Abstract
Droplet digital PCR (ddPCR), a method for measuring target nucleic acid sequence quantity, is useful for determining somatic mutation rates using TaqMan probes. In this study, the detection limit of copy numbers of test DNA by ddPCR was determined based on Poisson distribution. Peptide nucleic acid (PNA), which strongly hybridises to target lesions, can inhibit target amplification by PCR. Therefore, by combination of PCR with PNA and ddPCR (PNA–ddPCR), the detection limit could be lowered. We reanalysed a somatic GNAQ mutation (c.548G > A) in patients with Sturge–Weber syndrome (SWS) using ddPCR and PNA–ddPCR. Importantly, among three patients previously found to be mutation negative by next–generation sequencing, two patients had the GNAQ mutation with a mutant allele frequency of less than 1%. Furthermore, we were able to find the same mutation in blood leukocyte or saliva DNA derived from four out of 40 SWS patients. Vascular anomalies and blood leukocytes originate from endothelial cells and haemangioblasts, respectively, which are both of mesodermal origin. Therefore, blood leukocytes may harbour the GNAQ mutation, depending on the time when the somatic mutation is acquired. These data suggest the possibility of diagnosis using blood DNA in some patients with SWS.
Sturge–Weber syndrome (SWS, MIM#185300) is a neurocutaneous disorder characterised by the following manifestations: 1) cutaneous vascular malformations (port–wine stains), 2) ocular vascular malformations leading to choroidal vascular abnormalities, glaucoma, buphthalmia and hemianopia, and 3) intracranial vascular malformation resulting in neurological impairment including seizures and intellectual disability1,2,3,4,5. The prevalence is estimated at approximately 1/20,000–1/50,0001. It has been suggested that SWS is likely to be caused by somatic mutations because its occurrence is sporadic with no heritability6. Recently, a somatic c.548G > A mutation in GNAQ [encoding guanine nucleotide–binding protein, Q polypeptide (MIM600998)] was indeed identified in 88 and 92% of patients with SWS and those with non–syndromic port–wine stains, respectively3. We also confirmed the presence of low–prevalence somatic GNAQ mutations in 12 of 15 SWS samples using deep sequencing (80%), and no other possible somatic GNAQ mutations were found7. In these two reports, mutant allele frequencies in brain samples ranged from 1.0 to 11.15%7. Both studies adopted the 1% cut–off line to detect mutant alleles in total sequence reads to excluding possible errors of PCR and read misalignment/mapping. Therefore, there is a possibility that extreme low–prevalence (<1%) mutations could be overlooked.
Droplet digital PCR (ddPCR) is a sensitive method enabling the accurate quantification of a target nucleic acid sequence8,9. In this method, individual DNA molecules from a sample are captured within water–in–oil droplet partitions9. Droplets containing mutant or wild–type allele(s) are discriminated using two color–fluorescent TaqMan probes and the numbers of target DNA copies are counted at the end point of PCR8,10. Poisson distribution is used to assay DNA molecule concentration using numbers of accepted total amplified and un–amplified droplets9. Peptide nucleic acid (PNA) is a DNA/RNA mimic that can be hybridised to target sequences and prevent PCR amplification of target regions11,12. By combination of PCR with PNA and ddPCR (PNA–ddPCR), it may be possible to successfully detect low–prevalent mutant alleles more sensitively than with ddPCR alone as only mutant alleles are amplified.
We present an investigation of the detection limit of ddPCR and PNA–ddPCR using a target low–prevalence somatic GNAQ mutation (c.548G > A) in patients with SWS7 who were previously analysed only with next–generation sequencing (NGS)3,7.
Results
Detection limit of ddPCR
The detection limit of ddPCR was determined using serial dilutions of cloned mutant DNA (GNAQ c.548G > A) in non–mutant DNA at levels of 10, 5, 1, 0.5, 0.25 and 0.1%, the copy numbers of which were “300, 150, 30, 15 and 7.5” (in 3000), respectively (Table 1). However, mutant alleles at a frequency of <0.25% (7.5 copies) could not be consistently detected. Therefore, <0.25% is gray (under the detection limit) rather than completely negative. The mutation could be consistently detected at 10, 5, 1, 0.5 and 0.25% (Table 1). We evaluated the reliability of the detection limit using another statistical method based on the binominal distribution, supporting the above detection limit (see Supplementary Data and Table S1). This result also indicated that we were able to detect mutant DNA with confidence to 0.25% (see Supplementary Table S1). Therefore, the detection limit of ddPCR was defined as 0.25%. Fractional abundance (FA) (denoting the proportion of the mutant allele frequencies by QuantaSoft) of the 0.25% positive control actually indicated 0.26–0.42% (Table 1).
Table 1. Mutant clone ratios detected by ddPCR.
| Mut clone ratios | Mut copy number* | Mut drop number | Wt drop number | ACD | FA (%) | FA range (%) |
|---|---|---|---|---|---|---|
| 10% | 144 | 101 | 707 | 16444 | 12.3 | 14.5–10.0 |
| 10% | 166 | 114 | 754 | 16252 | 12.9 | 15.1–10.7 |
| 10% | 150 | 103 | 1046 | 16196 | 8.7 | 10.3–7.1 |
| 5% | 74 | 50 | 771 | 15951 | 6 | 7.6–4.3 |
| 5% | 74 | 49 | 714 | 15547 | 6.3 | 8.0–4.6 |
| 5% | 66 | 48 | 1050 | 16941 | 4.2 | 5.4–3.1 |
| 1% | 14.4 | 9 | 681 | 14684 | 1.3 | 2.1–0.4 |
| 1% | 10.2 | 7 | 729 | 16037 | 0.9 | 1.6–0.2 |
| 1% | 16 | 11 | 1182 | 16282 | 0.9 | 1.4–0.4 |
| 0.50% | 8.6 | 5 | 576 | 13723 | 0.8 | 1.6–0.1 |
| 0.50% | 4.6 | 3 | 552 | 15250 | 0.5 | 1.2–0 |
| 0.50% | 8.8 | 6 | 1031 | 16030 | 0.56 | 1.02–0.1 |
| 0.25% | 5 | 3 | 679 | 14038 | 0.43 | 0.95–0 |
| 0.25% | 3.6 | 2 | 631 | 12837 | 0.31 | 0.78–0 |
| 0.25% | 4.2 | 3 | 1104 | 16872 | 0.26 | 0.58–0 |
| 0.10% | 0 | 0 | 772 | 16230 | 0 | 0–0 |
| 0.10% | 1.4 | 1 | 788 | 16400 | 0.12 | 0.42–0 |
| 0.10% | 3.4 | 2 | 975 | 13931 | 0.2 | 0.50–0 |
*: Calculated copy number by Quantasoft™, Mut: mutant, Wt: wild–type, drop: droplet, ACD: accepted droplets, FA: fractional abundance, PFA: Poisson fractional abundance.
The GNAQ somatic mutation in patients with SWS detected by ddPCR
The GNAQ mutation (c.548G > A) was quantified by ddPCR in DNA of paired brain and either leukocytes or saliva samples from 15 SWS patients previously analysed by NGS3,7 (Table 2). Comparable results were produced by ddPCR and NGS in most samples. Of 18 of these samples (three from brain, 14 from blood and one from saliva) in which NGS had detected mutant allele frequencies of less than 1%, 17 showed mutant allele frequencies of less than 0.25% of FA (considered to be under the detection limit) while the remaining sample (the saliva DNA), showed a mutant allele frequency of 0.61% of FA (SWS8; Table 2).
Table 2. ddPCR and PNA–ddPCR results of 15 paired SWS patients.
| Sample | Sample type | Mut copy number | Mut drop | Wt drop | ACD | FA (%) | deep–seq | PNA-ddPCR |
|---|---|---|---|---|---|---|---|---|
| SWS1 | brain | 246.67 ( 9.29 ) | 138.33 ( 15.11 ) | 2188.00 ( 155.86 ) | 13208.33 ( 1009.30 ) | 5.50 ( 0.24 ) | 6.0 | + |
| blood | 7.80 ( 4.82 ) | 4.33 ( 2.87 ) | 2520.67 ( 88.51 ) | 12677.67 ( 740.68 ) | 0.15 ( 0.10 ) | 0.03 | +new | |
| SWS2 | brain | 80.67 ( 15.86 ) | 52.00 ( 8.60 ) | 1097.67 ( 13.47 ) | 15316.00 ( 839.64 ) | 4.37 ( 0.66 ) | 4.0 | + |
| blood | 8.87 ( 5.01 ) | 5.33 ( 3.09 ) | 4008.33 ( 2093.11 ) | 13836.00 ( 611.36 ) | 0.11 ( 0.06 ) | 0.05 | +new | |
| SWS3 | brain | 9.60 ( 3.37 ) | 4.67 ( 1.25 ) | 2876.67 ( 125.27 ) | 11946.33 ( 1546.93 ) | 0.14 ( 0.04 ) | 0.03 | +new |
| blood | 1.53 ( 1.23 ) | 1.00 ( 0.82 ) | 3066.33 ( 166.55 ) | 14435.33 ( 873.95 ) | 0.04 ( 0.02 ) | 0.03 | − | |
| SWS4 | brain | 11.33 ( 5.56 ) | 6.67 ( 3.86 ) | 2894.67 ( 128.23 ) | 13326.00 ( 1169.36 ) | 0.20 ( 0.11 ) | 0.16 | +new |
| blood | 2.87 ( 2.29 ) | 2.00 ( 1.63 ) | 2356.33 ( 450.70 ) | 14885.67 ( 1623.49 ) | 0.11 ( 0.02 ) | 0.03 | − | |
| SWS5 | brain | 222.67 ( 17.00 ) | 148.00 ( 15.12 ) | 2979.67 ( 215.29 ) | 15683.33 ( 423.06 ) | 4.33 ( 0.53 ) | 5.0 | NP |
| blood | 4.73 ( 2.46 ) | 3.00 ( 1.63 ) | 5104.00 ( 1862.52 ) | 14706.33 ( 645.52 ) | 0.05 ( 0.04 ) | 0.04 | − | |
| SWS6 | brain | 331.33 ( 10.50 ) | 207.67 ( 4.03 ) | 2772.33 ( 121.38 ) | 14858.67 ( 354.37 ) | 6.40 ( 0.22 ) | 6.0 | NP |
| blood | 3.60 ( 1.88 ) | 2.33 ( 1.25 ) | 3423.67 ( 513.43 ) | 15444.00 ( 1326.28 ) | 0.06 ( 0.03 ) | 0.04 | − | |
| SWS7 | brain | 177.33 ( 16.76 ) | 111.33 ( 13.89 ) | 2674.33 ( 173.12 ) | 14798.67 ( 634.96 ) | 3.63 ( 0.33 ) | 4.0 | NP |
| blood | 1.60 ( 1.40 ) | 1.00 ( 0.82 ) | 4894.00 ( 2358.55 ) | 15173.67 ( 1153.39 ) | 0.03 ( 0.02 ) | 0.04 | − | |
| SWS8 | brain | 295.33 ( 8.22 ) | 181.00 ( 7.12 ) | 2616.33 ( 124.93 ) | 14501.33 ( 320.86 ) | 5.97 ( 0.12 ) | 6.0 | + |
| saliva | 51.33 ( 5.25 ) | 35.33 ( 4.78 ) | 4856.33 ( 533.45 ) | 16400.33 ( 1350.59 ) | 0.61 ( 0.06 ) | 0.44 | + | |
| SWS9 | brain | 346.00 ( 17.05 ) | 178.00 ( 7.26 ) | 2339.00 ( 69.37 ) | 12196.67 ( 828.45 ) | 6.47 ( 0.09 ) | 8.0 | NP |
| blood | 1.07 ( 0.75 ) | 0.67 ( 0.47 ) | 3111.00 ( 107.34 ) | 14228.33 ( 241.84 ) | 0.02 ( 0.01 ) | 0.03 | − | |
| SWS10 | brain | 420.00 ( 41.09 ) | 232.00 ( 19.44 ) | 2565.00 ( 166.28 ) | 13152.67 ( 250.69 ) | 7.60 ( 0.08 ) | 8.0 | NP |
| blood | 3.40 ( 2.70 ) | 1.67 ( 1.25 ) | 2381.67 ( 237.10 ) | 12761.67 ( 1533.49 ) | 0.07 ( 0.05 ) | 0.04 | − | |
| SWS11 | brain | 4.60 ( 3.15 ) | 2.67 ( 1.70 ) | 2894.00 ( 226.81 ) | 14169.33 ( 1065.11 ) | 0.09 ( 0.06 ) | 0.03 | − |
| blood | 0.00 ( 0 ) | 0.00 ( 0 ) | 1078.33 ( 379.28 ) | 13129.00 ( 915.10 ) | 0.00 ( 0 ) | 0.03 | − | |
| SWS12 | brain | 136.67 ( 12.26 ) | 73.00 ( 20.22 ) | 2320.67 ( 629.80 ) | 12492.67 ( 2597.01 ) | 2.80 ( 0.08 ) | 4.0 | NP |
| blood | 0.87 ( 1.23 ) | 0.33 ( 0.47 ) | 1372.33 ( 544.09 ) | 10545.00 ( 2524.01 ) | 0.03 ( 0.04 ) | 0.04 | − | |
| SWS13 | brain | 170.67 ( 29.23 ) | 87.67 ( 10.62 ) | 2099.00 ( 302.03 ) | 12690.00 ( 3164.55 ) | 3.70 ( 0.08 ) | 4.0 | NP |
| blood | 3.67 ( 3.30 ) | 2.33 ( 2.05 ) | 2414.67 ( 78.47 ) | 14800.00 ( 921.06 ) | 0.09 ( 0.08 ) | 0.03 | − | |
| SWS14 | brain | 66.67 ( 4.11 ) | 43.00 ( 3.56 ) | 481.67 ( 24.64 ) | 15137.67 ( 854.48 ) | 8.07 ( 0.29 ) | 9.0 | NP |
| blood | 1.60 ( 1.23 ) | 1.00 ( 0.82 ) | 2086.67 ( 108.02 ) | 14751.33 ( 1099.24 ) | 0.04 ( 0.03 ) | 0.04 | − | |
| SWS15 | brain | 94.00 ( 21.42 ) | 55.33 ( 4.64 ) | 1485.67 ( 112.35 ) | 14378.33 ( 2276.07 ) | 3.43 ( 0.45 ) | 4.0 | NP |
| blood | 3.27 ( 1.23 ) | 2.00 ( 0.82 ) | 2560.33 ( 131.76 ) | 14101.00 ( 846.50 ) | 0.07 ( 0.02 ) | 0.03 | − |
Mut: mutant, Wt: wild–type, ACD: number of accepted droplets, FA: fractional abundance, PFA: Poisson fractional abundance, deep–seq: mutant ratio by deep sequencing (previously reported)7, NP: not performed, + : positive, −: negative, + new: positive only by PNA–ddPCR (not by the other methods). ddPCR was performed in triplicate. SD values are shown in parentheses after the mean values. PNA–ddPCR was performed in triplicate where “ +new” is shown.
Detection limit of PNA–ddPCR
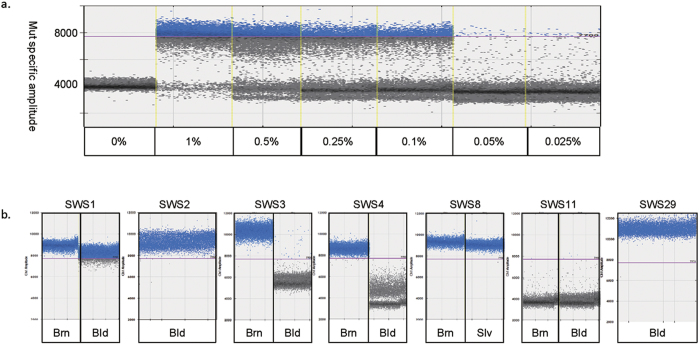
The sensitivity for PNA–ddPCR was determined using the same series of dilutions of mutant DNA. The mutation could be consistently detected at dilutions of 1, 0.5, 0.25 and 0.1% (Fig. 1a). However, 0.05–0.025% plasmid DNA showed inconsistently positive or negative results in each of triplicate experiments. The postulated reasons for this inconsistency for <0.1% plasmid DNA are as follows: 1) The mutant copy numbers at 0.1, 0.05 and 0.025% of 3000 copies would on average be 3, 1.5 and 0.75 copies. Therefore, the actual copy number of 0.025% can sometimes be one or zero (i.e. absent), depending on the aliquot used for PNA–ddPCR. 2) PCR contamination. PNA–ddPCR involves two rounds of PCR, so there is a risk of contamination. Based on the results of PNA–ddPCR using DNA from 30 healthy controls performed in triplicate, none of the results showed >1000 mutant droplets with >7700 signal intensity. Therefore, the criteria for positivity in detecting the mutant allele were set as >1000 mutant droplets with >7700 signal intensity. Considering the actual copy number of mutant DNA, 0.1% of loaded 1.5 × 10−5 ng clone DNA (equivalent to 3.0 × 103 copies) is approximately three copies. Therefore, to obtain a positive result by PNA–ddPCR, more than three copies per reaction is required.
Figure 1. PNA–ddPCR analyses in SWS patients.
Positive mutant–specific amplification was set to >1000 droplets with >7700 signal amplitude. (a) Detection limit of PNA–ddPCR. Mutant and wild–type clones were mixed serially in different ratios: 1, 0.5, 0.25, and 0.1% (mutant/mutant + wild–type). Ratios of 1–0.1% were positively amplified by PNA–ddPCR. (b) Mutation positivity was shown by PNA–ddPCR in 6 of 15 samples from SWS patients with <1% FA by ddPCR and in 1 of 25 samples from SWS patients for whom only blood samples were available in this study. Brn: Brain, Bld: Blood, Slv: saliva.
GNAQ mutation detection by PNA–ddPCR in SWS patients
Four of the 21 samples from 15 SWS patients previously analysed by NGS were positive for the GNAQ mutation by PNA–ddPCR. We especially focused on brain samples (n = 3) and blood leukocyte or saliva samples (n = 14) that were under the detection limit (<0.25% of FA) by ddPCR, as well as one saliva sample with 0.61% FA. PNA–ddPCR newly revealed that two blood leukocyte samples (SWS1 and SWS2) and two brain samples (SWS3 and SWS4) were mutant-positive, as was saliva of SWS8 (Fig. 1b and Table 2). Blood leukocyte samples were analysed by PNA–ddPCR from additional 25 SWS patients whose affected tissues were unavailable for mutation screening. Of note, one blood sample was positive for the GNAQ mutation (SWS29; Fig. 1b and see Supplementary Table S2).
Discussion
In this study, we have reached the conclusion that the detection limit of SNV by PNA–ddPCR was 3 copies in an aliquot. The data presented herein allow the determination of a hierarchy of sensitivity for novel mutation screening methods: 1% for NGS, 0.25% (7.5 copies) for ddPCR and 0.1% for PNA–ddPCR (3 copies). It thus follows that both 0.1% in 3.0 × 103 copies and 0.01% in 3.0 × 104 copies would produce a positive result because both scenarios would provide three copies in a single reaction. Importantly, therefore, if a greater overall quantity of DNA is used in PNA–ddPCR, the chance of detecting a low–prevalence mutation is increased. In practice, however, 3 × 104 copies of DNA is the machine limit in one–well analysis using the Droplet Digital PCR XQ200 system. As such, there is the trade–off between DNA quantity and the detection limit of ddPCR. Stahl et al. showed that the detection limit of ddPCR for the analysis of chimerism was 0.01% (1.95 copies)13. They used two types of marker (different loci) to distinguish donor cells and recipient cells for this chimerism analysis13. Our target is a single-nucleotide variation and discrimination of the mutant from the wild type is rather difficult.
One advantage of NGS over ddPCR and PNA–ddPCR is that while the latter two techniques target known genomic changes, NGS is capable of detecting a range of changes from single nucleotide variants to short insertions/deletions in all regions of the genome. NGS and ddPCR are quantitative methods, whereas PNA–ddPCR, though more sensitive is only qualitative. Among the original 15 SWS patients7, the GNAQ mutation (c.548G > A) in brain lesions was found in 14 patients (93%) using ddPCR and PNA–ddPCR. The mutation detection rate was therefore elevated from 80% (12/15) to 93% (14/15) by the use of ddPCR–based technologies.
We found the GNAQ mutation (c.548G > A) in brain lesions, blood leukocytes and saliva. Brain vascular system and lymphocytes in saliva and blood leukocytes are all of the mesodermal origin14. In embryogenesis, the mesoderm subsequently differentiates into blood islands and haemangioblasts in yolk sacs. In turn, haemangioblasts differentiate into endothelial cells and haemogenic endothelium14,15,16. Adult haematopoietic stem cells are differentiated from a particular type of haemogenic endothelium in aorta–gonad–mesonephros (AGM)15,16,17,18. These haematopoietic stem cells move to bone marrow after birth and continue haematopoiesis throughout life. Vascular malformations in SWS patients originate from endothelial cells. The GNAQ somatic mutation (c.548G > A) may occur in a haemangioblast or early endothelial cell. It is unknown whether the presence of a GNAQ mutation in blood or saliva is pathologically important. However, it is possible that cells harboring the mutation in blood leukocytes or saliva lymphocytes actively disseminate SWS lesions.
An extremely low–prevalence GNAQ mutation was found in two brain lesions and two blood samples in our 15 patients using PNA–ddPCR. Such low frequencies of mutant allele could be explained by an unusually low level of lesion infiltration contained in tissue sections used for DNA extraction. It is particularly important that we were able to find a GNAQ mutation in blood and saliva samples. A highly sensitive method of detecting the mutation in DNA of easily accessible tissues may be useful for developing a diagnostic method for SWS. The combination of two existing technologies described in this study could also be used for treatment monitoring or identification of relapse in some melanomas with the same GNAQ mutation19,20.
Methods
Patient and sample selection
This study was approved by the Institutional Review Board of Yokohama City University School of Medicine and Juntendo University Graduate School of Medicine. All experiments were carried out in accordance with the institutional guidelines. Written informed consent was obtained from patients or parents of pediatric patients. A total of 15 patients clinically diagnosed with SWS, who were previously analysed by NGS only7, were examined again in our study. Paired sets of peripheral blood leukocytes/saliva and surgically resected brain tissues were obtained from the 15 SWS patients7. In addition to the patients previously screened by NGS, we also included an additional cohort of 25 patients clinically diagnosed with SWS for whom only blood leukocytes were available and 30 normal controls in whom only blood leukocytes were available.
DNA extraction
Genomic DNA from RNAlater (Thermo Fisher Scientific, Waltham, MA, USA)–treated brain tissue blocks, peripheral blood leukocytes and saliva was extracted using Puregene Core Kit A (Qiagen, Valencia, CA, USA), PAXgene Blood DNA kit (Qiagen) and Oragene (DNA Genotek Inc., Ottawa, ON, Canada), respectively, according to the manufacturer’s instruction. DNA concentration was measured using the Qubit dsDNA BR assay kit (Thermo Fisher Scientific) according to the manufacturer’s instruction.
Control DNAs
The positive and negative control DNAs were necessary to set Tm values and minimise background for the ddPCR assay. A 178–bp fragment harboring c.548G > A in GNAQ or a corresponding wild–type allele was amplified using primers as shown in Table 3 and brain section DNA (as a template) from a SWS patient. The fragment was cloned in TOPO pCR2.1 Vector using a TOPO TA Cloning Kit (Life Technologies, Carlsbad, CA, USA). The presence of the inserted fragment in each clone was confirmed by Sanger sequencing.
Table 3. Sequences of primers and probes.
| Primer information | Primer and probe sequences |
|---|---|
| PCR primers amplifying a 178-bp fragment containing a GNAQ mutation which was cloned into TA vector. | 5′-ATTGTGTCTTCCCTCC−3′ (forward) 5′-GGTTTCATGGACTCAG−3′ (reverse) |
| ddPCR primers amplfying a 114-bp fragment containing a GNAQ mutation | 5′-CCTGCCTACGCAACAAGAT−3′ (forward) 5′-AGGTTTCATGGACTCAGTTACTAC-3′ (reverse) |
| LNA probes | 5′(HEX)−TGGGGAC + T + C + GAAC-3′(IABkFQ) (wild−type) |
| 5′(HEX)−TGGGG + AC + T + T + GAAC-3′(IABkFQ) (mutant) | |
| PNA probe (wild type) | 5′-GGGACTCGAACTCTA-3′ |
Confirmation of detection limit in ddPCR analysis
ddPCR was performed using a Droplet Digital PCR XQ200 system (Bio–Rad Laboratories, Hercules, CA, USA). The region–specific primers and customised locked nucleic acid (LNA) probes for wild–type and mutant alleles were purchased from Integrated DNA Technology (Coralville, IA, USA). Sequences of primers and probes were described in Table 3. We first tested ddPCR using serial dilutions of DNA of wild–type and mutant plasmids (each 4104 bp in size). One nanogram of plasmid DNA should contain 2.22 × 108 copies (one plasmid clone = 4.50 × 10−9 ng). Mutant and wild–type clones were mixed serially in different ratios: 10, 5, 1, 0.5, 0.25 and 0.1% (mutant/mutant + wild–type). Mixed plasmid DNA (1.5 × 10−5 ng equivalent to 3.0 × 103 copies) was added to a 20–μL PCR mixture containing 10 μL 2x ddPCR Supermix for probes (No dUTP; Bio–Rad), 900 nM target–specific PCR primers, 250 nM mutant–specific (FAM) and wild–type–specific (HEX) LNA probes. Twenty microliters of PCR mixture and 70 μL Droplet generation oil for Probes (Bio–Rad) were mixed and droplet generation was carried out using a QX100 Droplet Generator (Bio–Rad) according to the manufacturer’s manual. The droplet emulsion was thermally cycled in the following conditions: denaturing at 95 °C for 10 min, 40 cycles of PCR at 94 °C for 30 s and 57 °C for 2 min, and a final extension at 98 °C for 10 min. PCR amplification in droplets was confirmed using QX200 Droplet Reader (Bio–Rad). Threshold was determined by comparing wild–type and no–template ddPCR results. All data were evaluated above the threshold. All experiments were performed in triplicate.
Evaluation of a GNAQ mutation by ddPCR
The somatic GNAQ mutation (c.548G > A), which was previously evaluated using NGS in 15 patients with SWS7, was re–evaluated by ddPCR. Approximately 20 ng of genomic DNA (which is equivalent to 6.0 × 103 copies of a haploid genome) from paired brain and either blood leukocytes or saliva samples from SWS patients was similarly evaluated by ddPCR (Table 2 and Supplementary Table S2).
Qualitative evaluation using PNA–ddPCR
Wild–type–specific PNA corresponding to GNAQ c.548G was purchased from Fasmac (Atsugi, Kanagawa, Japan). Primer and probe sequences are described in Table 3.
To evaluate the sensitivity of PNA–ddPCR, the detection limit was determined in a similar way to ddPCR. Mutant and wild–type clones were mixed serially in different ratios: 1, 0.5, 0.25, 0.1, 0.05 and 0.025% (mutant/mutant + wild–type; Fig. 1a). Mixed plasmid DNA (1.5 × 10−5 ng, 3.0 × 103 copies) was used as a template in the first PCR mixture. The PCR mixture was prepared in a 10–μL volume containing 1 μL of 10x PCR Buffer (Takara, Otsu, Shiga, Japan), 0.8 μL of 2 mM dNTP Mixture (Takara), 0.05 μL of Takara Ex Taq HS (Takara), 1 μM forward and reverse primers, and 5 μM PNA together with mixed plasmid DNA. PCR conditions were as follows: denaturing at 94 °C for 2 min; 15 cycles at 94 °C for 30 s and 60 °C for 10 s; and extension at 72 °C for 3 min. Then, 1.5 μL PCR product was added to the ddPCR solution containing 10 μL 2x ddPCR Supermix for probes (No dUTP; Bio–Rad), 900 nM target–specific PCR primers and 250 nM mutant–specific (FAM) LNA probes. ddPCR was performed as described above. We also performed PNA–ddPCR using DNA from 30 healthy controls to confirm whether the threshold was appropriate.
The confirmation of detection limit of PNA–ddPCR assay
The GNAQ c.548G > A mutation was confirmed by PNA–ddPCR using genomic DNA of peripheral blood leukocytes from 40 SWS patients. Ten nanograms of genomic DNA (equivalent to 3.0 × 103 copies of a haploid genome) was added to the PCR mixture. PCR conditions were as follows: denaturing at 94 °C for 2 min; 20 cycles at 94 °C for 30 s and 60 °C for 10 s; and extension at 72 °C for 3 min. Each 1.5 μL of PCR product was evaluated by PNA–ddPCR.
Data analysis
The data was analysed using QuantaSoft (version 1.7.4.0917, Bio–Rad). Absolute quantification mode was used for all ddPCR measurements. Absolute quantification was based on the following underlined formula:
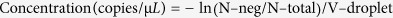 |
[N–neg = the number of total negative droplets in a well; N–total = the number of total droplets in a well; V–droplet = the volume of a droplet (0.85 nl, fixed)].
The threshold for positive amplification was determined based on the results of no template control, mutant and wild–type cloning plasmids and control genomic DNA. Fractional abundance (FA) was calculated as follows:
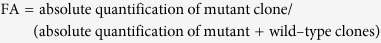 |
As for PNA–ddPCR, the threshold of positive amplification was set to more than 1,000 mutant droplet showing a mutant–specific signal amplitude > 7,700.
Additional Information
How to cite this article: Uchiyama, Y. et al. Ultra-sensitive droplet digital PCR for detecting a low-prevalence somatic GNAQ mutation in Sturge-Weber syndrome. Sci. Rep. 6, 22985; doi: 10.1038/srep22985 (2016).
Supplementary Material
Acknowledgments
We thank all patients and their families for their participation in this study. This work was supported in part by a grant for Research on Measures for Intractable Diseases, a grant for Comprehensive Research on Disability Health and Welfare, the Strategic Research Program for Brain Science, a grant for Initiative on Rare and Undiagnosed Diseases in Pediatrics and a grant for Initiative on Rare and Undiagnosed Diseases for Adults from the Japan Agency for Medical Research and Development; a Grant–in–Aid for Scientific Research on Innovative Areas (Transcription Cycle) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Grants–in–Aid for Scientific Research (A, B and C), and challenging Exploratory Research from the Japan Society for the Promotion of Science; the fund for Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program in the Project for Developing Innovation Systems from the Japan Science and Technology Agency; the Takeda Science Foundation; the Yokohama Foundation for Advancement of Medical Science; and the Hayashi Memorial Foundation for Female Natural Scientists.
Footnotes
Author Contributions Na.M. and K.Y. conceived the study. Na.M., M.N. and Y.U. designed the study. Y.U. performed ddPCR analyses. M.N. performed deep sequencing. S.W., H.M., A.K. and K.Y. conducted the studies. M.M. and A.H. evaluated patients and provided samples. M.T. supported the statistical analysis. Y.U. analyzed the data with the inputs from S.M., No.M. H.S., Y.U., M.N. and Na.M. wrote the manuscript. All authors reviewed the manuscript.
References
- Comi A. M. Update on Sturge-Weber syndrome: diagnosis, treatment, quantitative measures, and controversies. Lymphat. Res. Biol. 5, 257–264 (2007). [DOI] [PubMed] [Google Scholar]
- Dutkiewicz A. S. et al. A prospective study of risk for Sturge-Weber syndrome in children with upper facial port-wine stain. J. Am. Acad. Dermatol. 72, 473–480 (2015). [DOI] [PubMed] [Google Scholar]
- Shirley M. D. et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N. Engl. J. Med. 368, 1971–1979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslin J. S. et al. Sturge-Weber Syndrome (Encephalotrigeminal Angiomatosis): Recent Advances and Future Challenges. Asia Pac J Ophthalmol (Phila) 3, 361–367 (2014). [DOI] [PubMed] [Google Scholar]
- Lian C. G. et al. Novel genetic mutations in a sporadic port-wine stain. JAMA Dermatol 150, 1336–1340 (2014). [DOI] [PubMed] [Google Scholar]
- Huq A. H. et al. Evidence of somatic mosaicism in Sturge-Weber syndrome. Neurology 59, 780–782 (2002). [DOI] [PubMed] [Google Scholar]
- Nakashima M. et al. The somatic GNAQ mutation c.548G > A (p.R183Q) is consistently found in Sturge-Weber syndrome. J. Hum. Genet. 59, 691–693 (2014) [DOI] [PubMed] [Google Scholar]
- Vogelstein B. & Kinzler K. W. Digital PCR. Proc. Natl. Acad. Sci. USA 96, 9236–9241 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro L. B. et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 84, 1003–1011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. Digital PCR hits its stride. Nat Meth 9, 541–544 (2012). [Google Scholar]
- Kaihatsu K. et al. Recognition of chromosomal DNA by PNAs. Chem. Biol. 11, 749–758 (2004). [DOI] [PubMed] [Google Scholar]
- Paulasova P. & Pellestor F. The peptide nucleic acids (PNAs): a new generation of probes for genetic and cytogenetic analyses. Ann. Genet. 47, 349–358 (2004). [DOI] [PubMed] [Google Scholar]
- Stahl T. et al. Digital PCR to assess hematopoietic chimerism after allogeneic stem cell transplantation. Exp. Hematol. 43, 462–468 e461 (2015). [DOI] [PubMed] [Google Scholar]
- Fehling H. J. et al. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development 130, 4217–4227 (2003). [DOI] [PubMed] [Google Scholar]
- Forrai A. & Robb L. The hemangioblast–between blood and vessels. Cell Cycle 2, 86–90 (2003). [PubMed] [Google Scholar]
- Huber T. L. et al. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432, 625–630 (2004). [DOI] [PubMed] [Google Scholar]
- Huber T. L. Dissecting hematopoietic differentiation using the embryonic stem cell differentiation model. Int. J. Dev. Biol. 54, 991–1002 (2010). [DOI] [PubMed] [Google Scholar]
- Lam E. Y. et al. Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood 116, 909–914 (2010). [DOI] [PubMed] [Google Scholar]
- Wang Q. et al. Droplet Digital PCR for Absolute Quantification of EML4-ALK Gene Rearrangement in Lung Adenocarcinoma. J Mol Diagn 17, 515–520 (2015). [DOI] [PubMed] [Google Scholar]
- Mangolini A. et al. Diagnostic and prognostic microRNAs in the serum of breast cancer patients measured by droplet digital PCR. Biomark Res 3, 12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.