Highlight
LPA1 suppresses auxin signalling that interacts with C-22-hydroxylated and 6-deoxo brassinosteroids, which regulates lamina inclination independently of OsBRI1.
Key words: Auxin, C-22-hydroxylated BRs, lamina inclination, lamina joints, Loose Plant Architecture1 (LPA1), OsPINs, plant architecture, rice (Oryza sativa), 6-deoxos BRs.
Abstract
Lamina inclination is a key agronomical character that determines plant architecture and is sensitive to auxin and brassinosteroids (BRs). Loose Plant Architecture1 (LPA1) in rice (Oryza sativa) and its Arabidopsis homologues (SGR5/AtIDD15) have been reported to control plant architecture and auxin homeostasis. This study explores the role of LPA1 in determining lamina inclination in rice. LPA1 acts as a positive regulator to suppress lamina bending. Genetic and biochemical data indicate that LPA1 suppresses the auxin signalling that interacts with C-22-hydroxylated and 6-deoxo BRs, which regulates lamina inclination independently of OsBRI1. Mutant lpa1 plants are hypersensitive to indole-3-acetic acid (IAA) during the lamina inclination response, which is suppressed by the brassinazole (Brz) inhibitor of C-22 hydroxylase involved in BR synthesis. A strong synergic effect is detected between lpa1 and d2 (the defective mutant for catalysis of C-23-hydroxylated BRs) during IAA-mediated lamina inclination. No significant interaction between LPA1 and OsBRI1 was identified. The lpa1 mutant is sensitive to C-22-hydroxylated and 6-deoxo BRs in the d61-1 (rice BRI1 mutant) background. We present evidence verifying that two independent pathways function via either BRs or BRI1 to determine IAA-mediated lamina inclination in rice. RNA sequencing analysis and qRT-PCR indicate that LPA1 influences the expression of three OsPIN genes (OsPIN1a, OsPIN1c and OsPIN3a), which suggests that auxin flux might be an important factor in LPA1-mediated lamina inclination in rice.
Introduction
Rice (Oryza sativa) is a globally important cereal crop that provides carbohydrates for more than half of the world’s population. Planting density is a crucial factor for enhancing crop yields in a given area and the aerial plant structure or plant architecture affects crop yield. Rice leaf blades naturally bend downwards as plants become mature. Erect plant architecture is considered to be the ideal plant type and is continuously selected by farmers and breeders (Kovach et al., 2007).
Many genes have been reported to regulate leaf angle in rice and other plants. Rice Leaf Inclination2 (OsLC2) is involved in leaf bending by influencing cell division and hormone-responsive genes (Zhao et al., 2010). Rice Leaf and Tiller Angle Increased Controller (OsLIC) is an antagonistic factor of BRASSINAZOLE-RESISTANT1 (BZR1) and has an epistatic relationship with BR synthesis and BR signalling mutants (Wang et al., 2008; Zhang et al., 2012). Rice Increased Leaf Inclination1 (OsILI1) and its binding protein ILI1 Binding bHLH Protein1 (OsIBH1) function downstream of BZR1 in BR-mediated regulation of cell elongation (Zhang et al., 2009). Increased Leaf Angle1 (ILA1) affects rice leaf angle by regulating mechanical tissue formation at the leaf lamina joint (Ning et al., 2011). Wu et al. (2013) reported that LPA1 encodes an INDETERMINATE DOMAIN protein regulating tiller and leaf angle. The Arabidopsis OsLPA1 homologues (AtIDD14, AtIDD15 and AtIDD16) control branch angles by regulating the expression of auxin biosynthetic and transport genes (Cui et al., 2013).
Early studies employing excised lamina joints revealed the effects of auxin and BRs on lamina inclination (Maeda, 1965). Exposure of lamina joints to exogenous auxin or BRs induces strong lamina inclination (Wada et al., 1981). Such hormonal effects were later verified by genetic studies of mutants with altered lamina angles. ILI1, Brassinosterioid Upregulated1 (BU1) and OsLIC encode transcription factors that determine lamina bending via BR synthesis or signalling pathways (Wang et al., 2008; Zhang et al., 2009). In fact, BR synthesis and signalling mutants exhibit erect phenotypes (Sakamoto et al., 2006). Examination of rice Leaf Inclination1 (LC1), encoding an IAA-amido synthetase (OsGH3-1), revealed the effects of auxin on leaf inclination (Zhao et al., 2012), and the synergistic effects of BRs and auxin have been demonstrated (Takeno and Pharis, 1982; Mandava, 1988). A recent report proposed that there are two BR-mediated pathways that interact with auxin to regulate lamina joint bending: the C-22-hydroxylated and 6-deoxo BR-mediated pathway and an OsBRI1-mediated pathway (Nakamura et al., 2009).
Little information is available about the regulatory mechanisms that determine hormone-responsive lamina bending. In this study, we investigated the role of LPA1 in IAA-mediated lamina bending. We demonstrate that a main function of LPA1 in lamina inclination is to suppress the IAA signalling pathway that interacts with C-22-hydroxylated BRs, which is independent of OsBRI1. We provide genetic evidence verifying that two independent pathways, involving either BR compounds or the OsBRI1 receptor, function in IAA-mediated lamina inclination. In addition, RNA-seq analysis and qRT-PCR revealed that the expression levels of three OsPIN genes – OsPIN1a, OsPIN1c and OsPIN3a – are positively correlated with those of LPA1. This study increases our knowledge of the regulatory mechanisms underlying IAA- and BR-responsive bending of lamina joints.
Materials and methods
Plant materials and growth conditions
A new allele of lpa1 (lpa1-2) was isolated from gene trap Ds populations of the Dongjin japonica rice cultivar. For the wild types (WTs), either Dongjin or WT siblings from segregation populations were used. The d61 and d2 mutants (japonica type) were obtained from Dr Koh Hee Jong of Seoul National University. Following a previously published protocol (Chin et al., 1999), transgenic lines were generated from japonica rice cv. Dongjin. Plants were grown in paddy fields from June to October in South Korea and in the greenhouse during the winter.
Lamina inclination assay
Plants were grown on half-strength Murashige and Skoog (MS) medium in a growth chamber at 30°C under a 24h light regime. For the lamina inclination assays, 1 µl of ethanol containing a series of concentrations of BRs, IAA or Brz was spotted onto the tips of the lamina of 5- or 6-day-old seedlings. C-22-hydroxylated and 6-deoxo brassinosteroids were identical to those described by Nakamura et al. (2009). Treated seedlings were grown for two additional days, and lamina joints of the second leaves were photographed for lamina bending angle measurements.
Cloning, vector construction and transformation
Analysis of LPA1 cDNA from wild type and trap Ds line, vector construction, and transformation were performed following standard methods. See Supplementary Appendix S1, available at JXB online, for details.
Expression analysis
Total cellular RNA was isolated with TRIZOL reagent (MRC, http://www.mrcgene.com/) or an RNeasy Plant Mini Kit (Qiagen). For quantitative RT-PCR, the published methods of Je et al. (2010) were followed. The qRT-PCR products were quantified using CFX Manager software (Bio-Rad) and the values were normalized against 25S rRNA and Ubiquitin cDNA from the same samples.
RNA-seq analysis
Using an RNeasy Plant Mini Kit (Qiagen, http://www.qiagen.com/), total RNA was extracted from 1 cm-long leaf segments spanning the second lamina joints of 1-week-old seedlings of WT, mutants and overexpressors. Construction and sequencing of cDNA libraries for RNA sequencing and data analysis were described in Supplementary Appendix S2.
IAA treatments
Seeds were plated on half-strength MS agar medium and incubated in a growth chamber at 28°C under continuous light for 1 week. Uniform seedlings were selected and gently washed in dH2O to remove residual MS agar from their roots. For IAA treatments, seedling samples were transferred into 50ml Falcon tubes filled with distilled water. After acclimatization for 1 d, the samples were transferred to new Falcon tubes filled with 50ml of 20 µM IAA solution. After 3h of submergence, 1 cm-long segments that contained joints between the blades and sheaths of second leaves were sampled from 30 lpa1, e19 and WT plants. Using an RNeasy Plant Mini Kit (Qiagen, http://www.qiagen.com/), total RNA was extracted from leaf segments before and after 3h of IAA treatment.
GUS staining
GUS activity was detected by incubation in a 5-bromo-4-chloro-3-indoyl glucuronide solution, which was previously described (Chin et al., 1999). Samples were incubated at 37°C for 2 d in the dark and then dehydrated in a 30–70% graded ethanol series.
Microscopy
To detect GFP fluorescence, roots of LPA1:GFP transgenic plants were analysed using an Olympus confocal laser-scanning microscope (Fluoview FV 1000, http://www.olympus-global.com/). Fluorescence emission images were collected in the 480–540nm range. Propidium iodide (PI) staining was used for detection of nuclei.
For cellular measurement of lamina joint tissues, lamina joint tissues were collected from 1-week-old seedlings and fixed in 70% ethanol. The epidermal layers on the adaxial sides were imaged by microscopy (DP70; Olympus, Japan), and then the cell dimensions were measured by ImageJ.
Results
The identification of a new LPA1 allele, lpa1-2
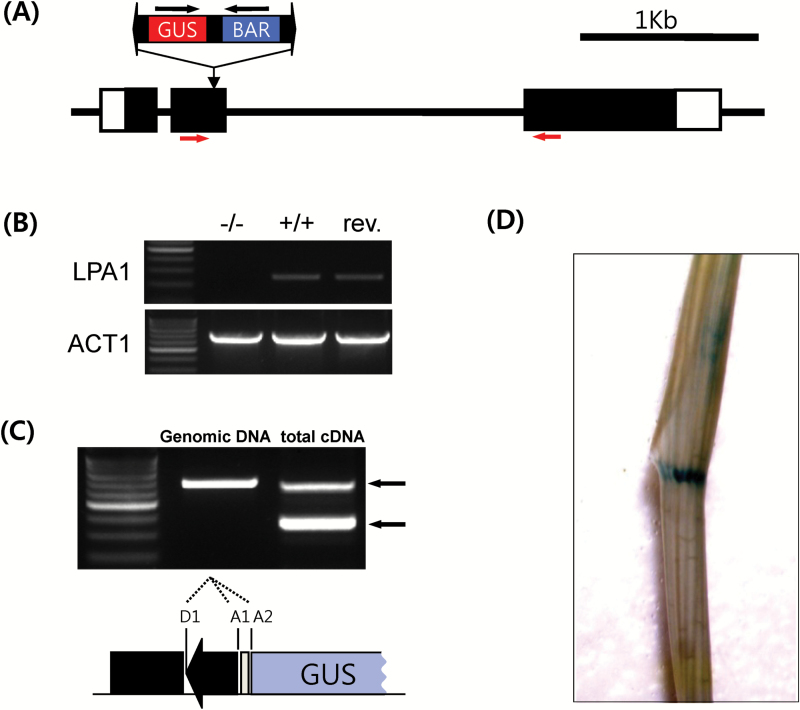
In 2007, we identified a mutant line with wide lamina and tiller angles in a Ds population field. This mutant arose from insertion of a gene trap Ds at the second exon of the gene LPA1 (Fig. 1A). RT-PCR showed that transcripts beyond the Ds insertion site were not detected in the mutants (Fig. 1B). Since a GUS coding region inside the gene trap Ds element was in the same orientation as the gene, GUS was to be transcribed with the 5ʹ truncated mRNA (Chin et al., 1999). We cloned the fusion transcripts by RT-PCR, and subsequent sequencing analysis revealed that two-thirds of the spliced transcripts were in-frame between the gene and GUS (Fig. 1C). New alleles including revertants were generated by remobilizing Ds via tissue culture. Of the more than 100 regenerated plants, four frame-shift alleles and three revertants were identified by sequencing Ds excision sites (Supplementary Table S1). LPA1 (Wu et al., 2013) is the same gene that was previously reported as OsIDD14 (Colasanti et al., 2006). Therefore, this Ds-tagged allele was designated as lpa1-2 and utilized for this study. LPA1/OsIDD14 has the highest sequence homology to SGR5/AtIDD15 of Arabidopsis (Morita et al., 2006), and it shares 42% identity and 54% similarity with SGR5/AtIDD15 at the amino acid level. LPA1 was highly expressed in lamina joints (Wu et al., 2013). To confirm the preferential expression of LPA1 at lamina joints, we examined GUS expression inside the gene trap Ds within LPA1 (LPA1::Ds). The GUS pattern of LPA1::Ds was consistent with the expression pattern detected by qRT-PCR. Strong GUS activity was detected in the lamina joint (Fig. 1D).
Fig. 1.
Analysis of the lpa1::Ds mutant allele and the expression pattern of LPA1. (A) The gene trap Ds carrying a GUS coding region and a bar gene was inserted at the second exon of LPA1 (LOC_Os03g13400). Open boxes indicate the untranslated region. Closed boxes indicate coding regions. Lines between boxes indicate introns. Black horizontal arrows indicate the orientation of transcription. (B) RT-PCR analysis was performed with RNAs from 2-week-old seedlings of lpa1, WT and a revertant. Primers recognized the second exon before the Ds insertion and the third exon. The locations of the primers are shown as red horizontal arrows in panel A. The Actin gene was used as a control. (C) GUS-fused transcripts were analysed by RT-PCR using primers from LPA1 and GUS. The upper arrow indicates cDNA from unspliced RNAs while the lower arrow indicates cDNA from spliced RNAs. The spliced cDNAs were sequenced to identify the joint sequences between the LPA1 and GUS coding regions. D1, A1 and A2 indicate splicing donor 1 and acceptor site 1 and 2, respectively, installed inside the element (Chin et al., 1999). (D) Leaf of 2-week-old lpa1:Ds plant incubated in GUS solution. Strong GUS staining was detected in the lamina joint. (This figure is available in colour at JXB online.)
LPA1 as a positive regulator suppressing lamina bending
We examined the mode of action of LPA1 on lamina bending by comparing the phenotypes among knockout, overexpression and repressor-fusion transgenic plants (Fig. 2). All of the mutants, overexpression and repressor-fusion transgenic lines were generated in the same rice japonica variety, Dongjin. The Ubiquitin promoter was used to overexpress LPA1 cDNA. To construct a vector expressing repressor-fused LPA1, the C-terminus of LPA1 was fused with the 24 amino acid fragment containing a conserved ERF-associated amphiphilic repression (EAR) motif of a gene (LOC_Os02g01090) orthologous to Arabidopsis SUPERMAN (At3g23130) (Hiratsu et al., 2004). The fusion gene was expressed under the control of the 2.4kb LPA1 promoter. We examined the expression levels of these lines by qRT-PCR.
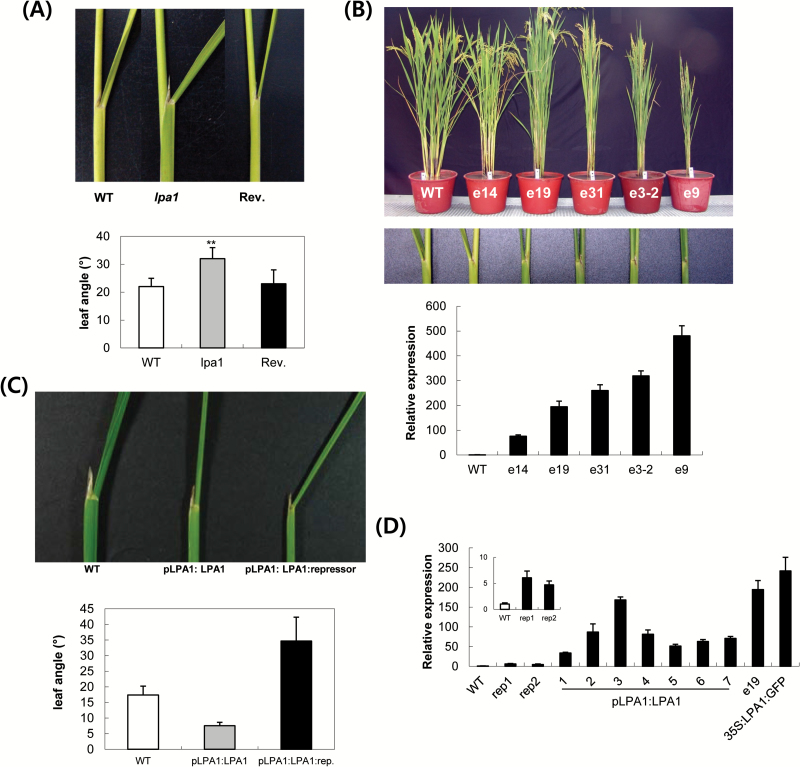
Fig. 2.
Phenotypic analysis of lpa1, LPA1 overexpressors (OX) and pLPA1:LPA1:repressor transgenic plants. (A) At the top, lamina joints of the leaves just below flag leaves at the reproductive stage are compared among WT, lpa1 and a revertant. At the bottom, measurements of lamina joint angles are shown in the diagrams. Data represent mean ±SE of more than ten plants analysed. **, P<0.001; P-value of lpa1 mutants was calculated with respect to one of WT or of revertant (B) OX lines (e14, e19, e31, e3-2 and e9) were aligned based on the expression levels of LPA1 mRNA. Lamina joint angles are shown at the reproductive stage. Using RNA from 10-day-old seedlings, qRT-PCR was employed to measure the expression levels of individual OX lines. Values were normalized against Ubiquitin cDNA from the same samples. (C) The third leaves from the reproductive stage plants are compared among Dongjin parental line (WT), pLPA1:LPA1 and pLPA1:LPA1:repressor. Since independent transgenic lines of the same constructs showed similar phenotypes of lamina bending, representative phenotypes are shown in the figure. At the bottom, measurements of lamina joint angles are shown in the diagrams. Data represent mean ±SD of more than ten plants from the lines analysed. (D) Levels of LPA1 mRNA of seven pLPA1:LPA1 (#1–7) and two pLPA1:LPA1:repressor (#1 and 2) lines measured by qRT-PCR using RNAs from 10-day-old seedlings. Values were normalized against Ubiquitin cDNA from the same samples. The transgenes were expressed under the control of the 2.4kb LPA1 promoter. (This figure is available in colour at JXB online.)
Lamina angles in LPA1-overexpressing plants and lpa1 mutants were not significantly different from those of WT plants for up to one month of growth, even under field conditions. After one month of growth, the Ds mutants exhibited exaggerated leaf angles, while the overexpressor lines exhibited an erect phenotype, with narrow leaf angles (Fig. 2A, B). The extent of the leaf angle was negatively correlated with the expression level of LPA1 mRNA in the overexpressors (Fig. 2B); plants expressing higher levels of mRNA exhibited more severe erect phenotypes. Higher LPA1 mRNA levels were associated with slightly shorter plant statures than those of WT plants, and these plants produced fewer tillers and narrower leaves. However, flowering time was not affected. Therefore, the function of LPA1 might be related to the suppression of lamina inclination. To further understand whether LPA1 acts as a positive or negative regulator in suppressing lamina inclination, we examined transgenic plants expressing repressor-fused LPA1 cDNA under the control of the 2.4kb LPA1 promoter. As a control, LPA1 cDNA was expressed under the control of the same LPA1 promoter. Figure 2C shows the lamina bending phenotype of the repressor-fusion LPA1 plants. The phenotype of the repressor-fused LPA1 plants exhibited the exaggerated lamina angles as observed in the knockout mutants (lpa1). By contrast, the pLPA1:LPA1 transgenic plants accumulated higher levels of LPA1 mRNA than normal plants (Fig. 2D) and developed the same erect phenotype as overexpressors under control of the Ubiquitin promoter. The data clearly show that the loss-of-function of LPA1 led to an increase in lamina joint bending angles. Therefore, LPA1 acts as a positive regulator that suppresses lamina bending.
The suppressive action of LPA1 on IAA-mediated lamina inclination is influenced by BR
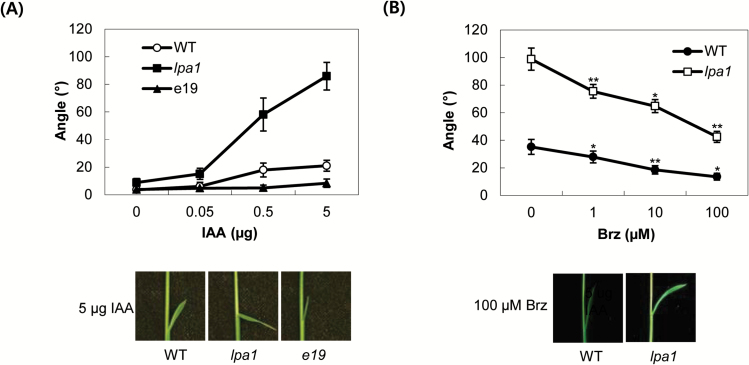
To examine whether LPA1 affects IAA-mediated lamina inclination, we measured the IAA sensitivity of lamina joints in the lpa1 mutant and an LPA1 overexpression line, e19. During the seedling stages, the angles of lamina joints were not obviously different among WT, mutant and overexpressor. We applied a series of different concentrations of IAA to the lamina joints of 5-day-old plants grown on half-strength MS medium (Fig. 3A and Supplementary Fig. S1A) and measured the downward inclination angles 2 d after treatment. Compared with WT plants, lpa1 plants were exceptionally sensitive to auxin. In the 5 µg IAA treatment group, WT sibling plants exhibited 20–40° angles in their lamina joints, while those of lpa1 plants were 100°. By contrast, the overexpressor was almost completely insensitive to auxin. These results imply that LPA1 exerts inhibitory action on auxin-mediated lamina inclination. To determine whether BRs influence the IAA-hypersensitivity of lpa1 plants, we pretreated these plants for 12h with a series of concentrations of brassinazole (Brz) prior to the application of 5 µg IAA. Brz inhibits C-22 hydroxylase in the BR synthesis pathway and blocks the production of C-22-hydroxylated BRs (Supplementary Fig. S7; Asami et al., 2001; Sakamoto et al., 2006). Figure 3B and Supplementary Fig. S1B show a comparison of the Brz sensitivity of lamina joints between lpa1 and the WT. As the concentration of Brz increased, auxin-mediated bending was suppressed in the lpa1 lamina joints. These data clearly show that BRs are involved in the hypersensitivity of lpa1 to IAA.
Fig. 3.
IAA-mediated lamina inclination and the effect of brassinazole on LPA1 mutants. (A) The indicated amounts of IAA in 1 µl of ethanol were spotted onto the tips of the second lamina of 5-day-old seedlings. After 2 d of incubation, the lamina joints were photographed for angle measurements; e19 is an LPA1 overexpressor line. A composite photograph of plants treated with aliquots of 5 µg IAA was taken. Mean values were derived from measurements of at least ten plants. Data represent the means ±SD of at least ten plants. (B) Five-day-old lpa1 and WT plants were treated with the indicated concentrations of brassinazole (Brz) on the tips of the second leaves 12h before 5 µg IAA treatment. Data represent the means ±SD of at least ten plants. *, P<0.05; **, P<0.001; P-values of samples treated with 1, 10 and 100 µM Brz were calculated with respect to those of the same samples treated with 0, 1 and 10 µM Brz, respectively. (This figure is available in colour at JXB online.)
Synergic effects of lpa1 with d2, but not OsBRI1, on auxin-mediated lamina inclination
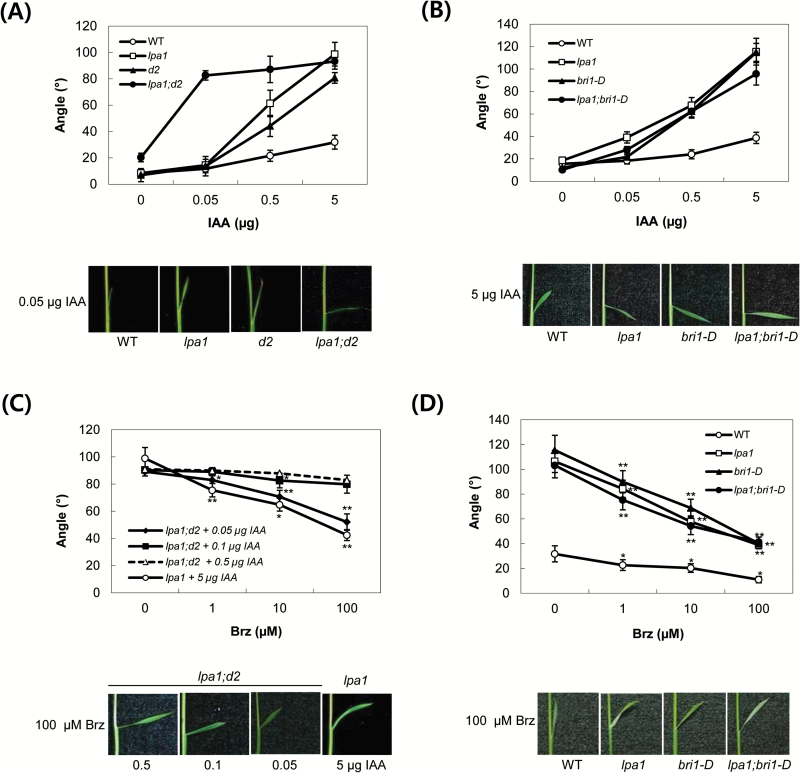
There are two possible explanations for the effect of Brz treatment on IAA-mediated inclination of lamina joints in lpa1. One explanation is that Brz treatment decreases the levels of endogenous BRs in the synthesis pathway, such as C-22-hydroxylated and 6-deoxo BRs, which are involved in IAA-mediated lamina inclination (Nakamura et al., 2009). The other explanation is that Brz treatment leads to a loss of active BR compounds, such as BL or castasterone (CS), which are required for the activity of OsBRI1. To distinguish between these two possibilities, d2 and bri1-D were crossed with lpa1 to generate double genetic combinations. Rice D2 encodes a CYP90D2 enzyme, which catalyses C-23 hydroxylation of various C-22-hydroxylated BRs [e.g. C-22-hydroxycampesterol (22-OHCR) and 6-deoxocathasterone (6-deoxoCT)] (Supplementary Fig. S7; Hong et al., 2003; Ohnishi et al., 2006; Nakamura et al., 2009). Therefore, d2 has accumulation of C-22-hydroxylated BRs and has been shown to enhance the IAA sensitivity of lamina joints (Nakamura et al., 2009). An OsBRI1 overexpression line (bri1-D) isolated from an activation T-DNA tagging population exhibits increased lamina joint angles (Jeong et al., 2002; Je et al., 2010). First, we compared the lamina inclination of WT, lpa1, lpa1;d2 and d2 in response to IAA treatment; all lines were derived from crosses between lpa1 and d2. Compared to lpa1 and d2, lpa1;d2 was extremely sensitive to IAA, achieving the maximal angles of lamina bending at only 0.05 µg IAA. For lpa1 and d2, the same maximal angle was observed at 5 µg IAA. IAA sensitivity was approximately 100-fold higher in lpa1;d2 than in each single mutant (Fig. 4A; Supplementary Fig. S2A). Second, to examine the action of OsBRI1 in the lamina joints of lpa1, we compared the IAA sensitivity in the double genetic combination lpa1;bri1-D with that of lpa1 and bri1-D. All lines were derived from crosses between lpa1 and bri1-D. As shown in Fig. 4B and Supplementary Fig. S2B, the magnitude of IAA-mediated lamina bending angles in lpa1;bri1-D was similar to that of lpa1 or bri1-D alone. These data indicate that the IAA-mediated lamina inclination of lpa1 is influenced by BRs (especially C-22-hydroxylated BRs) rather than OsBRI1.
Fig. 4.
IAA-mediated lamina inclination and the effect of brassinazole on lpa1;d2 and lpa1;bri1-D. (A, B) The indicated amounts of IAA in 1 µl of ethanol were spotted onto the tips of the second lamina of 5-day-old seedlings. The bending angles were measured after 2 d of incubation. A composite photograph of plants treated with 0.05 (A) and 5 µg IAA (B) was taken. Data represent the means ±SD of at least ten plants. (C) Five-day-old lpa1;d2 plants were treated with the indicated concentrations of brassinazole (Brz) on the tips of the second leaves 12h before IAA treatment; 0.05, 0.1 and 0.5 µg IAA were applied to Brz-treated plants, which were incubated for 2 d before measurement. For comparison, the effect of Brz on lpa1 plants treated with 5 µg IAA is shown. A composite photograph of plants treated with aliquots of 100 µM Brz was taken. (D) Five-day-old lpa1, lpa1;bri1-D and bri1-D plants were treated with the indicated concentrations of Brz on the tips of the second leaves 12h before 5 µg IAA treatment. A composite photograph of plants treated with aliquots of 100 µM Brz was taken. Data represent the means ±SD of at least ten plants. *, P<0.05; **, P<0.001; P-values of samples (C and D) treated with 1, 10 and 100 µM Brz were calculated with respect to those of the same samples treated with 0, 1 and 10 µM Brz, respectively. (This figure is available in colour at JXB online.)
In addition, we examined the effect of Brz on IAA-mediated lamina inclination in double genetic combinations lpa1;d2 and lpa1;bri1-D. For lpa1;d2, Brz treatment had much less of an effect on IAA-mediated inclination. To achieve the same magnitude of the Brz effect as observed in lpa1 and d2, lpa1;d2 plants required 100-fold lower IAA concentrations (Fig. 4C; Supplementary Fig. S2C). Such a reduction in the effect of Brz on lpa1;d2 may have been due to an interaction between lpa1 and d2 in the lamina joints that occurred prior to Brz treatment. By contrast, Brz had a similar effect on IAA-mediated inclination in lpa1;bri1-D to that in lpa1. Under increasing levels of Brz treatment, lpa1 and lpa1;bri1-D showed similar rates of inhibition of auxin-mediated bending of lamina joints (Fig. 4D; Supplementary Fig. S2D).
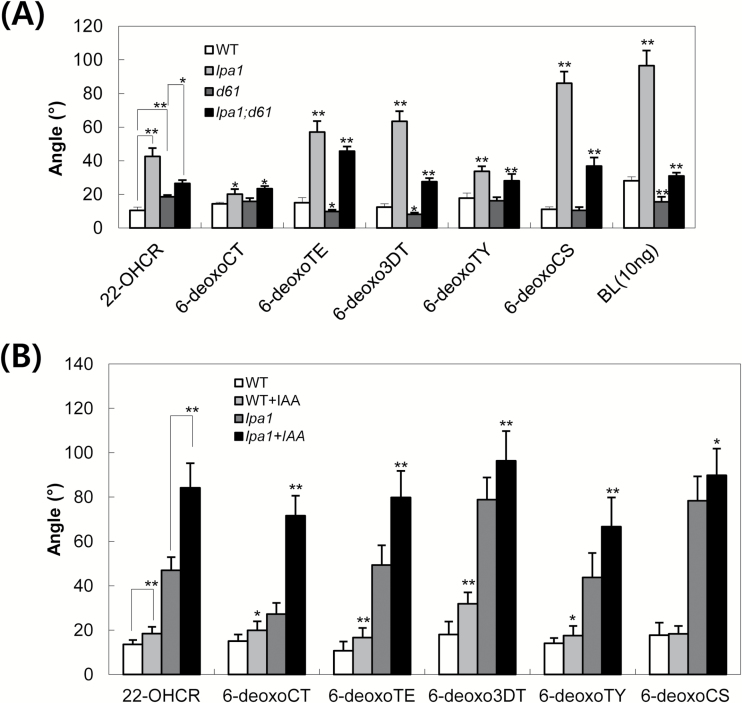
Sensitivity to C-22-hydroxylated and 6-deoxo BRs during lamina inclination
To verify that the IAA-mediated lamina inclination of lpa1 was enhanced in the presence of C-22-hydroxylated BRs, we examined the sensitivity of lamina joints to C-22-hydroxylated and 6-deoxo BRs in lpa1 and lpa1;d61-1 (Fig. 5A; Supplementary Fig. S3A). The following compounds were directly applied to lamina joints: 22-OHCR, 6-deoxoCT, 6-deoxoTE (teasterone), 6-deoxo3DT (3-dehydroteasterone), 6-deoxoTY (typhasterol) and 6-deoxoCS. Among these BRs, 22-OHCR and 6-deoxoCT are substrates for D2-encoded 23-hydroxylase (Supplementary Fig. S7). The compounds 6-deoxoTE, 6-deoxo3DT, 6-deoxoTY and 6-deoxoCS are converted to TE, 3DT, TY and CS, respectively, by CYP85A1 (BRD1 (BR-deficient Dwarf1)/OsDWARF1)-encoded C-6 oxidase (Supplementary Fig. S7; Hong et al., 2002). Due to the possibility that these C-22-hydroxylated or 6-deoxo BRs might be converted into the active BRs, CS or BL, it was important to compare the lamina sensitivity of lpa1 with lpa1;d61-1. All of the lines were derived from crosses between lpa1 and d61-1. As previously reported (Nakamura et al., 2009), lamina bending of WT and d61-1 plants was barely observed at a 100ng concentration of any of the BRs. However, lpa1 exhibited clear lamina bending under these conditions. Moreover, lpa1;d61-1 exhibited much stronger responses to all of the compounds than d61-1. Since lamina bending was more exaggerated in lpa1 than in lpa1;d61-1, it is possible that a portion of the exogenously supplied BRs was converted to active BRs, which would bind to OsBRI1 in the cells of lamina joints, thereby explaining the difference in lamina inclination between lpa1 and lpa1;d61-1. Taken together, these results suggest that LPA1 enhances the sensitivity of lamina joints not only to C-22-hydroxylated BRs, but also to deoxo BRs, which is independent of the action of OsBRI1.
Fig. 5.
Sensitivity of lpa1 and lpa1;d61-1 plants to C-22-hydroxylated and 6-deoxo brassinosteroids (BRs) and the combinatory effects with IAA on lpa1 mutants. (A) Aliquots of 100ng each of 22-OHCR, 6-deoxoCT, 6-deoxoTE, 6-deoxo3DT, 6-deoxoTY, 6-deoxoCS and 10ng BLs were applied to 5-day-old plants for 2 d and the resultant bending angle of the lamina joint was measured. Data represent the means ±SD of at least ten plants. *, P<0.05; **, P<0.001; P-values of lpa1and d61-1 were calculated with respect to the WT samples. The P-value of lpa1;d61-1 was calculated with respect to d61-1. (B) Mixtures of 0.05 µg IAA and 100ng aliquots each of 22-OHCR, 6-deoxoCT, 6-deoxoTE, 6-deoxo3DT, 6-deoxoTY and 6-deoxoCS were applied to 5-day-old plants for 2 d and the bending angles of lamina joints were measured. Data represent the means ±SD of at least ten plants. *, P<0.05; **, P<0.001; P-values of samples treated only with BR were calculated with respect to those treated with both IAA and the same BR. 22-OHCR, C-22-hydroxycampesterol; 6-deoxoCT, 6-deoxocathasterone; 6-deoxoTE, 6-deoxoteasterone; 6-deoxo3DT, 3-dehydro-6-deoxoteasterone; 6-deoxoTY, 6-deoxotyphasterol; 6-deoxoCS, 6-deoxocastasterone.
To estimate the effect of C-22-hydroxylated and 6-deoxo BRs on IAA-mediated lamina bending, we applied 100ng each of 22-OHCR, 6-deoxoCT, 6-deoxoTE, 6-deoxo3DT, 6-deoxoTY and 6-deoxoCS, along with 0.05 µg IAA, to lamina joints of lpa1 and WT plants. The IAA dosage of 0.05 µg was chosen since lamina bending was not evident in mutant or WT plants treated with 0.05 µg IAA (Fig. 3A). Figure 5B and Supplementary Fig. S3B show that significant interactions of these BRs with IAA were detected in lpa1. It is worth noting that the plants were more sensitive to 6-deoxo3DT, 6-deoxoTY and 6-deoxoCS than to 22-OHCR or 6-deoxoCT, which may be due to the fact that 6-deoxo3DT, 6-deoxoTY and 6-deoxoCS are substrates that are directly converted to CS or BL, which bind to OsBRI1. Since the maximal bending angle of lamina joints is approximately 100°, the combined effects of these 6-deoxo BRs and IAA on lamina bending were not as dramatic as expected. Taken together, these results suggest that LPA1 exerts a regulatory role in determining lamina bending via the interaction of IAA with C-22-hydroxylated and 6-deoxo BRs.
Relationship between LPA1 and OsBRI1 in determining lamina inclination
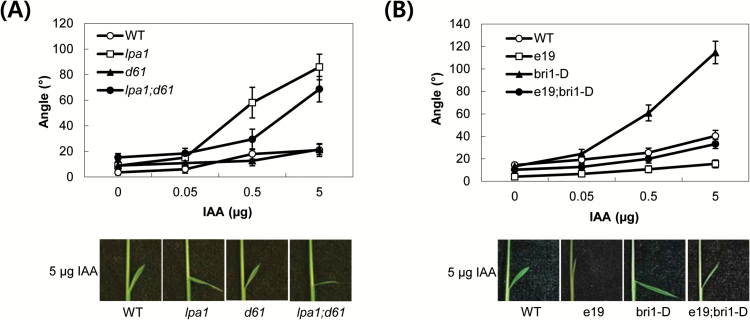
The data strongly suggest that LPA1 and OsBRI1 exert independent effects on IAA-mediated lamina inclination. As shown in Fig. 4B, both lpa1 and bri1-D were hypersensitive to IAA and showed similar bending angles in response to IAA in the lamina inclination assay. To further examine the relationship between LPA1 and OsBRI1 in auxin-mediated lamina inclination, two double genetic combinations of lpa1;d61-1 and e19;bri1-D were utilized. As shown in Fig. 6A and Supplementary Fig. S4A, the extent of lamina inclinations in lpa1;d61-1 was intermediate between that of lpa1 and d61-1. Similarly, the bending angles of e19;bri1-D were similar to those of the WT and were intermediate between those of e19 and bri1-D (Fig. 6B and Supplementary Fig. S4B). Therefore, LPA1 and OsBRI1 are independently required for IAA-mediated lamina inclination (Supplementary Fig. S5).
Fig. 6.
IAA-mediated lamina inclination of lpa1;d61-1 and e19;bri1-D. The indicated amounts of IAA in 1 µl of ethanol were spotted onto the tips of the second lamina of 5-day-old seedlings. All lines were derived from crosses of lpa1 with (A) d61-1 and (B) bri1-D. Data represent the means ±SD of at least ten plants. (This figure is available in colour at JXB online.)
Cellular measurements of the epidermal layer of lamina joints
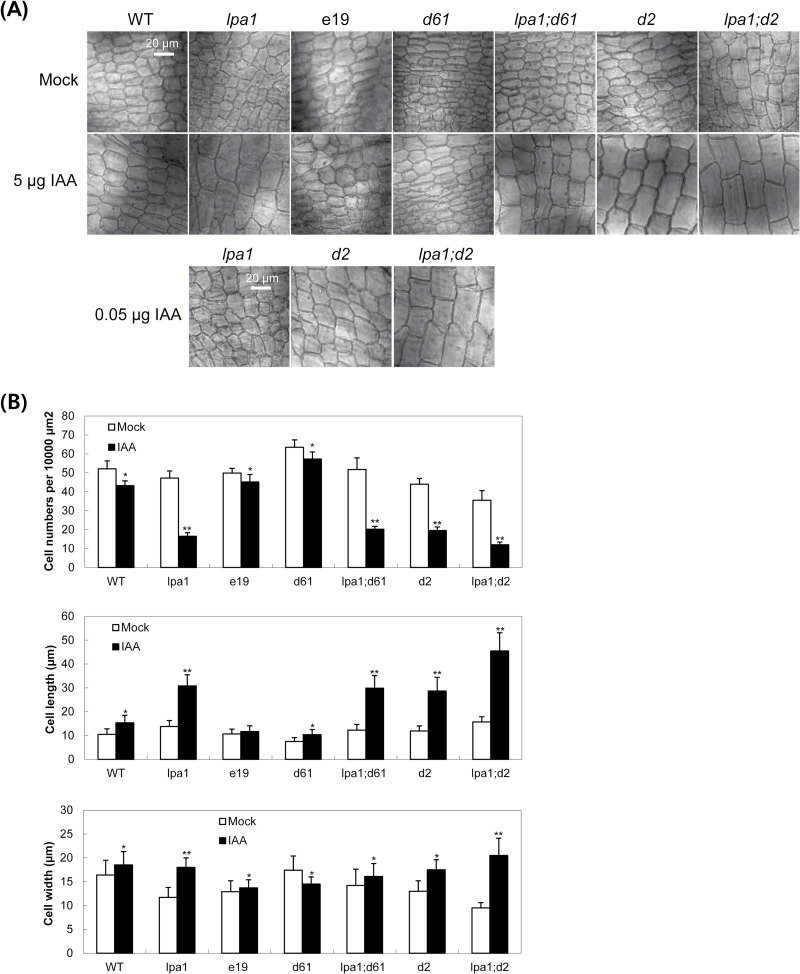
To obtain detailed information about the cellular responses of LPA1-affected lamina joints, we measured the cellular dimensions in the epidermal layers on the adaxial sides of these joints (Fig. 7A). We compared the lamina joints before and 2 d after 5 µg IAA treatment among lpa1, e19 (an LPA1 OX line), d61-1, lpa1;d61-1, lpa1;d2, d2 and WT siblings from the cross between lpa1 and d61-1. We also compared lpa1;d2 with lpa1 and d2 plants treated with 0.05 µg IAA. Figure 7B shows the cellular parameters of epidermal layers of the adaxial side. Cell numbers within given areas were dramatically reduced in lpa1, lpa1;d61-1, lpa1;d2 and d2 after the IAA treatments. Likewise, they showed significant increases in cell length in the same areas. Interestingly, lpa1 and lpa1;d2 exhibited enhanced expansion of cell width compared to lpa1;d61-1 and d2. Therefore, the dramatic lamina inclination observed in lpa1;d2 may have resulted from increases in both cell width and length.
Fig. 7.
Cellular measurements in the epidermal layers of lamina joints. (A) Epidermal cells of adaxial layers of the lamina joints are shown. The lamina joints of WT (siblings from lpa1;d61-1 segregation), lpa1, e19, d61-1, lpa1;d61-1, d2 and lpa1;d2 plants were treated with 1 µl ethanol (mock) or 1 µl ethanol containing 5 µg IAA. In addition, lpa1, d2 and lpa1;d2 plants treated with 0.05 µg IAA were compared. Bar, 20 µm. (B) The cell numbers, and length and width of epidermal cells of the lamina joints in these plants were measured by ImageJ after being imaged by microscopy. Data represent the means ±SD of at least four independent plants. *, P<0.05, **, P<0.001; student’s t-test. P-values of samples treated with IAA were calculated with respect to those of non-treated samples.
Possible mechanisms underlying the role of LPA1 in IAA-mediated lamina inclination
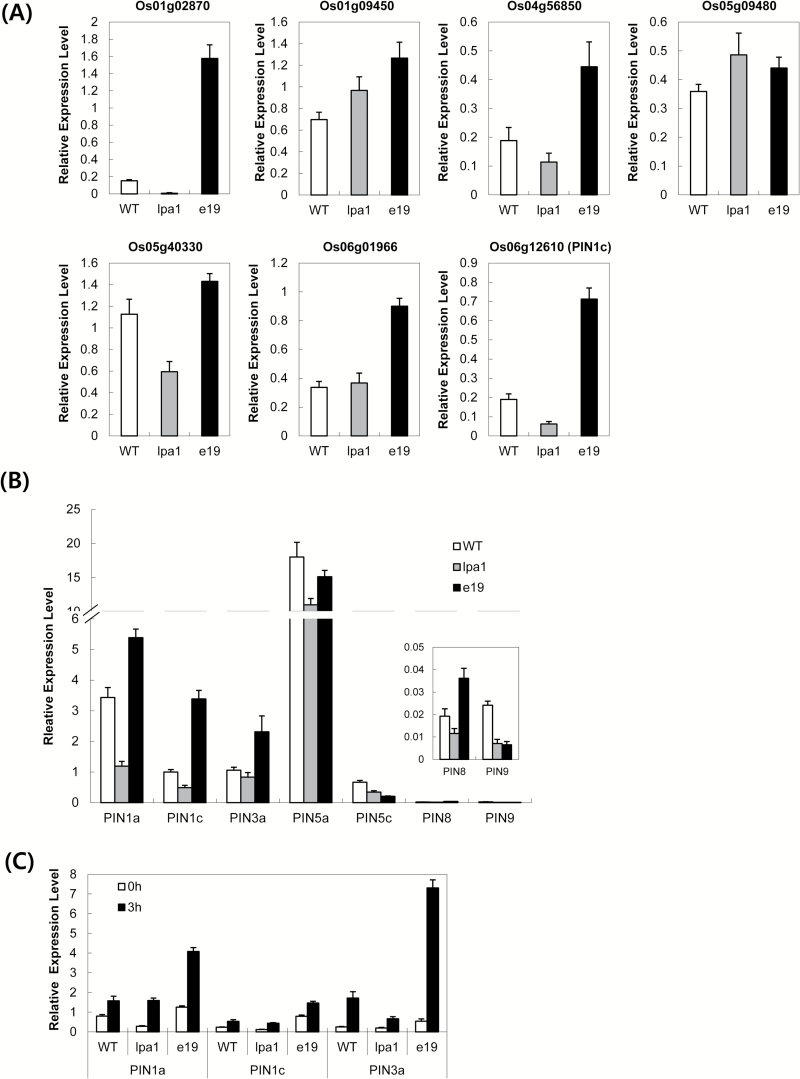
To explore the molecular mechanism underlying how LPA1 is involved in IAA-mediated lamina inclination, transcriptome profiling using Illumina RNA-seq was performed on leaf segments spanning lamina joints from lpa1 mutant, overexpressor (e19) and WT siblings. Total RNA was isolated from 1 cm-long leaf segments that spanned the lamina joints of 10-day-old plants. After analysis of RNA-seq reads, genes were identified that showed at least 1.5-fold differences in expression levels between WT sibling and lpa1 mutant or e19 overexpressor plants. Supplementary Table S3 lists the genes whose expression was 1.5-fold higher or lower in e19 or lpa1 than in the WT. Among these genes, seven genes whose functions are related to auxin signalling or action were further analysed using quantitative RT-PCR (Supplementary Table S4 and Fig. 8A). Among the auxin-related genes, four genes showed opposite expression patterns between lpa1 and e19. Compared to WT, these genes were downregulated in lpa1 and upregulated in e19. Particularly LOC_Os06g12610, which belongs to the OsPIN gene family and encodes the auxin efflux carrier OsPIN1c. To further evaluate the effect of LPA1 on the expression of OsPIN family genes, we examined RNA from leaf segments spanning lamina joints of lpa1, e19 and WT siblings using qRT-PCR (Fig. 8B). Twelve OsPIN genes are reported to be expressed in rice (Miyashita et al., 2010). The transcripts of seven genes were analysed by qRT-PCR using previously reported primers (Miyashita et al., 2010). Four OsPIN genes (OsPIN1a, OsPIN1c, OsPIN3a and OsPIN5) produced major RNA species in these leaf samples. Among these, three OsPIN genes (OsPIN1a, OsPIN1c and OsPIN3a) exhibited LPA1-dependent expression patterns; these genes were expressed at lower levels in lpa1 and higher levels in e19. To examine whether LPA1 influences the auxin sensitivity of these three OsPIN genes, we compared the RNA levels of OsPIN1a, OsPIN1c and OsPIN3a before and 3h after IAA treatment. Even after IAA treatment, the mutants maintained lower transcript levels of these three genes than the WT siblings, while the overexpressors produced higher transcript levels (Fig. 8C). These results indicate that the auxin-inducibility of these OsPIN genes is influenced by LPA1.
Fig. 8.
Quantitative RT-PCR analysis of auxin-related genes identified by RNA-seq analysis and OsPIN genes in WT, lpa1 and e19. RNA from 1 cm-long leaf segments spanning the lamina joints of WT, lpa1 and e19 was used for qRT-PCR analysis. (A) The expression of LOC_Os01g02870, LOC_Os06g12610, LOC_Os04g56850, LOC_Os05g09480, LOC_Os01g09450, LOC_Os06g01966, and LOC_Os05g40330, whose putative functions are listed in Supplementary Table S4, was re-examined by qRT-PCR. (B) The expression levels of seven OsPIN genes (OsPIN1a, OsPIN1c, OsPIN3a, OsPIN5a, OsPIN5c, OsPIN8 and OsPIN9) were measured by qRT-PCR. (C) Quantitative RT-PCR was performed to estimate the mRNA levels of OsPIN1a, OsPIN1c and OsPIN3a in WT, lpa1 and e19 before and 3h after IAA treatment; 25S rRNA was used as a control to normalize the expression data. Error bars are ±SD of the means of three qPCR replicates.
Discussion
The functions of LPA1 and its Arabidopsis homologues have been elucidated in previous reports. LPA1 and SGR5/AtIDD15 have defects in shoot gravity responses; therefore, they have been proposed to function during early shoot gravitropism responses by regulating amyloplast sedimentation rates (Morita et al., 2006; Wu et al., 2013). Subsequent work showed that AtIDD14, AtIDD15 and AtIDD16 gene products directly target auxin biosynthetic and transport genes, and influence organ development, which eventually controls plant architecture (Cui et al., 2013).
Lamina inclination is determined by cellular expansion of the lamina joint adaxial layers (Maeda, 1965). Lamina joints respond rapidly to extracellular hormones, which renders this system a suitable model for efficient analysis of cellular mechanisms regulating lamina inclination. The major objective of this study was to identify the role of LPA1 in hormone-dependent determination of lamina inclination in rice plants. We utilized BR synthetic and receptor mutants (d2, d61-1 and bri1-D) and treatment with IAA and BR compounds to elucidate the role of LPA1 in interacting with auxin and BR signalling, which determined lamina bending. Our studies revealed three primary observations. First, lamina joints in lpa1 mutants responded dramatically to IAA, which was suppressed by Brz inhibition of C-22 hydroxylase in the BR biosynthetic pathway. Second, IAA sensitivity of lpa1 mutants was strongly enhanced in genetic combination with d2 mutants, which are defective in CYP90D2 catalysis of C-22-hydroxylated BRs. However, no additive effect was observed in lpa1 plants in genetic combination with the BRI1 dominant mutation bri1-D. Third, lpa1 mutants exhibited sensitivity to C-22-hydroxylated and 6-deoxo BRs even in the absence of OsBRI1. These combined results indicate that LPA1 suppresses auxin signalling interacting with C-22-hydroxylated or 6-deoxo BRs, which regulates lamina inclination independently of OsBRI1. Supplementary Tables S5, S6 summarize the genetic and biochemical data on the auxin sensitivity of lamina bending.
Our work provides genetic evidence verifying the previously proposed hypothesis that BR interacts with auxin to determine lamina inclination via two different pathways. In one pathway, BRs interact directly with auxin. In the other pathway, the OsBRI1 receptor is required for BR interaction with auxin. Our study utilized two genetic combinations (e19 and bri1-D; lpa1 and d61-1) to investigate rice lamina inclination, and the results indicate that two pathways might exert independent effects on IAA-mediated lamina inclination. Currently, little is known about BR and IAA interactions in determining lamina inclination in rice, except for anatomical evidence that BR and IAA interaction results in cellular elongation and expansion (Fig. 7; Maeda, 1965; Nakamura et al., 2009). Further analysis of LPA1 would increase our understanding of lamina joint responses to auxin and BR.
Although the mechanisms underlying asymmetric responses of adaxial and abaxial cell layers to auxin during lamina bending are currently unknown, it is reasonable to speculate that differential sensitivity to auxin and asymmetrical auxin distribution are important factors in determining lamina inclination. Expression of Arabidopsis AtPIN1 was altered in gain-of-function and loss-of-function mutants of AtIDD14, AtIDD15 and AtIDD16, and IDD genes directly activated AtPIN1 expression (Cui et al., 2013). Our RNA-seq data showed that OsPIN expression depends on LPA1. This suggests that LPA1-dependent OsPIN expression might be responsible, at least in part, for the opposite auxin sensitivity during the lamina bending response between lpa1 and e19 plants. OsPIN2 overexpression in rice plants increases auxin transport from shoots to roots without altering auxin flux patterns (Chen et al., 2012). Therefore, it is reasonable to speculate that LPA1 overexpressors might have a greater capacity for transporting exogenous auxin than WT plants, and lpa1 mutants might exhibit less efficient auxin flux. To confirm the significance of auxin flux in LPA1-mediated lamina bending, lpa1 mutant and overexpressor e19 were treated with different concentrations of N-1-naphthylphthalamic acid (NPA) at lamina joints 12h before IAA application (Supplementary Fig. S6). NPA treatment increased IAA-induced lamina bending angles in e19-overexpressing plants but not in lpa1 mutants. These data support the proposal that auxin transport might be more active in LPA1-overexpressing plants than in lpa1 mutants. These combined results suggest that auxin flux and distribution might be important factors underlying the role of LPA1-mediated auxin sensitivity in lamina inclination.
Plant architecture is one of the major phenotypic parameters that crop breeders consider when developing high yielding varieties, and lamina angle is one of the major characters that determine plant type. Since overexpression of LPA1 leads to an erect phenotype, producing plants with the optimal expression pattern of LPA1 would lead to the production of ideal plant types. Therefore, LPA1 should serve as an important genetic tool that can contribute to breeding elite crop varieties. Indeed, we previously demonstrated the agricultural utility of LPA1/OsMPT1 (Han and Choe, 2010).
Supplementary data
Supplementary data is available at JXB online.
Appendix S1. Cloning, vector construction and transformation.
Appendix S2. RNA-seq analysis.
Figure S1. IAA-mediated lamina inclination and the effect of brassinazole on lpa1 mutants.
Figure S2. IAA-mediated lamina inclination and the effect of brassinazole on lpa1;d2 and lpa1;bri1-D.
Figure S3. Sensitivity of lpa1 and lpa1;d61-1 plants to C-22-hydroxylated and 6-deoxo brassinosteroids (BRs) and the combinatory effects with IAA on lpa1 mutants.
Figure S4. IAA-mediated lamina inclination of lpa1;d61-1 and e19;bri1-D.
Figure S5. Relationship between LPA1 and OsBRI1 in determining lamina inclination.
Figure S6. Effect of NPA on IAA-mediated lamina inclination in lpa1, overexpressor (e19) and WT.
Figure S7. Brassinosteroids (BRs) and brassinazole (Brz) utilized in this study and steps catalysed by D2 are shown in a BR biosynthetic pathway (adapted from Nakamura et al., 2009).
Table S1. Footprints after Ds excisions.
Table S2. Total number of reads mapped per sample.
Table S3. List of genes whose expression was 1.5-fold higher or lower in e19 or lpa1 than in the WT.
Table S4. List of auxin-related genes identified by RNA-seq.
Table S5. Auxin sensitivity of mutants for lamina inclination.
Table S6. Relative sensitivity of lpa1 and lpa1;d61-1 to BR compounds for lamina inclination.
Acknowledgements
This work was supported by grants from the Next-Generation BioGreen 21 Program (PJ01114601 and PJ01103901), the Rural Development Administration, Republic of Korea. JML, VK, and RAP are supported by scholarships from the BK21 program.
References
- Asami T, Mizutani M, Fujioka S, et al. 2001. Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. The Journal of Biological Chemistry 276, 25687–25691. [DOI] [PubMed] [Google Scholar]
- Chen YN, Fan XR, Song WJ, Zhang YL, Xu GH. 2012. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnology Journal 10, 139–149. [DOI] [PubMed] [Google Scholar]
- Chin HG, Choe MS, Lee SH, et al. 1999. Molecular analysis of rice plants harboring an Ac/Ds transposable element mediated gene trapping system. Plant Journal 19, 615–623. [DOI] [PubMed] [Google Scholar]
- Colasanti J, Tremblay R, Wong AYW, Coneva V, Kozaki A, Mable BK. 2006. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui DY, Zhao JB, Jing YJ, et al. 2013. The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genetics 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CD, Choe YD. 2010. OsMPT1 gene for modifying plant architecture and increasing yield, and uses thereof. Korean patent , no. 10-1144624. [Google Scholar]
- Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M. 2004. Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochemical Biophysical Research Communications 321, 172–178. [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, et al. 2002. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant Journal 32, 495–508. [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, et al. 2003. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. The Plant Cell 15, 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je BI, Piao HL, Park SJ, et al. 2010. RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. The Plant Cell 22, 1777–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G. 2002. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiology 130, 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR. 2007. New insights into the history of rice domestication. Trends in Genetics 23, 578–587. [DOI] [PubMed] [Google Scholar]
- Maeda E. 1965. Rate of lamina inclination in excised rice leaves. Physiologia Plantarum 18, 813–827. [Google Scholar]
- Mandava NB. 1988. Plant growth-promoting brassinosteroids. Annual Review of Plant Physiology and Plant Molecular Biology 39, 23–52. [Google Scholar]
- Miyashita Y, Takasugi T, Ito Y. 2010. Identification and expression analysis of PIN genes in rice. Plant Science 178, 424–428. [Google Scholar]
- Morita MT, Sakaguchi K, Kiyose S, Taira K, Kato T, Nakamura M, Tasaka M. 2006. A C2H2-type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. Plant Journal 47, 619–628. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Fujioka S, Takatsuto S, et al. 2009. Involvement of C-22-hydroxylated brassinosteroids in auxin-induced lamina joint bending in rice. Plant Cell Physiology 50, 1627–1635. [DOI] [PubMed] [Google Scholar]
- Ning J, Zhang B, Wang N, Zhou Y, Xiong L. 2011. Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the lamina joint of rice. The Plant Cell 23, 4334–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Szatmari AM, Watanabe B, et al. 2006. C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. The Plant Cell 18, 3275–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnish T, et al. 2006. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nature Biotechnology 24, 105–109. [DOI] [PubMed] [Google Scholar]
- Takeno K, Pharis RP. 1982. Brassinosteroid-induced bending of the leaf lamina of dwarf rice seedlings: an auxin-mediated phenomenon. Plant Cell Physiology 23, 1275–1281. [Google Scholar]
- Wada K, Marumo S, Ikekawa N, Morisaki M, Mori K. 1981. Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiology 22, 323–325. [Google Scholar]
- Wang L, Xu YY, Zhang C, et al. 2008. OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 3, e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XR, Tang D, Li M, Wang KJ, Cheng ZK. 2013. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiology 161, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xu YY, Guo SY, et al. 2012. Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genetics 8, e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, et al. 2009. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21, 3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SQ, Hu J, Guo LB, Qian Q, Xue HW. 2010. Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Research 20, 935–947. [DOI] [PubMed] [Google Scholar]
- Zhao SQ, Xiang JJ, Xue HW. 2012. Studies on the rice LEAF INCLINATION1 (LC1), an IAA–amido synthetase, reveal the effects of auxin in leaf inclination control. Molecular Plant 6, 174–187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.