Dear Editor,
Ebola virus disease (EVD), formerly known as Ebola hemorrhagic fever, is a severe infectious disease caused by Ebola virus (EBOV) species1,2. EBOV caused an epidemic in West Africa in 2013-20151,2. Early diagnosis of EVD is not only essential for implementation of effective interventions but also critical for prevention of the spread of infection, especially in regions with fragile and underfunded health system2. However, diagnosing EVD at an early stage is difficult, because EBOV causes symptoms observed in many other infections, including malaria, typhoid fever and influenza3, and some patients even develop illness without specific signs or symptoms. Current diagnostic methods to detect viral RNA or antigen in suspected patients are only effective at the late stage of illness. Lack of early biomarkers has led to diagnostic delays in current massive EVD outbreak4 and international spread of EBOV5,6. Recent studies by us and others have demonstrated that microRNAs (miRNAs), a class of small non-coding RNAs produced by eukaryotic cells and viruses, are present in human blood and other body fluids in highly stable, cell-free forms7,8. Circulating miRNAs can serve as non-invasive biomarkers for the early diagnosis of various diseases, including viral diseases7,8,9,10. Because EBOV is a negative-sense RNA virus, whether EBOV can produce miRNAs or miRNA-like RNA fragments remains unknown. In this study, we predicted and tested putative miRNA-like fragments generated by EBOV in the serum of EVD patients.
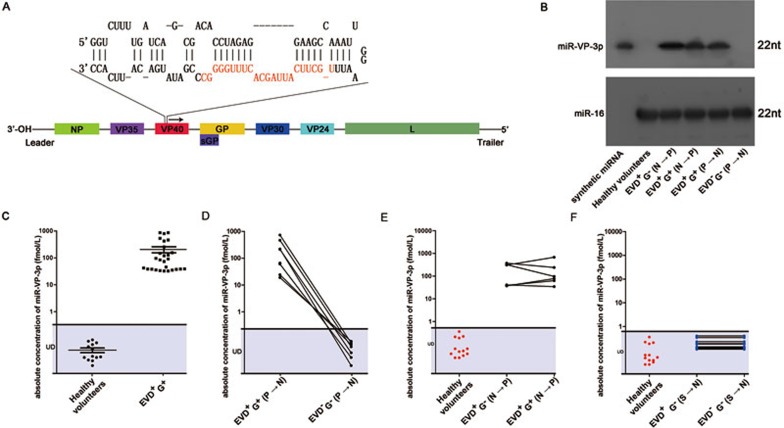
First, genome sequences of EBOV were used to predict hairpin structure of miRNA precursors. Three putative precursors (pre-miR-VP, pre-miR-NP and pre-miR-L) located in the VP40, NP and L gene of EBOV respectively were predicted (see Figure 1A for genomic organization of Ebola virus and pre-miR-VP). The mature miRNAs were next extrapolated, and three miRNA-like RNA fragments (miR-VP-3p, miR-NP-3p and miR-L-3p) in the 3′ arm of the precursors were identified (Figure 1A shows miR-VP-3p). Because miRNAs are generally highly conserved across species, the evolutionary conservation of the putative EBOV miRNAs was examined by aligning the precursor sequences with currently published full-length genomes of 2014 EBOV strains. Pre-miR-VP was highly conserved among 2014 EBOV strains (Supplementary information, Figure S1A), whereas pre-miR-L and pre-miR-NP were not. Furthermore, we performed NCBI Standard Nucleotide BLAST and confirmed that miR-VP-3p was unique to EBOV and had no sequence similarity to host genome or other virus genome. Thus, pre-miR-VP was retained for further analysis.
Figure 1.
Identification and detection of miRNA-like RNA fragment of EBOV in the sera from EVD patients. (A) The genomic position and the predicted stem-loop secondary structure of the EBOV-encoded pre-miR-VP. The mature miRNA derived from the 3′ arm of the stem–loop precursor is indicated in red. (B) Northern blotting analysis of the predicted miR-VP-3p in the sera of healthy volunteers, EVD patients before and after EBOV genomic RNA became detectable (denoted as EVD+G−(N→P) and EVD+G+(N→P)), and EBOV survivors during the acute phase (denoted as EVD+G+(P→N); positive for EBOV genomic RNA in a RT-PCR-based assay) and the convalescence phase (denoted as EVD−G−(P→N); negative for EBOV genomic RNA in a RT-PCR-based assay). EVD+ means that patients have Ebola-like symptoms; G+ means that patients have positive RT-PCR results for the EBOV genomic RNA. (C) Concentrations of miR-VP-3p in the serum samples from healthy volunteers and EVD+G+. (D) Concentrations of miR-VP-3p in the serum samples from EBOV survivors in the acute phase (denoted as EVD+G+(P→N)) and the convalescence phase (denoted as EVD−G−(P→N)). (E, F) Levels of miR-VP-3p were measured in the serum samples from 15 patients with suspected EVD via qRT-PCR. These patients exhibited EBOV infection symptoms but tested negative for EBOV genomic RNA based on RT-PCR at enrollment. During the prospective observation period, 6 patients (black dots, denoted as EVD+G−(N→P) and EVD+G+(N→P)) were confirmed to be EBOV positive (E); however, the other 9 patients (blue dots, denoted as EVD+G−(S→N) and EVD−G−(S→N)) remained EBOV negative (F). miR-VP-3p was detected in the 6 EBOV-positive suspected patients both at enrollment and during the observation; however, it was not detected in the other 9 EBOV-negative suspected patients.
To investigate whether putative EBOV miRNA can fold into a typical hairpin structure of miRNA precursor and can be further processed to a miRNA-like RNA fragment in cellular environment, the pre-miR-VP sequence was cloned into a plasmid and transfected into HEK293A cells (Supplementary information, Figure S1B and S1C). Northern blotting recovered both the mature form of miR-VP-3p (∼20-nt band) and the hairpin precursor of miR-VP-3p (∼80-nt band, corresponding to pre-miR-VP) (Supplementary information, Figure S1D). To confirm that the pre-miR-VP sequence is indispensable for the generation of miR-VP-3p, a mutation was introduced into the hairpin (Supplementary information, Figure S1B and S1C), which abolished the expression of miR-VP-3p (Supplementary information, Figure S1D), indicating that the mature miR-VP-3p was produced by specifically cutting the hairpin structure of pre-miR-VP.
To test whether the miRNA-like RNA fragments were actually produced by the cells, small RNA was isolated from sera of EBOV-infected patients and healthy volunteers. Northern blotting detected the presence of a miRNA-like RNA fragment corresponding to miR-VP-3p in sera of EVD patients but not in the sera of healthy volunteers (Figure 1B). Based on the sensitivity of northern blotting (Supplementary information, Figure S2A), we calculated that miR-VP-3p was present in the sera of EVD patients at a level higher than 100 fM. Furthermore, RT-PCR combined with TA-cloning and sequencing was performed to validate the presence of miRNA-like RNA fragment in EVD patients. Small RNA isolated from sera of EVD patients was subjected to RT-PCR to amplify putative EBOV miRNA, and then the amplified product was ligated into a TA-vector and sequenced. The sequences of the products from the EVD samples were completely identical to that of the predicted sequence of miR-VP-3p (Supplementary information, Figure S2B).
We next measured miR-VP-3p in the serum samples from 27 EBOV-infected individuals (denoted as EVD+G+) and 13 healthy volunteers using a TaqMan probe-based quantitative RT-PCR (qRT-PCR) assay. The patient information is listed in Supplementary information, Table S1A. To determine the dynamic range and sensitivity of the qRT-PCR assay for measuring miR-VP-3p, synthetic single-stranded miR-VP-3p was serially diluted and assessed using qRT-PCR to generate a standard curve, and a no-template control was used to determine the specificity of the primer set. The synthetic miR-VP-3p was efficiently amplified, and the no-template control was amplified at a much higher cycle range (Supplementary information, Figure S2C), indicating that the lower end of the dynamic quantification range for miR-VP-3p is 0.1 attomole. The measurements of miR-VP-3p in the serum samples from EVD+G+ and the healthy volunteers revealed that the CT values were consistently within the linear range for EVD+G+ but were outside the linear range for the healthy volunteers (Supplementary information, Figure S2D), suggesting that miR-VP-3p was detectable in the sera of EVD+G+ but absent in the sera of healthy volunteers. The concentration of miR-VP-3p was calculated according to the standard curve to be on average 205.3 fM in EVD+G+ sera (Figure 1C). As a control, the levels of miR-16 were also examined in the sera of EVD+G+ individuals and healthy volunteers and were found to be identical between the two groups (Supplementary information, Figure S2E-S2G).
Next, eight pairs of serum samples from EBOV survivors were collected and the levels of miR-VP-3p in these samples during the acute phase (denoted as EVD+G+(P→N); positive for EBOV genomic RNA based on RT-PCR) and the recovery phase (denoted as EVD−G−(P→N); negative for EBOV genomic RNA based on RT-PCR) were measured. The patient information is listed in Supplementary information, Table S1B. miR-VP-3p was present at high levels during the acute phase but disappeared in the recovery phase of the disease (Figure 1D and Supplementary information, Figure S2H). Northern blotting analysis confirmed that miR-VP-3p was absent in the sera of EVD survivors during the recovery phase (Figure 1B).
To determine whether miR-VP-3p can serve as potential biomarkers for the early diagnosis of EVD, a prospective observational study was conducted. Fifteen suspected patients with Ebola-like symptoms and negative RT-PCR-based results for EBOV genomic RNA (EBOV-negative) were included and prospectively observed. During the observational period, six patients became EBOV-positive (denoted as EVD+G−(N→P) or EVD+G+(N→P)), whereas the other 9 patients remained EBOV-negative (denoted as EVD+G−(S→N) or EVD−G−(S→N); Supplementary information, Figure S2I-S2K). The patient information is listed in Supplementary information, Table S1C. The levels of miR-VP-3p in the 15 patients with suspected EVD were measured via a qRT-PCR assay at the beginning and the end of the observation period. miR-VP-3p was undetectable in all 9 EBOV-negative suspected patients both at enrollment and at the end of the observation period (Figure 1F and Supplementary information, Figure S2M). Strikingly, a considerable level of miR-VP-3p was detected in the sera of the EVD+G−(N→P) patients, indicating that miR-VP-3p could be detected before the EBOV genomic RNA became positive (Figure 1E and Supplementary information, Figure S2L). Northern blotting analysis confirmed that miR-VP-3p was present in the suspected patients before the EBOV genomic RNA became detectable in serum (Figure 1B). One explanation of this observation may be that, during EBOV replication in the cells, some fraction of VP40 mRNA was processed to produce miR-VP-3p which was then released through exosomal pathway, even before the egress of viruses. That is, miR-VP-3p is released into the serum while virus is still replicating inside the cell, whereas genomic RNA makes into the serum only after viruses are released from the cells. In agreement with this hypothesis, miR-VP-3p was present mainly in the exosomal fractions but not the exosome-free fractions of the serum of EVD patients (Supplementary information, Figure S2N). Because miR-VP-3p was detected before EBOV genomic RNA was detected in the serum, this miRNA-like RNA fragment may represent a potential biomarker for early diagnosis of EVD before the development of detectable viremia in EBOV-positive patients.
Taken together, our study predicted several miRNA-like RNA fragments in EBOV genome and confirmed the presence of at least one such RNA fragment, miR-VP-3p, in the serum samples of EVD patients. This finding may be against the traditional concept that RNA viruses generally do not encode miRNA; however, it is supported by several recent reports that RNA viruses are capable of expressing miRNA-like small RNAs11,12,13,14. By characterizing the genome sequence and possible structure of these miRNA-like small RNAs, our results further suggest that formation of hairpin structure may be a key step in biogenesis of miR-VP-3p. Of course, more work is required to define the biogenesis of miR-VP-3p and determine the bioactivity of this miRNA-like RNA fragment during infection and host-virus interactions. Nevertheless, as miR-VP-3p is detected in EVD patients before development of viremia with detectable Ebola genomic RNA, it may serve as an early biomarker of EVD to advance the diagnosable window and reduce the difficulties in the isolation and treatment of patients who develop suspicious symptoms. Given that miR-VP-3p is highly conserved among different EBOV strains, it may serve as a tool to predict Ebola outbreaks in the future if Ebola arises again. Furthermore, future studies should investigate if similar miRNA-like small RNAs are produced in other viral infections.
Acknowledgments
We thank the Nanjing MicroMedMark Biotech Company (Nanjing, China) for the gift of the kits for RNA isolation. We thank Zheng Fu (Nanjing University), Zhe Zhou (Beijing Key Laboratory of New Molecular Diagnostics Technology), Wei Liu and Wenyi Zhang (Institute of Disease Control and Prevention, Academy of Military Medical Sciences), David Kargbo and Abdul Kamara (Sierra Leone Ministry of Health and Sanitation) for the technical support. This work was supported by grants from the National Basic Research Program of China (973 Program; 2014CB542300), the Chinese Science and Technology Major Project of China (2015ZX09102023-003), the Research Special Fund for Public Welfare Industry of Health (201302018), the National Natural Science Foundation of China (81550001), the National Key Program for Infectious Diseases of China (013ZX10004-203 and 2013ZX10004805-006).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
EBOV encoded a miRNA-like RNA fragment.
The levels of miR-VP-3p in EVD patients.
The demographic and clinical features of the EBOV-infected patients and healthy volunteers.
Materials and Methods
References
- World Health Organization. Ebola data and statistics — Situation summary; available at http://apps.who.int/gho/data/view.ebola-sitrep.ebola-summary-latest?lang=en.
- WHO Ebola Response Team. N Engl J Med 2014; 371:1481–1495. [DOI] [PMC free article] [PubMed]
- Kuhn JH, Becker S, Ebihara H, et al. Arch Virol 2010; 155:2083–2103. [DOI] [PMC free article] [PubMed]
- World Health Organization. Ebola virus disease fact sheet; available at http://www.who.int/mediacentre/factsheets/fs103/en.
- Lyon GM, Mehta AK, Varkey JB, et al. N Engl J Med 2014; 371:2402–2409. [DOI] [PubMed]
- Parra JM, Salmeron OJ, Velasco M. N Engl J Med 2014; 371:2439–2440. [DOI] [PubMed]
- Chen X, Ba Y, Ma L, et al. Cell Res 2008; 18:997–1006. [DOI] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Proc Natl Acad Sci USA 2008; 105:10513–10518. [DOI] [PMC free article] [PubMed]
- Zhang G, Zong J, Lin S, et al. Int J Cancer 2015; 136:E301–E312. [DOI] [PubMed]
- Li S, Zhu J, Zhang W, et al. Circulation 2011; 124:175–184. [DOI] [PubMed]
- Rosewick N, Momont M, Durkin K, et al. Proc Natl Acad Sci USA 2013; 110:2306–2311. [DOI] [PMC free article] [PubMed]
- Lu HJ, Qian J, Kargbo D, et al. Emerg Infect Dis 2015; 21:1921–1927. [DOI] [PMC free article] [PubMed]
- Gibb TR, Norwood DA Jr, Woollen N, et al. J Clin Microbiol 2001; 39:4125–4130. [DOI] [PMC free article] [PubMed]
- Rouha H, Thurner C, Mandl CW. Nucleic Acids Res 2010; 38:8328–8337. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EBOV encoded a miRNA-like RNA fragment.
The levels of miR-VP-3p in EVD patients.
The demographic and clinical features of the EBOV-infected patients and healthy volunteers.
Materials and Methods