Highlight
Phosphate-solubilizing bacteria associated with extraradical mycelia of Rhizophagus irregularis in the hyphosphere assimilated 13C-labeled plant photosynthates and enhanced the mineralization and turnover of soil organic P.
Key words: AM fungus, 13CO2 pulse labeling, hyphosphere, maize, organic phosphate, phosphate-solubilizing bacteria (PSB).
Abstract
This study used a [13C]DNA stable isotope probing (SIP) technique to elucidate a direct pathway for the translocation of 13C-labeled photoassimilate from maize plants to extraradical mycelium-associated phosphate-solubilizing bacteria (PSB) that mediate the mineralization and turnover of soil organic phosphorus (P) in the hyphosphere. Inoculation with PSB alone did not provide any benefit to maize plants but utilized the added phytate-P to their own advantage, while inoculation with Rhizophagus irregularis alone significantly promoted shoot biomass and P content compared with the control. However, compared with both sole inoculation treatments, combined inoculation with PSB and R. irregularis in the hyphosphere enhanced organic P mineralization and increased microbial biomass P in the soil. There was no extra benefit to plant P uptake but the hyphal growth of R. irregularis was reduced, suggesting that PSB benefited from the arbuscular mycorrhizal (AM) fungal mycelium and competed for soil P with the fungus. The combination of T-RFLP (terminal restriction fragment length polymorphism) analysis with a clone library revealed that one of the bacteria that actively assimilated carbon derived from pulse-labeled maize plants was Pseudomonas alcaligenes (Pseudomonadaceae) that was initially inoculated into the hyphosphere soil. These results provide the first in situ demonstration of the pathway underlying the carbon flux from plants to the AM mycelium-associated PSB, and the PSB assimilated the photosynthates exuded by the fungus and promoted mineralization and turnover of organic P in the soil.
Introduction
Phosphorus (P) security is emerging as one of the greatest global sustainability challenges of the 21st century (Cordell and White, 2014). Organic phosphates account for 20–80% of the total P in a wide range of soils (Dalal, 1977) and contribute substantially to the P nutrition of plants. Soil microorganisms play a key role in the major biogeochemical nutrient cycles, including those of organic carbon (C), nitrogen (N) and phosphorus (P), and in the control of nutrient availability in the soil–plant system (Stevenson and Cole, 1999). The main driving force of these microbe-mediated soil nutrient turnover and transformation processes originates from plant litter and rhizodeposition, which supply energy and nutrients for microbes (Fontaine et al., 2007; Kuzyakov, 2011; Sayer et al., 2011; Zhang et al., 2014). Understanding the C–P trade-off between plants and soil microbes has therefore been of considerable interest over many decades (Richardson and Simpson, 2011).
Physiological and molecular research has revealed that arbuscular mycorrhizal (AM) fungi represent an energetically efficient pathway for the acquisition of plant nutrients from soils (Smith et al., 2011). In a similar fashion to plant roots, the extraradical mycelium of AM fungi has been found to acquire organic N or P with attendant microbes that colonize the surface of AM fungal mycelium or the hyphae that are closely attached to the soil (defined as the hyphosphere) (Hodge and Fitter, 2010; Leigh et al., 2011; Wang et al., 2013; Zhang et al., 2014). The fungal hyphae-associated bacteria are able to stimulate the growth of extraradical mycelium, spore germination, and mycorrhizal formation (Xavier and Germida, 2003; Artursson et al., 2006; Frey-Klett et al., 2007), and accelerate the mineralization of organic N in the presence of AM fungi (Hodge et al., 2001; Leigh et al., 2009; Hodge and Fitter, 2010; Leigh et al., 2011; Herman et al., 2012).
Several studies have suggested that the extraradical hyphae of AM fungi exhibit a high C turnover rate (Staddon et al., 2003; Fitter et al., 2004; Godbold et al., 2006) and these hyphae are able to release a substantial amount of carbohydrates which stimulate bacterial growth and vitality (Filion et al., 1999; Mansfeld-Giese et al., 2002; Toljander et al., 2007) or even change the bacterial community composition under in vitro culture conditions (Toljander et al., 2007; Scheublin et al., 2010). The hyphosphere appears to be an environment that is C rich but deficient in available P for many microbes, which may stimulate their activity in mineralizing soil phytate-P and then incorporating the available P into microbial biomass P that is potentially available to AM fungal hyphae (Zhang et al., 2014). P locked in bacterial biomass can be released when C becomes limiting, soils undergo cycles of wetting and drying (Richardson and Simpson, 2011), or during processes of higher tropic-level predation such as bacterial grazing by nematodes (Becquer et al., 2014). The hyphosphere processes through which AM fungal hyphae provide active C derived from plant photosynthesis to microbes for soil P mobilization may be a win–win strategy among plant–AM fungi–fungal-associated bacteria in association with the soil ecosystem. Although there is increasing evidence that biological interactions occur between AM fungi and bacteria in nutrient transformation and turnover in hyphosphere soil, the underlying processes and ecological function are not understood. To date, however, no study has investigated whether fungal-associated bacteria assimilate carbohydrates derived from host plant photosynthesis and are deposited in the hyphosphere via the fungal hyphal exudates in exchange for mobilizing the unavailable nutrients from soil. The objective of the present study was to determine whether PSB that colonize the hyphosphere assimilate carbon from 13CO2-labeled maize through the extraradical hyphae of AM fungi and thereafter are involved in the mineralization and turnover of organic P in soil.
Materials and methods
Soil and microcosms
The soil used in this study was collected from Tai’an, Shandong Province, China and has the following physicochemical properties: pH (soil:H2O 1:5) 6.4, organic matter 5.19g kg−1, mineral N 7.2mg kg−1, Olsen-P [(NaHCO3) extractable] 4.9mg kg−1, and NH4Cl-exchangeable potassium 117.3mg kg−1. The soil was air-dried, sieved (2mm), and then irradiated before use to eliminate indigenous microorganisms (10 kGy, 60Co γ-rays, Beijing Radiation Application Research Center).
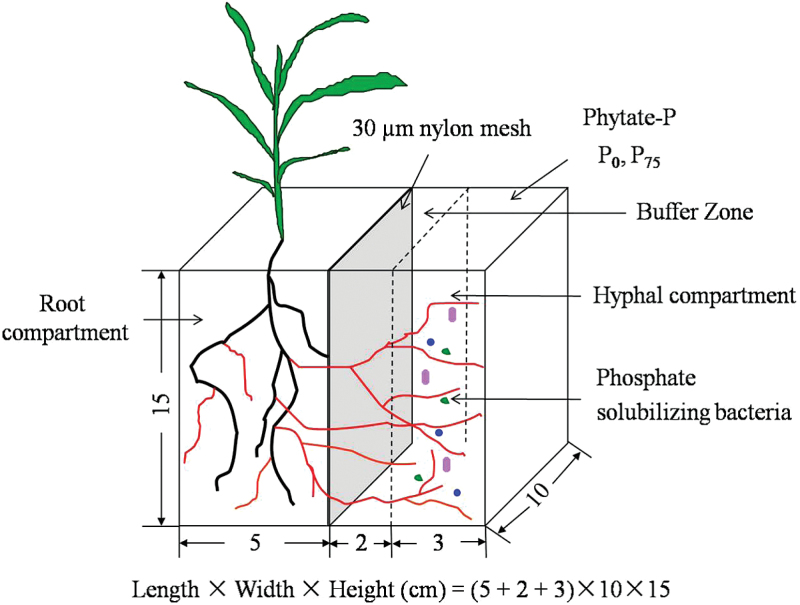
The microcosms used were acrylic rhizoboxes constructed to permit the spatial separation of soil zones for root and AM fungal hyphal growth. Each microcosm had two compartments, one for root growth including mycorrhizal structures (root compartment) and a second compartment (buffer zone and hyphal compartment) that was separated from the first compartment by 30 µm nylon mesh through which hyphae (but not roots) could pass (Fig. 1). There was no physical barrier between the buffer zone and the hyphal compartment. The soil density was adjusted to 1.2g cm−3 and the method was as follows. First, the volume of each compartment was calculated according to length, width, and height. Secondly, the soil subsamples to be transferred to each compartment were weighed separately based on the volume of the compartment and the target soil density. Thirdly, the soil was transferred very carefully to each compartment to maintain an equal soil density in all compartments. The rhizoboxes received the following amounts of soil: 800g in the root compartment, 320g in the buffer zone, and 480g in the hyphal compartment.
Fig. 1.
Schematic drawing of the two-compartment cultivation system (rhizobox) used in this study. The rhizoboxes were divided into a root compartment and a hyphal compartment separated by a 30 µm nylon mesh and a buffer zone. Overall dimensions were 10×10×15cm. (This figure is available in colour at JXB online.)
Host plants, and AM fungal and bacterial inoculants
Maize (Zea mays L., cv. Nongda 108) seeds were surface-sterilized with 10% H2O2 for 10min, thoroughly washed 5–8 times with distilled water, and then germinated on moist filter paper for 2 d at 26 °C in the dark. Two seeds were initially sown in each root compartment and thinned to one seedling after emergence.
The inoculum of Rhizophagus irregularis (formerly Glomus intraradices, BEG 141, kindly supplied by Professor Vivienne Gianinazzi-Pearson, INRA, France) was propagated on maize and clover and consisted of spores, mycelium, root fragments, and soil. Each mycorrhizal treatment received 15g of inoculum, and every non-mycorrhizal treatment received an equivalent amount of sterilized inoculum in combination with 5ml of filtrate of unsterilized inoculum to provide the same microflora with the exception of the absence of the AM fungus. The filtrate of inoculum was obtained by suspending 30g of unsterilized inoculum in 300ml of sterile water and filtering through five-layer quantitative filter paper (properties similar to Whatman Grade 43), which allowed passage of common soil microbes but efficiently retained spores and hyphae of mycorrhizal fungi. The inoculum was mixed uniformly with the soil of the root compartment before sowing of the germinated seeds.
The PSB Pseudomonas alcaligenes M20, Bacillus megaterium C4, and Rahnella aquatilis HX2 (kindly provided by Professor Sanfeng Chen of the College of Biological Sciences and Associate Professor Yanbin Guo of the College of Resources and Environmental Sciences, China Agricultural University) isolated from rice, maize, and grape rhizospheres, respectively, were used. The three bacterial species were previously tagged with the gfp gene encoding green fluorescent protein (GFP) and their capacity to mineralize organic phosphates (Supplementary Fig. S1 at JXB online) and colonize AM fungal hyphae (Supplementary Fig. S2) was also previously tested. Pseudomonas alcaligenes M20, which was transformed with the pGFP78 plasmid containing the gfp gene by electroporation, can release both monoester phosphatase and diester phosphatase to solubilize lecithin or phytate-P (Lv et al., 2005); B. megaterium C4, which was labeled with the pGFP4412 plasmid containing the gfp gene, can mineralize phytate-P (Zhang et al., 2012); and R. aquatilis HX2, which was tagged with the pSMC21 plasmid containing the gfp gene, can solubilize both Ca3(PO4)2 and phytate-P (Sun, 2012). All of the PSB strains were grown in liquid Luria–Bertani (LB) medium on an orbital shaker (180rpm) for 24h at 30 °C and then centrifuged at 6000rpm for 10min. The supernatant was discarded and the cells were re-suspended and diluted to 108 CFU ml−1 with sterile 155mM NaCl solution. After 30 d of plant growth, equivalent volumes of suspensions of the three bacterial species were mixed together and 10ml of the mixed bacterial suspension was then added to the hyphal compartment in the PSB treatments, whereas an equivalent amount of sterile bacterial suspension was added as a control to the non-inoculated PSB treatments.
Experimental design
The experiment was set up in a randomized block design with three factors: (i) two different organic P levels; (ii) two AM fungal levels, inoculated with R. irregularis or uninoculated; and (iii) two bacterial levels, inoculated with a mixed bacterial suspension or uninoculated. Phytate-P was added (0 or 75mg P kg−1 soil) as phytin (TCl, Tokyo, Japan) and was applied only to the hyphal compartment. The experiment was performed in triplicate, and the 24 rhizoboxes were arranged in a randomized block design in the glasshouse. The position of each rhizobox was re-randomized every week. Distilled water was supplied to all of the compartments to maintain the soil moisture level close to field capacity (~20% w/w) during the growth period.
All of the rhizoboxes received basal mineral nutrients which were mixed with the soil uniformly in each compartment at rates of 200mg kg−1 N as (NH4)2SO4, 200mg kg−1 K as K2SO4, 50mg kg−1 Mg as MgSO4·7H2O, 5mg kg−1 Zn as ZnSO4·7H2O, 5mg kg−1 Mn as MnSO4·H2O, and 2mg kg−1 Cu as CuSO4·5H2O. In addition, 10mg kg−1 P was applied as KH2PO4 to the root compartment to meet the minimum growth requirement of the plants. In order to enhance the solubilization of phytate-P in soil, (NH4)2SO4 as the N source was supplied to each compartment because AM fungal hyphae release protons to acidify the hyphosphere soil after absorbing ammonium (Wang et al., 2013). Accordingly, the nitrification inhibitor 3,4-dimethylpyrazole phosphate (DMPP; ‘ENTEC Flüssig’ produced by EuroChem Agro GmbH, Mannheim, Germany) was also added, at a rate of 1% (w/w) of the N applied to prevent nitrification of (NH4)2SO4 and maintain a higher NH4 + concentration in the soil.
13CO2 pulse labeling of plants
Treatment selection
To ensure the transfer of plant-derived C from mycorrhizal hyphae to soil microbes in the hyphosphere, 13CO2 pulse labeling was repeatedly conducted in the glasshouse over 2 years at the same seedling age and using the same labeling chambers, methods, labeling time period, and harvest time. On the first occasion, four microcosms were established as described above for 13C labeling experiments. Both duplicate mycorrhizal and non-mycorrhizal treatments, all with PSB inoculation in the hyphal compartment, were selected to perform 13C labeling. Then we found that the 13C was enriched in the hyphosphere soil in mycorrhizal treatments but did not appear in non-mycorrhizal treatments (Supplementary Table S1). This suggested that the 13C-labeled photosynthates were not enriched in the soil of the hyphal compartment in the absence of AM fungal hyphae. Therefore, in the present experiment, 13CO2 labeling was applied to the mycorrhizal treatments only together with PSB inoculation in duplicate. Unlabeled mycorrhiza combined with PSB treatment was needed to assess background 13C contamination during the two pulse labeling experiments.
Labeling chamber and procedure
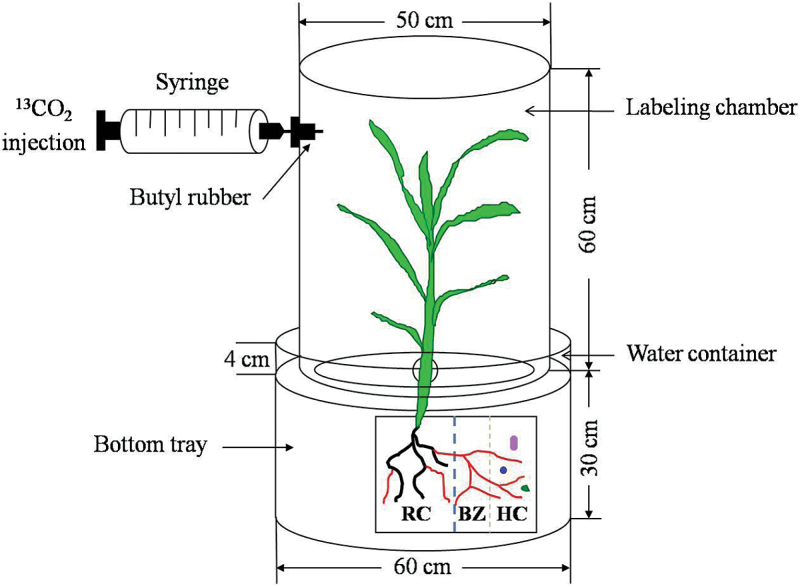
Fifty days after sowing, the maize plants were subjected to 13CO2 (99% of 13C atom) pulse labeling in an airtight Plexiglas growth chamber (Fig. 2). The chamber (volume 137 dm3) had a transparent hood and a bottom tray. The hood was equipped with one hole covered with a septum, the outer edge of the bottom tray was equipped with a water channel to prevent the exchange of 13C gas from ambient CO2 during the labeling period, and the center of the bottom tray was equipped with three holes fitted around the stem of each plant. The plant shoots protruded through the holes and the joins between stems and holes were sealed with silica gel to prevent exposure of the soil surface to the 13C gas. During pulse labeling, the plants were covered with the hood and the fan was fixed in the hood to circulate the air. The hood and bottom were then sealed with water. A 75ml aliquot of 13CO2 was injected through the septum using a gas-tight syringe every hour from 10:00h to 16:00h. The lid was removed after the last 13CO2 injection every day, when the CO2 concentration in the chamber had decreased to atmospheric level. The labeling was continued for 5 d. The temperature in the glasshouse was controlled at 25–28 °C. To remove the influence of vapor produced by plant evaporation during 13CO2 labeling on photosynthesis, three trays of CaCl2 (100g per tray) were placed in the chamber. The wet CaCl2 trays were removed and dried in a forced-air oven at 105 °C for 2h every day after the lid of the chamber was removed in the evening. Thus the CaCl2 was dried and re-used repeatedly.
Fig. 2.
Schematic drawing of pulse labeling of maize plants with 13CO2. A 75ml aliquot of 13CO2 (99 atom% 13C) was injected every hour from 10:00h to 16:00h (seven times a day) into the top chamber for a total of 5 d. (This figure is available in colour at JXB online.)
Harvest and sample analysis
The plants were harvested 8 weeks after sowing. The shoots were dried in a forced-air oven at 70 °C for 48h and weighed. The samples were then ground in a mill before elemental analysis. After the shoot harvest, the rhizoboxes were dismantled and separated into the root and hyphal compartments. To prevent the contamination of the surface soil in the hyphal compartment with exotic bacteria, we removed the top 2cm of soil to reduce the potential influence on hyphosphere soil samples. To obtain a thin slice of the hyphosphere soil, the soil block of the hyphal compartment was placed in an acrylic holder of similar dimensions and then lifted slightly by pushing an acrylic plate (10mm in thickness) underneath. The soil projecting from the holder was then sliced off with a sharp knife. The 10mm thick slice of soil was mixed well and regarded as hyphosphere soil for measuring phosphatase activity, hyphal density, extractable organic P, and microbial biomass P.
Analysis of plant samples
For determination of shoot P concentration, ground plant materials were digested in an H2SO4–H2O2 mixture at 370 °C for 2h. The plant P concentration was determined using the standard vanadomolybdate method (Hanson, 1950).
For determination of mycorrhizal colonization, the roots were washed with tap water and cut into 1cm long segments. The root segments were digested in 10% KOH at 90 °C for 60min, rinsed with water, acidified in 2% HCl at 90 °C for 5min, transferred (with no further rinsing) to 0.05% Trypan blue in lactic acid:glycerol (1:1, v/v) solution, and stained at 90°C for 30min. Finally, the roots were decolorized overnight in lactic acid:glycerol (1:1, v/v) solution at room temperature. The intensity of the mycorrhizal colonization (M%) in the roots was determined (Trouvelot et al., 1986) using the MYCOCALC program (http://www.dijon.inra.fr/mychintec/Mycocalc-prg/download.html).
Analysis of hyphosphere soil
Because all types of phosphatases are pH dependent, the phosphatase activity is usually measured at the standard pH to represent their general activity. However, this approach may not correctly reflect the actual phosphatase activity in environmental samples because the pH of the samples differs from the pH used in the assay buffer. We tested the actual acid phosphomonoesterase activity using buffers with pH adjustment according to the measured hyphosphere pH. The acid phosphomonoesterase activity in each hyphosphere soil was assayed using p-nitrophenyl phosphate (Sigma, St. Louis, MO, USA) as the substrate according to the method of Joner and Johansen (2000) with slight modifications. Fresh soil was weighed into an Eppendorf tube and incubated for 30min at 30 °C in a water bath with sodium acetate buffer (200mM, adjusted to the respective hyphosphere pH determined for each sample) and reaction substrate. The reaction was terminated by the addition of 0.5M NaOH and the mixture was then centrifuged for 5min at 1500 g. The production of p-nitrophenol was measured spectrophotometrically at 405nm.
The organic P concentration was measured by the method of Collos and Mornet (1993). Unlike NaOH-EDTA which extracts both soluble and non-soluble organic P that may not be available for soil phosphatase, the Collos and Mornet method extracts organic P with sodium bicarbonate (0.5M NaHCO3, pH 8.5); that is, P forms bioavailable for soil phosphatase. In brief, samples of air-dried hyphosphere soil were extracted with 0.5M NaHCO3, pH 8.5 (soil:extractant solution 1:20, w/v) at 25 °C for 30min on a reciprocal shaker (180rpm) and then filtered through the quantitative filter paper described above. The soil extract was divided into two parts, one of which was oxidized by acid potassium persulfate in an autoclave for 1h at 121 °C to measure the total P, and the other was used for the determination of inorganic P. The total P and inorganic P concentrations of soil extracts were determined by a modified molybdenum blue colorimetric method (Murphy and Riley, 1962). The extractable organic P was calculated by subtracting the concentration of inorganic P from the concentration of total P.
The microbial biomass phosphorus (MBP) was estimated using the chloroform fumigation–extraction method (Brookes et al., 1982). The weighed fresh hyphosphere soil was fumigated with alcohol-free liquid CHCl3 in desiccators. The samples were then extracted with 0.5M NaHCO3 (pH 8.5) for 30min on a reciprocal shaker (180rpm) at 25 °C. The extracts were filtered through quantitative filter paper. The Pi concentration in the soil extracts was measured colorimetrically by a modified molybdenum blue method (Murphy and Riley, 1962). MBP was calculated by subtracting the amount of inorganic P (Pi) extracted by 0.5M NaHCO3 (pH 8.5) from fresh unfumigated soil from the amount extracted from fumigated soil.
A 5g aliquot of soil in the hyphosphere was used for the estimation of the hyphal length density by a modified membrane filter technique as described previously (Jakobsen et al., 1992). The soil samples were carefully mixed and blended at high speed in a Waring Blender with 50ml of 0.25M sodium oxalate for 30s, and the blended suspension was poured through a 30 μm sieve to retain hyphae. The hyphae were rapidly transferred to a beaker with water and agitated vigorously with a magnetic stirrer at 1000rpm, and then left on the bench for 30s. The 5ml aliquots were pipetted onto 25mm millipore filters (0.45 μm pore size) in a filtration manifold. This procedure was repeated three times. The three filters from each sample were transferred to microscope slides, stained with lactoglycerol–Trypan blue for 5min, and then covered with coverslips. The intersections between blue-stained hyphae and a grid in the eyepiece were counted in 25 fields of view at ×200 magnification.
[13C]DNA stable isotope probing (SIP) analysis
Determination of carbon isotope ratios of plants and soil
–After the final 13C labeling assay, plants were immediately separated into shoots and roots for the labeled and unlabeled treatments. In the case of the hyphosphere soil, labeled and unlabeled samples were collected using the above-mentioned methods. Soil subsamples were frozen in liquid nitrogen immediately and stored at –80 °C until molecular analysis. Other soil subsamples were used for quantification of 13C enrichment that is reported as δ13C‰ (13C:12C ratio). Both soil and plant samples were oven-dried at 70 °C, ground, and sieved using an 80 μm mesh. The carbon isotope ratios of these shoot, root, and soil samples were determined using a DeltaPlusXP mass spectrometer (Thermo Scientific, Bremen, Germany) coupled with an elemental analyzer (FlashEA 1112; CE Instruments, Wigan, UK) in the continuous flow mode at the Stable Isotope Laboratory of the College of Resources and Environmental Sciences, China Agricultural University, Beijing, China. The elemental analyzer combustion temperature was 1020 °C. The carbon isotopic ratios are reported in the delta notation relative to the V-PDB (Vienna-Pee Dee Belemnite) standard using the following equation:
Where δ13C is the carbon isotope ratio of the sample in parts per million (‰), and Rsample and Rstandard are the 13C/12C ratios of the sample and standard, respectively. The SD for the δ13C measurements is <0.15‰.
DNA extraction, T-RFLP, and clone library construction
The DNA of hyphosphere soil in labeled mycorrhizal and unlabeled mycorrhizal treatments with PSB inoculation was extracted using the FastDNA SPIN Kit (MP Biomedicals LLC, Santa Ana, CA, USA) following the manufacturer’s instructions, and DNA fractionation was performed by cesium trifluoroacetate (CsTFA) equilibrium density gradient centrifugation (Lueders et al., 2004a). The centrifuged gradients were fractioned from bottom to top into 16 equal fractions. The buoyant density (BD) of DNA in the gradient fractions was determined using a digital refractometer (Reichert AR2000). The DNA fractions were then purified with isopropyl alcohol and 70% ethanol and stored at –20°C for further analysis.
For the T-RFLP (terminal restriction fragment length polymorphism) analysis, DNA fractions of different BDs were amplified using labeled forward primer 27f-FAM (5-carboxyfluorescein) and 907r (Lueders et al., 2004b) under the following reaction conditions: 3min of initial denaturation at 94 °C; 27 cycles of 30s of denaturation at 94 °C, 45s of annealing at 53 °C, and 1min of elongation at 72 °C; and 10min of final elongation at 72 °C. The FAM-labeled PCR products were purified using a TIANgel Midi Purification Kit (Tiangen, Beijing, China) and digested at 37 °C for 5h with MspI (TaKaRa, Shiga, Japan). The digestion products were purified by ethanol precipitation and were then size-separated using an ABI 3730xl DNA analyzer (Applied Biosystems, Waltham, MA, USA).
Unlabeled primers 27f and 907r were used for cloning analysis. PCR amplification was performed using the same reaction conditions as those described above. The PCR products were purified using a TIANgel Midi Purification Kit (Tiangen) and ligated into the pMD19-T vector (TaKaRa) according to the instructions supplied by the manufacturer. The competent cells (TaKaRa) were transformed with the ligation products and spread onto LB agar plates containing ampicillin (100 μg ml−1) with X-Gal/IPTG (TaKaRa). White colonies were randomly picked in each clone library and screened directly for inserts by performing colony PCR with the vector primers (M13-47 and RV-M). All of the clones were selected from each clone library and sequenced using an ABI 3730xl DNA analyzer (Applied Biosystems). The raw clone sequences were analyzed with ChromasPro software and then compared with sequences in the NCBI database using the Blast function. Clones that exhibited >98% sequence similarity were selected and a phylogenetic tree was constructed using the Neighbor–Joining algorithm with the MEGA6 software. The sequences obtained in this study were deposited in the GenBank database under the following accession numbers (KF830100–KF830127 and KJ909005–KJ909018).
Statistical analysis
Statistical analysis was performed with the Statistical Package for Social Sciences version 16.0 (SPSS Inc., Chicago, IL, USA). AM colonization data were arcsine transformed to normalize the distributions before statistical analysis. All statistical analysis data were checked for homogeneity of variances using Levene’s test (P>0.05). Different inoculation treatment data were analyzed using one-way ANOVA with organic P level as fixed factor for determining whether differences were significant, and mean values were compared with Duncan’s post-hoc multiple range test at P<0.05. Significant differences between zero phytate-P and phytate-P addition treatments for a given inoculation treatment were compared by t-test with 95% confidence intervals.
Results
AM colonization and external hyphal growth
Very little colonization (<0.35%) was observed in the non-mycorrhizal treatments, the plants were well colonized in the treatments inoculated with R. irregularis, and the proportion of root length colonization ranged from 55% to 65% (Table 1).
Table 1.
Mycorrhizal colonization and hyphal density in the hyphal compartment of maize plants inoculated with R. irregularis or uninoculated, and supplied with two organic P levels (0 or 75mg P kg−1) and with or without phosphate-solubilizing bacterial inoculation in the hyphosphere
| Root colonization (%) | Hyphal density (m g−1 soil) | ||||
|---|---|---|---|---|---|
| P0 | P75 | P0 | P75 | ||
| Control | 0.32±0.03 c A | 0.31±0.02 b A | 0 c A | 0 c A | |
| R. irregularis | 55.12±2.45 b B | 64.57±1.78 a A | 12.09±0.13 a B | 14.24±0.27 a A | |
| PSB | 0.33±0.03 c A | 0.29±0.03 b A | 0 c A | 0 c A | |
| R. irregularis+PSB | 60.92±0.63 a A | 64.83±2.27 a A | 9.93±0.31 b B | 11.79±0.21 b A | |
| Significance | |||||
| Mycorrhiza | *** | *** | |||
| PSB | NS | *** | |||
| Organic P level | ** | *** | |||
| Mycorrhiza×PSB | NS | *** | |||
| Mycorrhiza×organic P level | ** | *** | |||
| PSB×organic P level | NS | NS | |||
| Mycorrhiza×PSB×organic P level | NS | NS | |||
Data are presented as the mean (n=3) ±SE.
Values followed by different lower case letters in a column or upper case letters in a row are significantly different at the P<0.05 level. Significance of treatments and interactions was determined by ANOVA. NS indicates no significant difference, *P< 0.05, **P< 0.01, and ***P<0.001.
No AM fungal hyphae were detected in the hyphal compartment of the non-inoculated treatments, implying that the observed mycorrhizal colonization represents the soil background (Table 1). In contrast, the hyphae proliferated more extensively (9.93–14.24 m g−1 soil) into the hyphal compartment (P<0.001) in the presence of mycorrhizae with R. irregularis independently of phytate-P addition. Compared with the treatments inoculated solely with R. irregularis, co-inoculation with PSB and R. irregularis significantly reduced the hyphal length density by ~2.2 m g−1 soil regardless of phytate-P addition. The addition of phytate-P produced ~1.2 times greater hyphal length density than the P0 treatments independently of PSB inoculation.
Plant biomass and P uptake
Compared with the control, the shoot biomass and P uptake increased on average by 80% and 153%, respectively, when the plants were inoculated with R. irregularis. However, PSB inoculation alone did not show this effect regardless of phytate-P addition (Table 2). Compared with inoculation with R. irregularis, dual inoculation with PSB and R. irregularis did not produce significant effects on the shoot dry weight or P content regardless of phytate-P addition.
Table 2.
Shoot dry weight and P content of maize with or without R. irregularis inoculation, and supplied with two organic P levels (0 or 75mg P kg−1) and with or without phosphate-solubilizing bacterial inoculation in the hyphosphere
| Shoot biomass (g per plant) | Shoot P content (mg per plant) | |||||
|---|---|---|---|---|---|---|
| P0 | P75 | P0 | P75 | |||
| Control | 4.67±0.26 b A | 5.09±0.44 b A | 3.47±0.23 b A | 3.77±0.27 b A | ||
| R. irregularis | 8.29±0.53 a A | 9.32±0.12 a A | 7.88±0.64 a B | 10.52±0.27 a A | ||
| PSB | 4.52±0.46 b A | 5.22±0.18 b A | 3.96±0.52 b A | 3.77±0.09 b A | ||
| R. irregularis+PSB | 8.61±0.17 a A | 9.62±0.45 a A | 9.49±0.86 a A | 10.99±1.11 a A | ||
| Significance | ||||||
| Mycorrhiza | *** | *** | ||||
| PSB | NS | NS | ||||
| Organic P level | NS | * | ||||
| Mycorrhiza×PSB | NS | NS | ||||
| Mycorrhiza×organic P level | NS | * | ||||
| PSB×organic P level | NS | NS | ||||
| Mycorrhiza×PSB×organic P level | NS | NS | ||||
Data are presented as the mean (n=3) ±SE. Values followed by different lower case letters in a column or upper case letters in a row are significantly different at the P<0.05 level. Significance of treatments and interactions was determined by ANOVA. NS indicates no significant difference, *P<0.05, **P<0.01, and *** P < 0.001.
The addition of phytate-P significantly increased the P content (from 7.88mg to 10.52mg per plant) of the plants that were inoculated solely with R. irregularis (P<0.05), but there were no significant differences between the treatments with or without phytate-P addition when the plants were inoculated solely with PSB, dually inoculated with PSB and R. irregularis, or control (Table 2).
Phosphatase activity, soluble organic phosphorus, and microbial biomass phosphorus (MBP) in the hyphal compartment
Acid phosphomonoesterase activity was assayed using buffers with a pH in the measured pH range (pH 6.2–6.4). The acid phosphomonoesterase activities in the hyphal compartment of the soil were ~1.3 or 1.5 times higher after inoculation with R. irregularis or dual inoculation with R. irregularis and PSB compared with the control and after inoculation with PSB (Table 3). There was no significant difference between the control and the treatment with PSB inoculation only. The addition of phytate-P enhanced the actual acid phosphomonoesterase activities in the presence of mycorrhiza with R. irregularis, including sole inoculation and dual inoculation.
Table 3.
Soil pH and actual acid phosphomonoesterase phosphatase activity in the hyphosphere of maize with or without R. irregularis inoculation, and supplied with two organic P levels (0 or 75mg P kg−1) and with or without phosphate-solubilizing bacterial inoculation in the hyphosphere
| pH | Actual phosphatase activity (µg p-NPP g−1 DW soil min−1) |
|||
|---|---|---|---|---|
| P0 | P75 | P0 | P75 | |
| Control | 6.39±0.01 a A | 6.40±0.02 a A | 0.45±0.03 b A | 0.47±0.01 b A |
| R. irregularis | 6.20±0.01 c A | 6.20±0.02 b A | 0.64±0.01 a B | 0.70±0.01 a A |
| PSB | 6.37±0.01 a A | 6.39±0.01 a A | 0.49±0.02 b A | 0.48±0.04 b A |
| R. irregularis+PSB | 6.29±0.03 b A | 6.23±0.00 b A | 0.60±0.01 a B | 0.72±0.03 a A |
| Significance | ||||
| Mycorrhiza |
*** | *** | ||
| PSB | * | NS | ||
| Organic P level | NS | * | ||
| Mycorrhiza×PSB | ** | NS | ||
| Mycorrhiza×organic P level | * | * | ||
| PSB×organic P level | NS | NS | ||
| Mycorrhiza×PSB×organic P level | NS | NS | ||
Data are presented as the mean (n=3) ± SE. Values followed by different lower case letters in a column or upper case letters in a row are significantly different at the P < 0.05 level. Significance of treatments and interactions was determined by ANOVA. NS indicates no significant difference, *P<0.05, **P<0.01, and *** P<0.001.
Compared with the control, the NaHCO3-extractable organic P concentrations in the hyphal compartment of the soil decreased by 29% or 42% in the treatments of inoculation with PSB or dual inoculation with PSB and R. irregularis in the presence of phytate-P addition, whereas the organic P concentrations decreased by 32% under dual inoculation with PSB and R. irregularis treatment in the absence of phytate-P (Table 4).
Table 4.
NaHCO3-extracted organic P and microbial biomass phosphorus (MBP) in the hyphosphere of maize with or without R. irregularis inoculation, and supplied with two organic P levels (0 or 75mg P kg−1) and with or without phosphate-solubilizing bacterial inoculation in the hyphosphere
| Organic P concentration (mg kg−1) | MBP (mg kg−1) | ||||
|---|---|---|---|---|---|
| P0 | P75 | P0 | P75 | ||
| Control | 6.36±0.58 a B | 8.65±0.38 a A | 1.90±0.03 c B | 2.45±0.09 c A | |
| R. irregularis | 5.65±0.62 ab B | 7.61±0.55 a A | 2.08±0.02 c B | 2.59±0.02 c A | |
| PSB | 5.45±0.02 ab A | 6.12±0.22 b A | 2.85±0.07 b B | 3.44±0.07 b A | |
| R. irregularis+PSB | 4.32±0.59 b A | 5.05±0.32 b A | 4.45±0.10 a B | 4.87±0.09 a A | |
| Significance | |||||
| Mycorrhiza | ** | *** | |||
| PSB | *** | *** | |||
| Organic P level | *** | *** | |||
| Mycorrhiza×PSB | NS | *** | |||
| Mycorrhiza×organic P level | NS | NS | |||
| PSB×organic P level | * | NS | |||
| Mycorrhiza×PSB×organic P level | NS | NS | |||
Data are presented as the mean (n=3) ±SE. Values followed by different lower case letters in a column or upper case letters in a row are significantly different at the P<0.05 level. Significance of treatments and interactions was determined by ANOVA. NS indicates no significant difference, * P<0.05, ** P<0.01, and *** P<0.001.
The addition of phytate-P enhanced the MBP concentration in the hyphal compartment of the soil (P<0.001). Inoculation with PSB, regardless of whether it was combined with R. irregularis, significantly increased the MBP content per kilogram of soil (P<0.001) compared with the control or that obtained after inoculation with R. irregularis in the presence of both 0 and 75mg kg−1 phytate-P (Table 4).
Assimilation of 13C-labeled carbohydrate by active bacteria associated with extraradical hyphae
In unlabeled mycorrhizal treatments, the isotopic signature (δ13C) of shoots, roots, and hyphosphere soil was consistent with the atmospheric concentration, suggesting that 13C enrichment detected in the hyphosphere soil was not influenced by root exudates. After pulse labeling, the δ13C did differ between mycorrhizal and non-mycorrhizal treatments, independently of shoots, roots, and soil in the hyphal compartment (Supplementary Table S1). Similarly, in 13CO2 labeling experiment 2, the 13C enrichment of roots in the labeled treatments was ~100 times higher than in non-labeled treatments. Accordingly, the δ13C of hyphosphere soil in the absence of R. irregularis was consistent with a natural background of 13C, while R. irregularis inoculation led to an increase from –22.1‰ to +176.6‰ (Table 5). The isotopic signature (δ13C) in the two labeling experiments demonstrates that the shoots, roots, and soil of the hyphal compartment were enriched in 13C immediately after pulse labeling. Carbon translocation was especially rapid to the roots and then was transferred to the hyphosphere soil via the extraradical mycelia of R. irregularis, suggesting that microbes exploited a 13C-rich pool of recent photosynthates.
Table 5.
Carbon isotope ratios of plant roots and hyphosphere soil after 13CO2 application to maize inoculated with R. irregularis and phosphate-solubilizing bacteria in the hyphal compartment in labeling experiment 2
| Labeling status | Treatment | δ13C ‰ | |
|---|---|---|---|
| Root | Hyphosphere soil | ||
| Labeled samples | M+PSB | 2111.6±235.4 | 176.6±58.0 |
| Unlabeled samples | M+PSB | –21.4±0.4 | –22.1±0.3 |
Data are presented as the mean (n=2) ± SE.
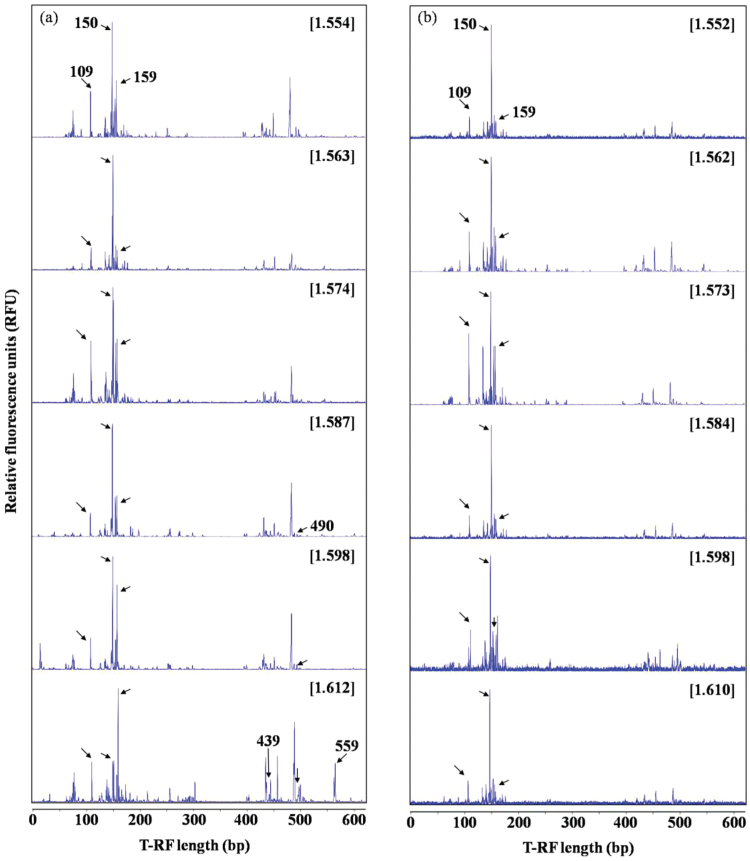
Maize plants were grown for 5 d with repeated pulses of 13CO2 (99 atom% 13C). DNA isolated from both the 13C-labeled and the non-labeled hyphosphere soil was subjected to density fractionation followed by PCR and T-RFLP fingerprinting of bacterial 16S rDNA. These DNA samples were analyzed across a BD gradient from 1.552g ml−1 to 1.612g ml−1. T-RFLP fingerprints showed that the bacterial community of the non-labeled soil did not change within any of the gradient fractions (Fig. 3b). In the case of the labeled soil, the T-RFLP patterns in ‘light’ DNA fractions (BD ≤1.574g ml−1) were similar to those of the unlabeled control soil (Fig. 3a). However, in the ‘heavy’ DNA fractions (BD ≥1.587g ml−1), a significant change in the bacterial community was found among the DNA fractions of different BDs. Specifically, the 109bp and 159bp T-RFs were markedly increased in the fractions of BD between 1.587g ml−1 and 1.612g ml−1. The T-RF with a length of 490bp appeared at a BD of 1.587g ml−1 and gradually increased with increasing BD. However, the 150bp T-RF showed a tendency to decrease with an increase in BD and the 439bp and 559bp T-RFs appeared only at a BD of 1.612g ml−1. The comparison of the T-RFLP fingerprints between labeled soil and non-labeled soil indicates that the bacterial populations with the characteristics of 109bp, 159bp, and 490bp T-RFs became labeled with 13C derived from maize photosynthates.
Fig. 3.
T-RFLP fingerprints of the bacterial 16S rRNA genes retrieved from density-resolved gradient fractions of 13C-labeled hyphosphere soil (a) and non-labeled control soil (b) in labeling experiment 2. The CsTFA buoyant densities (g ml−1) of the DNA fractions are shown in brackets. The number of base pairs and changes in T-RFs with DNA buoyant density are indicated by arrows.
The ‘heavy’ (BD 1.587g ml−1) and ‘light’ (BD 1.563g ml−1) DNA fractions were selected to construct clone libraries for bacterial 16S rRNA genes (Table 6). The dominant sequences, which accounted for 45%, were affiliated to Proteobacteria in the ‘heavy’ clone library and the remaining 55% belonged to Actinobacteria, Firmicutes, and uncultured bacteria. In the case of the ‘light’ clone library, Proteobacteria and Cyanobacteria each accounted for 21%, and the other 58% belonged to Actinobacteria, Firmicutes, and uncultured bacteria. Compared with the ‘light’ clone library, the frequency and diversity of the sequences affiliated with the Proteobacteria and Actinobacteria increased in the ‘heavy’ clone library. In contrast, no Cyanobacteria sequences were detected in the ‘heavy’ clone library but they were more abundant in the ‘light’ clone library.
Table 6.
Phylogenetic affiliations and clone number of bacterial 16S rRNA genes retrieved from ‘heavy’ (BD 1.587g ml−1) and ‘light’ (BD 1.563g ml−1) DNA fractions of maize hyphosphere soil in labeling experiment 2
| Phylogenetic group | Heavy | Light | T-RF (bp) |
|---|---|---|---|
| Alphaproteobacteria | |||
| Bradyrhizobiaceae | 4 | 150, 441 | |
| Methylobacteriaceae | 1 | 150 | |
| Uncultured | 10 | 3 | 150, 437 |
| Betaproteobacteria | 150 | ||
| Oxalobacteraceae | 2 | 109, 488 | |
| Uncultured | 7 | 4 | 452, 488, 492 |
| Gammaproteobacteria | |||
| Pseudomonadaceae | 1 | 490a | |
| Uncultured | 3 | 2 | 82, 141, 186 |
| Actinobacteria | |||
| Streptomycetaceae | 5 | 1 | 159 |
| Nocardioidaceae | 1 | 1 | 142, 161 |
| Micrococcaceae | 1 | 67 | |
| Uncultured | 3 | 1 | 161 |
| Firmicutes | 3 | 2 | 145, 153 |
| Cyanobacteria | 9 | 504 | |
| Uncultured | 21 | 20 | 82, 95, 113, 141, 153, 183, 287, 603 |
T-RFs with a relative abundance of >5% in total (105 clones) are indicated in bold.
a T-RF represents phosphate-solubilizing bacterium Pseudomonas alcaligenes added to the hyphal compartment in this study.
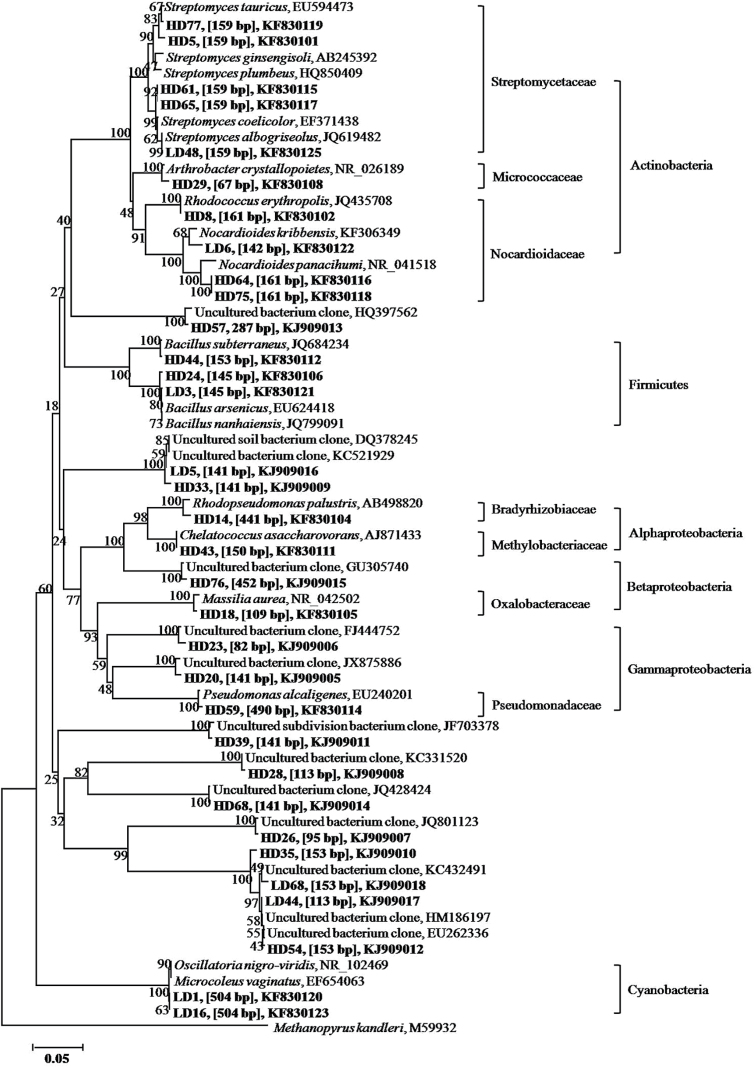
In silico analysis of the clone sequences showed that the Alphaproteobacteria group was mainly characterized by 150bp T-RF (Table 6; Fig. 4). One clone of Oxalobacteraceae in the Bateproteobacteria group and one clone of Pseudomonadaceae in the Gammaproteobacteria group showed 100% similarity to Massilia aurea and P. alcaligenes, and those characterized by T-RFs of 109bp and 490bp were detected only in the ‘heavy’ clone library with a minor abundance (Table 6; Fig. 4). Organisms related to Streptomycetaceae were characterized by a 159bp T-RF and showed higher abundance in the ‘heavy’ clone library. A comparison between T-RFLP fingerprints and the clone library indicates that an increase in the 159bp T-RF in the ‘heavy’ DNA fractions in the 13C-labeled hyphosphere soil (Fig. 3a) was consistent with the higher abundance of Streptomycetaceae (Table 6).
Fig. 4.
Phylogenetic relationship of bacterial 16S rRNA gene clone sequences derived from ‘heavy’ and ‘light’ DNA fractions of maize hyphosphere soil in labeling experiment 2. Sequences of the HD library represent the ‘heavy’ DNA fraction; those of the LD library represent the ‘light’ DNA fraction. The sequences obtained in this study are shown in bold, the in silico T-RF sizes of clone sequences digested with MspI are given in brackets, and the GenBank accession numbers of sequences are indicated. The scale bar represents 5% sequence divergence.
Discussion
Mineralization and turnover of phytate-P in the hyphosphere
Soil phosphatases are generally classified into acid and alkaline phosphatases based on their optimum pH (Vincent et al., 1992) and into phosphomonoesterases or phosphodiesterases based on the combined bond of the enzyme (Nannipieri et al., 2011). Phytase is one of the groups of phosphomonoesterases that are substrate specific for phytate mineralization and is generally classified into four subgroups (Lim et al., 2007). The overlapping of different groups is obtained after measuring phosphatase using a substrate that contains phosphomonoester bonds in the pH range. For example, purple acid phosphatases are a phosphomonoesterase and a phytase that has maximum activity at acidic pH (Tran et al., 2010). Therefore, we measured the mean activity of phosphomonoesterases that may be involved in the mineralization of phytate in the hyphosphere soil.
Phytate is the dominant form of organic P in soils (Turner et al., 2002). In order to distinguish the added phytate-P from the original soil P pool, we established control treatments that were not inoculated with AM fungi or did not receive phytate-P. We found that the AM fungi led to a positive response in plant biomass even when no additional phytate-P was added (Table 2), suggesting that the extraradical mycelia of R. irregularis absorbed P from the original soil P pool and delivered it to maize.
It is found that soil organic P will be overestimated when inorganic polyphosphates are present because polyphosphates do not react with the molybdate reagent during the measurement of inorganic P by molybdate blue colorimetry (Ron Vaz et al., 1993), but they may be hydrolyzed to inorganic P in the processes of oxidization (McKelvie, 2005) and therefore are included in the organic fraction (Haygarth et al., 1998). However, this drawback of the soil organic P measurement method will not influence the results of our present study. First, inorganic polyphosphates in soil occur in bacterial and fungal cells; their concentration is low compared with the soil organic P concentration (Turner et al., 2014) and thus they cannot be included in soil MBP. The highest MBP content in the dual inoculation treatment (Table 4) indicated that the treatment had a higher microbial biomass than the other three treatments. Secondly, the highest microbial biomass should potentially contain more inorganic polyphosphates if inorganic polyphosphates were to exist in the soil sample. We assume that if the soil organic P was to be mineralized with the same mineralization rate in all treatments, the detected organic P concentration should be the highest in the dual inoculation treatment. However, our results were completely opposite (Table 4), suggesting that organic P concentrations in this study were not overestimated, and the mineralization of organic P happened at different rates in different treatments.
Our previous study indicated that AM fungal hyphae release protons as they absorb ammonium and decrease the soil pH in the hyphosphere, which promotes the solubilization of phytate-P and enhances substrate availability of phytate-P to phosphatase (Wang et al., 2013). Therefore, we supplied (NH4)2SO4 as the N source to promote the acidification in hyphosphere soil. Additionally, a nitrification inhibitor named 3,4-dimethylpyrazole phosphate (DMPP) was also added to maintain a higher NH4 + concentration in the soil. The soil pH was significantly lower in the treatments inoculated with R. irregularis compared with the treatments without R. irregularis inoculation, and the lower soil pH supported the higher acid phosphomonoesterase activity (Table 3). Because the AM fungus and all of the PSB strains inoculated into the hyphal compartment are able to release phosphomonoesterase and mineralize phytate, we hypothesized that dual inoculation with both R. irregularis and PSB can enhance phytate mineralization and increase plant P uptake from phytate by enhancing phosphomonoesterase activities compared with that obtained after inoculation solely with R. irregularis or sole treatment with PSB. The results partly support our hypothesis. We found that the dual inoculation and sole R. irregularis treatments resulted in higher phosphomonoesterase activities compared with the treatments without R. irregularis inoculation (Table 3). The enhanced phosphomonoesterase activities reduced the soil organic P concentrations in the dual inoculation treatments compared with the inoculation with R. irregularis (Table 4). However, we did not find any enhancement of P uptake or shoot biomass in the dual inoculation treatment compared with the sole R. irregularis treatment (Table 2), but we found that the MBP contents increased in the dual inoculation treatment compared with the sole PSB or sole R. irregularis treatments (Table 4) and that the growth of the fungus (hyphal length density) was reduced (Table 1), suggesting that phytate-P mineralized by PSB was not delivered to the plants via the hyphae of R. irregularis but was incorporated into the MBP pool. This result is consistent with previous findings that the fungal-associated PSB are more competitive than the AM fungus in acquiring available P and the fungus loses out in the competition that inhibits fungal growth (Zhang et al., 2014).
In mycorrhizal research, the filtrates of inocula that may contain bacterial species and nutrient elements are added to the control treatments to maintain the same microbial flora with the exception of the AM fungus. Phytate-P supply enhanced the organic P concentration only in the control treatment and enhanced the MBP content in all four inoculated treatments. However, the organic P concentration in the control treatment was the highest but the MBP content was the lowest (Table 4). These results suggest that although the AM fungal inocula introduced some bacteria that may have the ability to mineralize phytate-P, their number and effects are markedly lower than the inoculated PSB. In addition, the findings support the aforementioned interpretation that the fungus is involved in the mineralization of phytate-P and then delivers it to the plants in the absence of a PSB competitor, whereas the mineralized phytate-P was preferentially incorporated into the MBP pool under conditions in which a PSB competitor is enriched based on their own advantage.
Carbon flow in the plant–AM fungi–PSB continuum
AM fungi are obligate biotrophs whose growth relies on carbon from a living host (Smith and Read, 2008). Mycorrhizal fungi have been estimated to receive up to 20% of the plant photosynthetic products, and are maintained only for 5–6 d followed by starting turnover (Staddon et al., 2003). Such a high rate of C turnover is significant even within the context of the global C balance (Szuromi, 2003). However, the pathways in which the photosynthates are consumed at such a rapid rate remain uncertain, although some studies have attributed this consumption to fungal respiration (Johnson et al., 2002, 2005). Recent studies showed that 13CO2-labeled photosynthates were rapidly transferred to AM fungi, followed by gradual release of the C to their associated microbial groups in the mycorrhizosphere and hyphosphere (Drigo et al., 2010; Kaiser et al., 2015). However, no study has investigated whether any functional bacterial group assimilates photosynthates derived from AM fungal structures and plays a significant role in soil nutrient turnover under soil conditions. In the present study, the C isotope signature of both plant materials (containing shoots and roots) and hyphosphere soil was detected after 13CO2 was fed to maize plants in the two 13CO2 labeling experiments (Supplementary Table S1; Table 5), suggesting that photosynthates flow from plant roots to hyphosphere soil via extraradical mycelia of R. irregularis. Although the results of bacterial T-RFLP fingerprinting and the clone library were not obtained in labeling experiment 1 due to technical problems, we obtained these data from labeling experiment 2. [13C]DNA-SIP analysis reveals that active bacterial groups in the hyphosphere soil incorporated C into their DNA that was sufficiently enriched in 13C to allow separation by density gradient centrifugation (Fig. 3). In this study the active bacteria in the hyphosphere were those closely related to Oxalobacteraceae within the Betaproteobacteria, Streptomycetaceae within the Actinobacteria, and Pseudomonadaceae within the Gammaproteobacteria (Table 6). The 109, 159, and 490bp T-RF characteristics for these organisms increased markedly in the ‘heavy’ fractions, but no change occurred in the ‘light’ fractions of the labeled soil (Fig. 3a) or in any of the fractions of the non-labeled soil (Fig. 3b). It would seem that the three groups of bacteria were highly active in assimilating the 13C-labeled substrates derived from extraradical mycelia of R. irregularis. In the Oxalobacteraceae group, the closest relative to our DNA sequence data is Massilia aurea (Fig. 4), which is a strictly aerobic bacterium and known to colonize the roots of many plant species in the rhizosphere (Ofek et al., 2012). Streptomyces spp., according to DNA sequence data, were apparently found to be the abundant population in the Streptomycetaceae group, and these organisms are the original source of many antibiotics. In the Pseudomonadaceae group, the DNA sequence data are exactly the same as for P. alcaligenes (Fig. 4), the bacterial strain that was inoculated into the hyphal compartment at the start of our experiment and was able to mineralize organic phosphate (Lv et al., 2005).
Although the soil was pre-sterilized, exotic bacteria in the air would have unavoidably contaminated the microcosms because the experiment was conducted in the open. Some environmental factors such as AM fungal inoculum and watering regime may cause bacterial contamination. Therefore, diverse T-RFLP fingerprints were detected in the soil in the hyphal compartment based on the 16S rRNA gene analysis (Figs 3, 4; Table 6).
In our current study, the fungal hyphae-associated bacteria can use two possible pathways to acquire the 13C-labeled photosynthates of maize plants via the AM fungal hyphae during the 5 d of labeling. One is from the hyphal exudates released by living hyphae, and the other is through the decay of dead hyphae. Although the processes of C flow from plants to the hyphosphere can be rapid (Johnson et al., 2002, 2005; Kaiser et al., 2015), particularly given that the half-life of some of the terminal hyphae is just a few days (Staddon et al., 2003), the fungal hyphae-associated bacteria are unlikely to be able to assimilate the dead debris of the fungus for the following reasons. First, the 13C-SIP method normally uses highly enriched 13CO2 (99%) and pulse labeling for several days to make the 13C-labeled DNA in rhizosphere bacteria detectable (Lu and Conrad, 2005). Secondly, the active cytoplasm is usually retracted to the backbone hyphae by forming septa upon death of the terminal hyphal branches (Bago et al., 1998; Logi et al., 1998), and there is not much 13C-labeled food left for the degraders (Jansa et al., 2013). Thirdly, the cell walls of AM fungal hyphae cannot to be degraded rapidly because they are composed of up to 40% of chitin, a carbohydrate that is recalcitrant to decomposition (Zhu and Miller, 2003). Moreover, a weighted mean residence time of chitin-derived products is 49±19 years (Gleixner et al., 2002). Our experiments involved harvesting 5 d after the beginning of 13C labeling. In such a short time, the fungal hyphae-associated PSB strains, particularly P. alcaligenes, are unlikely to be enriched by 13C by degrading the fungal cell wall, indicating that the PSB strain most probably assimilated 13C derived from the hyphal exudates. To the best of our knowledge, this study provides the first evidence that a PSB strain colonizing the hyphosphere assimilates photosynthates of the host plant from hyphal exudates of an AM fungus. In addition, the hyphal exudates prime the activity of the PSB, which further accelerates phytate-P turnover in the hyphosphere.
In summary, our findings indicate for the first time that a PSB strain in the hyphosphere, P. alcaligenes (Pseudomonadaceae), assimilated the photosynthates fixed by the plant and transferred them to R. irregularis, and they were subsequently exuded into the soil and taken up by the PSB. In combination with the AM fungus, the PSB promoted the processes of mineralization and turnover of phytate-P in hyphosphere soil. Thus, while the AM fungus and maize plants had a symbiotic relationship (i.e. they both benefited from the presence of each other), the PSB benefited from the plant and AM fungus but did not provide any reciprocal benefit to either in this experiment. Further studies are needed to link the interaction between AM fungi and the fungal-associated functional microbes to nutrient transformation and turnover in hyphosphere soil.
Supplementary data
Supplementary data are available at JXB online.
Figure S1 An obvious halo zone of phosphate solubilization on National Botanical Research Institute’s phosphate (NBRIP) agar plates produced by Pseudomonas alcaligenes M20 (a), Bacillus megaterium C4 (b), and Rahnella aquatilis HX2 (c) after 4 d growth at 30 °C.
Figure S2 Phosphate-solubilizing bacteria colonizing the surface of extraradical hyphae of Rhizophagus irregularis. (a) Pseudomonas alcaligenes M20, (b) Bacillus megaterium C4, and (c) Rahnella aquatilis HX2.
Table S1 Carbon isotope ratios of shoots, roots, and hyphosphere soils after 13CO2 application to maize inoculated with R. irregularis or uninoculated, and supplied with phosphate-solubilizing bacteria in the hyphal compartment in labeling experiment 1.
Acknowledgements
We thank Professor Peter Christie for help in revising the manuscript. This study was supported by the National Natural Science Foundation of China (31372139, U1403285), the PhD Programs Foundation of the Ministry of Education of China (20120008130001) and the Innovative Group Grant of the National Natural Science Foundation of China (31421092). We further thank State Key Laboratory of Desert and Oasis Ecology, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences for financially support in samples measurements. We would like to thank Professor Yahai Lu for technical assistance with SIP and valuable suggestions on the study.
References
- Artursson V, Finlay RD, Jansson JK. 2006. Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environmental Microbiology 8, 1–10. [DOI] [PubMed] [Google Scholar]
- Bago B, Azcon-Aguilar C, Goulet A, Piche Y. 1998. Branched absorbing structures (BAS): a feature of the extraradical mycelium of symbiotic arbuscular mycorrhizal fungi. New Phytologist 139, 375–388. [Google Scholar]
- Becquer A, Trap J, Irshad U, Ali MA, Claude P. 2014. From soil to plant, the journey of P through trophic relationships and ectomycorrhizal association. Frontiers in Plant Science 5, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes PC, Powlson DS, Jenkinson DS. 1982. Measurement of microbial biomass phosphorus in soil. Soil Biology and Biochemistry 14, 319–329. [Google Scholar]
- Collos Y, Mornet E. 1993. Automated procedure for determination of dissolved organic nitrogen and phosphorus in aquatic environments. Marine Biology 116, 685–688. [Google Scholar]
- Cordell D, White S. 2014. Life’s bottleneck: sustaining the world’s phosphorus for a food secure future. Annual Review of Environment and Resources 39, 161–188. [Google Scholar]
- Dalal RC. 1977. Soil organic phosphorus. Advances in Agronomy 29, 83–113. [Google Scholar]
- Drigo B, Pijl AS, Duyts H, et al. 2010. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2 . Proceedings of the National Academy of Sciences, USA 107, 10938–10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion M, ST-Arnaud M, Fortin JA. 1999. Direct interaction between the arbuscular mycorrhizal fungus Glomus intraradices and different rhizosphere microorganisms. New Phytologist 141, 525–533. [Google Scholar]
- Fitter AH, Heinemeyer A, Husband R, Olsen E, Ridgway KP, Staddon PL. 2004. Global environmental change and the biology of arbuscular mycorrhizas: gaps and challenges. Canadian Journal of Botany 82, 1133–1139. [Google Scholar]
- Fontaine S, Barot S, Barré P, Bdioui N, Mary B, Rumpel C. 2007. Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 450, 277–280. [DOI] [PubMed] [Google Scholar]
- Frey-Klett P, Garbaye J, Tarkka M. 2007. The mycorrhiza helper bacteria revisited. New Phytologist 176, 22–36. [DOI] [PubMed] [Google Scholar]
- Gleixner G, Poirier N, Bol R, Balesdent J. 2002. Molecular dynamics of organic matter in a cultivated soil. Organic Geochemistry 33, 357–366. [Google Scholar]
- Godbold DL, Hoosbeek MR, Lukac M, et al. 2006. Mycorrhizal hyphal turnover as a dominant process for carbon input into soil organic matter. Plant and Soil 281, 15–24. [Google Scholar]
- Hanson WC. 1950. The photometric determination of phosphorus in fertilizers using the phosphovanado-molybdate complex. Journal of the Science of Food and Agriculture 1, 172–173. [Google Scholar]
- Haygarth PM, Hepworth L, Jarvis SC. 1998. Forms of phosphorus transfer in hydrological pathways from soil under grazed grassland. European Journal of Soil Science 49, 65–72. [Google Scholar]
- Herman DJ, Firestone MK, Nuccio E, Hodge A. 2012. Interactions between an arbuscular mycorrhizal fungus and a soil microbial community mediating litter decomposition. FEMS Microbiology Ecology 80, 236–247. [DOI] [PubMed] [Google Scholar]
- Hodge A, Campbell CD, Fitter AH. 2001. An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature 413, 297–299. [DOI] [PubMed] [Google Scholar]
- Hodge A, Fitter AH. 2010. Substantial nitrogen acquisition by arbuscular mycorrhizal fungi from organic material has implications for N cycling. Proceedings of the National Academy of Sciences, USA 107, 13754–13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen I, Abbott LK, Robson AD. 1992. External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L. 1. Spread of hyphae and phosphorus inflow into roots. New Phytologist 120, 371–380. [Google Scholar]
- Jansa J, Bukovská P, Gryndler M. 2013. Mycorrhizal hyphae as ecological niche for highly specialized hypersymbionts—or just soil free-riders? Frontiers in Plant Science 4, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Krsek M, Wellington EMH, Stott AW, Cole L, Bardgett RD, Read DJ, Leake JR. 2005. Soil invertebrates disrupt carbon flow through fungal networks. Science 309, 1047. [DOI] [PubMed] [Google Scholar]
- Johnson D, Leake JR, Ostle N, Ineson P, Read DJ. 2002. In situ 13CO2 pulse-labelling of upland grassland demonstrates a rapid pathway of carbon flux from arbuscular mycorrhizal mycelia to the soil. New Phytologist 153, 327–334. [Google Scholar]
- Joner EJ, Johansen A. 2000. Phosphatase activity of external hyphae of two arbuscular mycorrhizal fungi. Mycological Research 104, 81–86. [Google Scholar]
- Kaiser C, Kilburn MR, Clode PL, Fuchslueger L, Koranda M, Cliff JB, Solaiman ZM, Murphy DV. 2015. Exploring the transfer of recent plant photosynthates to soil microbes: mycorrhizal pathway vs direct root exudation. New Phytologist 205, 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyakov Y. 2011. Ecology: prime time for microbes. Nature Climate Change 1, 295–297. [Google Scholar]
- Leigh J, Fitter AH, Hodge A. 2011. Growth and symbiotic effectiveness of an arbuscular mycorrhizal fungus in organic matter in competition with soil bacteria. FEMS Microbiology Ecology 76, 428–438. [DOI] [PubMed] [Google Scholar]
- Leigh J, Hodge A, Fitter AH. 2009. Arbuscular mycorrhizal fungi can transfer substantial amounts of nitrogen to their host plant from organic material. New Phytologist 181, 199–207. [DOI] [PubMed] [Google Scholar]
- Lim BL, Yeung P, Cheng C, Hill JE. 2007. Distribution and diversity of phytate-mineralizing bacteria. ISME Journal 1, 321–330. [DOI] [PubMed] [Google Scholar]
- Logi C, Sbrana C, Giovannetti M. 1998. Cellular events involved in survival of individual arbuscular mycorrhizal symbionts growing in the absence of the host. Applied and Environmental Microbiology 64, 3473–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Conrad R. 2005. In situ stable isotope probing of methanogenic archaea in the rice rhizosphere. Science 309, 1088–1090. [DOI] [PubMed] [Google Scholar]
- Lueders T, Manefield M, Friedrich MW. 2004a Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environmental Microbiology 6, 73–78. [DOI] [PubMed] [Google Scholar]
- Lueders T, Wagner B, Claus P, Friedrich MW. 2004b Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environmental Microbiology 6, 60–72. [DOI] [PubMed] [Google Scholar]
- Lv J, Li F, Chen S, Li J. 2005. The secretion of lecithinase was via type II secretion pathway in Pseudomonas alcaligenes S2. Chinese Science Bulletin 50, 1725–1730. [Google Scholar]
- Mansfeld-Giese K, Larsen J, Bodker L. 2002. Bacterial populations associated with mycelium of the arbuscular mycorrhizal fungus Glomus intraradices . FEMS Microbiology Ecology 41, 133–140. [DOI] [PubMed] [Google Scholar]
- McKelvie ID. 2005. Separation, preconcentration and speciation of organic phosphorus in environmental samples. In: Turner BL, Frossard E, Baldwin DS, eds. Organic phosphorus in the environment . Wallingford, UK: CABI. [Google Scholar]
- Murphy J, Riley JP. 1962. A modified single solution method for the determination of phosphate in natural waters. Analytica Chimica Acta 27, 31–36. [Google Scholar]
- Nannipieri P, Giagnoni L, Landi L, Renella G. 2011. Role of phosphatase enzymes in soil. In: Bunemann EK, Oberson A, Frossard E, eds. Phosphorus in action: biological processes in soil phosphorus cycling . New York: Springer, 215–243. [Google Scholar]
- Ofek M, Hadar Y, Minz D. 2012. Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS One 7, e40117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AE, Simpson RJ. 2011. Soil microorganisms mediating phosphorus availability. Plant Physiology 156, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayer EJ, Heard MS, Grant HK, Marthews TR, Tanner EVJ. 2011. Soil carbon release enhanced by increased tropical forest litterfall. Nature Climate Change 1, 304–307. [Google Scholar]
- Scheublin TR, Sanders IR, Keel C, van der Meer JR. 2010. Characterisation of microbial communities colonising the hyphal surfaces of arbuscular mycorrhizal fungi. ISME Journal 4, 752–763. [DOI] [PubMed] [Google Scholar]
- Smith SE, Jakobsen I, Grønlund M, Smith FA. 2011. Roles of arbuscular mycorrhizas in plant phosphorus nutrition: interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiology 156, 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read D. 2008. Mycorrhizal symbiosis , 3rd edn. New York: Academic Press. [Google Scholar]
- Staddon PL, Ramsey CB, Ostle N, Ineson P, Fitter AH. 2003. Rapid turnover of hyphae of mycorrhizal fungi determined by AMS microanalysis of 14C. Science 300, 1138–1140. [DOI] [PubMed] [Google Scholar]
- Stevenson FJ, Cole MA. 1999. Cycles of soils: carbon, nitrogen, phosphorus, sulfur, micronutrients . Canada: John Wiley & Sons, Inc. [Google Scholar]
- Sun Z. 2012. Colonization of Rahnella aquatilis HX2 on maize roots . China Agricultural University. [Google Scholar]
- Szuromi P. 2003. This week in Science . Science 300, 1049. [Google Scholar]
- Toljander JF, Lindahl BD, Paul LR, Elfstrand M, Finlay RD. 2007. Influence of arbuscular mycorrhizal mycelial exudates on soil bacterial growth and community structure. FEMS Microbiology Ecology 61, 295–304. [DOI] [PubMed] [Google Scholar]
- Tran HT, Hurley BA, Plaxton WC. 2010. Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Science 179, 14–27. [Google Scholar]
- Trouvelot A, Kough JL, Gianinazzi-Pearson V. 1986. Mesure du taux de mycorhization VA d’un système radiculaire. Recherche de méthodes d’estimation ayant une signification fonctionnelle. In: Gianinazzi-Pearson V, Gianinazzi S, eds. Physiological and genetical aspects of mycorrhizae . Paris: INRA Press, 217–221. [Google Scholar]
- Turner BL, Paphazy MJ, Haygarth PM, McKelvie ID. 2002. Inositol phosphates in the environment. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences 357, 449–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BL, Wells A, Condron LM. 2014. Soil organic phosphorus transformations along a coastal dune chronosequence under New Zealand temperate rain forest. Biogeochemistry 121, 595–611. [Google Scholar]
- Ron Vaz MD, Edwards AC, Shand CA, Cresser MS. 1993. Phosphorus fractions in soil solution: influence of soil acidity and fertiliser additions. Plant and Soil 148, 175–183. [Google Scholar]
- Vincent JB, Crowder MW, Averill BA. 1992. Hydrolysis of phosphate monoesters: a biological problem with multiple chemical solutions. Trends in Biochemical Sciences 17, 105–110. [DOI] [PubMed] [Google Scholar]
- Wang F, Jiang R, Kertesz MA, Zhang F, Feng G. 2013. Arbuscular mycorrhizal fungal hyphae mediating acidification can promote phytate mineralization in the hyphosphere of maize (Zea mays L.). Soil Biology and Biochemistry 65, 69–74. [Google Scholar]
- Xavier LJC, Germida JJ. 2003. Bacteria associated with Glomus clarum spores influence mycorrhizal activity. Soil Biology and Biochemistry 35, 471–478. [Google Scholar]
- Zhang L, Ding XD, Wang F, Tian ZY, Feng G. 2012. The effects of inoculation with phosphate solubilizing bacteria Bacillus megaterium C4 in the AM fungal hyphosphere on soil organic phosphorus mineralization and plant uptake. Acta Ecologica Sinica 32, 4079–4086. [Google Scholar]
- Zhang L, Fan J, Ding X, He X, Zhang F, Feng G. 2014. Hyphosphere interactions between an arbuscular mycorrhizal fungus and a phosphate solubilizing bacterium promote phytate mineralization in soil. Soil Biology and Biochemistry 74, 177–183. [Google Scholar]
- Zhu Y, Miller RM. 2003. Carbon cycling by arbuscular mycorrhizal fungi in soil–plant systems. Trends in Plant Science 8, 407–409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.