgrass and equal distribution in both cell types in C. gynandra, but strong enrichment in M cells in the two NADP-ME-type plants, S. viridis and maize. Strong enrichment in BS cells of genes involved in protein post-translational modification, protein glycosylation, and protein degradation mediated by AAA-type and ubiquitin proteases was observed in maize, whereas equal distribution in both cell types or moderate enrichment in M cells were found in switchgrass, S. viridis, and C. gynandra. In addition, genes involved in RNA regulation of transcripts were abundant in M cells of switchgrass, but are more abundant in BS cells of maize and S. viridis. This suggests that at both the post-transcriptional and translational levels, switchgrass, maize, and S. viridis may display different machineries for RNA transcriptional regulation and protein import, sorting, folding, assembly, and degradation that facilitate the preferential accumulation of proteins and accommodate protein homeostasis between M and BS cells. We suggest that the functional differentiation of post-transcriptional and translational regulatory mechanisms in M or BS cells of NAD-ME-type switchgrass, and NADP-ME-type maize and S. viridis, might be a result of their distinct evolutionary pathways, and/or may be associated with the accommodation of the different distribution of metabolites within M and BS cells of these two subtypes. For instance, NADP-ME-type plants require extra ATP produced by cyclic electron transport in BS cells to balance the production of NADPH for preventing photoinhibition or photodamage, and this is not required by NAD-ME-type plants (Nakamura et al., 2013; Walker et al., 2014).
Convergence and divergence of cell type-enriched transcription factors in C4 plants
Given that differentially expressed TFs will play a major role in regulatory networks for the functional differentiation of M and BS cell types (Li et al., 2010; Chang et al., 2012), we annotated and identified TFs in the M and BS cell transcriptomes in switchgrass based on their homologs to Arabidopsis and rice in the Plant TF Database v3.0 (Jin et al., 2014). Compared with the annotated TFs of S. viridis (John et al., 2014), maize (Chang et al., 2012), and C. gynandra (Aubry et al., 2014), 243 and 175, 232 and 296, 274 and 412, and 23 and 20 TFs were identified as enriched genes in M and BS cells of switchgrass, S. viridis, maize, and C. gynandra, respectively (Supplementary Table S8). These genes were assigned into 60 TF families according to the Plant TF Database v3.0 (Jin et al., 2014). Consistent with previous studies (Li et al., 2010; Chang et al., 2012), many TF families showed preferential cell type expression, indicating an overall differentiation in the regulatory networks underlying the functional differentiation of the two photosynthetic cell types in C4 leaves. However, similarities and differences were observed in the distributions of cell type preference of TF families within the four distinct C4 plants. Overall, maize and S. viridis shared more similarities in the distributions in TF families, resulting in Pearson’s correlation coefficients (r 2) of 0.70 and 0.89 in M and BS cells, respectively. In contrast, fewer similarities in distribution of cell type preference of TFs were shared between the monocot switchgrass and the dicot C. gynandra, although they belong to the same NAD-ME subtype.
To assess the potential co-option of TFs derived from a common ancestor, syntenic orthologs of TFs were determined between maize, S. viridis, and switchgrass. Switchgrass shared 34 and 13 TFs in M cells, and 44 and 18 TFs in BS cells as syntenic orthologs with S. viridis and maize, respectively. Among them, two and 10 syntenic ortholog TFs overlapped with M and BS cell-type enrichment, respectively, in the three C4 grass genomes. Interestingly, one and three syntenic ortholog TFs in maize with M and BS cell specificity, respectively, have been reported as regulatory candidates in differentiation of C4 Kranz anatomy (Wang et al., 2013; Tausta et al., 2014). For example, SCR is highly expressed in the BS in the three C4 grasses and has been proven to play an essential role in BS cell fate specification in plants (Wysocka-Diller et al., 2000; Slewinski et al., 2012; Cui et al., 2014). This suggests that TFs might be in convergent evolution from a common ancestor and recruited in parallel in distinct C4 linages.
To examine further possible candidates in BS establishment, homologous TFs with BS cell enrichment in four C4 plants were identified, and MYB59, which is reported to regulate the cell division process in Arabidopsis roots (Cominelli and Tonelli, 2009; Mu et al., 2009), was found to be significantly enriched in BS cells of all four C4 species. MYB59 in maize was listed as a candidate in regulating C4 Kranz formation in a previous study (Wang et al., 2013; Tausta et al., 2014). It is possible that MYB59 may play a role in controlling BS cell size in C4 species. In addition, transcriptional activators of lignification such as MYB42 (Hussey et al., 2013) were more abundantly expressed in the BS cells in the three monocot C4 grasses than in C. gynandra, and the transcriptional repressor of lignification MYB4 (Shen et al., 2012) was highly expressed in M cells in switchgrass. This is consistent with the thicker cell walls in the BS cells of monocot as compared with dicot plants (Majeran et al., 2005), and suggests that the C4 grasses might share similar regulatory mechanisms for secondary cell wall formation in BS cells.
Conclusions
The differentiation of M and BS cells with their specific accumulation of transcripts in C4 leaves is considered a hallmark of the C4 pathway (Burgess and Hibberd, 2015). Enzymatic and mechanical isolation or laser capture micro-dissection of M and BS cells have been applied in quantitative transcriptomics of specific cell types (Li et al., 2010; Chang et al., 2012; Aubry et al., 2014; John et al., 2014). Here we employed manual dissection of switchgrass M and BS cells, which is a simple and inexpensive means of obtaining a small specimen for further cell type-specific transcriptome analysis (Outlaw and Zhang, 2001). This has enabled, to the best of our knowledge, the first estimate of mRNA profiles of M and BS cells in a monocot NAD-ME subtype C4 plant.
A high degree of convergence in the distribution of C4 marker genes in M/BS cells was observed in the four distinct C4 species; monocot NAD-ME-type switchgrass, monocot NADP-ME-type S. viridis and maize, and dicot NAD-ME-type C. gynandra (Fig. 4A). Furthermore, these species shared a high degree of convergence in distribution of gene functional categories in M/BS cells based on analysis of the Pearson’s correlation coefficient (r 2) (Fig. 6; Supplementary Table S7). Genes grouped into the categories of TCA and major CHO metabolism were significantly or moderately enriched in BS cells in all four C4 species. A similar preferential expression of SCR and MYB59 TFs was found in C4 grasses and in all four C4 species, respectively (Supplementary Table S8). We conclude that the transcript abundance of core components in C4 photosynthesis, the distribution of gene function, and the mechanism of establishment of BS might be convergent in these four independent lineages of C4 plants.
The pathways of C4 photosynthesis are divided into three basic biochemical subtypes based on the major decarboxylating enzymes to release CO2: NAD-ME, NADP-ME, and PEPCK (Gowik and Westhoff, 2011). However, the differentiation observed in our comparative transcriptomes of NAD-ME and NADP-ME subtypes goes beyond the decarboxylating enzymes. Besides aminotransferase (Fig. 4A), the NAD-ME type differs from the NADP-ME type by the differential compartmentation of specific metabolite transporters, such as sodium:pyruvate and proton:pyruvate co-transporters (Supplementary Table S3), and by more general cell-specific functional enrichment, such as RNA regulation and protein biogenesis/homeostasis (Fig. 6). The abundance of transcripts functioning in protein synthesis, folding, and assembly is more prominent in BS and M cells of switchgrass and C. gynandra than in S. viridis and maize. C4 plants might have undergone convergent processes such as gene duplication, anatomical pre-conditioning, and compartmentalization of C4 core enzymes to evolve C4 photosynthesis (Sage, 2004), but may have undergone divergent processes for the optimization of M and BS cell co-ordination that are not directly associated with the C4 pathway.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Cross- and longitudinal sections of a switchgrass leaf.
Figure S2. Qualitative and quantitative analysis of total RNA from manually isolated BS and M cells using a Bioanalyzer.
Figure S3. Validation of C4-related gene expression in switchgrass by qRT–PCR.
Figure S4. Phylogenetic comparison of switchgrass NAD-ME with NAD-ME protein sequences in other species.
Figure S5. Phylogenetic comparison of switchgrass NADP-ME with NADP-ME protein sequences in other species.
Figure S6. Analysis of syntenic genes of NAD-ME and NADP-ME in grasses
Figure S7. Cell type-enriched functional groups in subcategories of protein degradation identified by Fisher exact test.
Table S1. Raw and normalized data.
Table S2. The fold change of C4 enzyme activity in age- and area-related experiments in switchgrass leaves.
Table S3. List of C4-related genes in the switchgrass M and BS transcriptomes.
Table S4. Sequences of primers used in the present work.
Table S5. Convergence in abundance of C4 marker genes in switchgass, S. viridis, maize, and C. gynandra.
Table S6. List of differentially expressed genes in M and BS cells from switchgrass, S. viridis, maize, and C. gynandra.
Table S7. Functional distribution of differentially expressed genes in M and BS cells from switchgass, S. viridis, maize, and C. gynandra.
Table S8. List of differentially expressed TFs in M and BS cells from switchgrass, S. viridis, maize, and C. gynandra.
Acknowledgements
This research was supported by grants to RAD from both The Advanced Research Projects Agency-Energy (ARPA-E) and the US Department of Energy Bioenergy Research Centers, through the Office of Biological and Environmental Research in the DOE Office of Science. Genome Panicum virgatum v1.1 sequence data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community. Computational resources were provided by UNT’s High Performance Computing Services. We thank Drs Rajeev Azad and Chenggang Liu for critical reading of the manuscript.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Kelly S, Kumpers BM, Smith-Unna RD, Hibberd JM. 2014. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genetics 10, e1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, et al. 2012. Reference genome sequence of the model plant Setaria. Nature Biotechnology 30, 555–561. [DOI] [PubMed] [Google Scholar]
- Bouton JH. 2007. Molecular breeding of switchgrass for use as a biofuel crop. Current Opinion in Genetics and Development 17, 553–558. [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brautigam A, Kajala K, Wullenweber J, et al. 2011. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology 155, 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam A, Schliesky S, Kulahoglu C, Osborne CP, Weber AP. 2014. Towards an integrative model of C4 photosynthetic subtypes: insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. Journal of Experimental Botany 65, 3579–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell TP, Wang L, Swartwood K, Goldschmidt A, Jackson D, Zhu XG, Kellogg E, van Eck J. 2010. Setaria viridis: a model for C4 photosynthesis. The Plant Cell 22, 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SJ, Hibberd JM. 2015. Insights into C4 metabolism from comparative deep sequencing. Current Opinion in Plant Biology 25, 138–144. [DOI] [PubMed] [Google Scholar]
- Chang YM, Liu WY, Shih AC, et al. 2012. Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiology 160, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Kellogg EA, Vicentini A, Besnard G. 2009. Integrating phylogeny into studies of C4 variation in the grasses. Plant Physiology 149, 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Tonelli C. 2009. A new role for plant R2R3-MYB transcription factors in cell cycle regulation. Cell Research 19, 1231–1232. [DOI] [PubMed] [Google Scholar]
- Cui H, Kong D, Liu X, Hao Y. 2014. SCARECROW, SCR-LIKE 23 and SHORT-ROOT control bundle sheath cell fate and function in Arabidopsis thaliana. The Plant Journal 78, 319–327. [DOI] [PubMed] [Google Scholar]
- Edwards G, Walker DA. 1983. C3 C4: mechanisms, and cellular and environmental regulation, of photosynthesis . Oxford: Blackwell Scientific Publications. [Google Scholar]
- Edwards GE, Franceschi VR, Ku MS, Voznesenskaya EV, Pyankov VI, Andreo CS. 2001. Compartmentation of photosynthesis in cells and tissues of C4 plants. Journal of Experimental Botany 52, 577–590. [PubMed] [Google Scholar]
- Fermani S, Trivelli X, Sparla F, Thumiger A, Calvaresi M, Marri L, Falini G, Zerbetto F, Trost P. 2012. Conformational selection and folding-upon-binding of intrinsically disordered protein CP12 regulate photosynthetic enzymes assembly. Journal of Biological Chemistry 287, 21372–21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT. 2011. Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? Journal of Experimental Botany 62, 3103–3108. [DOI] [PubMed] [Google Scholar]
- Furbank R, Hatch M, Jenkins CD. 2004. C4 photosynthesis: mechanism and regulation. In: Leegood R, Sharkey T, Caemmerer S, eds. Photosynthesis . Dordrecht: Springer, 435–457. [Google Scholar]
- Furbank RT, Taylor WC. 1995. Regulation of photosynthesis in C3 and C4 plants: a molecular approach. The Plant Cell 7, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumoto T, Yamaguchi T, Ohshima-Ichie Y, et al. 2011. A plastidial sodium-dependent pyruvate transporter. Nature 476, 472–475. [DOI] [PubMed] [Google Scholar]
- Gimeno J, Eattock N, Van Deynze A, Blumwald E. 2014. Selection and validation of reference genes for gene expression analysis in switchgrass (Panicum virgatum) using quantitative real-time RT–PCR. PLoS One 9, e91474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Westhoff P. 2011. The path from C3 to C4 photosynthesis. Plant Physiology 155, 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groben R, Kaloudas D, Raines CA, Offmann B, Maberly SC, Gontero B. 2010. Comparative sequence analysis of CP12, a small protein involved in the formation of a Calvin cycle complex in photosynthetic organisms. Photosynthesis Research 103, 183–194. [DOI] [PubMed] [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta 895, 81–106. [Google Scholar]
- Hussey SG, Mizrachi E, Creux NM, Myburg AA. 2013. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Frontiers in Plant Science 4, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Wan X, Zhu W, Sun D, Zheng C, Liu P, Huang J. 2015. Overexpression of mitochondrial phosphate transporter 3 severely hampers plant development through regulating mitochondrial function in Arabidopsis. PLoS One 10, e0129717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J. 2014. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research 42, D1182–D1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John CR, Smith-Unna RD, Woodfield H, Covshoff S, Hibberd JM. 2014. Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiology 165, 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita H, Nagasaki J, Yoshikawa N, et al. 2011. The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. The Plant Journal 65, 15–26. [DOI] [PubMed] [Google Scholar]
- Knappe S, Flugge UI, Fischer K. 2003. Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiology 131, 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, et al. 2010. The developmental dynamics of the maize leaf transcriptome. Nature Genetics 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- Liu H, Osborne CP. 2015. Water relations traits of C4 grasses depend on phylogenetic lineage, photosynthetic pathway, and habitat water availability. Journal of Experimental Botany 66, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Calcagno PE, Howard TP, Raines CA. 2014. The CP12 protein family: a thioredoxin-mediated metabolic switch? Frontiers in Plant Science 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, et al. 2008. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with Rosids. Plant Physiology 148, 1772–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A, Zell MB, Maurino VG. 2011. Malate decarboxylases: evolution and roles of NAD(P)-ME isoforms in species performing C4 and C3 photosynthesis. Journal of Experimental Botany 62, 3061–3069. [DOI] [PubMed] [Google Scholar]
- Majeran W, Cai Y, Sun Q, van Wijk KJ. 2005. Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. The Plant Cell 17, 3111–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, van Wijk KJ. 2009. Cell-type-specific differentiation of chloroplasts in C4 plants. Trends in Plant Science 14, 100–109. [DOI] [PubMed] [Google Scholar]
- McLaughlin SB, Kszos LA. 2005. Development of switchgrass (Panicum virgatum) as a bioenergy feedstock in the United States. Biomass and Bioenergy 28, 515–535. [Google Scholar]
- Meyer E, Aspinwall MJ, Lowry DB, Palacio-Mejia JD, Logan TL, Fay PA, Juenger TE. 2014. Integrating transcriptional, metabolomic, and physiological responses to drought stress and recovery in switchgrass (Panicum virgatum L.). BMC Genomics 15, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Moser LE, Vogel KP, Waller SS, Johnson BE, Pedersen JF. 1991. Describing and quantifying growth stages of perennial forage grasses. Agronomy Journal 83, 1073–1077. [Google Scholar]
- Mu RL, Cao YR, Liu YF, et al. 2009. An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Research 19, 1291–1304. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Iwano M, Havaux M, Yokota A, Munekage YN. 2013. Promotion of cyclic electron transport around photosystem I during the evolution of NADP-malic enzyme-type C4 photosynthesis in the genus Flaveria. New Phytologist 199, 832–842. [DOI] [PubMed] [Google Scholar]
- Outlaw WH, Jr, Zhang S. 2001. Single-cell dissection and microdroplet chemistry. Journal of Experimental Botany 52, 605–614. [PubMed] [Google Scholar]
- Palmer NA, Donze-Reiner T, Horvath D, Heng-Moss T, Waters B, Tobias C, Sarath G. 2015. Switchgrass (Panicum virgatum L) flag leaf transcriptomes reveal molecular signatures of leaf development, senescence, and mineral dynamics. Functional and Integrative Genomics 15, 1–16. [DOI] [PubMed] [Google Scholar]
- Pick TR, Brautigam A, Schluter U, et al. 2011. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. The Plant Cell 23, 4208–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast H, Hattersley P, Stone N. 1987. New structural/biochemical associations in leaf blades of C4 grasses (Poaceae). Functional Plant Biology 14, 403–420. [Google Scholar]
- Rao X, Krom N, Tang Y, Widiez T, Havkin-Frenkel D, Belanger FC, Dixon RA, Chen F. 2014. A deep transcriptomic analysis of pod development in the vanilla orchid (Vanilla planifolia). BMC Genomics 15, 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. 2004. The evolution of C4 photosynthesis. New Phytologist 161, 341–370. [DOI] [PubMed] [Google Scholar]
- Saigo M, Bologna FP, Maurino VG, Detarsio E, Andreo CS, Drincovich MF. 2004. Maize recombinant non-C4 NADP-malic enzyme: a novel dimeric malic enzyme with high specific activity. Plant Molecular Biology 55, 97–107. [DOI] [PubMed] [Google Scholar]
- Schnable JC, Freeling M, Lyons E. 2012. Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biology and Evolution 4, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MK, Sharma R, Cao P, et al. 2012. A genome-wide survey of switchgrass genome structure and organization. PLoS One 7, e33892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. 1999. C4 gene expression. Annual Review of Plant Physiology and Plant Molecular Biology 50, 187–217. [DOI] [PubMed] [Google Scholar]
- Shen H, He X, Poovaiah CR, et al. 2012. Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytologist 193, 121–136. [DOI] [PubMed] [Google Scholar]
- Slewinski TL, Anderson AA, Zhang C, Turgeon R. 2012. Scarecrow plays a role in establishing Kranz anatomy in maize leaves. Plant and Cell Physiology 53, 2030–2037. [DOI] [PubMed] [Google Scholar]
- Sommer M, Brautigam A, Weber AP. 2012. The dicotyledonous NAD malic enzyme C4 plant Cleome gynandra displays age-dependent plasticity of C4 decarboxylation biochemistry. Plant Biology 14, 621–629. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Palanichelvam K, Ma J, Steele J, Blancaflor E, Tang Y. 2010. Collection and analysis of expressed sequence tags derived from laser capture microdissected switchgrass (Panicum virgatum L. Alamo) vascular tissues. BioEnergy Research 3, 278–294. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausta SL, Li P, Si Y, Gandotra N, Liu P, Sun Q, Brutnell TP, Nelson T. 2014. Developmental dynamics of Kranz cell transcriptional specificity in maize leaf reveals early onset of C4-related processes. Journal of Experimental Botany 65, 3543–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, et al. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnology 31, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theoretical Bioscience 131, 281–285. [DOI] [PubMed] [Google Scholar]
- Walker BJ, Strand DD, Kramer DM, Cousins AB. 2014. The response of cyclic electron flow around photosystem I to changes in photorespiration and nitrate assimilation. Plant Physiology 165, 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Kelly S, Fouracre JP, Langdale JA. 2013. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. The Plant Journal 75, 656–670. [DOI] [PubMed] [Google Scholar]
- Wang Y, Brautigam A, Weber AP, Zhu XG. 2014. Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. Journal of Experimental Botany 65, 3567–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zeng X, Iyer NJ, Bryant DW, Mockler TC, Mahalingam R. 2012. Exploring the switchgrass transcriptome using second-generation sequencing technology. PLoS One 7, e34225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DA, Ku MS, Edwards GE. 1987. Photosynthesis, leaf anatomy, and cellular constituents in the polyploid C4 grass Panicum virgatum . Plant Physiology 84, 461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn JD, Schnable JC, Davidse G, Pires JC. 2015. Phylogeny and photosynthesis of the grass tribe Paniceae. American Journal of Botany 102, 1493–1505. [DOI] [PubMed] [Google Scholar]
- Weber AP, von Caemmerer S. 2010. Plastid transport and metabolism of C3 and C4 plants—comparative analysis and possible biotechnological exploitation. Current Opinion in Plant Biology 13, 257–265. [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. 2000. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127, 595–603. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Lee YC, Torres-Jerez I, et al. 2013. Development of an integrated transcript sequence database and a gene expression atlas for gene discovery and analysis in switchgrass (Panicum virgatum L.). The Plant Journal 74, 160–173. [DOI] [PubMed] [Google Scholar]
- Zhou C, Han L, Hou C, Metelli A, Qi L, Tadege M, Mysore KS, Wang ZY. 2011. Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. The Plant Cell 23, 2106–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.