Abstract
Chemotaxis and cell migration are fundamental, universal eukaryotic processes essential for biological functions such as embryogenesis, immunity, cell renewal, and wound healing, as well as for pathogenesis of many diseases including cancer metastasis and chronic inflammation. To identify novel chemotaxis inhibitors as probes for mechanistic studies and leads for development of new therapeutics, we developed a unique, unbiased phenotypic chemotaxis-dependent Dictyostelium aggregation assay for high-throughput screening using rapid, laser-scanning cytometry. Under defined conditions, individual Dictyostelium secrete chemoattractants, migrate, and aggregate. Chemotaxis is quantified by laser-scanning cytometry with a GFP marker expressed only in cells after chemotaxis/multi-cell aggregation. We applied the assay to screen 1,280 known compounds in a 1536-well plate format and identified two chemotaxis inhibitors. The chemotaxis inhibitory activities of both compounds were confirmed in both Dictyostelium and in human neutrophils in a directed EZ-TAXIscan chemotaxis assay. The compounds were also shown to inhibit migration of two human cancer cell lines in monolayer scratch assays. This test screen demonstrated that the miniaturized assay is extremely suited for high-throughput screening of very large libraries of small molecules to identify novel classes of chemotaxis/migratory inhibitors for drug development and research tools for targeting chemotactic pathways universal to humans and other systems.
Chemotaxis is a fundamental process whereby cells sense and migrate in chemoattractant gradients. Chemotaxis and cell migration play pivotal roles in embryogenesis, inflammation, wound healing, and renewal of skin and intestinal cells. They also mediate development of chronic inflammatory diseases, such as asthma, chronic obstructive pulmonary disease, rheumatoid arthritis, and atherosclerosis, as well as, cancer, angiogenesis, and metastasis1. Chemotaxis is regulated by G protein coupled receptors (GPCRs) and heterotrimeric G proteins that transduce chemotactic signals to the cytoskeleton to dynamically polarize migratory cells. Such biased polarizations may help re-localize the intracellular machinery for basal cell motility toward directed movement2. The molecular mechanisms that regulate chemotaxis/migration under different biological and pathological conditions are complex, and discovering novel small molecule probes of these pathways is important to analyze mechanistic functions and to develop new therapeutics1,3,4,5,6,7,8. Pathways involving ligand sensing, signal transduction, and basal cell mobility may all be targets for inhibition. Previous in vitro drug screens for anti-inflammatory and anti-metastasis potential have often focused on chemokine receptors, adhesion molecules, and limited downstream pathways4,5,6,7,8. However, the compounds identified from molecular target-based screens generally have poor activity in vivo and only few of them have been advanced to clinical trials.
Several cell-based migration assays are being optimized for more high-throughput image screening9,10,11,12,13,14,15,16,17,18,19,20, but they are not yet compatible to screen 1000s of compounds across a broad range of concentrations. Microfluidic devices are easily automated and show promise, but have been difficult to scale beyond 96 format arrays11,12,18,21. Magnetically labeled cell groups can be cultured in 3D, disrupted, and allowed to coalesce in a ring closure assay that assesses cell motility. The assay can be performed in 96-well plates, but requires magnetic and mechanical manipulations14. Boyden chambers have been re-configured to 96-well formats, but whole single-plate image analysis is limited and cell migration efficiency can be restricted to ~20%9,10.
Here, we report a simple, phenotypic, fluorescent chemotaxis-dependent aggregation assay in a 1536-well plate format that utilizes the unique chemotactic properties of Dictyostelium. Chemotaxis has been comprehensively studied in Dictyostelium discoideum, a model organism for eukaryote development, cell differentiation, and chemotaxis1,22,23,24,25,26, that shares similar chemotactic mechanisms with mammalian migratory cells1,26. Dictyostelium has also proven to be a highly sensitive system to evaluate the effects of various compounds on chemotaxis27,28,29,30.
Dictyostelium has a unique life cycle involving unicellular growth and multicellular development. Dictyostelium cells grow individually under nutrient abundant conditions, but upon starvation, they enter a developmental program and secrete the chemoattractant cAMP, which directs cell-to-cell chemotactic migration, formation of tight multi-cell aggregates, and multicellular development, with the terminal differentiation of distinct cell classes1,22,23,24,25. A GFP reporter, which is only expressed upon chemotactic-mediated aggregation31, is used to assess chemotaxis-dependent aggregation (Fig. 1A and Supplementary Movie 1). The assay was miniaturized and automated to a 1536-well plate format, where GFP fluorescence can be rapidly quantified (Fig. 1). A viability counter screen was also developed and incorporated, to eliminate cytotoxic false positives.
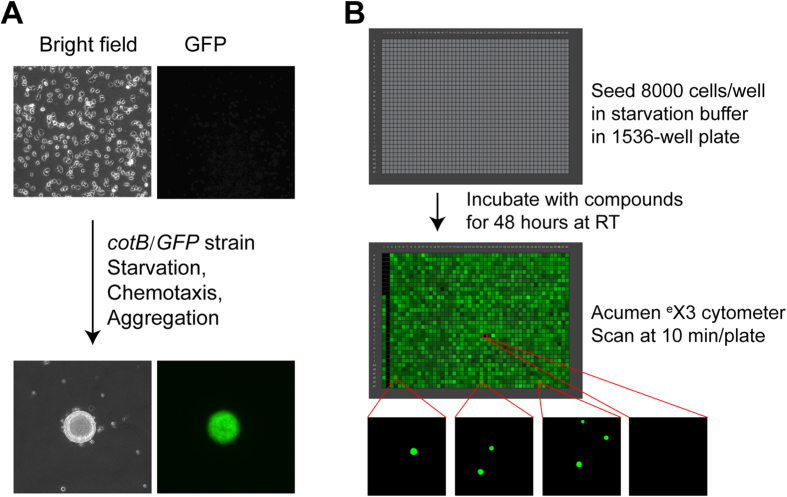
Figure 1. Imaging GFP reporter-based Dictyostelium chemotaxis-dependent aggregation assay for chemotaxis inhibitor screening.
(A) Principle of the screening assay. Top: Single cells before aggregation (bright field microscopy) do not express the cotB/GFP reporter. Bottom: Under starvation conditions, Dictyostelium secrete the chemoattractant cAMP, chemotax, and form multicellular aggregates, which express GFP after 24 hr. (B) Protocol for compound screening. Top: 1536-well Acumen eX3 GFP image at the start of the experiment. In general, wells contained 8,000 Dictyostelium cells. The left-most vertical row of cells has decreasing concentrations of latrunculin A (see Fig. 3A). The second (from left) vertical row of cells has the highest concentration of latrunculin A in all wells, as a negative signal (i.e. a full inhibitory) control. The third and fourth vertical rows of cells have 0.38% DMSO, as positive signal controls (i.e. without any inhibition). All other wells contain different small molecule compounds. Middle: An entire, 1536-well Acumen eX3 GFP image after 48 hr incubation. Bottom: Selected Acumen eX3 GFP images of individual wells. GFP positive, usually form 1–3 multicellular Dictyostelium aggregates within each well of a 1536-well plate format, while no aggregates form when chemotaxis is inhibited. Criteria are set to quantify GFP signals in objects >30 μM diameter; non-aggregated cells have a zero GFP signal. Each image is an entire well, 1.53 mm × 1.53 mm.
While the described Dictyostelium chemotaxis-dependent aggregation assay system offers unique advantage for HTS, we recognize that the readout is indirect. Still, compounds that affect development independently of chemotaxis, signal-response, and motility are easily eliminated in directed screens. Further, we demonstrate successful proof of application, using mammalian cells as readouts.
In a test screen of 1280 small molecules, at 7 (i.e. >4-log) concentrations each, using the described Dictyostelium chemotaxis-dependent aggregation assay system, we identified two compounds that inhibit chemotaxis in mammalian cells. The inhibitory activity of both compounds was independently confirmed using an EZ-TAXIScan chemotaxis assay13,32, applied to Dictyostelium and human neutrophils, and a scratch migration assay33, applied to human migratory cancer cells. The Dictyostelium aggregation assay is a first assay suitable for HTS of chemotaxis/migration inhibitors in large libraries of small molecules, and we have described a novel technological approach to identify lead compounds in chemotaxis for mechanistic studies and new therapeutics.
Results
Principles of Assay and Signal Detection
Upon nutrient depletion, Dictyostelium initiate a multicellular developmental program involving secretion of the chemoattractant cAMP that directs chemotaxis and tight multi-cell aggregation. This morphological transition from unicellular to multicellular state is easily observed (Fig. 1A). Chemotaxis/aggregation is quantified using Dictyostelium carrying a stable GFP reporter (cotB/GFP)31 that is only expressed in multicellular aggregates (Fig. 1A). We miniaturized and automated the assay (Supplementary Table 1) to a 1536-well plate format that allows the formation of only 1–3 (GFP-positive) aggregates per well in a limited surface area (2.3 mm2 and 6 μl media per well; Fig. 1B).
We evaluated three different instruments to rapidly quantify GFP signals following chemotaxis/aggregation in 1536-well plates. The ViewLux CCD imager based fluorescence plate reader was not sufficiently sensitive to detect the GFP signals. The 1 to 3 GFP-aggregates per well were frequently localized outside the detection field of the EnVision photomultiplier tube based fluorescence plate reader. In contrast, GFP-aggregation fluorescent signals were readily detected by the Acumen eX3 laser scanning based plate cytometer, with full data collection of an entire 1536-well plate within ~10 min.
Scanning threshold was set to capture signals from objects greater than 30 μm in diameter (>3 cell equivalents) and record numbers of aggregates, total fluorescence intensity, and peak fluorescence intensity of aggregates within each well (Fig. 1B). The GFP peak intensity per well proved to be the most consistent, on well-to-well and plate-to-plate bases, and was chosen as the primary readout.
In addition to high throughput and quick data acquisition, the phenotypic screening approach provides further advantages. Since GFP is only expressed in multicellular aggregates (not individual cells, see Fig. 1A), fluorescent signals are highly concentrated to the limited multi-cellular foci (aggregates) within each well (Fig. 1 and Supplementary Movie 1). GFP-positive aggregates are readily distinguished from non-chemotaxing, GFP-negative individual cells. Because there is no background GFP signal below the 30 μm diameter detection threshold, compounds inhibiting chemotaxis would ablate strong positive GFP signals.
Assay Automation, Miniaturization, and Optimization
Time course
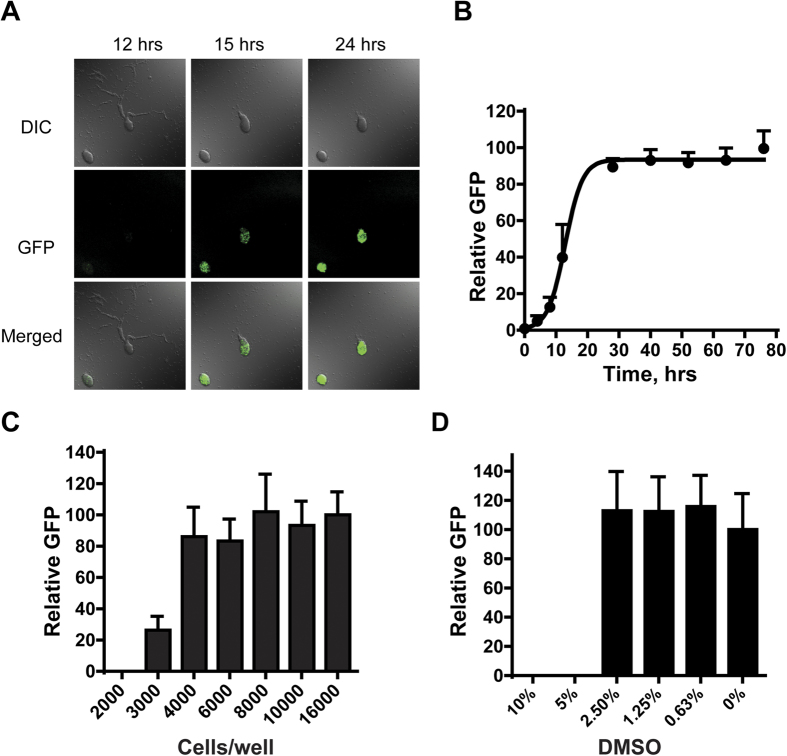
To evaluate timing, we seeded Dictyostelium in starvation/differentiation buffer in 1536-well plates, at 6 μl/well, and incubated at ~20 °C (Supplementary Table 1). Chemoattractant (cAMP) signaling was initiated endogenously after 8 hr and resulted in cells chemotaxing and aligning in a head-to-tail mode (i.e. cell-to-cell streaming; Fig. 2A and Supplementary Movie 1)34. GFP expression was observed in early aggregated cells and fluorescence intensity increased with incubation times (Fig. 2A,B and Supplementary Movie 1). The maximal GFP expression in the 1536-well format was observed after ~30 hr incubation and remained constant for an additional 2 days (Fig. 2B). We chose a 48-hr incubation time to ensure that all aggregates of entire plates attained peak GFP signals.
Figure 2. Optimization of Dictyostelium chemotaxis-dependent aggregation assays.
(A) Time course of reporter GFP expression during Dictyostelium chemotaxis and aggregation. Cells were incubated in starvation buffer for various times and visualized by differential interference contrast (DIC) and fluorescent confocal (GFP) microscopy (see Supplementary Movie 1). (B) For measuring time course of GFP expression in 1536-well plate format, cells were seeded into plates and assayed at times indicated for GFP signal intensity (with standard deviations) with the Acumen eX3 laser scanning cytometer. The data are representative of two independent experiments. (C) For cell density optimization, cells were deposited in 1536-well plates at different densities and GFP intensity (with standard deviations) measured after 48 hr. (D) For evaluation of DMSO effects, 8,000 cells per well were incubated with different concentrations of DMSO and GFP intensity (with standard deviations) was measured after 48 hr.
Cell density
To minimize well-to-well variation and ensure reproducibility, we optimized cell density for chemotaxis-dependent aggregation in small surface areas of 1536-plate wells. Cells at low density (<2,000 cells/well) failed to aggregate or express GFP (Fig. 2C). The peak GFP signal intensity increased from 3,000 to 8,000 cells/well, and plateaued (Fig. 2C). Our data also indicated that at cell densities >12,000 cells/well, aggregation and GFP expression could occur, even with poor chemotaxis. Thus, 8,000 cells/well (~3.5 × 105 cells/cm2) in 6 μl buffer was chosen as optimal for the 1536-well plate format (Fig. 2C and Supplementary Table 1). This cell density was proportionally similar to that previously developed for 12-well plates (~1.5 × 105 cells/cm2)35. Approximately, 1–3 aggregates with variable sizes form in each well of a 1536-well plate under optimal conditions (Fig. 1B).
Temperature sensitivity
Multi-cell aggregate formation was robust and reproducible at 20–23.5 °C. Our results were consistent with the reported optimum growth temperature for Dictyostelium at ~22 °C. Dictyostelium undergo heat-shock at 28 °C36; chemotaxis is significantly delayed at <15 °C.
Assay verification
Since DMSO was used as the universal solvent for all compounds, we examined DMSO effects on aggregation in 1536-well plate formats. Neither aggregation nor GFP expression was affected at final DMSO concentrations <2.5% (Fig. 2D). The DMSO concentration in our compound screens is 0.38% (Supplementary Table 1).
Since the GFP-expressing multicellular aggregates formed within each well have different sizes and numbers, the total fluorescence intensity signals can vary [coefficient of variation (CV) = 27%)] from well-to-well and plate-to-plate, with a Z’ factor of 0.47. Importantly, the parameters of the laser scanning plate cytometer are set to only quantify GFP fluorescence presented in multicellular aggregates. The design of the assay results in a “zero” signal when aggregation is completely inhibited (see Fig. 1) and, consequently, in non-conventional signal-to-basal ratio calculations. This high throughput assay is extremely robust, sensitive, and reliable for the identification of novel chemotaxis inhibitors within small molecule libraries.
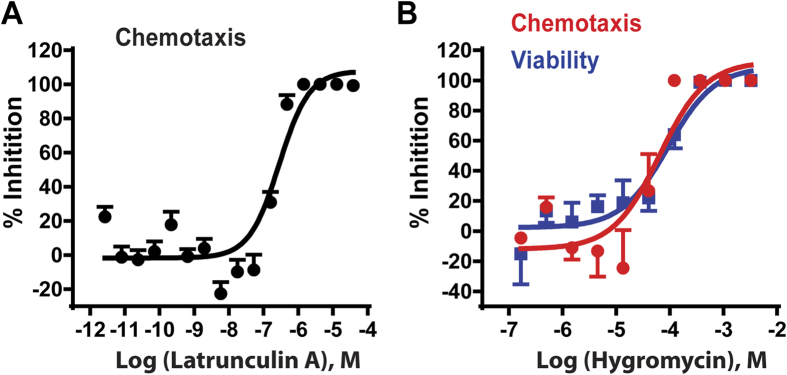
Latrunculin A is a natural product, which binds actin monomers at 1:1 with a Kd of 200 nM in vitro37. Inhibition of actin polymerization by latrunculin A disrupts actin filament formation, cytoskeletal organization, cell migration, and chemotaxis15,38. We evaluated inhibitory activity of latrunculin A on chemotaxis-dependent aggregation in the 1536-well format. Latrunculin A inhibited chemotaxis-dependent aggregation with an IC50 of 282 nM (95% confidence interval, 193–412 nM; Fig. 3A), similar to that of mammalian cells. Results demonstrated that the Dictyostelium assay is an effective alternate approach for compound screening to identify chemotaxis inhibitors. While latrunculin A may cause lethality of oncogenic mammalian cells, it is not toxic to Dictyostelium in our assay. When Dictyostelium are seeded in starvation buffer, growth is arrested and non-dividing cells survive without actin-dependent processes for external nutrient capture, cell division, or motility; cellular energy, etc. derive from autophagy. When we simultaneously monitored dose response effects of latrunculin A on aggregation and cell viability, complete inhibition of aggregation is confirmed at 1 μM, whereas cells remained viable to 93+/−2.3%, as assayed by vital dye staining39. Latrunculin A is more cytotoxic at 10 μM (~30% viability).
Figure 3. Validation of the Dictyostelium chemotaxis-dependent aggregation assay for inhibitor screens.
(A) Effects of latrunculin A on chemotaxis-dependent aggregation (GFP expression). 8,000 cells per well were incubated with different concentrations of the chemotaxis inhibitor latrunculin A in the 1536-well plate. GFP intensity (with standard deviations) was measured after 48 hr. (B) ATP content assay as a counter screen for cytotoxicity. Parallel wells with different concentrations of the cytotoxic compound hygromycin B were assayed for viability (500 cells/well) after 3 days and chemotaxis-dependent aggregation (8,000 cells/well) after 2 days. Inhibition of cell viability was assayed by loss of cellular ATP and inhibition of aggregation was assayed by loss relative GFP intensity. The viability and aggregation inhibition curves (with standard deviations) superimpose.
We do not observe partial inhibition of GFP expression by lactrunculin A across a broad concentration range (Fig. 3A), as might have been anticipated; in addition, smaller aggregates are not a characteristic of inhibition. The non-linearity of the detected GFP signal response is intrinsic to this unique phenotypic assay and also provides significant advantage. First, cell-cell communication (signal-relay) between individual Dictyostelium is essential to chemotaxis-dependent aggregation and both signal-relay and chemotaxis are regulated by many of the same intracellular pathways22,23,24,26,40. Thus, inhibitory effects can be amplified if they impact both chemotaxis and signal-relay, as does latrunculin A34,41.
In addition, since the assay proceeds for 48 hrs, we lessen minimal inhibitory effects on chemotaxis-dependent aggregation and GFP expression. Finally, GFP is only detected in objects >30 μM, which further emphasizes positive/negative contrasts. We suggest that these criteria polarize effects toward either non-inhibitory or highly inhibitory signals (Figs 1B and 3A). Since we test compounds across a wide concentration range, these data enhance discrimination of true inhibitory molecules from false positives.
Counter screen to eliminate toxic compounds
Since compounds that cause cell lethality will appear as inhibitory positives in our assay, we adapted a cell-based ATP content assay to evaluate compound cytotoxicity to eliminate those false positives from a primary screen42. Since Dictyostelium cells within aggregates were not easily lysed with the assay-kit buffer, we established an ATP content assay (Supplementary Table 2) using 500 cells/well in which aggregation was not formed due to non-permissive numbers of cells (Fig. 2C). Hygromycin B was used as a positive control for cytotoxicity that exhibited similar IC50 values of 92.8 μM and 64.5 μM in the ATP content and aggregation assays, respectively (Fig. 3B). Our results indicate that 3-day incubation is optimal for measurement of compound cytotoxicity using the ATP content assay. Hygromycin B was then used as a parallel cytotoxic control in all experiments. The signal-to-noise ratio was 22-fold, CV was 12%, and Z’ factor was 0.6 in this Dictyostelium cytotoxicity assay.
LOPAC collection screening
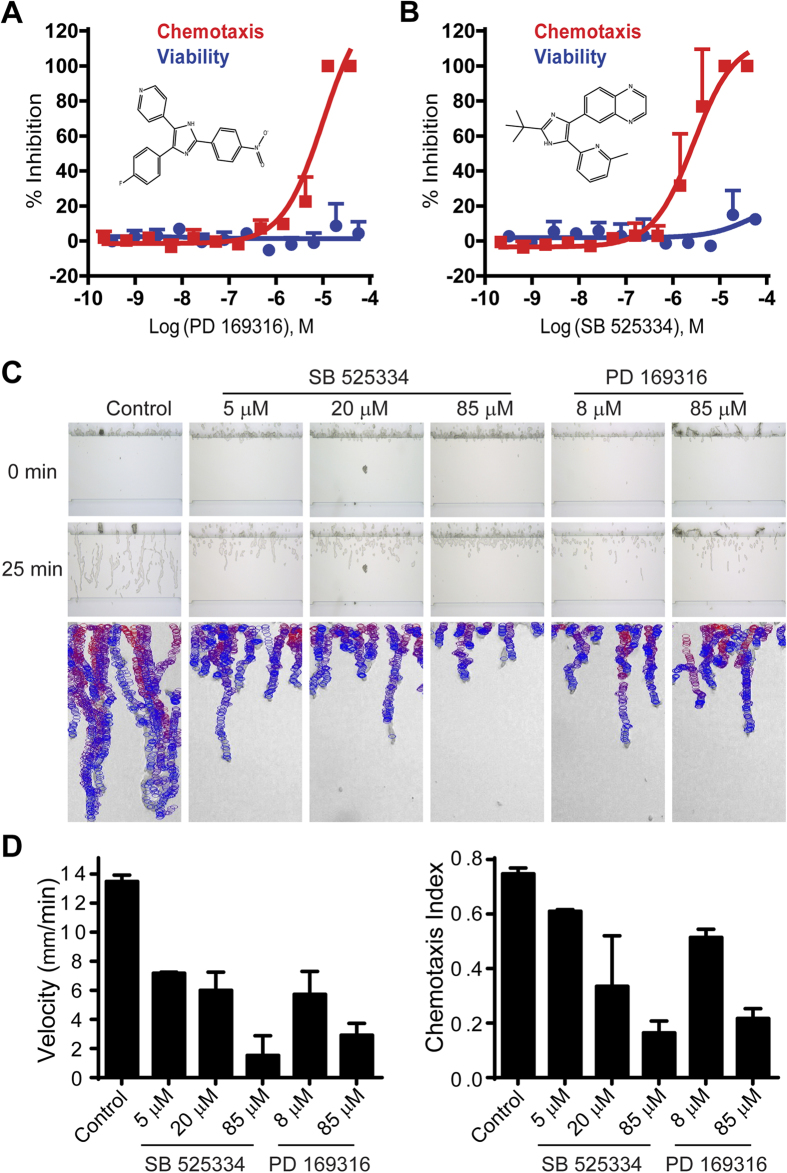
The LOPAC (Library of Pharmacologically Active Compounds) collection contains 1280 compounds commonly used to test and validate screen assays. We performed a primary Dictyostelium chemotaxis-dependent aggregation assay with all 1280 LOPAC compounds in serial dilution (from ~2 nM to ~50 μM) in parallel with an ATP content cytotoxicity assay. Based on calculation of IC50, three compounds fully inhibited chemotaxis/aggregation at ≤1.5 μM, but all three were also cytotoxic; at ≤7.7 μM, 22 compounds were inhibitory, with 20 also cytotoxic (see Figure S1A,B). The inhibitory effects on aggregate formation of the two non-toxic compounds PD 169316 and SB 525334 (Fig. 4A,B) were confirmed by microscopic examination; PD 169316 and SB 525334 did not simply suppress GFP expression, but also inhibited chemotaxis-dependent aggregation.
Figure 4. Identification and confirmation of Dictyostelium chemotaxis inhibitors PD 169316 and SB 525334 from LOPAC screening.
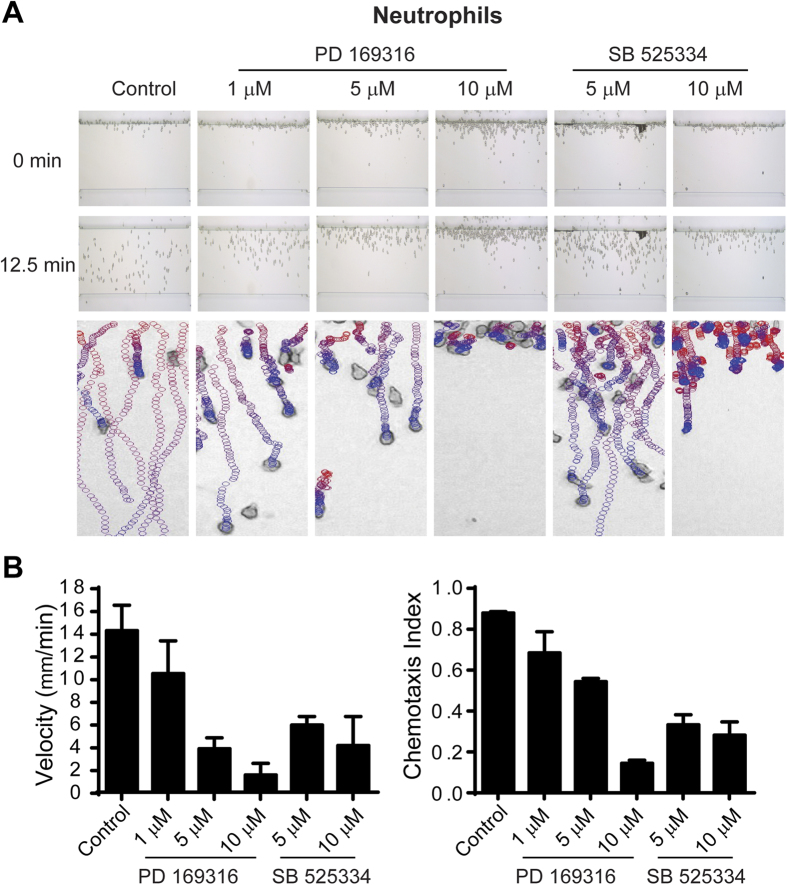
(A,B) Concentration response of PD 169316 (A) and SB 525334 (B) with strong inhibitory effects on chemotaxis-dependent aggregation (GFP), without obvious effect on cell viability (ATP). The experiments were performed in triplicate. Error bars represent standard deviations. Structures of PD 169316 and SB 525334 are embedded in the graphs. (C) Validation of inhibitory effects of compounds PD 169316 and SB 525334 on Dictyostelium chemotaxis in EZ-TAXIScan assays. Developed Dictyostelium were treated with various doses of PD 169316 and SB 525334 and assayed for chemotaxis within a cAMP gradient by EZ-TAXIScan. Top and middle panels: the images show the positions of cells at 0 and 25 min of chemotaxis; at 25 min, control cells approach the end of the gradient (see Figure S2A). Bottom panels: the images show migratory paths of individual Dictyostelium during an entire chemotaxis assay. Cells are depicted in migratory tracts as circles, progressing from red to blue during increasing times of the assay. For clarity, only cells that moved in ten consecutive frames are shown. Non-moving (e.g. stationary) features are ignored by the algorithm, which compares objects among multiple images. Data are representative of three independent experiments. (See Supplementary Movie 2). (D) Chemotaxis parameters were calculated using data obtained from (C). The chemotaxis velocity was computed as the total chemotaxis path length divided by time. The chemotaxis index was computed as the ratio of net path length toward cAMP compared to the total path length.
Finally, while 123 other compounds were inhibitory at only the single highest dose tested, 38.3 μM, 111 of these compounds were also cytotoxic; we did not pursue the remaining 12 compounds, rather focusing on the more active compounds PD 169316 and SB 525334.
PD 169316 and SB 525334 were individually re-assayed for dose-response inhibition of aggregation and cytotoxicity over a wider concentration range (Fig. 4A,B). PD 169316 (Fig. 4A) inhibited Dictyostelium aggregation with an IC50 of 10 μM (95% confidence interval, 5.1–19.7 μM) with no detectable cytotoxicity. SB 525334 (Fig. 4B) also strongly inhibited aggregation (IC50 2.8 μM, 95% confidence interval, 1.5–5.1 μM). The LOPAC test screen demonstrated the suitability of the Dictyostelium chemotaxis-dependent aggregation assay in 1536-well plate formats for HTS to identify potential inhibitors of chemotaxis.
PD 169316 and SB 525334 inhibit chemotaxis of both Dictyostelium and human neutrophils
To confirm the chemotactic inhibitory properties of both PD 169316 and SB 525334, we examined direct effects on chemotaxis of Dictyostelium and human neutrophils in specific linear chemoattractant gradients. Dictyostelium were applied to an EZ-TAXIScan channel, in a 0–0.1 μM gradient of cAMP at room temperature, in the presence of varying doses of PD 169316 or SB 525334 (Fig. 4C, Figure S2A, and Supplementary Movie 2). A similar assay of PD 169316 and SB 525334 was performed using fresh human neutrophils, under a 0–0.1 μM gradient of fMLP at 37 °C (Fig. 5A, Figure S2B, and Supplementary Movie 3). Dictyostelium and neutrophils rapidly polarized and migrated efficiently in control chambers in the presence of appropriate chemotactic agents. PD 169316 and SB 525334 inhibited chemotaxis velocity and directional index of both cells in dose-dependent manners, albeit with different potencies (Figs 4C,D and 5A,B). Notably, the IC50 values of PD 169316 and SB 525334 for Dictyostelium chemotaxis were similar to that determined in the original Dictyostelium assay screen (Fig. 4). In neutrophils, chemotaxis inhibition by PD 169316 was relatively more potent than by SB 525334 (Fig. 5B).
Figure 5. Validation of inhibitory effects of compounds PD 169316 and SB 525334 on human neutrophil chemotaxis in EZ-TAXIScan assays.
(A) Human neutrophils were treated with various doses of PD 169316 and SB 525334 and assayed for chemotaxis within an fMLP gradient by EZ-TAXIScan. Top and middle panels: the images show the positions of cells at 0 and 12.5 min of chemotaxis; at 12.5 min, control cells approach the end of the gradient (see Figure S2B). Bottom panels: The images show migratory paths of individual neutrophils during an entire (30 min) chemotaxis assay. Cells are depicted in migratory tracts as circles, progressing from red to blue during increasing times of the assay. For clarity, only cells that moved in ten consecutive frames are shown. Non-moving (e.g. stationary) features are ignored by the algorithm, which compares objects among multiple images. Data are representative of three independent experiments. (See Supplementary Movie 3). (B) Chemotaxis parameters were calculated using data obtained from (A). The chemotaxis velocity was computed as the total chemotaxis path length divided by time. The chemotaxis index was computed as the ratio of net path length toward fMLP compared to the total path length.
To assess effects of PD 169316 and SB 525334 on Dictyostelium chemotaxis in the absence of development, we monitored response of growing cells to folate using EZ-TAXIScan. Growing Dictyostelium are responsive to folate, whereas both PD 169316 and SB 525334 at 50 μM significantly impair chemotaxis (Figure S3). Stimulation of growing Dictyostelium with either folate or cAMP leads to the rapid, but transient, activation of mTORC2 and consequent phosphorylation of PKBR1. These responses were not impaired by either PD 169316 or SB 525334 (Figure S4). Thus, these compounds inhibit the chemotaxis of Dictyostelium and not simply a developmental response to cAMP.
Although compounds SB 525334 and PD 169316 are used to preferentially target (<100 nM) TGFβ receptors and p38 MAPK43,44, respectively, their specificities are not absolute43,45. Furthermore, Dictyostelium does not appear to have TGFβ-signaling, and although there are two ERKs in Dictyostelium, their normalized activations by folate or cAMP are not significantly (+/−10%) affected by either SB 525334 or PD 169316 (Figure S5).
PD 169316 and SB 525334 inhibit human cancer cells in scratch migration assays
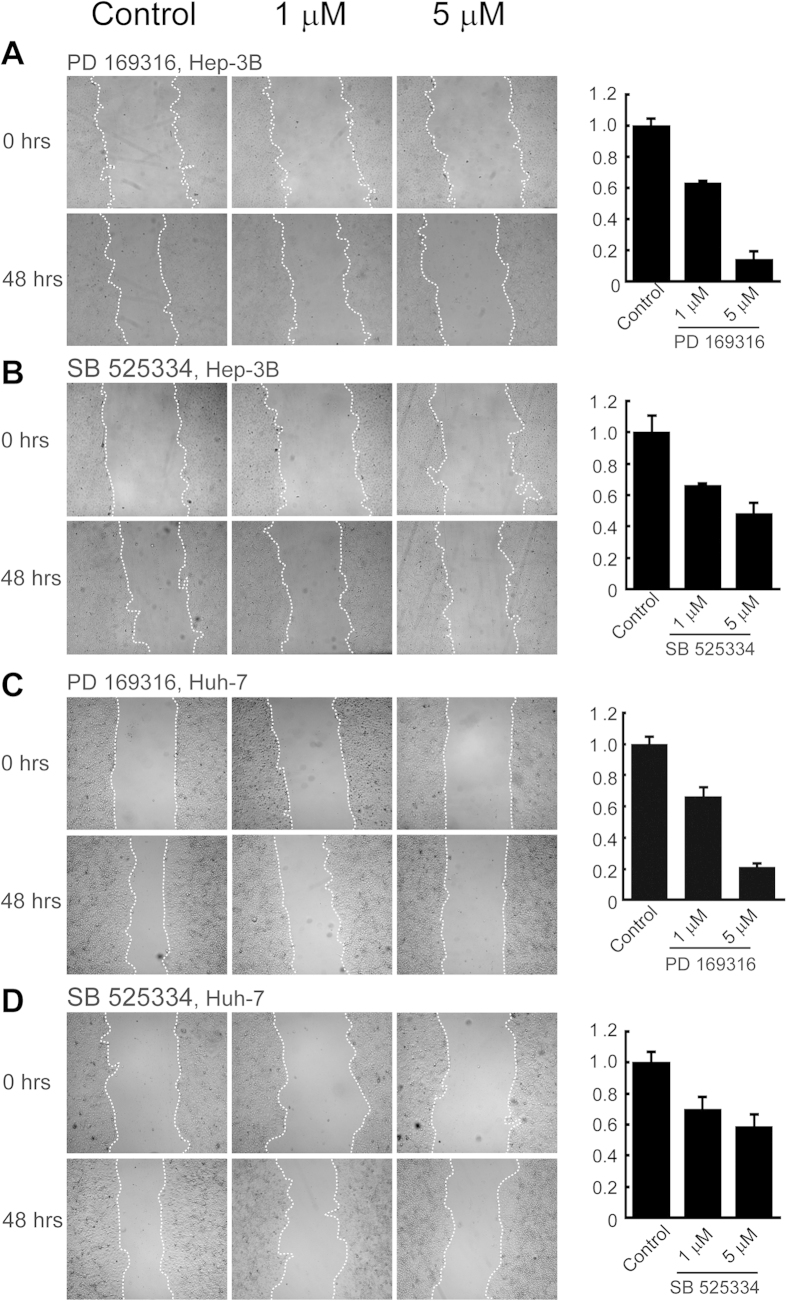
To additionally assess the validity of the compounds identified, and, thus, the application of our method, we assayed the effects of PD 169316 and SB 525334 on human cancer cells during migration in monolayer scratch assays33. Hep 3B2.1–7 and HuH-7 Hepatoma cells were cultured in monolayer to ~95% confluence and treated with mitomycin C to arrest growth. Scratches were introduced into the cell monolayers, floating cells were removed, and fresh media was added with or without various doses of PD 169316 or SB 525334. Cell migration into scratch areas was recorded and quantified. Both PD 169316 and SB 525334 inhibited cell migration in dose dependent manners (Fig. 6). As with chemotaxis inhibition of neutrophils (see Fig. 5), cells migration in these scratch assays was more sensitive to inhibition by PD 169316 than by SB 525334.
Figure 6. Validation of inhibitory effects of compounds PD 169316 and SB 525334 on migration of human cells in monolayer scratch assays.
Hep 3B2.1–7 (A,B) and HuH-7 (C,D) hepatoma cells were grown to ~95% confluency in monolayer and straight line scratches were made across each monolayer (19). Cells were incubated with 0 (control), 1, and 5 μM PD 169316 (A,C) or SB 525334 (B,D). The exact scratch areas were recorded immediately and after two days. Invaded areas were quantified by ImageJ software. 8 Scratches were assayed in each monolayer and experiments were performed in duplicate. Error bars represent standard deviations.
We conclude that the HTS Dictyostelium assay is able to identify compounds that similarly inhibit chemotaxis/migration of human cells.
Discussion
We have developed and validated a chemotaxis-dependent aggregation assay using Dictyostelium as a model chemotactic system that has capability for HTS. The phenotypic assay approach to identify small molecule inhibitors of chemotaxis in large complex libraries has unique advantages in comparison to other chemotactic assays9,10,11,12,13,14,15,16,17,18,19,20. The use of a laser scanning cytometer plate reader enabled a miniaturized assay format in 1536-well plates for the rapid detection of GFP-aggregates that quantifies chemotactic success. The assay provides a simple phenotypic approach to identify chemotaxis inhibitors of complex chemotactic processes.
Since there are large variations in the size and number of GFP aggregates per well, the 27% coefficient of variation (CV) for GFP expression may seem high. However, this is not a confounding issue. The detection threshold of the laser scanning plate cytometer is set to only capture GFP fluorescence signals from multicellular structures; the background fluorescence intensity in non-aggregated cells is zero (Figs 1 and 2A). This null background signal contrasts sharply with the several thousand GFP fluorescence units detected from the GFP aggregates formed after chemotaxis. Thus, results were in a unique format that precludes the calculation of conventional Z’ factors or signal-to-basal ratios, which approach infinity. Potent inhibitory compounds that fully suppress GFP expression/aggregate formation are readily identified from a primary screen, even though signal-to-basal ratios cannot be calculated by conventional means. In addition, for every LOPAC screening plate, we include DMSO, as negative control wells, and concentration ranges of latrunculin A, as positive inhibitory control wells. When the hundreds of these wells are classified in a binary fashion, with zero (no GFP expression) values as positive and non-zero (GFP expression) values as negative, there was perfect correlation between inhibitory and non-inhibitory effects, emphasizing the robustness of the assay.
It is interesting that partial inhibition of the GFP signal was not routinely observed. To assess this further, we monitored aggregation by direct microscopic examination of the mini-wells. The data were similar; cells either completed aggregation and expressed GFP or remained as individual cells and did not express GFP. This is not unlike the effects of cell density on the aggregation of WT Dictyostelium, where a two-fold cell dilution in monolayer can separate robust chemotaxis and aggregation from non-responsiveness (see Fig. 2C). These observations can be partly explained by the characteristics of Dictyostelium signal-response. During WT aggregation, cells must both release and respond to secreted cAMP, in a positive feedback loop1,22,23,24,26,40. There appears a density threshold, below which cells are unable to relay the signal-response and aggregation cannot proceed46.
The non-linearity of the assay is reflected in the latrunculin A data (see Fig. 3A). Similarly, while PD 169316 and SB 525334 inhibit Dictyostelium chemotaxis velocity and directionality (see Fig. 4D) in a dose-dependent manner, partial inhibition of GFP expression is often not observed. While both PD 169316 and SB 525334 were very strongly inhibitory in the screening assay, with IC50 values similar to that observed in the EZ-TAXIScan assay, we did not anticipate that they would inhibit chemotaxis to 100% in the directed assays. There are at least 6 intracellular signaling pathway arms that regulate chemotaxis in Dictyostelium1,22,23,24,26,40 and although they cross-interact, genetic ablation of several in an individual cell does not completely suppress chemotaxis23,47. Indeed, partial chemotaxis of both Dictyostelium and neutrophils in EZ-TAXIScan assays is observed at the highest compound concentrations tested (Figs 4C,D and 5).
Although there is a significant false positive rate due to the large number of cytotoxic compounds (e.g. 20 out of 22 primary hits), these false positives are readily recognized and eliminated using a parallel counter-screen with a highly sensitive ATP content assay for cell viability. Other false positives (e.g. inhibitors of cAMP synthesis, starvation sensing, GFP) are easily screened in direct EZ-TAXIScan or other chemotaxis assays. Whereas, not every compound identified in our initial screen is expected to be equally effective on mammalian chemotaxis, our results indicate that both of the active compounds identified from the Dictyostelium aggregation assay also inhibited chemotaxis/migration of human neutrophils and cancer cells. Indeed, Dictyostelium and mammalian cells share many pathways required for chemoattractant/chemokine responses (e.g. GPCRs, G proteins, ERKs, PKA, PI3K, TORC2, AKT, PLA2, PDK1, cyclases)1,22,23,24,26,40, although we recognize that not all compounds that can inhibit chemotaxis will be detected in our screen. LY-294002 may be considered such a false negative. LY-294002 is in the LOPAC library, can inhibit chemotaxis, but was not selected in our screen. At the highest concentration tested (~40 μM), LY-294002 was only partially (<50%) inhibitory in our system, consistent with LY-294002 concentrations of >100 μM required to inhibit all chemotaxtic functions (e.g. mTORC2)35.
The described Dictyostelium assay is, nonetheless, a reliable model system to identify active compounds that regulate chemotaxis, in general, and that can be further confirmed and followed in experiments with mammalian cells. Dictyostelium strains with specific genetic deficiencies in various chemotactic components are available and represent useful tools to dissect the mechanism of action and protein targets for the inhibitors of chemotaxis found in the phenotypic screens1,22,23,24,48,49.
In addition, this assay may also be applicable to screen for enhancers of chemotaxis. We observed that dozens of LOPAC compounds increased the number of aggregates per well (up to 18 aggregates; unpublished observations). Compounds that induce aggregation and GFP expression, under conditions normally non-permissive for Dictyostelium chemotaxis and aggregation [e.g. <2,000 cells/well (see Fig. 2C)] may be excellent candidates.
Conclusions
We have developed and optimized a novel assay for high-throughput screening to identify novel inhibitors of chemotaxis. The assay uses a rapid laser-scanning cytometry plate reader for the detection of GFP-aggregates formed during Dictyostelium chemotaxis and a parallel cell viability counter-screen to eliminate cytotoxic compounds. The assay allowed the HTS of 1200 compounds, at 7 concentrations + controls, in ~50,000 wells with 32 (1536-well) plates. We further demonstrated that the 2 compounds identified as inhibitors of Dictyostelium chemotaxis are also capable of inhibiting chemotaxis/migration of human neutrophils and cancer cells. This new compound screening approach, using a phenotypic Dictyostelium chemotaxis-dependent aggregation assay, enables the rapid identification of lead compounds that inhibit chemotaxis in human and other cells for drug development and research tools.
Methods
Materials
Dictyostelium strain DBS0236466 (AX4 cells expressing GFP under the control of the prespore cotB promoter) was kindly provided by Dr. William Loomis (UCSD, La Jolla, CA)31. Hepatoma cell lines, Hep 3B2.1–7 and HuH-7, were purchased from Cell Bank, Chinese Academy of Sciences. One liter of D3-T media for Dictyostelium growth culture was composed of 7.15 g of peptone (BD Bioscience, #211693), 7.15 g of thiotone (BD Bioscience, #211921), 7.15 g of yeast extract (BD Bioscience, #212750), 15.4 g of D-glucose, 0.48 g of KH2PO4 and 0.525 g of Na2HPO4•7H2O. DB starvation buffer at pH 6.5 was composed of 7.4 mM NaH2PO4-H2O, 4 mM Na2HPO4•7H2O, 2 mM MgCl2 and 0.2 mM CaCl2. The ATPlite assay kit for measuring ATP content in cells as a cytotoxicity indicator was purchased from Perkin Elmer (#6016731). Latrunculin A was purchased from Sigma-Aldrich (#L5163). Hygromycin B and the Library of Pharmacologically Active Compounds (LOPAC) containing 1280 compounds were also purchased from Sigma-Aldrich. MEM media were composed of MEM (GIBCO, #41500034), 1.5 g/L of NaHCO3, 0.11 g/L of Sodium Pyruvate and 10% of fetal bovine serum (GIBCO, #10437-028). DMEM media was composed of DMEM (GIBCO, #12800017), 1.5 g/L of NaHCO3 and 10% of fetal bovine serum.
Cell Growth and Starvation
Dictyostelium cotB/GFP cells31 were grown to confluence in nutrient abundant D3-T media in stationary flasks at 20 °C. Cells were diluted into fresh D3-T media at 2 × 105 cells/ml and cultured in shaking flasks for 2 days at 220 rpm, 20 °C). Cells at log-phase (1–3 × 106 cells/ml) were harvested by centrifugation at 1000 × g for 5 min, washed twice with DB, and resuspended in DB at appropriate cell densities.
Hepatoma cell lines, Hep 3B2.1–7 and HuH-7, were cultured with MEM and DMEM media in a humidified 5% CO2-95% air atmosphere incubator at 37 °C.
Preparation of Compound Plates
All of the compounds were dissolved in 100% DMSO at 10 mM stock solutions. For the LOPAC library, the compounds were serially diluted with DMSO in 384-well plates at a 1:5 titration ratio over 7 concentrations and then formatted to 1536-well compound plates using an Evolution P3 system (PerkinElmer, Wellesley, MA). The stock solutions of control compounds and the selected compounds were diluted with DMSO in 384-well plates at a 1:3 titration ratio over 12 concentrations, and then formatted to 1536-well compound plates50. The final concentrations of each compound ranged from 2.46 nM to 38.3 μM in the chemotaxis-dependent aggregation assay (0.38% DMSO in all wells) and from 3.68 nM to 57.5 μM in the ATP content assay (0.58% DMSO in all wells).
Data Analyses
Screening data were normalized and concentration-effect relationships derived using NCGC-developed software51. IC50 values were obtained by data fit using a residual error minimization algorithm with automatic outlier determination to a 4-parameter Hill. Concentration-effect relationships (CERs) were categorized by fit quality (r2), response magnitude, and degree of measured activity51. For validation, data were analyzed using GraphPad Prism software. Percent inhibition was derived from normalization of the Logarithms of positive and negative controls within the same experimental set. IC50 values were obtained from nonlinear curve fits using a sigmoidal dose−response model.
EZ-TAXIScan Chemotaxis Assay
The EZ-TAXIScan chemotaxis assays are modifications32. For Dictyostelium chemotaxis, log phase cells (1–3 × 106 cells/ml) were harvested, washed twice and resuspended in DB starvation buffer at 2 × 107 cells/ml. Cells were shaken at 125 rpm at 20 °C and pulsed with 75 nM final concentration of cAMP every 6 min. After one hour of cAMP pulsing, various doses of SB 525334, PD 169316, or an equivalent amount of DMSO as a control was added to the cells and pulsing was continued for additional four and half hours. Cells were then applied onto the EZ-TAXIScan (Effector Cell Institute, Tokyo, Japan) channel with 0–0.1 μM gradient of cAMP in the chemoattractant well at room temperature.
Blood samples were obtained from paid healthy volunteers who gave written informed consent to participate in an IRB-approved study for the collection of blood samples for in vitro research use. The protocol is designed to protect subjects from research risks as defined in 45CFR46 and to abide by all NIH guidelines for human subjects research (protocol number 99-CC-0168). Neutrophils were isolated from heparinized human whole blood by dextran sedimentation and differential centrifugation over Histopaque 1077 (Sigma-Aldrich) as described previously32. Residual erythrocytes were removed by hypotonic lysis. The fresh-isolated neutrophils were treated with PD 169316, SB 525334, or an equivalent amount of DMSO as a control was added to the cells for 4 hrs. Cells were then applied onto the EZ-TAXIScan channel with 0–0.1 μM gradient of fMLP in the chemoattractant well at 37 °C.
Migratory Dictyostelium or neutrophil cells were recorded every 15 sec for 30 min32. Cell velocity and chemotaxis index were calculated with MATLAB software, as described32.
For Dictyostelium chemotaxis to folate, log phase cells (1–3 × 106 cells/ml) were incubated for 30 min. with 50 μM SB 525334, 50 μM PD 169316, or an equivalent amount of DMSO (0.16%) as a control. Cells were then applied onto the EZ-TAXIScan channel with 0–100 μM gradient of folate in the chemoattractant well at room temperature.
Monolayer Scratch Migration Assay
The assay is modified from a previous report33. Hep 3B2.1–7 and HuH-7 Hepatoma cells were grown in monolayer, 6-well plates to ~95% confluency and treated with mitomycin C to arrest cell growth. Two straight scratches were made with a pipette tip across each cell monolayer. Floating cells were removed by washing with PBS and fresh media, with varying doses of PD 169316, SB 525334, or an equivalent amount of DMSO, were added into each well. 8 areas of scratches lines were marked for each cell monolayer and immediately recorded by camera; after two days, the exact scratch areas were again recorded. The original and resulting areas absent of cells were quantified by ImageJ software. The 8 areas of each well were averaged and variation further assessed with duplicate wells for each treatment. Normalization was relative to % control invasion, set to 1.0.
Phosphorylation assays
cAMP and folate stimulations of PKBR1 and ERK phosphorylation were assayed by immunoblot, as previously described52,53.
Trypan blue staining
Control and latrunculin A treated cells were incubated with 0.1% trypan blue (Lonza) for 10 min at room temperature39 and cell viability quantified using a Nexcelom Cellometer and light microscopy.
Additional Information
How to cite this article: Liao, X.-H. et al. A High-Throughput, Multi-Cell Phenotype Assay for the Identification of Novel Inhibitors of Chemotaxis/Migration. Sci. Rep. 6, 22273; doi: 10.1038/srep22273 (2016).
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Programs of The National Institutes of Health, The National Cancer Institute, The National Center for Advancing Translational Sciences, The National Institute of Diabetes and Digestive and Kidney Diseases, and The Molecular Libraries Initiative of the NIH Roadmap for Medical Research. The authors thank Amy Melpolder and the National Institutes of Health Blood Bank for providing human blood from healthy volunteers. The authors also thank the compound management group in NCATS/NIH for providing the compounds used in this study. Finally, we wish to thank Dr. Pundrik Jaiswal for comments, interest, and assistance and Dr. Aleksandra Nita-Lazar and colleagues for access to EZ-TAXIScan.
Footnotes
Author Contributions X.-H.L. and A.R.K. developed the concepts and approaches. X.-H.L., N.M., L.L., M.S., A.L.Z., M.P., J.J.X. and A.R.K. performed experiments. X.-H.L., N.M., N.S., L.L., M.S., A.L.Z., J.J.X., T.J., C.A.P., W.Z. and A.R.K. analyzed the data. X.-H.L., C.A.P., W.Z. and A.R.K. prepared the manuscript prior to submission.
References
- Jin T., Xu X. & Hereld D. Chemotaxis, chemokine receptors and human disease. Cytokine 44, 1–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A. T. et al. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol 178, 185–91 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G. & Mooney D. J. New materials for tissue engineering: towards greater control over the biological response. Trends Biotechnol 26, 382–92 (2008). [DOI] [PubMed] [Google Scholar]
- Mackay C. R. Moving targets: cell migration inhibitors as new anti-inflammatory therapies. Nat Immunol 9, 988–98 (2008). [DOI] [PubMed] [Google Scholar]
- Djekic U. V., Gaggar A. & Weathington N. M. Attacking the multi-tiered proteolytic pathology of COPD: new insights from basic and translational studies. Pharmacol Ther 121, 132–46 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Lee V. C., Chevalier E. & Hwang S. T. Chemokine receptors as targets for cancer therapy. Curr Pharm Des 15, 742–57 (2009). [DOI] [PubMed] [Google Scholar]
- Su W., Chen Q. & Frohman M. A. Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future Oncol 5, 1477–86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamdar G. S., Madhunapantula S. V. & Robertson G. P. Targeting the MAPK pathway in melanoma: why some approaches succeed and other fail. Biochem Pharmacol 80, 624–37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastyugin V., McWhinnie E., Labow M. & Buxton F. A quantitative high-throughput endothelial cell migration assay. J Biomol Screen 9, 712–8 (2004). [DOI] [PubMed] [Google Scholar]
- Hulkower K. I. & Herber R. L. Cell migration and invasion assays as tools for drug discovery. Pharmaceutics 3, 107–24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthier E. et al. Low-volume toolbox for the discovery of immunosuppressive fungal secondary metabolites. PLoS Pathog 9, e1003289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guckenberger D. J., Berthier E. & Beebe D. J. High-Density Self-Contained Microfluidic KOALA Kits for Use by Everyone. J Lab Autom (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. et al. [Eosinophil chemotaxis assay using novel device EZ-TAXIScan]. Arerugi 57, 1317–24 (2008). [PubMed] [Google Scholar]
- Timm D. M. et al. A high-throughput three-dimensional cell migration assay for toxicity screening with mobile device-based macroscopic image analysis. Sci Rep 3, 3000 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltaninassab S. R. et al. A Novel High Throughput-Compatible Cell Migration Screening Assay. BioTek Application Note Rev. 8/12, 1–4 (2008). [Google Scholar]
- Ponath P. D., Wang J. & Heath H. Transwell chemotaxis. Methods Mol Biol 138, 113–20 (2000). [DOI] [PubMed] [Google Scholar]
- Hattori H. et al. Small-molecule screen identifies reactive oxygen species as key regulators of neutrophil chemotaxis. Proc Natl Acad Sci USA 107, 3546–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wu X. & Lin F. Recent developments in microfluidics-based chemotaxis studies. Lab Chip 13, 2484–99 (2013). [DOI] [PubMed] [Google Scholar]
- Collins S. R. et al. Using light to shape chemical gradients for parallel and automated analysis of chemotaxis. Mol Syst Biol 11, 804 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai S., Guan X. J., Kuspa A. & Cox E. C. High-throughput analysis of spatio-temporal dynamics in Dictyostelium. Genome Biol 8, R144 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia D. & Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr Biol (Camb) 1, 506–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney K. F., Huang C. H. & Devreotes P. N. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys 39, 265–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert P. J. & Veltman D. M. Chemotaxis: navigating by multiple signaling pathways. Sci STKE 2007, pe40 (2007). [DOI] [PubMed] [Google Scholar]
- McMains V. C., Liao X. H. & Kimmel A. R. Oscillatory signaling and network responses during the development of Dictyostelium discoideum. Ageing Res Rev 7, 234–48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G. Dictyostelium finds new roles to model. Genetics 185, 717–26 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artemenko Y., Lampert T. J. & Devreotes P. N. Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell Mol Life Sci 71, 3711–47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. S. et al. The mood stabiliser lithium suppresses PIP3 signalling in Dictyostelium and human cells. Dis Model Mech 2, 306–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade K. J., Nakajima A., Ilacqua A. N., Shimada N. & Sawai S. The green tea catechin epigallocatechin gallate (EGCG) blocks cell motility, chemotaxis and development in Dictyostelium discoideum. PLoS One 8, e59275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garige M. & Walters E. Curcumin inhibits development and cell adhesion in Dictyostelium discoideum: Implications for YakA signaling and GST enzyme function. Biochem Biophys Res Commun 467, 275–81 (2015). [DOI] [PubMed] [Google Scholar]
- Kuwayama H. & Kubohara Y. Differentiation-inducing factor-1 and -2 function also as modulators for Dictyostelium chemotaxis. PLoS One 4, e6658 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol A., Rappel W., Levine H. & Loomis W. F. Cell-sorting in aggregates of Dictyostelium discoideum. J Cell Sci 112(Pt 22), 3923–9 (1999). [DOI] [PubMed] [Google Scholar]
- Liu L., Das S., Losert W. & Parent C. A. mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev Cell 19, 845–57 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C. C., Park A. Y. & Guan J. L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2, 329–33 (2007). [DOI] [PubMed] [Google Scholar]
- Kriebel P. W., Barr V. A., Rericha E. C., Zhang G. & Parent C. A. Collective cell migration requires vesicular trafficking for chemoattractant delivery at the trailing edge. J Cell Biol 183, 949–61 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. H., Buggey J. & Kimmel A. R. Chemotactic activation of Dictyostelium AGC-family kinases AKT and PKBR1 requires separate but coordinated functions of PDK1 and TORC2. J Cell Sci 123, 983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis W. F. & Wheeler S. Heat shock response of Dictyostelium. Dev Biol 79, 399–408 (1980). [DOI] [PubMed] [Google Scholar]
- Coue M., Brenner S. L., Spector I. & Korn E. D. Inhibition of actin polymerization by latrunculin A. FEBS Lett 213, 316–8 (1987). [DOI] [PubMed] [Google Scholar]
- Pring M., Cassimeris L. & Zigmond S. H. An unexplained sequestration of latrunculin A is required in neutrophils for inhibition of actin polymerization. Cell Motil Cytoskeleton 52, 122–30 (2002). [DOI] [PubMed] [Google Scholar]
- Giusti C. et al. Analysis of autophagic and necrotic cell death in Dictyostelium. Methods Enzymol 446, 1–15 (2008). [DOI] [PubMed] [Google Scholar]
- Jin T. Gradient sensing during chemotaxis. Curr Opin Cell Biol 25, 532–7 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang S., Charest P. G. & Firtel R. A. Spatiotemporal regulation of Ras activity provides directional sensing. Curr Biol 18, 1587–93 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miret S., De Groene E. M. & Klaffke W. Comparison of in vitro assays of cellular toxicity in the human hepatic cell line HepG2. J Biomol Screen 11, 184–93 (2006). [DOI] [PubMed] [Google Scholar]
- Grygielko E. T. et al. Inhibition of gene markers of fibrosis with a novel inhibitor of transforming growth factor-beta type I receptor kinase in puromycin-induced nephritis. J Pharmacol Exp Ther 313, 943–51 (2005). [DOI] [PubMed] [Google Scholar]
- Kummer J. L., Rao P. K. & Heidenreich K. A. Apoptosis induced by withdrawal of trophic factors is mediated by p38 mitogen-activated protein kinase. J Biol Chem 272, 20490–4 (1997). [DOI] [PubMed] [Google Scholar]
- Fu Y., O’Connor L. M., Shepherd T. G. & Nachtigal M. W. The p38 MAPK inhibitor, PD169316, inhibits transforming growth factor beta-induced Smad signaling in human ovarian cancer cells. Biochem Biophys Res Commun 310, 391–7 (2003). [DOI] [PubMed] [Google Scholar]
- Gregor T., Fujimoto K., Masaki N. & Sawai S. The onset of collective behavior in social amoebae. Science 328, 1021–5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman D. M., Keizer-Gunnik I. & Van Haastert P. J. Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J Cell Biol 180, 747–53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert P. J. Analysis of signal transduction: formation of cAMP, cGMP, and Ins(1,4,5)P3 in vivo and in vitro. Methods Mol Biol 346, 369–92 (2006). [DOI] [PubMed] [Google Scholar]
- Mendoza M. C. & Firtel R. A. Assaying chemotaxis of Dictyostelium cells. Methods Mol Biol 346, 393–405 (2006). [DOI] [PubMed] [Google Scholar]
- Inglese J. et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci USA 103, 11473–8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall N., Jadhav A., Huang R., Nguyen T. & Wang Y. Enabling the Large Scale Analysis of Quantitative High Throughput Screening Data, 442–463 (Taylor and Francis, New York, 2009). [Google Scholar]
- Liao X. H. & Kimmel A. R. Biochemical responses to chemoattractants in Dictyostelium: ligand-receptor interactions and downstream kinase activation. Methods Mol Biol 571, 271–81 (2009). [DOI] [PubMed] [Google Scholar]
- Meena N. & Kimmel A. R. Biochemical responses to chemically distinct chemoattractants during the growth and development of Dictyostelium. Methods Mol Biol (in press) (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.