Abstract
Pathogenic bacteria have evolved multiple mechanisms to capture iron or iron-containing heme from host tissues or blood. In response, organisms have developed defense mechanisms to keep iron from pathogens. Very little of the body's iron store is available as free heme; rather nearly all body iron is complexed with heme or other proteins. The feline leukemia virus, subgroup C (FeLV-C) receptor, FLVCR, exports heme from cells. It was unknown whether FLVCR regulates heme-iron availability after infection, but given that other heme regulatory proteins are upregulated in macrophages in response to bacterial infection, we hypothesized that macrophages dynamically regulate FLVCR. We stimulated murine primary macrophages or macrophage cell lines with LPS and found that Flvcr is rapidly downregulated in a TLR4/MD2-dependent manner; TLR1/2 and TLR3 stimulation also decreased Flvcr expression. We identified several candidate TLR-activated transcription factors that can bind to the Flvcr promoter. Macrophages must balance the need to sequester iron from systemic circulating or intracellular pathogens with the macrophage requirement for heme and iron to produce reactive oxygen species. Our findings underscore the complexity of this regulation and point to a new role for FLVCR and heme export in macrophages responses to infection and inflammation.
1. Introduction
Nearly all organisms require iron because of its ability to catalyze redox reactions, and humans have evolved mechanisms to recycle almost all the iron contained within hemoglobin and cellular enzymes with only minimal daily loss through the gastrointestinal tract [1]. Nevertheless, iron-deficiency anemia and anemia of chronic disease (caused in part by iron-restricted erythropoiesis) together are responsible for the majority of anemia cases worldwide [2], and while these conditions cause morbidity and mortality, it has become clear that limiting iron availability is in fact an innate immune strategy against microbes. Indeed studies in humans and mice have shown that oral iron supplementation leads to increased mortality due to infection [3, 4]. Pathogens that enter and proliferate within a host must acquire iron from the host and have evolved a large and diverse number of mechanisms to accomplish this [5], and in response, mammals have developed complex mechanisms to keep iron from pathogens [6, 7].
Over the last two decades, there has been much effort aimed at understanding the pathophysiology of anemia associated with chronic inflammation/disease, characterized by hypoferremia. This led to the finding that a small peptide, hepcidin, initially identified as an antimicrobial peptide [8], is a master regulator of systemic iron stores. Hepcidin is produced mainly by the liver and its production is regulated by inflammation/iron levels, hypoxia, and erythropoiesis; hepcidin is elevated in anemia of chronic inflammation [9]. Hepcidin can bind to ferroportin 1 (FPN1), an iron export protein found on macrophages, enterocytes, and other cell types, and in doing so causes FPN1 internalization and degradation [10]. This results in sequestration of iron within macrophages and decreased intestinal absorption. The vast majority of bodily iron stores is complexed with heme in hemoglobin in red cells, and senescent red cells are broken down and phagocytosed by specialized macrophages in the spleen, which are highly efficient in recycling iron from hemoglobin. Nevertheless, pathogens have evolved complex mechanisms to obtain heme from blood or tissues as an alternate source of iron [11]. While the heme synthesis pathway has been well characterized in mammals, much less is known about how heme is regulated or trafficked within cells or systemically [12]. We identified the feline leukemia virus subgroup C receptor (FLVCR), a 12-transmembrane domain protein and member of the major facilitator superfamily, as a heme exporter in mammalian cells [13, 14]. Heme exported from cells through FLVCR is rapidly bound by plasma proteins including hemopexin and albumin, which can then transport the heme to other sites for utilization [15]. We found that FLVCR is required for normal erythroid [16] and T lymphocyte development [17]. Although macrophages express high levels of FLVCR, consistent with a role for macrophages in recycling heme/heme iron from phagocytosed senescent red cells [16], the role of FLVCR in regulating heme-iron after infection remains unexplored. Macrophages upregulate heme oxygenase-1 (HMOX1), a heme-degrading enzyme, in response to inflammation or infection [18]; therefore we hypothesized that macrophages dynamically regulate FLVCR in response to inflammation or infection.
We analyzed Flvcr (also referred to as Mfsd7b) mRNA levels as well as that of the other key heme and iron regulatory genes, Fpn1 (also referred to as Slc40a1), Hmox1, and ferritin light chain (Ftl) in macrophages treated with LPS. We found that Flvcr mRNA decreases quickly upon LPS stimulation, similar to Fpn1, before recovering to baseline 24–48 hours later. The return to baseline Flvcr expression coincides with the major increase in Hmox1 and Ftl1 expression, suggesting an initial need for increased heme in macrophages after infection accomplished by FLVCR downregulation, then at later time points, heme is degraded by HMOX1 and iron is sequestered in ferritin. While macrophage sequestration of heme and iron may be one aspect of antimicrobial defense, macrophages need heme and iron for the reactive oxygen (ROS) production and bacterial killing [19, 20]. These observations suggest that, upon infection, macrophages initially transiently increase intracellular heme and iron in order to kill bacteria before shifting to a strategy focused on iron sequestration.
2. Materials and Methods
2.1. Macrophage Cell Culture
The J774A.1 cell line (henceforth referred to as J774) was obtained from ATCC and maintained in cDMEM: DMEM medium (Gibco) supplemented with 10% inactivated FBS, penicillin/streptomycin/L-glutamine (1 unit/mL, 1 μg/mL, and 2 mM; Gibco), HEPES (10 mM; Gibco), β-mercaptoethanol (0.05 M; Sigma), and MEM-non-essential amino acids (0.1 mM; Gibco). Bone-marrow-derived macrophages (BMDM) were prepared by euthanizing 6–8-week-old male C57BL/6 mice and sterilely dissecting both femurs. The bone marrow was flushed from the femur with HBSS, homogenized into single cell suspension by pipetting, and centrifuged at 400 g × 4 minutes (4°C) and supernatant aspirated. The cells were then resuspended at 5 × 106 cells/mL in BMDM media: RPMI1640 (Gibco) supplemented with 20% inactivated FBS, 30% L929-conditioned media (LCM), penicillin/streptomycin/L-glutamine (1 unit/mL, 1 μg/mL, 2 mM; Gibco), β-mercaptoethanol (0.05 M; Sigma), and 10 mL plated/10 cm sterile nontissue culture-treated Petri dish (Corning). After 4-5 days, the media containing nonadherent cells were removed and replaced with fresh media. BMDM were harvested by trypsin/versene treatment and replated at lower density as below for stimulation assays. BMDM were used 6–8 days after harvesting.
2.2. Mice
C57BL/6 mice were purchased from The Jackson Laboratory. Tlr4 −/−; Ly96 −/− mice were previously described [21]. All mice were bred and maintained in a specific pathogen-free barrier facility at the University of Washington. Experiments were performed in compliance with the University of Washington Institutional Animal Care and Use Committee regulations.
2.3. Macrophage Stimulation
J774 or BMDM were plated at 5 × 105 cells/well in 12-well tissue culture plates in cDMEM (J774) or BMDM media and allowed to adhere overnight. The following day, the media were exchanged for fresh cDMEM with varying concentrations of hemin or LPS (E. coli O111:B4; Sigma) for varying durations. For later experiments, including those using Tlr4 −/−; Ly96 −/− BMDM, O111:B4 ultrapure LPS from InvivoGen was used. Pam3Csk4 was obtained from EMC Microcollections and polyinosinic:polycytidylic acid (pIC) from Amersham.
2.4. mRNA Isolation and Quantitative RT-PCR
At the appropriate time points, media were aspirated from stimulation wells and macrophages lysed with RLT buffer (Qiagen) and stored at −80°C. RNA was then purified from lysate using RNeasy Plus Mini Kit (Qiagen) and reverse-transcribed using iScript reverse transcriptase (BioRad). Multiplex quantitative real-time PCR (qPCR) was performed on cDNA using the KAPA ProbeFast BioRad iCycler reaction mix (KAPA Biosystems) with gene-specific primers obtained from Integrated DNA Technology (IDT). Primer sequences: β-actin (F 5′-ACCTTCTACAATGAGCTGCG-3′, R 5′-CTGGATGGCTACGTACATGG-3′, 5′-/5Cy5/TCTGGGTCATCTTTTCACGGTTGGC/3IAbRQsp/-3′; Flvcr (F 5′-ATCTGGAACCTGTGCAGAAACA-3′, R 5′-ATTGAATAAAATGCTCCAGTCATGAT-3′, Probe 5′/HEX/CCCCTTTGTTCTCCTGCTGGTCAGTTATG/IABkFQ/-3′); Hmox1 (F 5′-CTGCTAGCCTGGTGCAAGATACT-3′, R 5′-GTCTGGGATGAGCTAGTGCTGAT-3′, Probe 5′-/FAM/AGACACCCCGAGGGAAACCCCA/IABkFQ/-3′); Fpn1 (F 5′-CCAACCGGAAATAAAACCACAG-3′), (R 5′-AGGAGAAAACAGGAGCAGATTAG-3′), (Probe 5′-/FAM/CCAACCGGAAATAAAACCACAG/IABkFQ/-3′); and Ftl1 (F 5′-CAGCCATGACCTCTCAGATTC-3′), (R 5-CCACGTCATCCCGATCAAAA-3′), (Probe 5′-/HEX/CGCCTGGTCAACTTGCACCTG/IABkFQ/-3′). Flvcr, Hmox1, and β-actin primer sets were run together, and Fpn1, Ftl1, and β-actin primer sets were run together. Gene expression (mRNA RQ) was quantified as fold-change expression using the Pffafl method [22]; β-actin was the reference gene and untreated (0 ng/mL) cells were the reference sample. A dilution series of untreated C57BL/6 macrophage cDNA was run for every assay to determine the reaction efficiency for the Pfaffl calculation, to ensure that amplification was linear, and to ensure that the samples being assayed were within the linear range of the assay.
2.5. Macrophage Polarization
Polarization of BMDM to the M1 and M2 states was performed using established protocols [23] as follows. BMDM were prepared and 1 × 106 BMDM plated per well of 6-well plates. The following day, either IFNγ/LPS (100 U/mL; 100 ng/mL), IL4 (100 U/mL), or nothing was added and cells were lysed at various times afterward as above for RNA isolation, cDNA production, and qRT-PCR. IFNγ and IL4 were obtained from eBioscience.
2.6. Transcription Factor (TF) Binding to Flvcr Promoter
The Promoter Binding TF Profiling Plate Array (Signosis) was used to assess TF binding to the murine Flvcr promoter. In brief, this 96-well plate-based competition assay utilizes an array of biotinylated oligos specific for 48 transcription factors (in duplicate). Nuclear extracts from the cell of interest are incubated with purified, PCR-amplified Flvcr promoter. The purified promoter competes with biotinylated TF-specific oligos for binding to TFs present in the nuclear extract. If there is no competition, each TF-bound oligo can hybridize to its specific complementary DNA on the 96-well plate and be detected through luminescence. TF that are present in the extract and bind to the Flvcr promoter will show decreased signal in the presence of the promoter compared to no promoter. Nuclear extracts were prepared from 1 × 106 BMDM using the Nuclear Extraction Kit (Signosis), and the protein content was determined by the Bradford assay. 10 μg of BMDM nuclear extract was incubated with or without 15 pmol of purified PCR-amplified murine Flvcr promoter, processed, and hybridized to the TF array plate following the manufacturer's protocol. Bound probe was detected by chemiluminescence.
3. Results
3.1. Flvcr mRNA Levels in Macrophages Do Not Change in Response to Heme
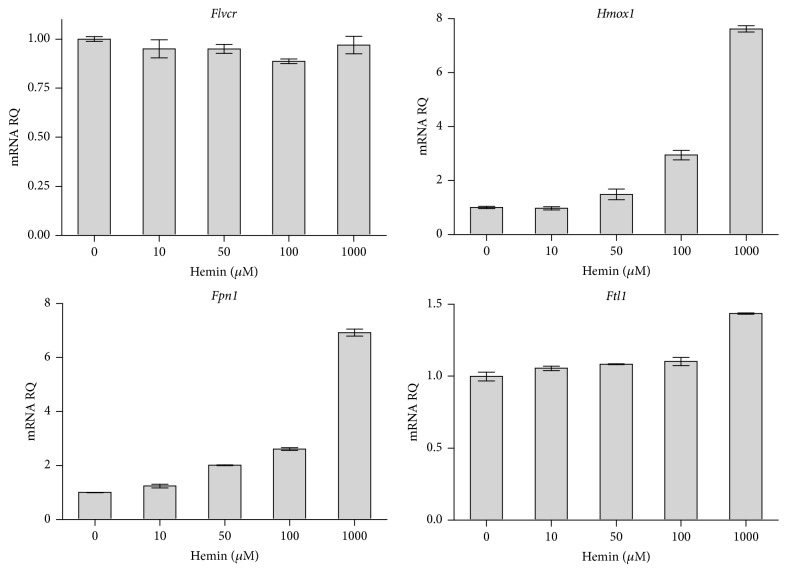
Previously, we observed that macrophages expressed high levels of FLVCR, consistent with a role for macrophages in recycling heme/heme iron from phagocytosed senescent red cells [16]. To determine whether Flvcr expression in macrophages is regulated by heme, J774 macrophages were exposed to increasing doses of hemin for different times and then Flvcr, Hmox1, Fpn1, and Ftl1 mRNA levels were determined by multiplex quantitative RT-PCR (qPCR). While the Hmox1, Fpn1, and Ftl1 mRNA showed a dose-responsive increase to hemin exposure for 10 hours, Flvcr mRNA levels did not change (Figure 1). Similar results were seen at 3 and 24 hours (data not shown). This led us to ask whether FLVCR in macrophages might have another role aside from heme regulation after erythrophagocytosis. Macrophages are key regulators of systemic iron balance that maintain organismal iron supply while sequestering iron from pathogens [1]. Given that much of the body iron store is found in heme, we next tested whether macrophages modulate FLVCR expression in response to infection.
Figure 1.
Macrophages do not regulate Flvcr in response to heme treatment. J774 macrophages were treated with hemin and mRNA levels of Flvcr, Hmox1, Fpn1, and Ftl1 at 10 hours are shown. There was no change in macrophage Flvcr expression with hemin treatment, in contrast to Hmox1, Fpn1, and Ftl1, which increased. Similar trends were seen at 3 and 24 hours (data not shown). Expression levels are shown as mRNA relative quantity (RQ) of treated cells relative to nontreated cells and normalized to β-actin expression. The mean and range of duplicate samples are shown. The data is representative of 2 independent experiments.
3.2. Macrophages Downregulate Flvcr Expression in Response to LPS Stimulation
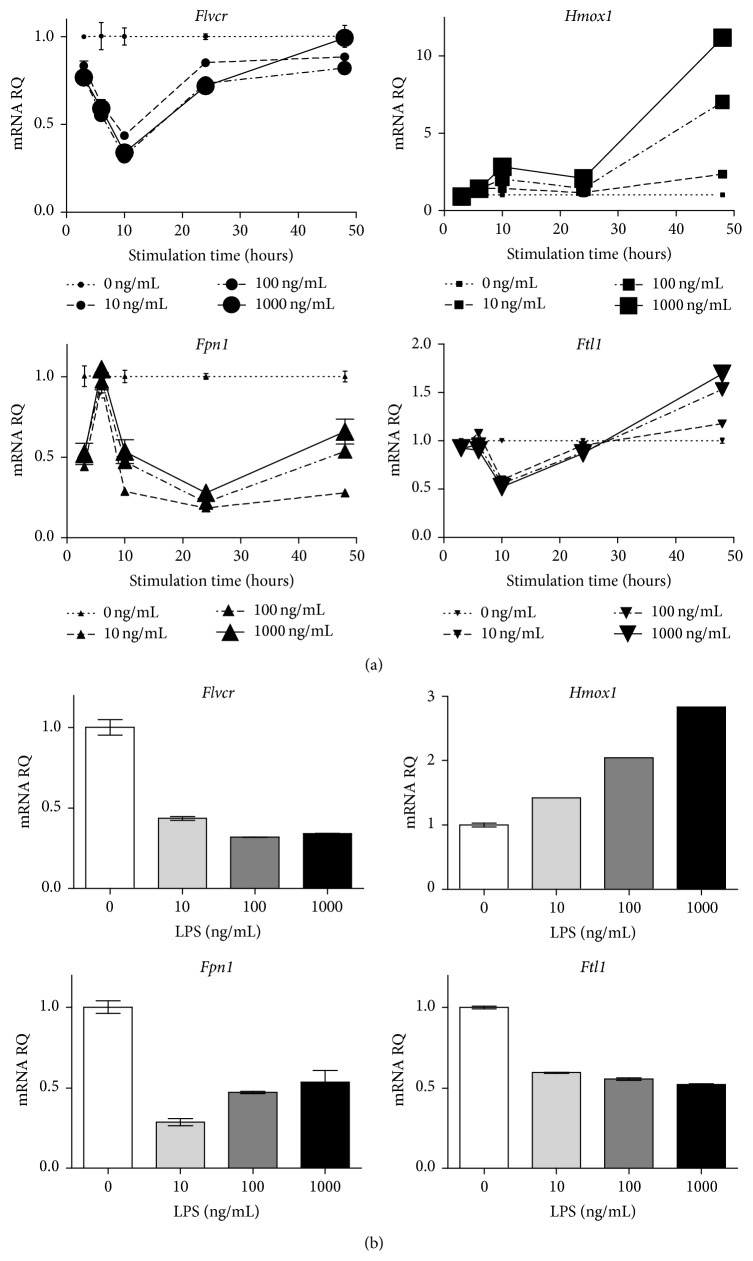
Fpn1 transcription is downregulated in macrophages stimulated by LPS [24, 25]; therefore we stimulated J774 with varying concentrations of LPS for different durations and quantified mRNA levels of Flvcr and key heme/iron regulatory genes. We found that Flvcr expression decreased rapidly upon LPS stimulation before recovering to baseline at 24–48 hours (Figure 2(a)). Fpn1 increased rapidly and transiently before then decreasing over the first 24 hours; Fpn1 recovery was slower and not complete by 48 hours (Figure 2(a)). This transient increase in Fpn1 expression prior to downregulation has not been previously described. As expected, Hmox1 expression increased with time. Ftl1 kinetics were similar to those of Flvcr though the initial decrease in expression was not as marked as Flvcr and at later time points Ftl1 increased above baseline (Figure 2(a)).
Figure 2.
Macrophages downregulate Flvcr in response to LPS stimulation. (a) J774 cells were stimulated with LPS at varying doses and times. Multiplex qPCR was then performed to assess mRNA levels of Flvcr, Fpn1, Hmox1, and Ftl1. The maximal decrease in Flvcr expression occurred at 10 hours and then recovered to baseline. (b) mRNA levels from the 10-hour time point in (a) are shown as bar graphs to demonstrate that Flvcr downregulation was LPS dose-responsive. Expression levels are shown as mRNA relative quantity (RQ) of LPS-treated cells relative to nontreated cells and normalized to β-actin expression. The mean and range of duplicate samples are shown. The data is representative of 3 independent experiments.
The decrease in Flvcr expression was dose-responsive between 0 and 100 ng with no further decrease at higher LPS doses, as seen in Figure 2(b), which shows heme and iron regulatory gene expression at 10 hours. Both Flvcr and Hmox1 show a dose-responsive decrease and increase in mRNA expression, respectively, whereas Fpn1 and Ftl1 had the maximal drop in expression at the lowest dose of LPS, 10 ng/mL (Figure 2(b)).
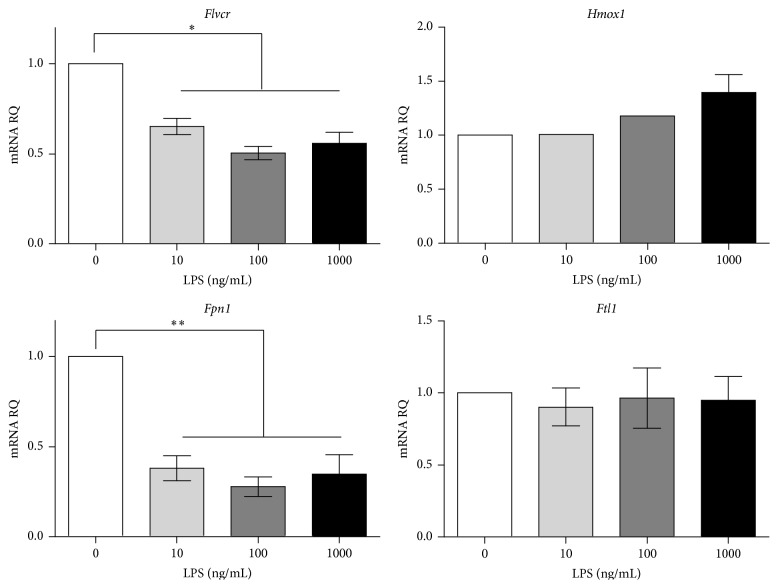
We next assessed the effect of LPS stimulation on primary murine bone-marrow-derived macrophages (BMDM) and again observed a dose-responsive decrease in Flvcr similar to the decrease in Fpn1 expression (Figure 3). The kinetics of Flvcr and Fpn1 downregulation in response to LPS were similar to that seen in J774 (data not shown). Thus, both primary BMDM and macrophage cell lines respond to LPS signaling by downregulating heme and iron export.
Figure 3.
Primary bone-marrow-derived murine macrophages (BMDM) downregulate Flvcr in response to LPS stimulation. Primary macrophages were treated with varying doses and duration of LPS and showed a dose-responsive decrease in both Flvcr and Fpn1 levels in response to 24 hours of LPS treatment. The mean and SEM are shown (n = 3 mice). The data is representative of 3 independent experiments. ∗ p < 0.001. ∗∗ p < 0.003.
3.3. Flvcr Expression is Differentially Regulated by Macrophage Polarization
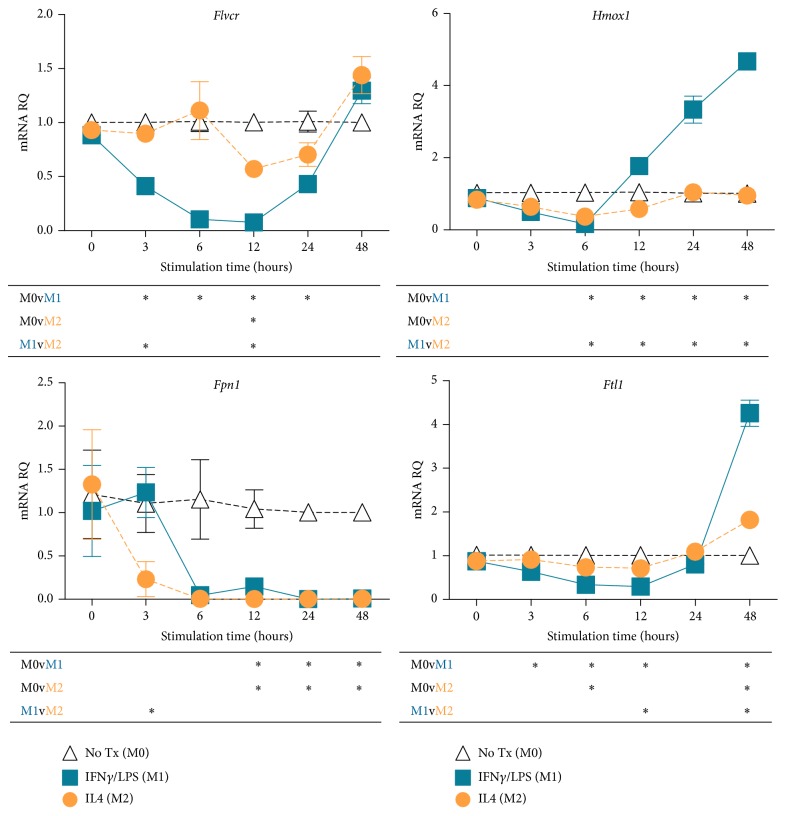
Activation of macrophages in vitro or in vivo by various stimuli leads to distinct gene expression patterns, a process referred to as macrophage polarization [23]. LPS and IFNγ treatment leads to M1 macrophage polarization, suited for combating infection and acute inflammation, whereas IL4 treatment leads to M2 polarization. M2 macrophages promote tissue regeneration and the return to baseline homeostasis. Most studies on macrophage regulation of iron balance have been done on M1 macrophages, but one study found that in contrast to M1 macrophages, M2-polarized human macrophages do not sequester iron but rather release iron to the surrounding tissues, promoting proliferation [26]. To determine whether Flvcr was differentially regulated in M1- versus M2-polarized macrophages, we generated primary murine undifferentiated macrophages (M0) and then treated with either LPS and IFNγ or IL4 to generate M1- and M2-polarized macrophages. Similar to previous reports, we found that Hmox1 and Ftl1 expression was not upregulated in M2 macrophages (Figure 4). However, both M1 and M2 macrophages downregulated Fpn1 mRNA, and in contrast to LPS treatment alone, Fpn1 mRNA remained suppressed at 48 hours. Interestingly, Flvcr expression decreased later and to a much lower extent in M2 versus M1 macrophages. Thus, M2 macrophages maintain Flvcr expression, possibly to export heme to cells in regenerating tissues.
Figure 4.
Macrophages downregulate Flvcr and Fpn1 expression in response to M1 and M2 polarization. Primary macrophages (M0) were treated with LPS/IFNγ or IL4 to polarize to M1 or M2 state, respectively. Both M1- and M2-polarized macrophages downregulated Fpn1 to a similar extent as compared to M0 macrophages. Interestingly, Flvcr downregulation in M2 macrophages occurred later and was less pronounced than in M1 macrophages. The mean and SEM are shown (n = 3 mice). Below each graph is a table summarizing statistical significance for all pairwise comparisons at each time point. ∗ indicates p ≤ 0.05. The data is representative of 2 independent experiments.
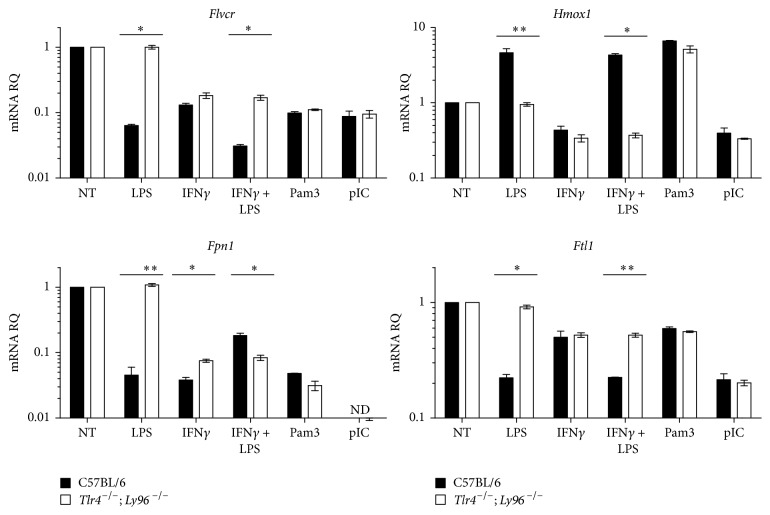
3.4. LPS-Induced Flvcr Downregulation Is TLR4-Dependent
LPS binds to the TLR4 receptor complex on macrophages to activate multiple downstream signaling pathways [27]. To confirm that LPS-induced Flvcr downregulation was mediated by the TLR4 pathway, we stimulated BMDM from Tlr4 −/−; Ly96 −/− mice or controls with LPS or LPS/IFNγ. Ly96 encodes MD2, a coreceptor with TLR4 required for LPS signaling. Loss of TLR4 and MD2 completely reversed LPS-induced downregulation of Flvcr and Fpn1 (Figure 5). To demonstrate specificity, we stimulated wild-type and Tlr4 −/−; Ly96 −/− BMDM with the TLR1/2 agonist Pam3Csk4 (Pam3) and TLR3 agonist polyinosinic:polycytidylic acid (pIC). We found that both Pam3 and pIC treatment led to Flvcr and Fpn1 downregulation in both wild-type and Tlr4 −/−; Ly96 −/− BMDM (Figure 5). Another group recently showed that TLR2 and TLR6 agonists cause Fpn1 mRNA downregulation in macrophages [28]. Thus, decreased transcription of the genes encoding both heme and iron export proteins is a common pathway following TLR activation in macrophages. Interestingly, Hmox1 was upregulated by LPS and Pam3, but not by pIC (Figure 5), suggesting distinct regulation of Flvcr as compared to other heme regulatory genes.
Figure 5.
Macrophages downregulate Flvcr expression in response to multiple TLR agonists, and the downregulation is TLR-dependent. Primary BMDM were generated from control C57BL/6 or Tlr4 −/− and Ly96 −/− mice and stimulated with various TLR agonists and IFNγ. Flvcr and Fpn1 downregulation in response to LPS was abrogated in Tlr4 −/− and Ly96 −/− macrophages. Stimulation with TLR1/2 agonist Pam3Csk4 (Pam3) and the TLR3 agonist polyinosinic:polycytidylic acid (pIC) also caused a decrease in Flvcr and Fpn1 expression in both wild-type and Tlr4 −/−; Ly96 −/− macrophages. The mean and SEM are shown (n = 3 mice). ND: not detected, as amplification was below the lower limit of detection. The data are representative of 2 independent experiments. ∗ p ≤ 0.01. ∗∗ p ≤ 0.001.
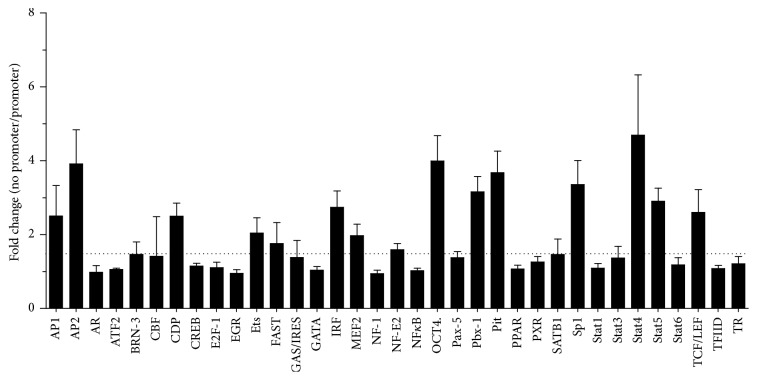
3.5. Transcription Factors Activated by LPS/TLR Signaling Bind to the Flvcr Promoter
To explore the connection between LPS stimulation and Flvcr mRNA transcription, we used the EPDnew eukaryotic promoter database [29] to identify the human Flvcr promoter sequence and then queried the promoter sequence for transcription factor (TF) binding sites using PROMO [30, 31]. This analysis revealed many potential binding sites motifs for transcription factors known to be expressed in macrophages such as NF-κB, IRF-1, and C/EBPβ [27] (see Supplemental Figure 1 of the Supplementary Material available online at http://dx.doi.org/10.1155/2016/4039038). We next used a multiplex TF binding assay to identify TF present in murine BMDM that can bind to the murine Flvcr promoter. Several of the TF with highest in vitro Flvcr promoter binding activity (Figure 6) such as STAT4, AP2, SP-1, and IRF-1 had predicted binding sites with the human Flvcr promoter (Supplemental Figure 1). Moreover, STAT4 [32] and IRF-1 [33] are known downstream mediators of LPS/TLR4 signaling in macrophages.
Figure 6.
LPS-activated transcription factors in macrophages bind to Flvcr promoter. Nuclear extracts were prepared from primary murine BMDM and incubated with or without purified Flvcr promoter DNA. Transcription factor (TF) activity was measured using the Signosis Promoter Binding Transcription Factor Profiling Array 1 and results are presented as the amount of TF activity without promoter over TF activity with promoter, reflecting promoter binding. The dotted line indicates 1.5-fold change. Several of the TF with highest Flvcr promoter binding (STAT4, AP2, SP-1, and IRF-1) had predicted binding sites with the human Flvcr promoter (Supplemental Figure 1), and STAT4 and IRF-1 are known downstream mediators of LPS/TLR4 signaling in macrophages. The mean and SEM are shown (n = 3 mice).
4. Discussion
In this study, we found that primary and immortalized murine macrophages downregulate Flvcr mRNA levels upon LPS stimulation, similar to the downregulation of Fpn1 expression previously described [24, 25]. Macrophages also downregulated Flvcr and Fpn1 in response to TLR1/2 and TLR3 agonists, suggesting that heme and iron sequestration in macrophages is a general response to inflammatory/infectious stimuli. It was previously reported that M1 polarization causes human macrophages to sequester iron through FPN1 downregulation while M2 polarization leads to increased FPN1 protein expression [26]. A more recent study [34] found that Fpn1 expression was decreased in murine M0 macrophages polarized to both the M1 and M2 states. Interestingly, this study also found that heme (in the form of RBC or free heme) polarizes M0 macrophages to the M1 state [34]. We found that, in murine macrophages, both M1 and M2 polarization caused decreased expression of Fpn1 and Flvcr, though notably the decrease in Flvcr expression occurred later and was less marked under M2 polarizing conditions. The hypothesis is that macrophages sequester iron in response to infection (M1) and export iron and heme under M2 conditions in which tissue regeneration and proliferating cells have higher demand for heme and iron. Future studies could explore Flvcr and Fpn1 expression in vivo under physiologic conditions of M1 (acute infection) and M2 polarization (late-stage wound healing) or investigate how tumor-associated macrophages regulate heme and iron.
Surprisingly, Flvcr mRNA levels in J774 macrophages did not change significantly in response to heme treatment. While free heme may not alter Flvcr mRNA expression in macrophages, it is possible that Flvcr expression may change in response to macrophage erythrophagocytosis, especially in splenic macrophages specialized to take up senescent RBC. Heme has been shown to regulate Fpn1 transcription through binding the transcriptional factors Btb and Cnc Homology 1 (BACH1) and Nuclear Factor Erythroid 2-like (NRF2), which associate with a conserved Maf Recognition Element (MARE)/Antioxidant Response Element (ARE) in the Fpn1 promoter [35]. We did not identify any MARE/ARE elements in the Flvcr promoter.
Thus, while HMOX1 and FPN1 in macrophages may respond to changes in intracellular heme and iron levels in addition to inflammatory/infectious signals, the primary role of FLVCR in macrophages may be the regulation of heme in response to infection/inflammation. It is notable that the decrease in Flvcr expression occurs rapidly within 10 hours after LPS stimulation, while Fpn1 expression had an initially small increase followed by a slower decrease in expression (Figures 2(a) and 4). Whether this transient Fpn1 increase prior to the subsequent downregulation is functionally important is not known. The different expression kinetics suggest that Flvcr and Fpn1 are regulated differently. FPN1 protein expression is also regulated posttranslationally through inflammation-induced hepcidin [6]. It is not known whether FLVCR is also posttranslationally regulated, though one study using transfected cell lines found that FLVCR had a long half-life (>16 hours) [36]. It has been difficult to study the localization and regulation of FLVCR protein in vivo or from ex vivo murine cells because there is no antibody available. Once alternative strategies (such as genetic knock-in of epitope tag sequences) make in vivo and ex vivo of murine FLVCR detection and localization feasible, it will be important to study the kinetics and trafficking of FLVCR and FPN1 protein after LPS treatment.
The decrease in Flvcr expression precedes the major increase in Hmox1 expression, at which point Flvcr expression has returned to baseline. This suggests that the decrease in FLVCR is functionally important in the first hours after infection, and as other mechanisms for systemic iron regulation (hepcidin-mediated FPN1 degradation, increased Ferritin expression) are initiated, FLVCR returns to baseline. One explanation is that early FLVCR downregulation might be important for macrophage killing of intracellular pathogens. Heme and iron-containing enzymes produce the reactive oxygen species and other compounds needed for intracellular killing of bacteria [19]. Macrophages that have just encountered and/or endocytosed bacteria may downregulate FLVCR in order to increase heme available for cytolytic enzymes. A recent review highlighted the “macrophage paradox,” that is the finding that many pathogens preferentially replicate inside macrophages in spite of their specialized killing function [37], and different bacteria have different intracellular niches within macrophages. Given that heme and iron are also trafficked and regulated differently in intracellular compartments [12, 38–40], there are likely several layers of regulation of heme and iron regulatory proteins at transcriptional, posttranscriptional, translational, and posttranslational (including trafficking) levels required to meet the challenges presented by specific pathogens. This is supported by the recent finding that the survival of Listeria monocytogenes localized to different intracellular compartments in macrophages was differentially altered by FPN1 expression [41]. Moreover, macrophages are not a uniform population, but rather there are several subtypes and macrophage differentiation states which serve specific functions in different tissues and depending on the conditions [23]. Macrophages that are specialized for erythrophagocytosis may regulate heme and iron regulatory proteins in response to heme and iron levels [40] rather than inflammatory signals as we observed here. Our finding that macrophages dynamically regulate Flvcr expression in response to TLR signaling points to a new potential role for FLVCR and heme export in macrophages during infection and inflammation. Future studies aimed at elucidating the transcriptional and posttranscriptional regulation of FLVCR in response to TLR and inflammatory signaling will improve our understanding of the complex interplay cell and tissue-localized demands for heme and iron and systemic heme/iron homeostasis.
Supplementary Material
Supplemental Figure 1: The EPDnew (1) eukaryotic promoter database was used to identify the Flvcr promoter sequence from -499 to +100 base pairs relative to the transcription start site. This sequence was queried for human transcription factor (TF) binding sites using PROMO (2) and the resulting output is shown above. Of the TF binding sites identified, many are known to be expressed in macrophages and activated by LPS signaling.
Acknowledgments
The authors thank Drs. Raymond Doty, Andrea Schietinger, and Peter Lauer for helpful discussion and Alicia Brasfield for technical support. This work was supported by National Institutes of Health Grants K08 CA158069 (to Mary Philip), R01 HL031823 (to Janis L. Abkowitz), and R01 DK085146 (to Janis L. Abkowitz) and by an American Society of Hematology Basic Science Fellow Award (to Mary Philip) and Trainee Research Award (to Edison Y. Chiu). Mary Philip was also supported by National Institutes of Health Grant T32 CA009515.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Cassat J. E., Skaar E. P. Iron in infection and immunity. Cell Host and Microbe. 2013;13(5):509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kassebaum N. J., Jasrasaria R., Naghavi M., et al. A systematic analysis of global anemia burden from 1990 to 2010. Blood. 2014;123(5):615–624. doi: 10.1182/blood-2013-06-508325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sazawal S., Black R. E., Ramsan M., et al. Effects of routine prophylactic supplementation with iron and folic acid on admission to hospital and mortality in preschool children in a high malaria transmission setting: community-based, randomised, placebo-controlled trial. The Lancet. 2006;367(9505):133–143. doi: 10.1016/s0140-6736(06)67962-2. [DOI] [PubMed] [Google Scholar]
- 4.Kortman G. A., Mulder M. L., Richters T. J., et al. Low dietary iron intake restrains the intestinal inflammatory response and pathology of enteric infection by food-borne bacterial pathogens. European Journal of Immunology. 2015;45(9):2553–2567. doi: 10.1002/eji.201545642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheldon J. R., Heinrichs D. E. Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS Microbiology Reviews. 2015;39(4):592–630. doi: 10.1093/femsre/fuv009. [DOI] [PubMed] [Google Scholar]
- 6.Drakesmith H., Prentice A. M. Hepcidin and the iron-infection axis. Science. 2012;338(6108):768–772. doi: 10.1126/science.1224577. [DOI] [PubMed] [Google Scholar]
- 7.Nairz M., Haschka D., Demetz E., Weiss G. Iron at the interface of immunity and infection. Frontiers in Pharmacology. 2014;5, article 152 doi: 10.3389/fphar.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krause A., Neitz S., Mägert H.-J., et al. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Letters. 2000;480(2-3):147–150. doi: 10.1016/S0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 9.Ganz T. Systemic iron homeostasis. Physiological Reviews. 2013;93(4):1721–1741. doi: 10.1152/physrev.00008.2013. [DOI] [PubMed] [Google Scholar]
- 10.Nemeth E., Tuttle M. S., Powelson J., et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306(5704):2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 11.Contreras H., Chim N., Credali A., Goulding C. W. Heme uptake in bacterial pathogens. Current Opinion in Chemical Biology. 2014;19(1):34–41. doi: 10.1016/j.cbpa.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan A. A., Quigley J. G. Control of intracellular heme levels: heme transporters and heme oxygenases. Biochimica et Biophysica Acta. 2011;1813(5):668–682. doi: 10.1016/j.bbamcr.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quigley J. G., Burns C. C., Anderson M. M., et al. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood. 2000;95(3):1093–1099. [PubMed] [Google Scholar]
- 14.Quigley J. G., Yang Z., Worthington M. T., et al. Identification of a human heme exporter that is essential for erythropoiesis. Cell. 2004;118(6):757–766. doi: 10.1016/j.cell.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z., Philips J. D., Doty R. T., et al. Kinetics and specificity of feline leukemia virus subgroup c receptor (flvcr) export function and its dependence on hemopexin. The Journal of Biological Chemistry. 2010;285(37):28874–28882. doi: 10.1074/jbc.m110.119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keel S. B., Doty R. T., Yang Z., et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319(5864):825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- 17.Philip M., Funkhouser S. A., Chiu E. Y., et al. Heme exporter FLVCR is required for T cell development and peripheral survival. The Journal of Immunology. 2015;194(4):1677–1685. doi: 10.4049/jimmunol.1402172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizzardini M., Terao M., Falciani F., Cantoni L. Cytokine induction of haem oxygenase mRNA in mouse liver. Interleukin 1 transcriptionally activates the haem oxygenase gene. Biochemical Journal. 1993;290(2):343–347. doi: 10.1042/bj2900343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West A. P., Shadel G. S., Ghosh S. Mitochondria in innate immune responses. Nature Reviews Immunology. 2011;11(6):389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmström K. M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nature Reviews Molecular Cell Biology. 2014;15(6):411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 21.Hajjar A. M., Ernst R. K., Fortuno E. S., III, et al. Humanized TLR4/MD-2 mice reveal LPS recognition differentially impacts susceptibility to Yersinia pestis and Salmonella enterica . PLoS Pathogens. 2012;8(10) doi: 10.1371/journal.ppat.1002963.e1002963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(9, article e45) doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray P., Allen J., Biswas S., et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X.-B., Nguyen N.-B. H., Marquess K. D., Yang F., Haile D. J. Regulation of hepcidin and ferroportin expression by lipopolysaccharide in splenic macrophages. Blood Cells, Molecules, and Diseases. 2005;35(1):47–56. doi: 10.1016/j.bcmd.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Yang F., Liu X.-B., Quinones M., Melby P. C., Ghio A., Haile D. J. Regulation of reticuloendothelial iron transporter MTP1 (Slc11a3) by inflammation. The Journal of Biological Chemistry. 2002;277(42):39786–39791. doi: 10.1074/jbc.m201485200. [DOI] [PubMed] [Google Scholar]
- 26.Recalcati S., Locati M., Marini A., et al. Differential regulation of iron homeostasis during human macrophage polarized activation. European Journal of Immunology. 2010;40(3):824–835. doi: 10.1002/eji.200939889. [DOI] [PubMed] [Google Scholar]
- 27.Medzhitov R., Horng T. Transcriptional control of the inflammatory response. Nature Reviews Immunology. 2009;9(10):692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 28.Guida C., Altamura S., Klein F. A., et al. A novel inflammatory pathway mediating rapid hepcidin-independent hypoferremia. Blood. 2015;125(14):2265–2275. doi: 10.1182/blood-2014-08-595256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dreos R., Ambrosini G., Périer R. C., Bucher P. EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Research. 2013;41(1):D157–D164. doi: 10.1093/nar/gks1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messeguer X., Escudero R., Farré D., Núñez O., Martínez J., Albà M. M. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18(2):333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 31.Farré D., Roset R., Huerta M., et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Research. 2003;31(13):3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuroda E., Kito T., Yamashita U. Reduced expression of STAT4 and IFN-gamma in macrophages from BALB/c mice. The Journal of Immunology. 2002;168:5477–5482. doi: 10.4049/jimmunol.168.11.5477. [DOI] [PubMed] [Google Scholar]
- 33.Barber S. A., Fultz M. J., Salkowski C. A., Vogel S. N. Differential expression of interferon regulatory factor 1 (IRF-1), IRF-2, and interferon consensus sequence binding protein genes in lipopolysaccharide (LPS)-responsive and LPS-hyporesponsive macrophages. Infection and Immunity. 1995;63(2):601–608. doi: 10.1128/iai.63.2.601-608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinchi F., Costa da Silva M., Ingoglia G., et al. Hemopexin therapy reverts heme-induced proinflammatory phenotypic switching of macrophages in a mouse model of sickle cell disease. Blood. 2016;127(4):473–486. doi: 10.1182/blood-2015-08-663245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marro S., Chiabrando D., Messana E., et al. Heme controls ferroportin1 (FPN1) transcription involving Bach1, Nrf2 and a MARE/ARE sequence motif at position—7007 of the FPN1 promoter. Haematologica. 2010;95(8):1261–1268. doi: 10.3324/haematol.2009.020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanatori I., Yasui Y., Miura K., Kishi F. Mutations of FLVCR1 in posterior column ataxia and retinitis pigmentosa result in the loss of heme export activity. Blood Cells, Molecules, and Diseases. 2012;49(1):60–66. doi: 10.1016/j.bcmd.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Price J. V., Vance R. E. The macrophage paradox. Immunity. 2014;41(5):685–693. doi: 10.1016/j.immuni.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 38.Chiabrando D., Vinchi F., Fiorito V., Mercurio S., Tolosano E. Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes. Frontiers in Pharmacology. 2014;5, article 61 doi: 10.3389/fphar.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evstatiev R., Gasche C. Iron sensing and signalling. Gut. 2012;61(6):933–952. doi: 10.1136/gut.2010.214312. [DOI] [PubMed] [Google Scholar]
- 40.Delaby C., Rondeau C., Pouzet C., et al. Subcellular localization of iron and heme metabolism related proteins at early stages of erythrophagocytosis. PLoS ONE. 2012;7(7) doi: 10.1371/journal.pone.0042199.e42199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haschka D., Nairz M., Demetz E., Wienerroither S., Decker T., Weiss G. Contrasting regulation of macrophage iron homeostasis in response to infection with Listeria monocytogenes depending on localization of bacteria. Metallomics. 2015;7(6):1036–1045. doi: 10.1039/C4MT00328D. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: The EPDnew (1) eukaryotic promoter database was used to identify the Flvcr promoter sequence from -499 to +100 base pairs relative to the transcription start site. This sequence was queried for human transcription factor (TF) binding sites using PROMO (2) and the resulting output is shown above. Of the TF binding sites identified, many are known to be expressed in macrophages and activated by LPS signaling.