Abstract
We previously found that deletion of the multifunctional factor ANP32B (a.k.a. SSP29, APRIL, PAL31, PHAPI2) resulted in a severe but strain-specific defect resulting in perinatal lethality. The difficulty in generating an adult cohort of ANP32B-deficient animals limited our ability to examine adult phenotypes, particularly cancer-related phenotypes. We bred the Anp32b-null allele into the BALB/c and FVB/N genetic background. The BALB/c, but not the FVB/N, background provided sufficient frequency of adult Anp32b-null (Anp32b−/−) animals. From these, we found no apparent oncogenic role for this protein in mammary tumorigenesis contrary to what was predicted based on human data. We also found runtism, pathologies in various organ systems, and an unusual clinical chemistry signature in the adult Anp32b−/− mice. Intriguingly, genome-wide single-nucleotide polymorphism analysis suggested that our colony retained an unlinked C57BL/6J locus at high frequency. Breeding this locus to homozygosity demonstrated that it had a strong effect on Anp32b−/− viability indicating that this locus contains a modifier gene of Anp32b with respect to development. This suggests a functionally important genetic interaction with one of a limited number of candidate genes, foremost among them being the variant histone gene H2afv. Using congenic breeding strategies, we have generated a viable ANP32B-deficient animal in a mostly pure background. We have used this animal to reliably exclude mouse ANP32B as an important oncogene in mammary tumorigenesis. Our further phenotyping strengthens the evidence that ANP32B is a widespread regulator of gene expression. These studies may also impact the choice of subsequent groups with respect to congenic breeding versus de novo zygote targeting strategies for background analyses in mouse genetics.
Keywords: chromatin regulators, congenic breeding, modifier locus, strain-dependent SNPs
Introduction
Although phenotypes in mice provide significant insight into the role of particular genes in human development and disease etiology, the genes are commonly examined in only one or two genetic backgrounds [10, 24]. Strain-dependent phenotypes have long been known to complicate developmental genetics [2, 5, 8, 11,12,13]. As new gene-ablation systems provide for fast and efficient gene ablations in zygotes [25, 30], the mouse-genetics community must plan to utilize strain-dependent phenotypes for maximal benefit, particularly with respect to identification of genetic interactions. Novel zygote targeting strategies in purebred animals will facilitate hypothesis-driven research, but it may also limit opportunities for genetic interaction discovery. Regardless, the BALB/c strain, being distantly related to the most commonly used C57BL/6J strain [19] and the second most frequently cited strain for analyses in mice [7], is an excellent candidate strain for examining genetic background effects on phenotypes.
The acidic (leucine-rich) nuclear phosphoprotein family member B (ANP32B) is a member of the ANP32 family of proteins that are composed of an amino terminal leucine-rich repeat domain and a carboxy-terminal low-complexity acidic region [reviewed in 22]. The conservation of these factors combined with their overlapping biochemical activities suggest that they fulfill important activities, potentially with some redundancy.
ANP32B (also known as SSP29, APRIL, PAL31, PHAPI2) has reported roles in apoptosome activation [6], ELAVL1/HuR-assisted mRNA transport [3, 4], Caspase-3 inhibition [23, 27], and KLF5-mediated transcriptional repression by means of chromatin remodelling [15]. ANP32B is expressed in proliferating tissue [1, 26] and its mRNA expression is also a negative prognostic indicator for human breast cancer [20] although ANP32B may also function as a tumor-suppressor protein based on genome-wide mutation mapping [29].
In contrast to ANP32A and ANP32E, which do not demonstrate apparent knockout phenotypes [18, 21, 30], we previously showed that ANP32B is important for normal mouse development [20]. The Anp32b-null (Anp32b−/−) phenotype was strain dependent with almost fully penetrant lethality in the C57BL/6J congenic background and semi-penetrant perinatal lethality in the mixed C57BL/6J-129P2/OlaHsd background, hereafter referred to as the “mixed background”. In the mixed background the ANP32B deficiency was characterized by runtism, premature aging, various pathologies and anatomical defects including eustachian tube defects [20].
Here, we describe the phenotyping of the ANP32B deficiency in the FVB/N and BALB/c strains. We show that, as was seen in the C57BL/6J background, FVB/N-congenic Anp32b−/− mice occur very infrequently. In contrast, a large percentage of six-generation BALB/c-congenic ANP32B-deficient mice were viable. In addition to presenting cancer and developmental data on this background, we present the discovery a modifier locus of the Anp32b−/− viability phenotype in the BALB/c background and suggest that the modifier gene may be the variant histone gene H2afv.
Methods and Materials
Animals
Mice containing the Anp32b-null allele in the congenic C57BL/6J background [20] were bred with BALB/c and FVB/N wild-type mice (InVivos Pte Ltd., Singapore). All mice were provided 5% irradiated chow and water ad libitum and housed under SPF conditions in individually ventilated cages. Genotyping was performed on tail biopsies by PCR according to a previously reported protocol [20]. Animals were maintained in social housing except in cases where males demonstrated aggression toward cocaged mice. In such cases environmental differences were minimized by biweekly exchange of bedding between paired animals. Euthanasia was carried out by carbon dioxide asphyxiation followed by cervical dislocation.
For aging experiments, pairs of animals were examined daily for signs of morbidity including signs of body weight loss; inability to eat or drink; behavioral abnormalities such as hunched posture, shivering, decreased activity, or immobility; or clinical symptomatology such as ruffled fur-coat, lameness, paralysis, dyspnea, edema, not eating or drinking, and abnormal discharge. Evidence of any of these was indicative of humane endpoint.
Chemical cancer induction
Female experimental pairs were provided the polycyclic hydrocarbon 7,12-dimethylbenz (a) anthracene (DMBA) per oral as described in [9] with small modifications. To facilitate orogastric gavage, mice were maintained without chow for 30 min prior to gavage and DMBA was dosed using 0.05 ml of 20 mg/ml DMBA in tricaprylin (Tokyo Chemical Industry Co., Ltd.). Animals were examined daily for signs of morbidity including signs of body weight loss; inability to eat or drink, behavioural abnormalities such as hunched posture, shivering, decreased activity, or immobility; or clinical symptomatology such as ruffled fur coat, lameness, paralysis, dyspnea, edema, not eating or drinking, and abnormal discharge. Evidence of any of these or evidence of a tumor of 2 cm in diameter was indicative of humane endpoint.
Clinical chemistry
After CO2 asphyxiation, blood was collected by cardiac puncture from paired animals in K2-EDTA tubes (cat # 365973, Becton, Dickenson and Co., Franklin Lakes, NJ, USA). Plasma was separated from leukocytes by centrifugation (4°C, 4,250 × g), snap frozen in liquid nitrogen, and stored at −20°C. Samples were shipped frozen for expanded toxicity analysis (cat # 60514, IDEXX BioResearch, Columbia, MO, USA).
Immunohistochemistry
Tumors from DMBA-treated mice were stained with anti-cytokeratin 5 (Covance) and anti-cytokeratin 8. The TROMA-1 antibody, against cytokeratin 8, developed by Rolf Kemler and Philip Brulet was obtained from the Developmental Studies Hybridoma Bank, created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) of the National Institutes of Health (NIH) and maintained at the University of Iowa.
Genome analysis
Four random heterozygous mice were selected from separate BALB/c-congenic breeding pairs for strain-specific single-nucleotide polymorphism (SNP) analysis (Harlan Labs). Targeted rs6190775 genotyping was performed using an SNP genotyping assay (Invitrogen). Custom exome sequencing was performed (Genomax Technologies, Singapore) on a separate cohort of mice that were selected from rs6190775-homozygous Anp32b-heterozygous stocks.
Ethics statement
All animal care and experiments were performed under protocol 2012/SHS/725, which was approved by the SingHealth Institutional Animal Care and Use Committee (IACUC). The approval process required agreement to daily animal health checks for animals put at risk due to aging and tumorigenesis studies. The IACUC recognized that the study aims were not aided by spontaneous animal deaths and, therefore, the authors had incentive to minimize spontaneous deaths by daily monitoring.
Results
BALB/c-congenic ANP32B-deficient animals are partially viable
The Anp32b−/− phenotype is complex and background dependent. In an attempt to identify a genetically pure strain that provides viable Anp32b−/− mice, we backcrossed the Anp32b-null allele from a congenic C57BL/6J background into FVB/N and BALB/c mice for six generations each, to generate two congenic strains. For these six generations the Anp32b-null allele was maintained at heterozygosity.
After breeding six generations, heterozygotes (Anp32b+/−) of each strain were crossed to generate congenic Anp32b−/− animals. In order to have unequivocal parentage, all heterozygote breedings were performed using one male and one female per cage. Consistent with previous analyses, we found that viability was strain dependent (Table 1). The congenic FVB/N strain produced very few Anp32b−/− mice that survived until typing at postnatal day 14 (Table 1A). Strangely, three homozygous null mice that survived were born during one week within two litters. This suggests that unknown environmental factors may be playing a role in the survival of these mice. Ultimately, no environmental factors could be identified to explain the clustered survival of these mice.
Table 1. Strain-dependent survival of Anp32b-deficient mice. Mendelian ratios of Anp32b heterozygote intercrosses from six-generation (A) FVB/N and (B) BALB/c congenic backgrounds.
| (A) FVB/N Congenic | (B) BALB/c Congenic | ||||||
|---|---|---|---|---|---|---|---|
| Anp32b genotype | Anp32b genotype | ||||||
| +/+ | +/– | –/– | +/+ | +/– | –/– | ||
| Expected | 62 | 123 | 62 | 83 | 167 | 83 | |
| Observed | 92 | 148 | 6 | 102 | 181 | 50 | |
(A) Chi-square test: P < 10-15, (B) Chi-square test: P < 10-4.
In contrast, the BALB/c backcrossed Anp32b+/− breedings produced a significant number of Anp32b−/− mice (Table 1B). This varied from Mendelian ratios but not to the degree evident in FVB/N or C57BL/6J [20]. These results suggest that the BALB/c background can be used as a tool for examining the Anp32b−/− mice in a commonly used strain. We then examined male and female fertility of the BALB/c-congenic ANP32B-deficient animals by allowing three male and three female null mice to breed with heterozygotes. We found that all of the BALB/c-congenic Anp32b−/− animals were able to produce litters. Thus, the BALB/c-congenic ANP32B-deficient animals of both sexes are fertile.
Phenotype of the BALB/c congenic Anp32b−/− mice
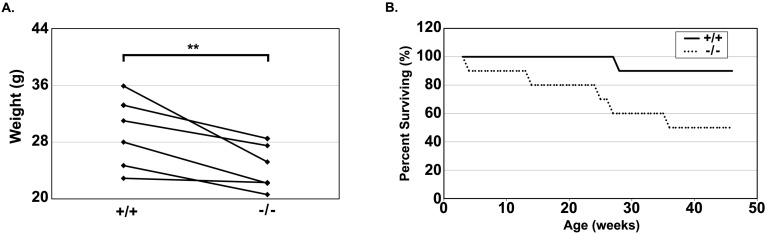
With sufficient BALB/c congenic Anp32b−/− mice surviving to weaning, we set aside 10 pairs (6 male, 4 female) of sex-matched littermate wild-type and null mice for analysis of a potential aging phenotype. Runtism in the Anp32b−/− mice is one of the most prominent phenotypes in the mixed background Anp32b−/− mouse. In order to examine whether runtism was evident in the BALB/c congenic Anp32b−/− mice, we measured weights of 6 pairs of littermates that survived beyond 24 weeks and found that an average 15.9% (± 3.7% SEM) reduction in weight in the null mice compared to the co-caged wild-type mice (P<0.01 by paired t-test; Fig. 1A). Thus, runtism is a common Anp32b−/− phenotype in both the mixed background and the BALB/c congenic animals.
Fig. 1.
BALB/c congenic Anp32b−/− mice demonstrate growth and survival defects. (A) Weights of cocaged, sex-matched littermate pairs of adult BALB/c-congenic wild-type (+/+) and Anp32b-null (−/−) mice. **, P<0.01 by Student’s t-test; (B) Kaplan-Meier survival curve of adult BALB/c-congenic wild-type (+/+) and Anp32b-null (−/−) mice. Survival was determined by absence of defined humane endpoints. Results were statistically significant by log-rank analysis (P<0.05).
To examine any potential pathologies, the animals were allowed to age to 46 weeks. Mice were examined daily for signs of pathology in a genotype-blinded manner until humane endpoints were reached. In this analysis, 1/10 Anp32b+/+ mice and 4/10 Anp32b−/− mice died prior to humane endpoint criteria being evident. Genotypes of animals at humane endpoint or death were recorded and necropsies were performed. Figure 1B shows the Kaplan-Meier curve for untreated BALB/c congenic ANP32B-competent and ANP32B-deficient animals. With 50% of Anp32b−/− animals reaching endpoint by 36 weeks of age, the loss of ANP32B has a statistically significant effect on survival of these mice (P<0.05 by log-rank analysis). Thus, consistent with what was seen in the mixed background, ANP32B deficiency compromises adult viability in the BALB/c congenic animals.
Unfortunately, among the mice analyzed, and consistent with the findings of the mixed genetic background, the defects in the aged BALB/c congenic ANP32B-deficient mice were irregular. Individual incidents of hydrocephalus, pulmonary adenoma, lymphoid aggregates on lungs, hyperplastic salivary gland, hamartoma of cardiomyocytes in the lung, segmental aplasia of the uterine horn, cardiomegaly, and dilated pancreatic ducts were noted in these mice. Also consistent with what was seen in the mixed-bred background, two separate incidents of hemorrhage were noted: one ovarian and one subpleural. The lack of consistent anatomical defects in the context of high penetrance of defects and overall reduced viability associated with ANP32B deficiency suggests that ANP32B is important in a wide range of tissues in the body.
In addition to examining anatomical differences in these mice, we also tested clinical chemistry parameters of blood from adult pairs of ANP32B-competent and ANP32B-deficient mice. As shown in Table 2, we identified that each of the parameters of total protein, total cholesterol, and total bilirubin were lower in the ANP32B-deficient mouse to a highly statistically significant degree (P<0.001 by Wilcoxon signed-rank test). High-density lipoprotein cholesterol was also reduced in the null mice to a lesser extent. No other parameters examined were altered in a statistically significant manner between the mice (Supplemental Table S1).
Table 2. Changes in clinical chemistry in adult Anp32b −/− mice.
| Parameter | ||||
|---|---|---|---|---|
| Total protein (g/dl) | Total bilirubin (mg/dl) | Total cholesterol (mg/dl) | HDL cholesterol (mg/dl) | |
| +/+ average | 5.525 ± 0.09 | 0.2 ± 0.02 | 118.25 ± 3.3 | 60.63 ± 4.1 |
| –/– average | 5.212 ± 0.13 | 0.125 ± 0.02 | 100.38 ± 6.9 | 54.75 ± 3.0 |
| Average pairwise reduction | 5.54% ± 1.1% | 35.42% ± 8% | 14.03% ± 3.5% | 10.11% ± 3.5% |
| Statistical test | Signed-rank | Signed-rank | Signed-rank | Student’s t-test |
| P-value | < 0.001 | < 0.001 | < 0.001 | < 0.05 |
Indicated parameters demonstrated statistically significant changes between adult paired +/+ and −/− mice (n=8). Statistical tests were chosen based on Anderson-Darling normality.
Anp32b deficiency does not protect against mammary tumorigenesis
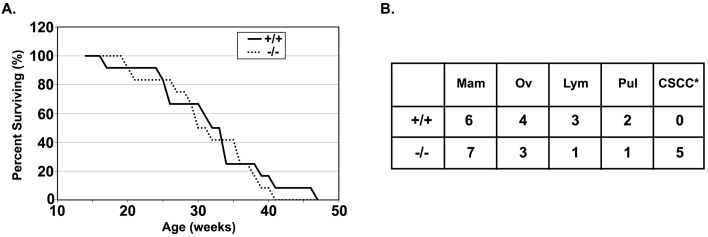
Earlier studies suggested that ANP32B might be a prognostic factor in breast cancer in humans [20]. With a viable ANP32B-deficient mouse in the congenic BALB/c background, we were able to study mammary tumorigenesis directly in the mouse using a chemical induction strategy involving DMBA [9]. In this analysis, 5/12 Anp32b+/+ mice and 3/12 Anp32b−/− mice died prior to humane endpoint criteria being evident. We found that ANP32B-competent and ANP32B-deficient mice reached humane endpoint or died at similar times after treatment (Fig. 2A). These data demonstrated no statistically significant effect of genotype either in isolation (P=0.76) or accounting for spontaneous deaths of the Anp32b−/− mice (P=0.31) by log-rank testing. Furthermore, the incidences of mammary, ovarian, and lymphoid tumors were not statistically different between the different genotypes (Fig. 2B). One evident difference was the incidence of cutaneous squamous cell carcinoma (CSCC) in the Anp32b−/− group (P<0.05 by chi-square test), occurring either biphasic with mammary tumors or independently. This suggests that ANP32B may have a tumor-suppressive function in cutaneous squamous epithelia.
Fig. 2.
Cancer induction in the BALB/c-congenic ANP32B-deficient mouse. (A) Kaplan-Meier survival curve of DMBA-treated BALB/c-congenic wild-type (+/+) and Anp32b-null (−/−) mice. Survival was determined by absence of defined humane endpoints. No genotype effect was evident at statistically significant levels. (B) Incidence of different tumor types in DMBA-treated mice. Mam, mammary; Ov, ovarian; Lym, lymphoid; Pul, pulmonary; CSCC, cutaneous squamous cell carcinoma. *, P<0.05 by Chi-square test.
Congenic BALB/c ANP32-deficient mice occur at variable frequencies
In our examination of the frequency of ANP32B-deficient BALB/c-congenic mice, we noted discrepancies in obtaining these null mice over time. The frequency of obtaining ANP32B-deficient mice from heterozygous breedings varied from 18% attainment in the first 10 litters to 12% among litters 51–60 (Supplementary Table S2A). As a similar effect was noted but not recorded for the earliest generations of the mixed background, we wanted to more closely examine this effect. One hypothesis to explain this finding was that ANP32B deficiency in the maturing oocyte might specifically reduce viability of these oocytes prior to oestrus. A corollary to this hypothesis is that, since oocytes carry only one allele, such an effect would also skew Anp32b+/− to Anp32b+/+ ratios. Whereas the Anp32b+/− to Anp32b+/+ ratios were commonly below the expected Mendelian ratios, there was no evident association with rates of Anp32b−/− acquisition. Thus it is unlikely that reduced oocyte viability is the cause of the reduced number of Anp32b−/− progeny. This hypothesis would also predict that older dams would produce fewer Anp32b−/− pups. When we stratified the litters based on age of the dam at time of litter birth we did not find an association (Supplementary Table S2B), again suggesting that oocyte development in the dam is not the primary reason for variable frequencies of Anp32b−/− mice occurring over time in the congenic BALB/c background.
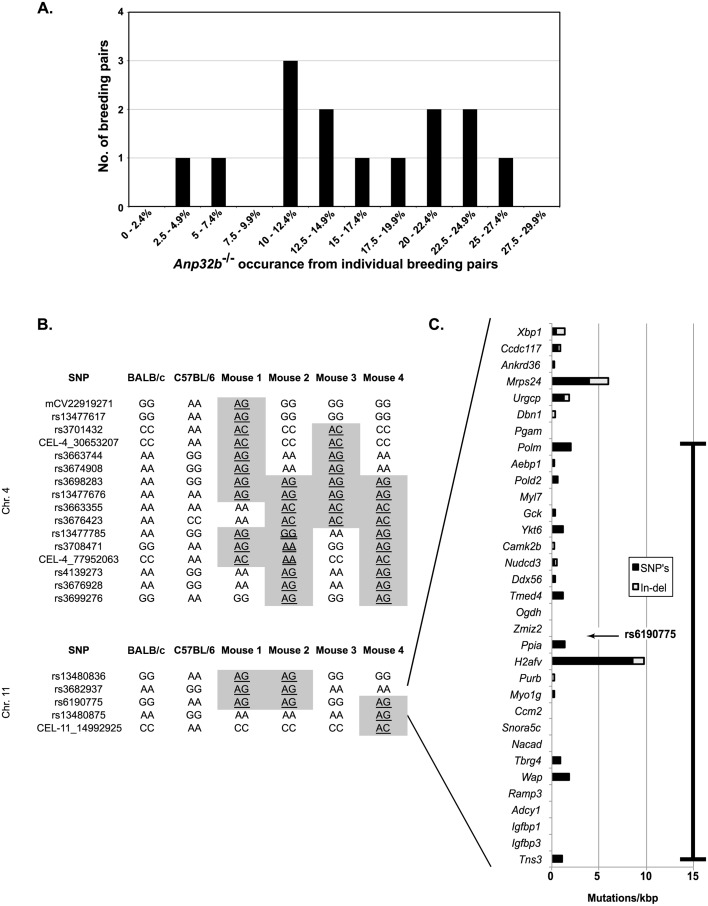
We next examined whether there might be genetic variability within our population by examining the frequency at which nullizygous mice occurred from individual breeding pairs. When we plotted a histogram of the different percentages of Anp32b−/− mice arising from breeding pairs a possible trimodal distribution was noted with peaks of Anp32b−/− attainment around 5%, around 12.5%, and around 22.5% (Fig. 3A). This suggested that a strong, non-linked, modifier gene of Anp32b might exist in our colony.
Fig. 3.
Indication and identification of a retained unlinked locus in the BALB/c-congenic ANP32B-deficient heterozygotes. (A) Histogram of Anp32b−/− mice provision from individual breeding pairs of six-generation BALB/c-congenic colony. (B) Strain-specific SNP mapping of representative Anp32b heterozygotes indicates two regions of C57BL/6J retention on chromosomes 4 and 11 in the six-generation BALB/c-congenic mice. The retained locus on chromosome 4 surrounds Anp32b. (C) Genes in the region of the retained C57BL/6J SNP rs6190775 demonstrated different degrees of exonic mutation between the strains. The heavily bolded line indicates the region of C57BL/6 retention for animals used in exome sequencing as part of SNP homozygosity breeding. SNP, single-nucleotide polymorphism; Indel, insertion/deletion mutations.
An unlinked C57BL/6J locus is conserved in Anp32b heterozygotes
In order to directly examine this possibility, four Anp32b+/− progeny from different breeding pairs were examined by strain-specific SNP analysis to examine possible retention of C57BL/6J loci through the 6-generation backcrosses. The mean interval between strain-specific SNPs in this analysis was 1.75 Mbp ± 0.03 Mbp (mean ± SEM). As expected for six-generation backcrossing, each of the mice demonstrated greater than 97.5% BALB/c-specific SNP content. As shown in Fig. 3B, two regions retained C57BL/6J genomic signatures in the majority of the mice sampled. Not unexpectedly, one encompassed the Anp32b gene on chromosome 4, for which heterozygosity was selected by direct genotyping throughout breeding. The second region was present on chromosome 11 wherein SNP rs6190775 was conserved in a heterozygous state in three of four animals examined. In addition to having no apparent linkage to the selected Anp32b locus, the rs6190775 SNP conservation was the product of different neighboring recombination events in at least two animals (Fig. 3B). For an unlinked locus, we calculated the probability of this retention as being less than 10−6 without selective pressure, which is highly statistically significant even with Bonferroni adjustment of normal significance levels (Supplemental Table S3). We conclude, therefore, that this C57BL/6J sequence was likely retained by virtue of some conferred survival advantage in the Anp32b heterozygotes. Furthermore, the selection of this locus was potentially the cause of the observed variability in the occurrence of Anp32b−/− mice.
Neither of the examined SNPs neighboring rs6190775 maintained C57BL/6J conservation in the majority of animals tested. Thus, a gene within the 2.88Mbp interval between the SNPs surrounding rs6190775 is likely a modifier of Anp32b heterozygous survival. In order to examine which of the genes in the region of rs6190775 may be altered between C57BL/6J and BALB/c mice, we examined a database of sequenced mouse genomes. Fig. 3C shows the frequency of nucleotide substitutions and insertions/deletions for all the annotated genes surrounding this SNP, normalized by mRNA length. Two genes of high mutation density were evident. One was proximal to rs6190775, within H2afv, and one was more distal, within Mrps24.
Modification of the Anp32b phenotype linked to rs6190775
The Anp32b−/− phenotype is complex but reduced viability is its primary characteristic (Table 1) [20]. In order to examine whether there was a phenotypic modification of Anp32b−/− associated with the SNP conserved in C57BL/6J, we generated two categories of Anp32b+/− breedings with homozygosity in the rs6190775 locus. The “SNP A” strain conserves the C57BL/6J sequence for the rs6190775 locus whereas the “SNP G” strain conserved the BALB/c sequence.
When we examined the progeny of these screenings we found a striking effect (Table 3). Homozygous SNP G mice produced near Mendelian ratios of Anp32b−/− mice (17.9%). Indeed, chi-square analysis of the Mendelian ratios did not demonstrate a statistically significant effect associated with the Anp32b null allele in the SNP G homozygous background. In contrast, Anp32b-heterozygote breedings in the context of SNP A homozygosity produce only 4.4% null mice, far below the expected Mendelian ratios. These data indicate that an Anp32b-modifying gene is closely linked to the rs6190775 locus.
Table 3. rs6190775 modifies the ANP32B viability phenotype.
| (A) BALB/c Congenic + SNP G/G | (B) BALB/c Congenic + SNP A/A | ||||||
|---|---|---|---|---|---|---|---|
| Anp32b genotype | Anp32b genotype | ||||||
| +/+ | +/– | –/– | +/+ | +/– | –/– | ||
| Expected | 24 | 48 | 24 | 17 | 34 | 17 | |
| Observed | 32 | 46 | 17 | 28 | 37 | 3 | |
Mendelian ratios of Anp32b heterozygote intercrosses from (A) rs6190775 G/G and (B) rs6190775 A/A homozygous backgrounds in the six-generation BALB/c congenic strain. SNP, ingle-nucleotide polymorphism. (A) Chi-square test: P = 0.09, (B) Chi-square test: P < 10-4.
In order to directly examine the extent of the varied locus in these breedings, we performed directed exome sequencing for the genes surrounding rs6190775 in the SNP A and SNP G homozygous breeders. Sampling three Anp32b+/− mice from each of the SNP homozygous breeding colonies, we found that rs6190775 mimicked known strain-specific mutations between the Polm and Tns3 genes, outside of which BALB/c polymorphisms were homozygous in all mice. This suggests a very limited number of genes that may be responsible for this modification phenotype. In comparison with a mouse genome sequencing resource [8], we noted two aberrations with respect to reported strain differences (Supplementary Table S4) although the functionality of these or any other mutations in the relevant genes remains to be determined.
Discussion
Breeding the murine ANP32B deficiency into alternate genetic backgrounds has provided a viable congenic model allowing studies in the adult, reaching near Mendelian ratios. The BALB/c-congenic background has reaffirmed the complex and strain-dependent phenotype found with ANP32B in the mixed background. The pleiotropic effects suggest that ANP32B functions in gene expression in many tissues, consistent with other broadly expressed chromatin regulators. Contrary to expectation based on clinical data, we found no apparent effect of ANP32B on mammary carcinoma induction or prognosis. Due to the improbable retention of a non-linked C57BL/6J chromosomal segment, our study also revealed a modifier locus with a phenotypic difference between C57BL/6J and BALB/c strains.
We previously described the Anp32b−/− phenotype as strain dependent and complex, consistent with its widespread involvement in gene expression during development [20]. The current study reinforced that finding. Our data present a partially penetrant lethality in the six-generations congenic BALB/c background with various pathologies arising into adulthood. This generally resembles the reported phenotype in the mixed genetic background. Our attempts to find genes whose loss may resemble this phenotype, particularly in clinical chemistry, led to a number of genes that have defect that include but are not limited to the changes seen in the ANP32B deficiency. These include chromatin modifiers Chd7 and Mysm1, signaling genes Akap9, Dbn1, and Ltbp1, peroxisomal biogenesis gene Pex3, as well as a gene of unknown function Fam73b. It is unknown how much the variability of the phenotype relies on the variability of this modifying locus both within the six-generation BALB/c congenic and the mixed background. Since BALB/c is unique among the examined strains for the polymorphism in the region of rs6190775, it is unlikely that genes in this region are modifying the phenotype in the mixed background.
It is perplexing that the C57BL/6J version of the modifying locus is retained in the heterozygous breeding but does not support improved viability of the ANP32B-deficient mice. Indeed, the C57BL/6J version is clearly detrimental to homozygosity. We cannot exclude that heterozygosity of this locus is preferred, irrespective of Anp32b status. In light of this, we would suggest the rs6190775 to be closely examined in any BALB/c congenic breeding from a C57BL/6J precursor.
Among the list of genes surrounding this SNP, only one gene with an indirect relationship to a known ANP32 function was identified, namely H2afv. H2afv encodes H2A.V (also known as H2A.Z.2), which is a variant H2A histone likely involved in transcriptional start site accessibility [16]. ANP32E was identified as a histone chaperone specific to H2A.Z-containing nucleosomes [17]. Furthermore, H2afv carries a high number of strain-dependent differences albeit in its 3′ untranslated region. Therefore, we consider H2afv to be a strong candidate for being the modifying gene at this locus. Although H2A.Z and H2A.V have long been believed to be functionally interchangeable, H2A.V was recently shown to have functions distinct from those of H2A.Z in cell-cycle progression and cellular drug resistance [28]. No biochemical evidence currently exists to suggest that ANP32B interacts with variant histones although ANP32B has been implicated in promoter-specific nucleosomal placement at the transcriptional start site [15] where H2A.Z-containing nucleosomes are normally present. It is possible that a genetic interaction exists whereby histone H2A.V and ANP32B have opposing activities in transcription without direct interaction.
Cancer induction in the BALB/c-congenic Anp32b-mutant mouse does not indicate a role for ANP32B in mammary cancer induction in the mouse. Furthermore, viability of the carcinogen-treated ANP32B-deficient mice is not enhanced as may be predicted from human prognostic data [20]. We did note incidents of CSCC in the Anp32b−/− animals that were not seen in Anp32b+/+ animals suggesting that ANP32B may have a tumor-suppressive function in CSCC. Notably however, Anp32b is closely linked to a major CSCC-related gene Xpa [14] on mouse chromosome 4. In the context of our experiment, strain-dependent differences in Xpa, including a reported missense mutation, are likely carried with the ANP32B deletion and this should be viewed as a major caveat with respect to any role of ANP32B in cutaneous squamous cell carcinoma. Here, since Xpa and Anp32b are within 1 Mb of each other and would likely cosegregate even after ten-generation backcrossing, this complication could best be resolved by zygote targeting in the BALB/c strain.
ANP32A is the most closely related gene to ANP32B. Based on the prevalence of ANP32A in biochemical isolations, it was somewhat surprising that the ANP32A-deficient mouse did not manifest a strong phenotype. In contrast, the ANP32B deficiency had a prominent effect on development and viability in the mixed-bred and C57BL/6J background. The strong strain dependency of ANP32B suggests that there may be differential requirement of ANP32 proteins depending on genetic background. The capacity of BALB/c mice to tolerate loss of ANP32B suggests that ANP32A may be more important in terms of redundant functions in this background.
This study of genetic background impact on ANP32B phenotype can be viewed as a tale of caution as well as one of serendipity. The improbable retention of the chromosome 11 locus has complicated the phenotypic analysis of the BALB/c-congenic mouse. It is unlikely, based on the degree of retention after six generations, that additional backcrosses would have eliminated the C57BL/6J locus. Certainly, global SNP mapping at each generation, as per speed congenics services, may have made the generation of a clean congenic animal more likely. Conversely, this would have abrogated our opportunity to identify the modifying locus in the null mice and to identify candidate genes that can now be tested for genetic interaction. Hence, we have found a benefit from conventional backcrossing.
Supplementary Material
Acknowledgments
We would like to thank Ms. Teo Wei Ling, Mr. Clint Menendez, Mr. Bryan Buenaflor, and Dr. Sebastian David for their technical assistance. This study was financially supported by grants from the National Cancer Centre Research Foundation. The funding body had no role in study design.
References
- 1.Amasaki H., Ogawa M., Nagasao J., Mutoh K., Ichihara N., Asari M., Shiota K.2003. Distributional changes of BrdU, PCNA, E2F1 and PAL31 molecules in developing murine palatal rugae. Ann. Anat. 185: 517–523. doi: 10.1016/S0940-9602(03)80116-4 [DOI] [PubMed] [Google Scholar]
- 2.Becker D.J., Myers J.T., Ruff M.M., Smith P.L., Gillespie B.W., Ginsburg D.W., Lowe J.B.2003. Strain-specific modification of lethality in fucose-deficient mice. Mammalian Genome 14: 130–139. [DOI] [PubMed]
- 3.Brennan C.M., Gallouzi I.E., Steitz J.A.2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151: 1–14. doi: 10.1083/jcb.151.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fries B., Heukeshoven J., Hauber I., Grüttner C., Stocking C., Kehlenbach R.H., Hauber J., Chemnitz J.2007. Analysis of nucleocytoplasmic trafficking of the HuR ligand APRIL and its influence on CD83 expression. J. Biol. Chem. 282: 4504–4515. doi: 10.1074/jbc.M608849200 [DOI] [PubMed] [Google Scholar]
- 5.Ince-Dunn G., Okano H.J., Jensen K.B., Park W.Y., Zhong R., Ule J., Mele A., Fak J.J., Yang C., Zhang C., Yoo J., Herre M., Okano H., Noebels J.L., Darnell R.B.2012. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron 75: 1067–1080. doi: 10.1016/j.neuron.2012.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X., Kim H.E., Shu H., Zhao Y., Zhang H., Kofron J., Donnelly J., Burns D., Ng S.C., Rosenberg S., Wang X.2003. Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science 299: 223–226. doi: 10.1126/science.1076807 [DOI] [PubMed] [Google Scholar]
- 7.Johnson M.2012. Laboratory Mice and Rats. Labome: Materials and Methods 2.
- 8.Keane T.M., Goodstadt L., Danecek P., White M.A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M., Furlotte N.A., Eskin E., Nellåker C., Whitley H., Cleak J., Janowitz D., Hernandez-Pliego P., Edwards A., Belgard T.G., Oliver P.L., McIntyre R.E., Bhomra A., Nicod J., Gan X., Yuan W., van der Weyden L., Steward C.A., Bala S., Stalker J., Mott R., Durbin R., Jackson I.J., Czechanski A., Guerra-Assunção J.A., Donahue L.R., Reinholdt L.G., Payseur B.A., Ponting C.P., Birney E., Flint J., Adams D.J.2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. doi: 10.1038/nature10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S., Roopra A., Alexander C.M.2012. A phenotypic mouse model of basaloid breast tumors. PLoS ONE 7: e30979. doi: 10.1371/journal.pone.0030979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraev A.2014. Parallel universes of Black Six biology. Biol. Direct 9: 18. doi: 10.1186/1745-6150-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krauskopf A., Eriksson O., Craigen W.J., Forte M.A., Bernardi P.2006. Properties of the permeability transition in VDAC1(-/-) mitochondria. Biochim. Biophys. Acta 1757: 590–595. doi: 10.1016/j.bbabio.2006.02.007 [DOI] [PubMed] [Google Scholar]
- 12.LeCouter J.E., Kablar B., Whyte P.F., Ying C., Rudnicki M.A.1998. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development 125: 4669–4679. [DOI] [PubMed] [Google Scholar]
- 13.Leonard J.R., Klocke B.J., D’Sa C., Flavell R.A., Roth K.A.2002. Strain-dependent neurodevelopmental abnormalities in caspase-3-deficient mice. J. Neuropathol. Exp. Neurol. 61: 673–677. [DOI] [PubMed] [Google Scholar]
- 14.Miller K.L., Karagas M.R., Kraft P., Hunter D.J., Catalano P.J., Byler S.H., Nelson H.H.2006. XPA, haplotypes, and risk of basal and squamous cell carcinoma. Carcinogenesis 27: 1670–1675. doi: 10.1093/carcin/bgi376 [DOI] [PubMed] [Google Scholar]
- 15.Munemasa Y., Suzuki T., Aizawa K., Miyamoto S., Imai Y., Matsumura T., Horikoshi M., Nagai R.2008. Promoter region-specific histone incorporation by the novel histone chaperone ANP32B and DNA-binding factor KLF5. Mol. Cell. Biol. 28: 1171–1181. doi: 10.1128/MCB.01396-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nekrasov M., Soboleva T.A., Jack C., Tremethick D.J.2013. Histone variant selectivity at the transcription start site: H2A.Z or H2A.Lap1. Nucleus 4: 431–438. doi: 10.4161/nucl.26862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obri A., Ouararhni K., Papin C., Diebold M.L., Padmanabhan K., Marek M., Stoll I., Roy L., Reilly P.T., Mak T.W., Dimitrov S., Romier C., Hamiche A.2014. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature. [DOI] [PubMed]
- 18.Opal P., Garcia J.J., McCall A.E., Xu B., Weeber E.J., Sweatt J.D., Orr H.T., Zoghbi H.Y.2004. Generation and characterization of LANP/pp32 null mice. Mol. Cell. Biol. 24: 3140–3149. doi: 10.1128/MCB.24.8.3140-3149.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petkov P.M., Ding Y., Cassell M.A., Zhang W., Wagner G., Sargent E.E., Asquith S., Crew V., Johnson K.A., Robinson P., Scott V.E., Wiles M.V.2004. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 14: 1806–1811. doi: 10.1101/gr.2825804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reilly P.T., Afzal S., Gorrini C., Lui K., Bukhman Y.V., Wakeham A., Haight J., Ling T.W., Cheung C.C., Elia A.J., Turner P.V., Mak T.W.2011. Acidic nuclear phosphoprotein 32kDa (ANP32)B-deficient mouse reveals a hierarchy of ANP32 importance in mammalian development. Proc. Natl. Acad. Sci. USA 108: 10243–10248. doi: 10.1073/pnas.1106211108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reilly P.T., Afzal S., Wakeham A., Haight J., You-Ten A., Zaugg K., Dembowy J., Young A., Mak T.W.2010. Generation and characterization of the Anp32e-deficient mouse. PLoS ONE 5: e13597. doi: 10.1371/journal.pone.0013597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reilly P.T., Yu Y., Hamiche A., Wang L.2014. Cracking the ANP32 whips: important functions, unequal requirement, and hints at disease implications. BioEssays: news and reviews in molecular, cellular and developmental biology 36: 1062–1071. [DOI] [PMC free article] [PubMed]
- 23.Shen S.M., Yu Y., Wu Y.L., Cheng J.K., Wang L.S., Chen G.Q.2010. Downregulation of ANP32B, a novel substrate of caspase-3, enhances caspase-3 activation and apoptosis induction in myeloid leukemic cells. Carcinogenesis 31: 419–426. doi: 10.1093/carcin/bgp320 [DOI] [PubMed] [Google Scholar]
- 24.Sigmund C.D.2000. Viewpoint: are studies in genetically altered mice out of control? Arterioscler. Thromb. Vasc. Biol. 20: 1425–1429. doi: 10.1161/01.ATV.20.6.1425 [DOI] [PubMed] [Google Scholar]
- 25.Singh P., Schimenti J.C., Bolcun-Filas E.2015. A Mouse Geneticist’s Practical Guide to CRISPR Applications. Genetics. 199: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun W., Hattori N., Mutai H., Toyoshima Y., Kimura H., Tanaka S., Shiota K.2001. PAL31, a nuclear protein required for progression to the S phase. Biochem. Biophys. Res. Commun. 280: 1048–1054. doi: 10.1006/bbrc.2000.4244 [DOI] [PubMed] [Google Scholar]
- 27.Sun W., Kimura H., Hattori N., Tanaka S., Matsuyama S., Shiota K.2006. Proliferation related acidic leucine-rich protein PAL31 functions as a caspase-3 inhibitor. Biochem. Biophys. Res. Commun. 342: 817–823. doi: 10.1016/j.bbrc.2006.02.026 [DOI] [PubMed] [Google Scholar]
- 28.Vardabasso C., Gaspar-Maia A., Hasson D., Punzeler S., Valle-Garcia D., Straub T., Keilhauer E.C., Strub T., Dong J., Panda T., Chung C.Y., Yao J.L., Singh R., Segura M.F., Fontanals-Cirera B., Verma A., Mann M., Hernando E., Hake S.B., Bernstein E. Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Molecular Cell 59: 75–88. doi: 10.1016/j.molcel.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volinia S., Mascellani N., Marchesini J., Veronese A., Ormondroyd E., Alder H., Palatini J., Negrini M., Croce C.M.2008. Genome wide identification of recessive cancer genes by combinatorial mutation analysis. PLoS ONE 3: e3380. doi: 10.1371/journal.pone.0003380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R.2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. doi: 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.