Summary
Cellular lipid requirements are achieved through a combination of biosynthesis and import programs. Using isotope tracer analysis, we show that type I interferon (IFN) signaling shifts the balance of these programs by decreasing synthesis and increasing import of cholesterol and long chain fatty acids. Genetically enforcing this metabolic shift in macrophages is sufficient to render mice resistant to viral challenge, demonstrating the importance of reprogramming the balance of these two metabolic pathways in vivo. Unexpectedly, mechanistic studies reveal that limiting flux through the cholesterol biosynthetic pathway spontaneously engages a type I IFN response in a STING-dependent manner. The upregulation of type I IFNs was traced to a decrease in the pool size of synthesized cholesterol, and could be inhibited by replenishing cells with free cholesterol. Taken together, these studies delineate a metabolic-inflammatory circuit that links perturbations in cholesterol biosynthesis with activation of innate immunity.
Introduction
Evidence indicates an intimate relationship exists between host lipid metabolism and intracellular pathogens (Baek et al., 2011; Bard et al., 2005; Coppens, 2013; Griffin et al., 2012). Perturbations in host lipid homeostasis are observed in viral and microbial infections (Cui et al., 2014; Greseth and Traktman, 2014; Herker and Ott, 2011). While it remains mechanistically unclear as to how each invading pathogen subverts host lipid metabolism, it has been proposed that co-opting of host metabolism is a general strategy employed by pathogens to meet the anabolic requirements of pathogen lifecycle, and facilitate evasion from host defense (Cui et al., 2014; Greseth and Traktman, 2014; Heaton et al., 2010; Herker and Ott, 2011; Kapadia and Chisari, 2005; Ouellet et al., 2011). Consistent with this concept, genetic or pharmacologic inhibition of host lipid metabolism has been shown to attenuate pathogenesis of both viral and microbial infections in a number of model systems (Gilbert et al., 2005; Herker and Ott, 2011; Liu et al., 2008; Munger et al., 2008; Parihar et al., 2014; Petersen et al., 2014; Rzouq et al., 2014).
Cumulatively, these studies suggest that co-opting host lipid metabolism facilitates microbial or viral pathogenesis, and leads to the hypothesis that host defense pathways should attempt to overwrite the metabolic changes induced by invading pathogens. In support of this concept, recent studies have demonstrated that components of host responses to pathogens (e.g., TLR3/4 and type I interferon (IFN) signaling) specifically rewire components of the lipid metabolic program by downregulating de novo cholesterol biosynthesis at the genetic level (Blanc et al., 2013; Blanc et al., 2011; Liu et al., 2013; Reboldi et al., 2014). Thus, it has been suggested that the influence of type I IFN signaling on the cholesterol homeostasis would serve to limit the availability of lipid metabolites for intracellular organisms. In contrast, other studies have shown that these same inflammatory signals (e.g., TLR3/4 and type I IFNs) increase lipid uptake from environmental sources, resulting in the accumulation of neutral lipids and ultimately facilitating foam cell formation (Dushkin and Kovshik, 2013; Funk et al., 1993; Huang et al., 2014; Keyel et al., 2012). Thus, it remains unclear if the purpose of type I IFN-mediated metabolic reprogramming is to specifically limit the availability of lipid metabolites (e.g., cholesterol) for pathogens as been proposed, or if there are alternative reasons for the selective reprogramming of flux through the cholesterol biosynthetic pathway.
Herein, we examine this question and delineate a lipid metabolic-inflammatory circuit that links the induction of type I IFN-mediated inflammation with perturbations in the pool size of synthesized cholesterol. Using stable isotope enrichment analysis, we demonstrate that type I IFN signaling shifts the balance of lipid metabolism away from de novo synthesis to favor lipid import, without limiting total long chain fatty acid and cholesterol content in macrophages. Strikingly, we find that genetically enforcing this shift in lipid homeostasis in macrophages alone is sufficient to protect mice from viral challenge, demonstrating the importance of reprogramming the “set point” between synthesis and import in host defense. Unexpectedly, we find that limiting flux through the cholesterol biosynthetic pathway spontaneously induces a type I IFN response that primes cells for heightened anti-viral immunity in both an autocrine and paracrine manner. Mechanistic studies indicate that the STING (TMEM173) signaling axis is required to link the induction of a type I IFN response with alterations in flux through the mevalonate pathway. Replenishing free cholesterol was able to normalize type I IFN levels by limiting STING signaling in cells lacking enzymes of the mevalonate pathway, establishing the importance of cholesterol homeostasis in type I-mediated inflammation.
Results
Reprogramming the balance of lipid synthesis and import in macrophages
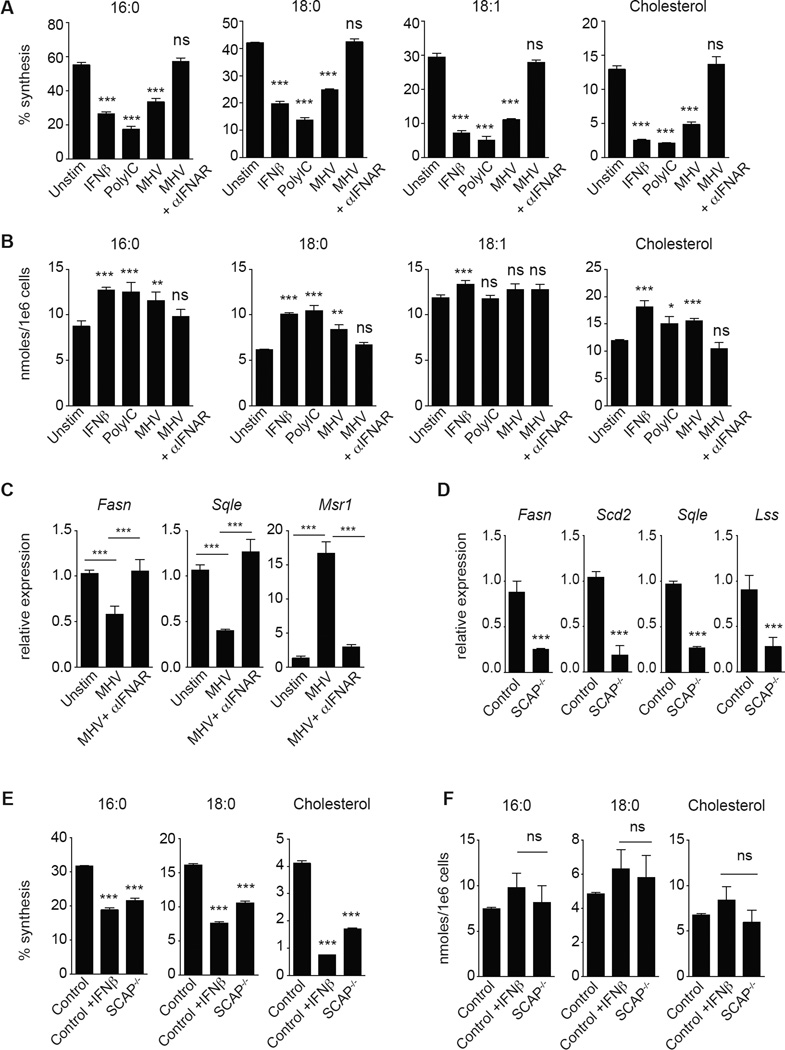
To better understand how type I IFN signaling modulates the balance of lipid synthesis and import, we employed gas chromatography-mass spectrometry (GC-MS) paired with 13C-stable isotope enrichment studies to measure total and synthesized cholesterol and long chain fatty acid levels in mouse bone marrow-derived macrophages (BMDMs) stimulated with IFNβ, Poly:IC (TLR3 agonist) or infected with murine gammaherpesvirus-68 (MHV-68). Isotopic Spectral Analysis (ISA) (Williams et al., 2013) was applied to the mass isotopomer distributions to derive the contribution of de novo synthesis to the total cholesterol and long chain fatty acid pools over the labeling period. Treatment of BMDMs with either IFNβ or Poly:IC significantly decreased the synthesis of saturated long chain fatty acids (16:0, 18:0), unsaturated long chain fatty acids (18:1) and cholesterol (Figure 1A, S1A). However, total fatty acid (16:0, 18:0, 18:1 and 18:2) and cholesterol content increased on a per cell basis (Figure 1B, S1B), suggesting that these cells increase lipid import. Consistent with our MS studies, we observed a significant increase in the uptake of amount of dil-acetylated LDL in response to Poly:IC or IFNβ stimulation in BMDMs (Figure S1C). Importantly, infection of BMDMs with MHV-68 drove a similar shift in the balance of lipid synthesis and import in an IFNAR-dependent manner, indicating that type I IFNs mediate lipid metabolic reprogramming (Figure 1A, 1B, S1A, S1B). We also considered the possibility that the availability of environmental lipids would influence IFN-mediated reprogramming. To address this, we cultured BMDMs with increasing percentages of serum (5, 10 and 20%) before stimulation with IFNβ. ISA modeling indicated that type I IFN treatment consistently decreased de novo synthesis of cholesterol and increased import to nearly the same extent in all serum conditions (Figure S1D).
Figure 1. Type I interferon signaling shifts the balance of lipid synthesis and import.
(A) Percent synthesis of palmitic acid (16:0), stearic acid (18:0), oleic acid (18:1) and cholesterol as measured by Isotopic Spectral Analysis (ISA) on C75BL/6 BMDMs with or without interferonβ (1000U/mL IFNβ), Poly:IC (2ug/mL) or MHV-68 (MOI=0.5) +/− 10ug/mL IFNAR neutralizing antibody for 48h. (B) Total cellular palmitic acid (16:0), stearic acid (18:0), oleic acid (18:1) and cholesterol from BMDMs stimulated as in (A). (C) qPCR analysis of Fasn, Sqle, and Msr1 gene expression in BMDMs infected with MHV-68 (MOI=0.5) +/− 10ug/mL IFNAR neutralizing antibody for 48h. (D) qPCR analysis of indicated lipid synthesis genes in quiescent LysM Cre+/− control (designated Control) or LysM Cre+/− SCAPfl/fl (SCAP−/−). (E) Percent synthesis of palmitic acid (16:0), stearic acid (18:0) and cholesterol as measured by ISA of Control or SCAP−/− BMDMs unstimulated or stimulated with IFNβ as above for 24h. (F) Total cellular palmitic acid (16:0), stearic acid (18:0) and cholesterol from Control or SCAP−/− BMDMs unstimulated or stimulated with IFNβ as above for 24h. All mass spec and gene expression experiments are expressed as means ± SD from three independent experiments. *P < 0.05; **P < 0.01, ***P < 0.005 (two-tailed unpaired Student’s t test). See also Figures S1 and S2.
Next, we examined the expression pattern of genes involved in cholesterol and fatty acid synthesis and import in MHV-infected BMDMs. We observed that infection decreased expression of genes encoding enzymes of the de novo cholesterol and fatty acid biosynthetic pathways (Figure 1C), similar to previous studies on cholesterol synthesis using cytomegalovirus (Blanc et al., 2013). MHV infection also significantly increased expression of genes involved in lipid import including Macrophage Scavenger Receptor (Msr1) and the fatty acid translocase Cd36 (Figure 1C and S1E). Stimulation of BMDMs with IFNβ or Poly:IC for 24h revealed a similar reprogramming at the gene and protein expression level (Figure. S1F, S1G). Importantly, blocking IFNAR signaling abrogated reprogramming of lipid metabolism driven by either MHV-68 infection or type I IFN stimulation (Figure 1C and S1G). Thus, we conclude that type I IFN signals reprogram the balance of lipid synthesis and import within a macrophage, but do not appear to specifically limit cholesterol and long chain fatty acid availability.
Loss of SREBP activity shifts the balance of lipid synthesis and import
The results of our isotope labeling studies led us to ask if this shift in the balance of lipid homeostasis driven by type I IFN is an important component of host defense. To directly address this would require a genetic model where the “set point” controlling the balance of lipid synthesis and scavenging was shifted independent of TLR and IFNAR signaling. To accomplish this, we generated mice with macrophage-specific deletion of the SREBP cleavage-activating protein (SCAP) using the LysM-Cre model. SCAP is an endoplasmic reticulum (ER) sterol-sensing protein required to chaperone the sterol regulatory element binding proteins (SREBP1 and SREBP2) (Horton et al., 2002). In the absence of SCAP protein, SREBP1 and 2 transcriptional activities are significantly attenuated, resulting in markedly reduced expression of cholesterol and fatty acid biosynthesis genes (see Figure S2A for schematic) (Kidani et al., 2013).
As expected, gene expression analysis of BMDMs confirmed that genetic deletion of SCAP markedly decreases expression of the genes encoding enzymes of the de novo cholesterol and fatty acid biosynthetic programs (Figure 1D). However, loss-of-SCAP does not appear to influence the in vitro differentiation of BMDMs (Figure S2B), nor impact the frequency of macrophages or other immune cell populations in LN, spleen and bone marrow (Figure S2C). Next, we performed 13C-isotope enrichment and ISA to determine if loss-of-SCAP altered lipid biosynthetic capacity. We observed that, the contribution of synthesis to total lipid pools in SCAP−/− BMDMs was decreased compared to controls (Figure 1E, S2D). Total cholesterol and fatty acid pool sizes were similar between IFNβ-treated control and SCAP−/− BMDMs (Figure 1F), demonstrating that loss-of-SCAP does not significantly decrease the total amounts of these lipids in BMDMs. Taken together, these data demonstrate that SCAP-deficiency phenocopies the shift in lipid homeostasis observed in response to IFNβ stimulation.
Loss of SCAP protects from viral inflection
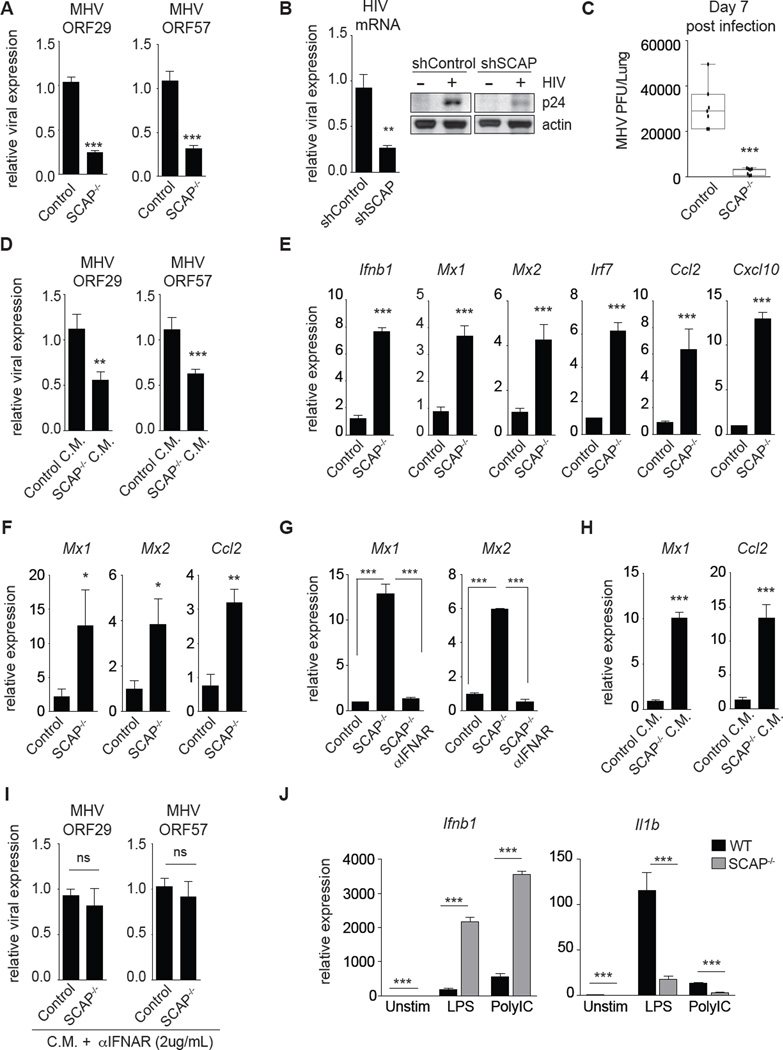
Next, we sought to determine if genetically enforcing this metabolic shift in macrophages influenced host defense. To begin investigating this, control and SCAP−/− BMDMs were infected with MHV-68. Examination of viral gene expression revealed that SCAP−/− BMDMs contained significantly lower viral transcripts (Figure 2A), indicative of lower viral burden. Similarly, stable knockdown of SCAP in human THP1 macrophages rendered cells resistance to HIV-1, decreasing HIV-1 RNA and p24 protein expression by approximately 70% (Figure 2B). To determine if macrophage-specific perturbations in the lipid biosynthetic program could influence viral pathogenesis in vivo, LysM-Cre+/− and LysM-Cre+/−/Scapfl/fl mice were challenged intranasally with MHV-68. Similar to our in vitro results, we found that the loss-of-SCAP in macrophages alone was sufficient to reduce viral load in lungs by over 90% (Figure 2C). Taken together, our data indicates that genetic deletion of SCAP can be protective in viral inflections, and supports the concept that lipid metabolic reprogramming can be a protective component of host response to viral infections.
Figure 2. Genetic inhibition of the lipid biosynthetic program primes antiviral immunity.
(A) qPCR analysis of murine gammaherpesvirus-68 (MHV-68) genes in Control or SCAP−/− BMDMs infected with MHV-68 (MOI=0.5) for 48h. (B) qPCR analysis of HIV-1 mRNA (left) and representative immunoblots of HIV-1 p24 protein levels (right) from shControl or shSCAP THP1 macrophages 96h after infection. (C) MHV-68 titers from the lungs of LysM Cre+/− control (Control) or LysM Cre+/− SCAPfl/fl (SCAP−/−) mice on d.7 post intranasal infection (N=7; data shown is the combined results from two separate infection experiments of n=3 per group and n=4 per group). (D) qPCR analysis of indicated MHV-68 genes in WT BMDMs pretreated for 4h with Control or SCAP−/− conditioned media (C.M.) before MHV-68 infection (MOI=0.5) for 48h. C.M. from Control or SCAP−/− BMDMs was collected on d.7 post-differentiation. (E) qPCR analysis of Ifnb1 and representative interferon stimulated genes (ISGs) in unstimulated control or SCAP−/− BMDMs on d.8 of differentiation. (F) qPCR analysis of ISGs in ex vivo alveolar macrophage isolated from bronchoalveolar lavage from LysM Cre+/− control (Control) or LysM Cre+/− SCAPfl/fl (SCAP−/−) (3 mice/group). (G) qPCR analysis of indicated ISGs in unstimulated control or SCAP−/− BMDMs on d.9 of differentiation +/− 5ug/mL IFNAR blocking antibody for last 48h. (H) qPCR analysis of ISGs in WT BMDMs treated for 4h with Control or SCAP−/− conditioned media (C.M.). (I) MHV-68 ORF29 and ORF57 gene expression in WT BMDMs pretreated for 4h with Control or SCAP−/− conditioned media (CM) + 2ug/mL IFNAR blocking antibody before MHV-68 infection (MOI=0.5) for 48h. (J) qPCR analysis of Ifnb1 and Il1b expression in Control or SCAP−/− BMDMs unstimulated or stimulated with LPS (50ng/mL) or Poly:IC (1ug/mL) for 1h on d.8 of differentiation. All experiments are reported as means ± SD from three independent experiments, unless noted otherwise. *P < 0.05; **P < 0.01, ***P < 0.005 (two-tailed unpaired Student’s t test). See also Figure S3.
Loss of SCAP primes a type I IFN response
MHV replicates in a number of cell types including macrophages, type 1 and type 2 pneumocytes (Hughes et al., 2010), leading us to hypothesize that viral resistance conferred by loss-of-SCAP in macrophages may be transferable to neighboring cells. To directly test this, conditioned media (C.M.) from naïve control or SCAP−/− macrophages was collected and transferred to wildtype (C57BL/6) BMDM cultures before challenge with MHV-68 (see Figure S3A for schematic). Remarkably, transfer of SCAP−/− C.M. to WT BMDM cultures was able to reduce MHV-68 viral burden by approximately 50% (Figure 2D). These results indicate that the viral resistance phenotype observed in SCAP-deficient macrophages occurs, at least in part, through the production of secreted factors that then prime cells for anti-viral immunity.
Type I IFNs are secreted proteins that are important anti-viral immunity, leading us to ask if genetic deletion of SCAP influenced type I IFN responses. Gene expression studies on unstimulated BMDMs revealed that loss-of-SCAP results in the spontaneous induction of a type I IFN-inflammatory response characterized by heightened Ifnb1 and interferon-stimulated genes (ISGs, e.g., Mx1, Mx2, Irf7, Ccl2, Cxcl10) (Figure 2E). Heightened ISG expression was also observed in alveolar macrophages from LysM-Cre+/−/Scapfl/fl (Figure 2F, S3b), indicating that loss-of-SCAP drives a type I IFN signature in vivo. Addition of IFNAR blocking antibody to BMDM cultures was able to abrogate ISG expression (Figure 2G), suggesting that SCAP−/− macrophage spontaneously produce more type I IFNs. In support of this, transfer of culture supernatants from unstimulated SCAP−/− BMDMs to quiescent WT macrophages was able to induce an ISG signature (Figure 2H). We also observed the upregulation of a type I IFN signature in human THP1 macrophages in response to either stable or transient silencing of SCAP (Figure S3C, S3D). Importantly, addition of IFNAR blocking antibody abrogated antiviral capacity of C.M. from SCAP−/− BMDMs (Figure 2I), supporting the concept that the anti-viral effect of SCAP-deficiency was, in large part, mediated through production of type I IFNs.
Next, we assessed the ability of SCAP-deficient macrophage to respond to TLR3/4 and IFNAR signaling. We observed that both human and mouse SCAP-deficient macrophages have significantly heightened induction of Ifnb1 and ISGs in response to both LPS and Poly:IC, as well as type I IFN treatment (Figure 2J left, S3E–G). Interestingly, not all inflammatory responses were upregulated in SCAP-deficient macrophages. We found that induction of Il1b, a canonical pro-inflammatory factor downstream of TLR4 activation, was significantly attenuated in response to LPS (Figure 2J, right). IFNAR blockade was able to partially restore upregulation of Il1b expression in TLR4 stimulated SCAP-deficient macrophages (Figure S3H), indicating that type IFN signaling contributes, in part, to repression of Il1b in LPS stimulated cells. Taken together, these data indicate that reprogramming of lipid metabolism can prime macrophages for heightened IFN responses to PAMPs or cytokine signals, however this type of metabolic reprogramming also entrains specific inflammatory responses at the expense of other inflammatory programs.
Induction of a type I IFN signature segregates with loss of SREBP2
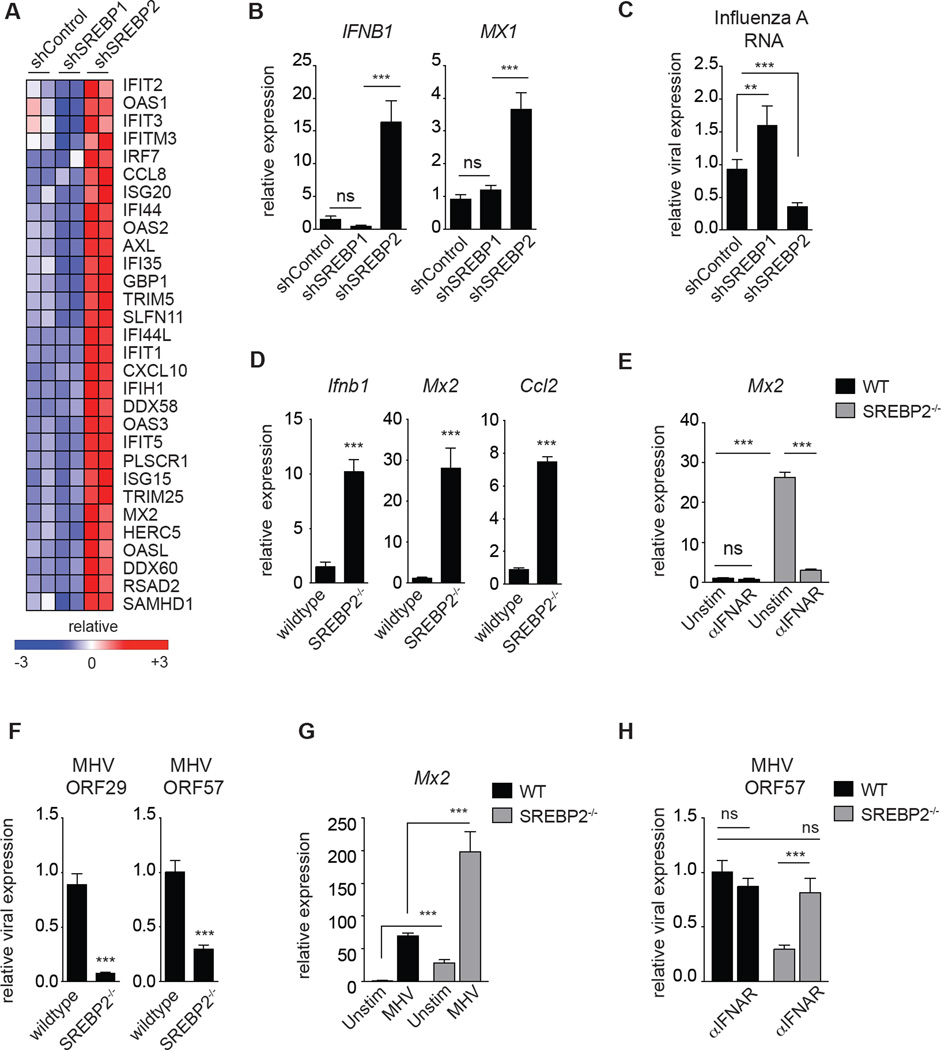
SCAP is required for activation of all isoforms of the SREBP transcription factors. In mammals there are two SREBP genes that express three SREBP proteins. SREBP1a and SREBP1c are produced via alternative transcriptional start sites on SREBF1, whereas the SREBF2 gene encodes SREBP2 (Horton et al., 2002). In general, SREBP1 activates transcription of genes involved in fatty acid synthesis, while SREBP2 is responsible for transcriptionally activating genes required for cholesterol synthesis (schematic Figure S2A). To determine if the ISG signature observed in SCAP-deficient macrophages was dependent on loss of the SREBP1 or SREBP2 transcriptional axis, we generated THP1 macrophages stably expressing shRNA targeting either SREBF1 or SREBF2 (designated herein as shSREBP1 or shSREBP2, respectively) and performed RNA-sequencing studies on unstimulated macrophages. As expected, knockdown of SREBP1 attenuated expression of genes involved in fatty acid synthesis, whereas loss of SREBP2 decreased expression of cholesterol biosynthesis genes (Figure S4A, S4B). Bioinformatic analysis of RNA-seq data indicted that heightened expression of ISGs, and enrichment of pathways involved in host defense to viral infection, segregated entirely with the loss of SREBP2 expression (Figure 3A and S4C). qPCR analysis confirmed that silencing of SREBP2 in primary human PBMC-derived macrophages or THP1 cells resulted in heightened basal expression of and an ISG signature (Figure 3B, S4D, S4E). In line with this pro-inflammatory phenotype, SREBP2-deficient macrophages were significantly resistant to Influenza A and HIV-1 challenge (Figure 3C, S4F). In contrast, a type I IFN signature was not observed in shSREBP1 macrophages (Figure 3A, 3B and S4E), indicating that the spontaneous induction of type I interferon-mediated inflammation occurs specifically in response to genetically perturbing the SCAP/SREBP2 pathway.
Figure 3. Inhibiting the SREBP2 transcriptional pathway engages a type I IFN inflammatory response.
(A) RNA-seq data from unstimulated shControl, shSREBP1 and shSREBP2 THP1 cells 72 h after PMA-differentiation into macrophages. Data shown are biologic replicates of each genotype. (B) qPCR analysis of IFNB1 and MX1 in unstimulated shControl, shSREBP1 and shSREBP2 primary PBMC-derived human macrophages. (C) qPCR analysis of Influenza A RNA from shControl, shSREBP1 or shSREBP2 THP1 cells 72h post infection. (D) qPCR analysis of Ifnb1 and indicated ISGs expression in WT control or SREBP2−/− MEFs cultured in DMEM containing 1% FBS for 24h. (E) qPCR analysis of Mx2 expression in WT or SREBP2−/− MEFs cultured in 1% FBS for 24h +/− 5ug/mL IFNAR blocking antibody as indicated (F) qPCR analysis of indicated MHV-68 gene expression in WT or SREBP2−/− MEFs infected with MHV-68 (MOI=1.0) for 24h. (G) qPCR analysis of Mx2 in WT or SREBP2−/− MEFs +/− MHV infection (MOI=1.0) for 24h. (H) qPCR analysis of MHV-68 ORF57 expression in WT or SREBP2−/− MEFs infected with MHV-68 (MOI=1.0) for 24h +/− 24h pretreatment with 5ug/mL IFNAR blocking antibody. All experiments are reported as means ± SD from three independent experiments. *P < 0.05; **P < 0.01, ***P < 0.005 (two-tailed unpaired Student’s t test). See also Figure S4.
Nearly every cell in the body has the ability to produce type I IFNs, thus we sought to determine if genetic deletion of SREBP2 in non-immune cells would also result in the spontaneous induction of a type I IFN and resistance to viral challenge. To that end, we examined the basal expression levels of ISGs from SREBP2-null mouse embryonic fibroblasts (designated SREBP2−/− MEFs). Similar to our studies in macrophages, we observed that SREBP2−/− MEFs constitutively expressed heightened levels of Ifnb1 and ISGs (Figure 3D), and IFNAR blockade significantly reduced ISG expression to that of control MEFs (Figure 3E). Correspondingly, SREBP2−/− MEFs were significantly resistance to MHV-68 challenge (Figure 3F), and produced heightened ISGs in response to MHV infection (Figure 3G). Importantly, pretreatment with IFNAR-neutralizing antibodies was sufficient to abrogate the heightened anti-viral immunity observed in SREBP2−/− MEFs (Figure 3H), indicating that the anti-viral immunity seen in SREBP2-deficient cells is largely dependent on IFNAR signaling.
Limiting flux through the mevalonate pathway engages a type I IFN response
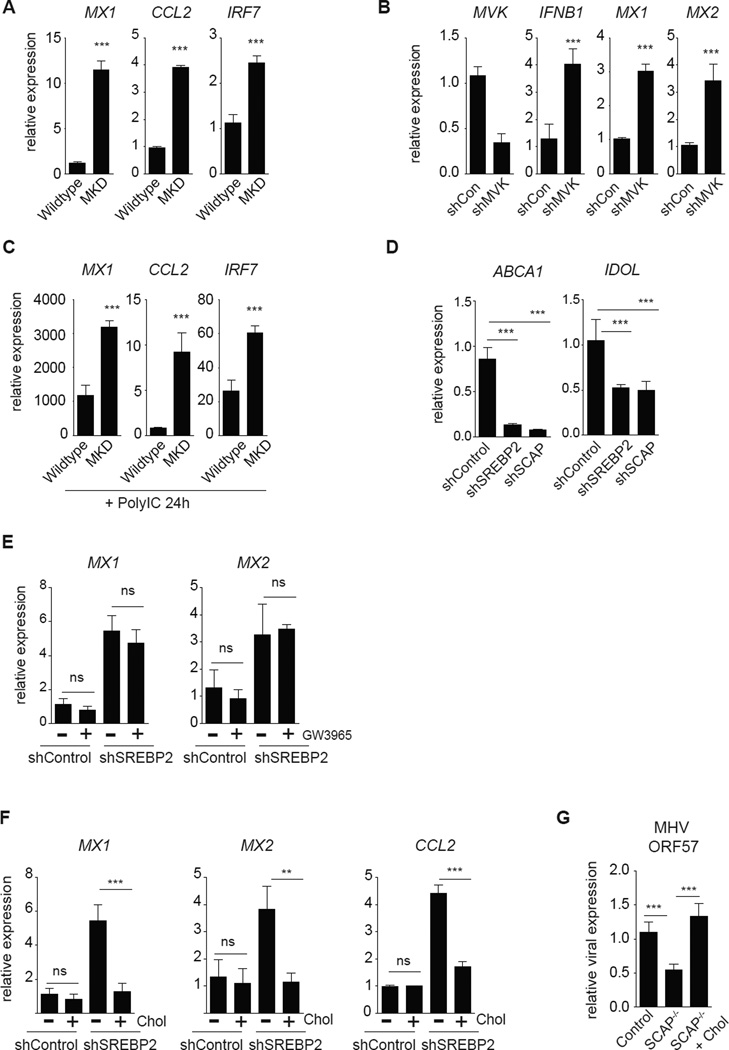
Mevalonate Kinase Deficiency (MKD) is an autosomal recessive disease caused by significantly decreased enzymatic activity of mevalonate kinase (MVK) (Esposito et al., 2014; Santos et al., 2014). Individuals with moderate residual enzymatic activity (~20% of wildtype) are characterized by recurrent episodic fevers, persistent inflammatory responses and hyper IgD production (termed Hyperimmunoglobulinemia D with Periodic Fever Syndrome or HIDS). SREBP2 has a well-defined role in transcriptionally regulating the enzymes involved in the mevalonate pathway (Horton et al., 2002), leading us to ask if MKD results in the spontaneous generation of type I IFN-mediated inflammation. Examination of primary fibroblasts from an individual with MKD revealed heightened basal ISG expression when compared to population controls (Figure 4A). Genetic silencing of MVK or HMGCR (HMG-CoA reductase), the rate-limiting enzyme in cholesterol biosynthesis, in macrophages also resulted in the upregulation of IFNB1 and ISGs (Figure 4B, S5A, S5B). In contrast, silencing of stearoyl-CoA desaturase 1 (SCD1), a key enzyme involved in flux through the long-chain fatty acid biosynthetic pathway (Williams et al., 2013), did not induce ISGs (Figure S5A, S5C). Importantly, we also find that MKD fibroblasts are primed for exaggerated type I IFN-mediated inflammatory responses. Activation of MKD fibroblasts with Poly:IC resulted in a significantly heightened ISG signature (Figure 4C). Taken together, these data indicate that decreasing flux through the mevalonate pathway results in the upregulation of a type I IFN response, and provides a potential mechanistic explanation for the inflammatory symptoms and immune dysregulation observed in individuals with MKD.
Figure 4. Limiting flux throught the cholesterol biosynthetic pathway engages a type I IFN response.
(A) qPCR analysis of ISG expression in population control (Wildtype) primary human fibroblasts or fibroblasts from an individual with MVK-deficiency (MKD; ~2% MVK activity compared to population controls). (B) qPCR analysis of MVK, IFNB1 and ISG expression in shControl or shMVK THP1 cells after 72h of PMA-differentiation (C) qPCR analysis of ISG expression in population control (Wildtype) primary human fibroblasts or fibroblasts from an individual with MVK-deficiency (MKD) primed with 2ug/mL Poly:IC for 24h. (D) qPCR analysis of ABCA1 and IDOL expression in shControl, shSREBP2 and shSCAP THP1 macrophages differentiated for 72h. (E) qPCR analysis of indicated ISG expression in THP1 shControl or shSREBP2 cells differentiated as above and stimulated with vehicle (DMSO) or LXR ligand (1uM GW3965) for 72h. (F) qPCR analysis of indicated ISG expression of differentiated THP1 shControl or shSREBP2 macrophages in media supplemented with 0.1mg/mL methyl beta cyclodextrin (MβCD)-cholesterol (“Chol”) as indicated for 72h. G. qPCR analysis of MHV-68 ORF57 expression in Control or SCAP−/− BMDMs +/− 0.25mg/mL MβCD-cholesterol (Chol) for 48h, then infected with MHV-68 (MOI=0.5) for 24h. All experiments are reported as means ± SD from three independent experiments. *P < 0.05; **P < 0.01, ***P < 0.005 (two-tailed unpaired Student’s t test). See also Figure S5.
Intermediary metabolites of cholesterol biosynthesis have been shown to activate the Liver X Receptors (LXRα and β), nuclear receptors with a clear role in repressing lipid-driven metabolic inflammation (Glass and Witztum, 2001; Shibata and Glass, 2009; Spann et al., 2012; Yang et al., 2006). Consistent with this, we found that LXR target genes ABCA1 and IDOL were significantly decreased in both SREBP2- and SCAP-deficient macrophages (Figure 4D). Thus, we considered the possibility that the upregulation of IFNB1 and downstream ISGs resulted from decreased LXR signaling. However, treatment of SREBP2-deficient macrophages with LXR ligand GW3965, which restored LXR function (Figure S5D), failed to reduce expression of ISGs (Figure 4E). These findings indicate that LXRs do not repress type I IFN-mediated inflammation, and are in agreement with previous work (Castrillo et al., 2003).
Replenishing free cholesterol attenuates the type IFN-signature in SREBP2-null cells
In combination, our data demonstrate that decreasing flux through the mevalonate pathway induces type I IFN-mediated inflammation, leading us to ask if replenishing cholesterol could attenuate this inflammatory program. To this end, control and SREBP2-deficient macrophage cultures were treated with methyl-beta-cyclodextrin conjugated cholesterol (MβCD-cholesterol) to supplement free cholesterol levels in loss-of-function cells. Supplementing SREBP2-deficient macrophages with MβCD-cholesterol significantly reduced the expression of ISGs (Figure 4F). Thus, we conclude that restoring cholesterol homeostasis alone is sufficient to abrogate the induction of a type I IFN signature in cells that have the mevalonate pathway perturbed. Consistent with this, replenishing cholesterol in SCAP−/− BMDMs abrogated the anti-viral phenotype (Figure 4G). In combination, these data suggest that acutely decreasing synthesized cholesterol appears to provide a novel “danger” signal that activates a type I IFN-mediated anti-viral response.
IRF3 links perturbations in cholesterol homeostasis to Ifnb1 transcription
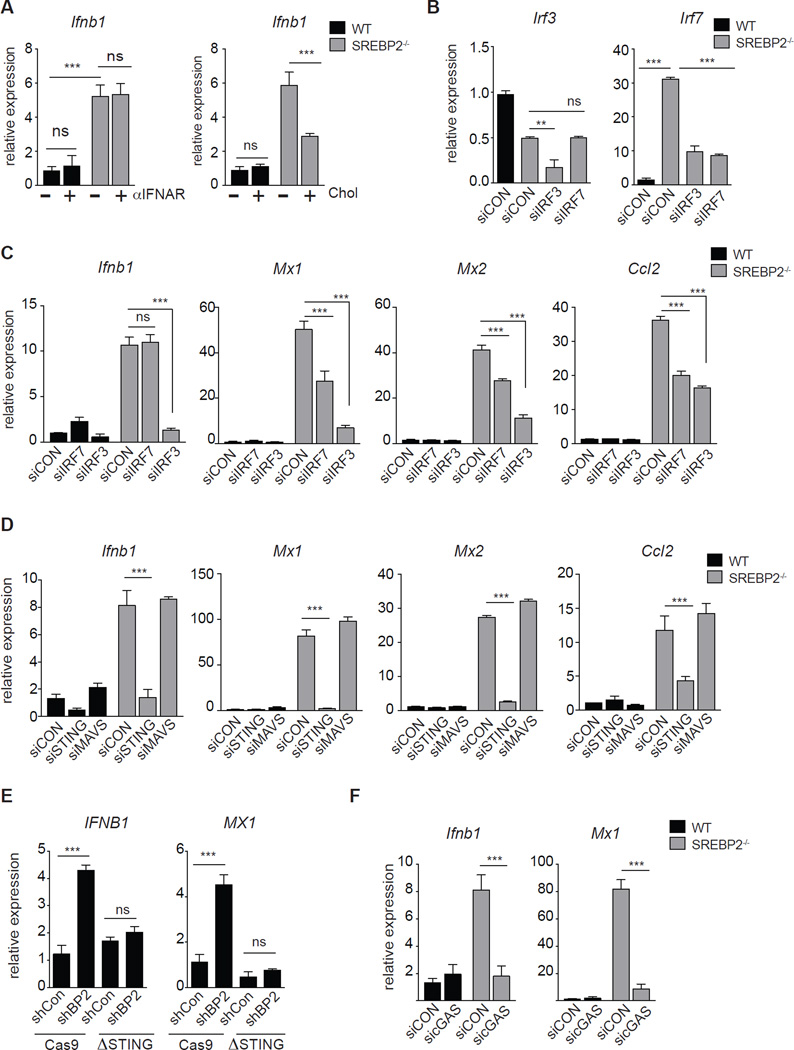
Finally, we sought to define the molecular mechanism linking perturbations in cholesterol biosynthesis with the spontaneous induction of type I interferon-mediated inflammation. We had observed that IFNAR blockade reduced ISG expression (Figure 2G, 3E), but was not able to decrease Ifnb1 gene expression in both SCAP- and SREBP2-deficient cells (Figure 5A left, S6A). However, addition of MβCD-cholesterol was able to reduce the expression level of Ifnb1 (Figure 5A right, S6B), supporting a model where induction of inflammation in SREBP2- or SCAP-deficient cells is dependent on transcriptional upregulation of Ifnb1. Interferon Regulatory Factor 3 and 7 (IRF3 and IRF7) are transcription factors with well-defined roles in the transcriptional regulation of Ifnb1 (Fitzgerald et al., 2003; Honda et al., 2005). We observed that SREBP2-deficeint cells had significantly elevated expression of IRF7 (Figure 2E, 3A, 5B), leading us to ask if IRF7 was important in mediating inflammation. To address this, we transiently silenced Irf3 or Irf7 in SREBP2−/− MEFs (Figure 5B). We observed that silencing of Irf7 was able to partially reduce ISG expression (Figure 5C), but had no influence on Ifnb1 expression (Figure 5C, left). In contrast, silencing of Irf3 completely abrogated Ifnb1 expression and correspondingly attenuated ISG expression (Figure 5C). Taken together, these data indicate that IRF3 is required to drive the spontaneous upregulation of Ifnb1 and ISGs observed in SREBP2-deficeint cells, and that IRF7 serves to amplify the ISG program.
Figure 5. IRF3/STING link changes in cholesterol biosynthesis with type I IFN production.
(A) Left: qPCR analysis of Ifnb1 in WT or SREBP2−/− MEFs cultured in 1% FBS for 24h +/− 5ug/mL IFNAR blocking antibody as indicated. Right: qPCR analysis of Ifnb1 in WT and SREBP2−/− MEFs treated 0.075mg/mL MβCD-cholesterol (Chol) as indicated for 72h. (B) qPCR analysis of Irf3 and Irf7 in WT or SREBP2−/− MEFs transfected with control siRNA, siIRF3 or siIRF7 (C) qPCR analysis of Ifnb1, Mx1, Mx2 and Ccl2 in WT or SREBP2−/− MEFs transfected with control siRNA, siIRF3 or siIRF7 (D) qPCR analysis of Ifnb1, Mx1, Mx2 and Ccl2 in WT or SREBP2−/− MEFs transfected with control siRNA, siMAVS or siSTING as indicated (E) qPCR of IFNB1 and MX1 gene expression from control (Cas9) or CRISPR/Cas9-edited STING (ΔSTING) THP1 cells stably transduced with shControl or shSREBP2. (F) qPCR analysis of Ifnb1 and Mx1 in WT or SREBP2−/− MEFs transfected with control siRNA or siRNA to cGAS. All experiments are reported as means ± SD from three independent experiments. *P < 0.05; **P < 0.01, ***P < 0.005 (two-tailed unpaired Student’s t test). See also Figure S6.
STING links changes in cholesterol synthesis with IFNβ production
Stimulator of Interferon Genes (STING; TMEM173) and Mitochondrial Antiviral-Signaling (MAVS) pathways are known to facilitate IRF3 nuclear translocation and Ifnb1 transcription in response to cytosolic dsDNA or ssRNA, respectively (Ishikawa and Barber, 2008; Seth et al., 2005; Sun et al., 2013; Tanaka and Chen, 2012). To determine if these pathways were contributing to IRF3 activation and Ifnb1 expression, we silenced MAVS or STING in SREBP2-deficient cells. Silencing of MAVS had no effect on heightened Ifnb1 and ISG expression in SREBP2-deficient MEFs (Figure 5D, S6C). In contrast, silencing of STING completely abrogated heightened Ifnb1 and ISG expression in MEFs and macrophages (Figure 5D, 5E S6C–G). STING ligands (cyclic di-nucleotides) can be generated by the enzymatic activity of cGAMP synthase (cGAS; Mb21d1). Gene expression studies revealed that SREBP2-deficeint cells have basally heightened expression of cGAS (Figure S6C), which is partially dependent on STING, IRF3 and IFNAR signaling (Figure S6C, S6H, S6I). Transiently silencing cGAS reduced the expression of Ifnb1 and ISGs in SREBP2-deficient cells to that of controls, indicating that heightened STING activity observed in SREBP2−/− cells is dependent on generation of endogenous STING ligands (Figure 5F).
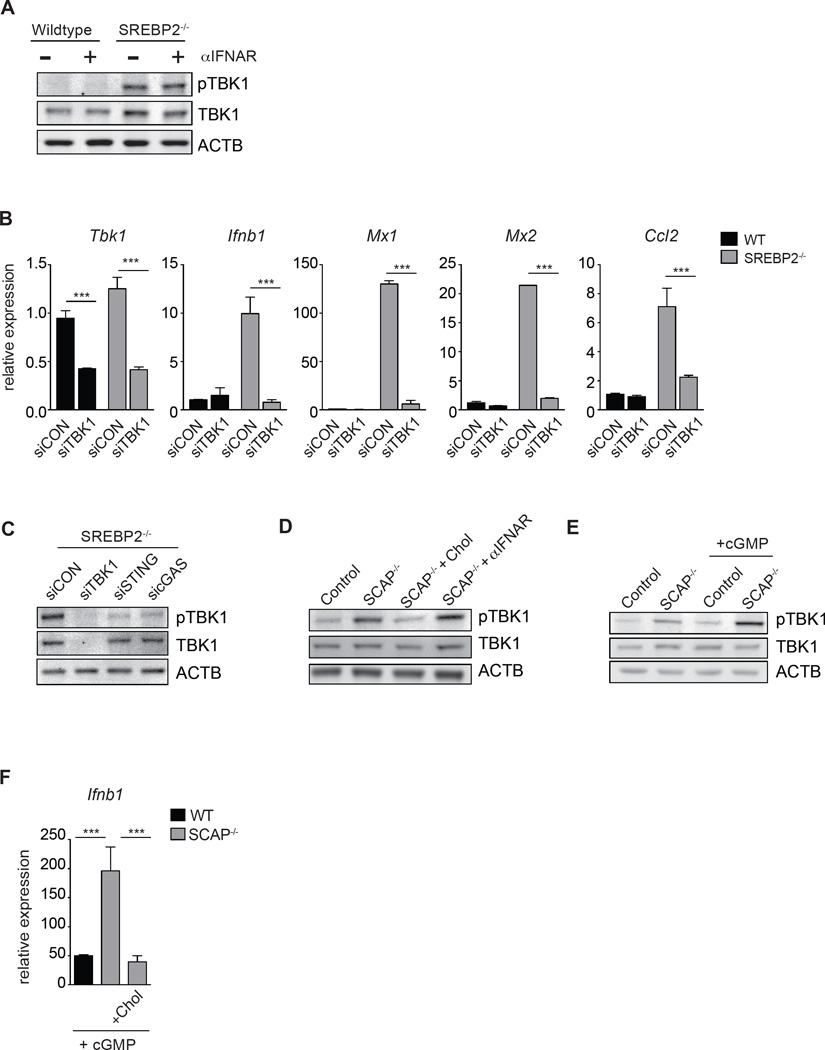
STING is an ER-resident protein that promotes phosphorylation of TANK-binding kinase 1 (TBK1) and IRF3 in response to cyclic di-nucleotides (Tanaka and Chen, 2012). Western blot analysis demonstrated increased basal phospho-TBK1 in SREBP2−/− MEFs (Figure 6A). Silencing of Tbk1 alone was sufficient to reduce expression of Ifnb1 and ISGs (Figure 6B) in SREBP2−/− cells. Likewise, silencing of STING or cGAS attenuated heightened pTBK1 (Figure 6C). Taken together, these data confirm a requirement for the cGAS/STING/TBK1/IRF3 signaling axis in linking changes in cholesterol metabolism with induction of Ifnb1.
Figure 6. Perturbations in cholesterol homeostasis alters STING sensitivity to di-cyclic nucleotides.
(A) Western blot analysis of phospho-TBK1 (pTBK1) and total TBK1 from whole cell lysates in WT or SREBP2−/− MEFs cultured in 1% FBS for 24h +/− 5ug/mL IFNAR blocking antibody as indicated. (B) qPCR analysis of Tbk1, Ifnb1, Mx1, Mx2 and Ccl2 in WT or SREBP2−/− MEFs transfected with control siRNA, or siTBK1. (C) Representative western blot analysis of phospho-TBK1 (pTBK1) and total TBK1 from whole cell lysates of SREBP2−/− MEFs transfected with control siRNA, siSTING or sicGAS as indicated. (D) Western blot analysis of pTBK1 and TBK1 total in Control or SCAP−/− BMDMs +/− 0.25mg/mL MβCD-cholesterol (Chol) for 48h or 10ug/mL IFNAR neutralizing antibody. (E) Western blot analysis of pTBK1 and TBK1 total in Control or SCAP−/− BMDMs +/− 5ug/mL c-di-GMP (cGMP) for 45 min. (F) qPCR analysis of Ifnb1 expression in Control or SCAP−/− BMDMs cultured +/− 0.25mg/mL MβCD-cholesterol for 48h, then stimulated with 5ug/mL cGMP for 1h.
We posited that addition of exogenous cholesterol should reduce signaling activity through the STING/TBK1 pathway. Replenishing cholesterol to SCAP−/− BMDMs significantly reduced phospho-TBK1 levels (Figure 6D), whereas IFNAR blockade had no effect on pTBK1 (Figure 6A, 6D). These data suggest that cholesterol levels directly influence the ability of STING to transduce signals to TBK1. To address this, control and SCAP−/− BMDMs were treated with a low dose of exogenous STING ligand (herein denoted as “cGMP”). We observed that cGMP treatment significantly increased pTBK1 levels in SCAP−/− BMDMs compared to that of controls (Figure 6E). Importantly, addition of cholesterol to these cells markedly diminished cGMP induced Ifnb1 expression (Figure 6F). Taken together, these data support a model where perturbations in cholesterol biosynthetic flux intrinsically influence responsiveness of STING/TBK1 axis to cyclic di-nucleotides, and provides a molecular mechanism linking cholesterol homeostasis with type I IFN-mediated inflammation.
Discussion
In this study, we identify a metabolic-inflammatory circuit that is an important component of host defense. We initially show that decreasing de novo cholesterol and fatty acid biosynthesis is a physiologic response to viral infection via IFNAR signaling pathway,, in agreement with other recently published gene expression studies (Blanc et al., 2011; Reboldi et al., 2014). One potential explanation for this is that decreasing lipid synthesis may serve to limit the amount of lipids available to invading pathogens. However, our studies also indicate that these same pro-inflammatory signals maintain or increase the cellular pool size of cholesterol and long chain fatty acids, suggesting that type I IFN-mediated reprogramming of lipid metabolism is to alter the balance between lipid synthesis and scavenging, rather than to decrease lipid pool sizes. These data also suggest that limiting intracellular lipid metabolite availability (e.g., cholesterol) for pathogen utilization is a less likely explanation for why IFN signals downregulate de novo lipid synthesis. Rather, our data indicates that selectively decreasing flux through the cholesterol biosynthetic pathway engages a type I IFN response and primes cells for heightened anti-viral immunity. In this way, IFN signaling decreases cholesterol biosynthesis and in a reciprocal manner, acutely decreasing cholesterol biosynthesis drives type I IFN responses. Therefore, we conclude that the mevalonate pathway and IFN-signaling pathway are part of a metabolic-inflammatory circuit that ensures any changes in the activity of one pathway are sensed by the other pathway.
One striking implication of our data is that synthesized lipids convey unique information about cellular status despite being chemically indistinguishable from their imported counterparts. A simple explanation for this observation is that de novo synthesized lipids are initially partitioned into the ER bilayer before being distributed to subcellular pools or the plasma membrane (Horton et al., 2002). In contrast, lipid import occurs at the plasma membrane and must transit through a number of cellular processing/transport pathways before reaching the ER or other organelles (Das et al., 2014; Wang et al., 2010). As such, subcellular concentration of a specific lipid species would convey information regarding cellular metabolic homeostasis. Indeed, this is the operating principle underlying the tight regulation of the SREBP pathway, where ER sterol content is monitored by the SCAP and INSIG proteins (Radhakrishnan et al., 2008; Sun et al., 2007). STING protein is in also embedded in the ER membrane (Ishikawa and Barber, 2008), thus we propose a model where the STING signaling axis is activated in response to decreasing the ER bilayer cholesterol pool size. Moreover, given that all nucleated cells have the ability to synthesize cholesterol and that STING is expressed in a wide variety of cell types, we predict that this metabolic inflammatory circuit is likely to exist in a broad array of tissues.
As to how perturbations in the pool size of synthesized cholesterol directly engages the STING pathway remains unclear at this time. We considered the possibility that perturbing flux through the cholesterol biosynthetic pathway could significantly increase intracellular levels of STING ligands (Sun et al., 2013). However, cGAMP levels in both control and SREBP2-deficient cells remained below our limits of detection by LC-MS (~240 fmole; data not shown (West et al., 2015; White et al., 2014)). Thus, we believe it unlikely that changes in cholesterol homeostasis is significantly altering the levels of endogenous STING ligands in cells. Rather,, we propose a model where decreasing cholesterol levels in the ER membrane facilitate STING/TBK1 interactions. In this system, STING from SREBP2-deficient cells would respond more efficiently to the same amount of cGAS-produced ligand, explaining the requirement for cGAS for enhanced IFNβ production in SREBP2-deficient cells. Of course, it remains possible that cholesterol is also regulating the function of other components of this signaling axis (e.g., cGAS or TBK1) and it will be important in future studies to mechanistically address these questions.
Finally, our observation that cholesterol pool size can be monitored by host defense machinery raises the intriguing possibility that other classes of lipids are also monitored through yet to be defined sensor-signaling pathways. Indeed, our data indicates that IFNAR signaling can also limit flux through the de novo fatty acid biosynthetic program, but the immunological consequences of this observation still remain unclear. Further experiments will be required to elucidate the signaling pathways involved in shaping the lipid “codes” enforced by innate immune receptors and to fully appreciate the role of lipid metabolic reprogramming during the innate immune response. In conclusion, the studies presented herein provide mechanistic insights as to how perturbations in cholesterol homeostasis engage inflammatory pathways, and advance our understanding of the crosstalk between lipid metabolism and host defense.
Experimental Procedures
Mouse Strains
C57BL/6, B6;129-Scaptm1Mbjg/J and LysM-Cre+/− were purchased from Jackson Labs. B6;129-Scaptm1Mbjg/J were crossed with LysM-Cre+/− mice to obtain LysM-Cre+/−/SCAPfl/fl.s
Human Cell lines
THP1 cells cultured in RPMI media with 10% FBS (Omega Scientific) with penicillin/streptomycin (Gibco). THP1 cells were differentiated with phorbol 12-myristate 13-acetate (PMA) at 50ng/mL (EMD Millipore) for 48–96hrs depending on the experiment. For viral infection studies, THP1 cells were differentiated for 72h prior to infection.
Primary Human cells
Human monocyte-derived macrophage: Peripheral blood mononuclear cells (PBMCs) were isolated from leukopacks using standard ficoll procedures. Monocytes were separated from PBMCs via plastic adherence. Monocytes were differentiated into macrophage with 10ng/mL human M-CSF (Biolegend) in RPMI (Gibco) media with 10% FBS for 7 days prior to experimental use. Human primary fibroblasts from MVK-deficient (Cat# GM12014) or population controls (Cat# ND38530) were obtained from Coriell Institute for Medical Research and cultured under recommended conditions.
Mouse cells
Bone marrow was differentiated into macrophages in DMEM containing 20% FBS (Omega), 5% M-CSF conditioned media, 1% pen/strep (Gibco), 1% glutamine (Invitrogen) 0.5% sodium pyruvate (Invitrogen) for 7–9 days prior to experimental use. WT and SREBP2−/− MEFs were obtained from Dr. Karen Reue (UCLA). MEFs were cultured in DMEM plus 10% FBS with 1% Pen/Strep unless noted otherwise.
Viruses
HIV1 was produced by transfecting 293T cells with pHIV89.6 using lipofectamin 2000 (Life Technology) according to the manufacturer’s protocol. HIV189.6 was harvested 2 d after transfection and purified with 0.22um filter and quantified by p24 ELISA. Influenza A/WSN/33 (H1N1) was produced by transfecting 293T cells with the 8-plasmid reverse genetics system (Neumann et al., 1999). Wild-type MHV-68 was purchased from ATCC. For all in vitro infection experiments, cells were infected with virus for 3 hours, then changed into fresh media.
RNA-seq
RNA was purified using Qiagen RNeasy Kit and submitted to the UCLA Clinical Microarray Core for RNA-seq analysis (see supplemental methods for details of methods and analysis). Data available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73942
U13C-glucose labeling of cells and Isotope Enrichment Analysis
Day 8 differentiated BMDMs were labeled with U13C-glucose and Isotopomer Spectral Analysis (ISA) was performed as previously described (Williams et al., 2013). Please see supplemental experimental procedure for detailed methods.
Supplementary Material
Acknowledgements
We thank Drs. Peter Tontonoz, Stephen T. Smale, Jake Lusis, Caius Radu, Thotsophon “Todd” Taechariyakul, Thuc Le and Genhong Cheng (UCLA) for kindly providing reagents and thoughtful analysis. We thank the CFAR Virology Core Lab & Tissue Culture/PCR Facility at the UCLA AIDS Institute for their help with PBMC collection. We thank Dr. Robert Damoiseaux at the UCLA Molecular Screening Shared Resource (a core facility supported by the NCI Cancer Center Support Grant P3016042-35 to Judy Gasson). AGY was supported in part by California HIV/AIDS Research Program Training Fellowship (D12-LA-351) and UCLA Dissertation Year Fellowship. JPA was supported in part by a NIH CBI Grant (T32 GM008469) and UCLA Dissertation Year Fellowship. NCW. was supported in part by UCLA Molecular Biology Philip Whitcome Pre-Doctoral Fellowship and UCLA Dissertation Year Fellowship. G.B. was supported by the Interdisciplinary Training in Virology and Gene Therapy Training Grant (NIH T32 AI 060567). EEG was supported in part by Cancer Research Institute Fellowship Program. EJT was supported in part by NIH HL 116181 and UCLA CTSI-DRC UL1TR000124. DGB was supported by R01 AI085043. RS and TTW were supported by NCI RO1 CA177322 and NIDCR RO1 DE023519. KR and LV were supported in part by NIH P01 HL028481, NIH NCRR S10RR026744, and NIH P01 HL090553. SJB was supported by NIH AI093768, HL126556 and the Lupus Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
AGY designed/implemented experiments, analyzed data, conceptualized this project, and constructed the manuscript. JPA, KJW, GB, LV, EEG, NCW, AZ, DHY, CRC, EJT, DHG, AKY and ESW designed/implemented experiments and analyzed data. MQW and DC provided formal analysis. TTW, RS, SGK, DGB, KR, and DBS provided scientific advice. SJB provided resources & supervision, conceptualized this project, analyzed data and constructed the manuscript.
References
- Baek SH, Li AH, Sassetti CM. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS biology. 2011;9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard M, Sturm AM, Pierson CA, Brown S, Rogers KM, Nabinger S, Eckstein J, Barbuch R, Lees ND, Howell SA, et al. Sterol uptake in Candida glabrata: rescue of sterol auxotrophic strains. Diagnostic microbiology and infectious disease. 2005;52:285–293. doi: 10.1016/j.diagmicrobio.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Blanc M, Hsieh WY, Robertson KA, Kropp KA, Forster T, Shui G, Lacaze P, Watterson S, Griffiths SJ, Spann NJ, et al. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity. 2013;38:106–118. doi: 10.1016/j.immuni.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc M, Hsieh WY, Robertson KA, Watterson S, Shui G, Lacaze P, Khondoker M, Dickinson P, Sing G, Rodriguez-Martin S, et al. Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS biology. 2011;9:e1000598. doi: 10.1371/journal.pbio.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo A, Joseph SB, Vaidya SA, Haberland M, Fogelman AM, Cheng G, Tontonoz P. Crosstalk between LXR and toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism. Molecular cell. 2003;12:805–816. doi: 10.1016/s1097-2765(03)00384-8. [DOI] [PubMed] [Google Scholar]
- Coppens I. Targeting lipid biosynthesis and salvage in apicomplexan parasites for improved chemotherapies. Nature reviews Microbiology. 2013;11:823–835. doi: 10.1038/nrmicro3139. [DOI] [PubMed] [Google Scholar]
- Cui HL, Ditiatkovski M, Kesani R, Bobryshev YV, Liu Y, Geyer M, Mukhamedova N, Bukrinsky M, Sviridov D. HIV protein Nef causes dyslipidemia and formation of foam cells in mouse models of atherosclerosis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014;28:2828–2839. doi: 10.1096/fj.13-246876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Brown MS, Anderson DD, Goldstein JL, Radhakrishnan A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife. 2014;3 doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dushkin MI, Kovshik GG. Effect of toll-like receptor agonists on the formation of macrophage/foam cells upon acute peritonitis in mice. Bulletin of experimental biology and medicine. 2013;156:49–52. doi: 10.1007/s10517-013-2275-1. [DOI] [PubMed] [Google Scholar]
- Esposito S, Ascolese B, Senatore L, Bosis S, Verrecchia E, Cantarini L, Rigante D. Current advances in the understanding and treatment of mevalonate kinase deficiency. International journal of immunopathology and pharmacology. 2014;27:491–498. doi: 10.1177/039463201402700404. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Funk JL, Feingold KR, Moser AH, Grunfeld C. Lipopolysaccharide stimulation of RAW 264.7 macrophages induces lipid accumulation and foam cell formation. Atherosclerosis. 1993;98:67–82. doi: 10.1016/0021-9150(93)90224-i. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Bergeron M, Methot S, Giguere JF, Tremblay MJ. Statins could be used to control replication of some viruses, including HIV-1. Viral immunology. 2005;18:474–489. doi: 10.1089/vim.2005.18.474. [DOI] [PubMed] [Google Scholar]
- Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- Greseth MD, Traktman P. De novo fatty acid biosynthesis contributes significantly to establishment of a bioenergetically favorable environment for vaccinia virus infection. PLoS Pathog. 2014;10:e1004021. doi: 10.1371/journal.ppat.1004021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chemistry & biology. 2012;19:218–227. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E, Ott M. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol Metab. 2011;22:241–248. doi: 10.1016/j.tem.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YL, Morales-Rosado J, Ray J, Myers TG, Kho T, Lu M, Munford RS. Toll-like receptor agonists promote prolonged triglyceride storage in macrophages. J Biol Chem. 2014;289:3001–3012. doi: 10.1074/jbc.M113.524587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DJ, Kipar A, Sample JT, Stewart JP. Pathogenesis of a model gammaherpesvirus in a natural host. Journal of virology. 2010;84:3949–3961. doi: 10.1128/JVI.02085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci U S A. 2005;102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyel PA, Tkacheva OA, Larregina AT, Salter RD. Coordinate stimulation of macrophages by microparticles and TLR ligands induces foam cell formation. Journal of immunology. 2012;189:4621–4629. doi: 10.4049/jimmunol.1200828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CI, Liu GY, Song Y, Yin F, Hensler ME, Jeng WY, Nizet V, Wang AH, Oldfield E. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319:1391–1394. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nature biotechnology. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet H, Johnston JB, de Montellano PR. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends in microbiology. 2011;19:530–539. doi: 10.1016/j.tim.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parihar SP, Guler R, Khutlang R, Lang DM, Hurdayal R, Mhlanga MM, Suzuki H, Marais AD, Brombacher F. Statin therapy reduces the mycobacterium tuberculosis burden in human macrophages and in mice by enhancing autophagy and phagosome maturation. The Journal of infectious diseases. 2014;209:754–763. doi: 10.1093/infdis/jit550. [DOI] [PubMed] [Google Scholar]
- Petersen J, Drake MJ, Bruce EA, Riblett AM, Didigu CA, Wilen CB, Malani N, Male F, Lee FH, Bushman FD, et al. The major cellular sterol regulatory pathway is required for Andes virus infection. PLoS Pathog. 2014;10:e1003911. doi: 10.1371/journal.ppat.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell metabolism. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A, Dang EV, McDonald JG, Liang G, Russell DW, Cyster JG. Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science. 2014;345:679–684. doi: 10.1126/science.1254790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzouq F, Alahdab F, Olyaee M. Statins and hepatitis C virus infection: an old therapy with new scope. The American journal of the medical sciences. 2014;348:426–430. doi: 10.1097/MAJ.0000000000000291. [DOI] [PubMed] [Google Scholar]
- Santos JA, Arostegui JI, Brito MJ, Neves C, Conde M. Hyper-IgD and periodic fever syndrome: a new MVK mutation (p.R277G) associated with a severe phenotype. Gene. 2014;542:217–220. doi: 10.1016/j.gene.2014.03.031. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shibata N, Glass CK. Regulation of macrophage function in inflammation and atherosclerosis. J Lipid Res. 2009;50(Suppl):S277–S281. doi: 10.1194/jlr.R800063-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci U S A. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Science signaling. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ML, Motamed M, Infante RE, Abi-Mosleh L, Kwon HJ, Brown MS, Goldstein JL. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell metabolism. 2010;12:166–173. doi: 10.1016/j.cmet.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015 doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KJ, Argus JP, Zhu Y, Wilks MQ, Marbois BN, York AG, Kidani Y, Pourzia AL, Akhavan D, Lisiero DN, et al. An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer research. 2013;73:2850–2862. doi: 10.1158/0008-5472.CAN-13-0382-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, McDonald JG, Patel A, Zhang Y, Umetani M, Xu F, Westover EJ, Covey DF, Mangelsdorf DJ, Cohen JC, et al. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J Biol Chem. 2006;281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.