Abstract
Fructose is a major component of dietary sugar and its overconsumption exacerbates key pathological features of metabolic syndrome. The central fructose-metabolising enzyme is ketohexokinase (KHK), which exists in two isoforms: KHK-A and KHK-C, generated through mutually exclusive alternative splicing of KHK pre-mRNAs. KHK-C displays superior affinity for fructose compared with KHK-A and is produced primarily in the liver, thus restricting fructose metabolism almost exclusively to this organ. Here we show that myocardial hypoxia actuates fructose metabolism in human and mouse models of pathological cardiac hypertrophy through hypoxia-inducible factor 1α (HIF1α) activation of SF3B1 and SF3B1-mediated splice switching of KHK-A to KHK-C. Heart-specific depletion of SF3B1 orgenetic ablation of Khk, but not Khk-A alone, in mice, suppresses pathological stress-induced fructose metabolism, growth and contractile dysfunction, thus defining signalling components and molecular underpinnings of a fructose metabolism regulatory system crucial for pathological growth.
Heart failure is a leading cause of death worldwide. It is often preceded by hypertrophic growth of the organ as it strains to maintain function under conditions of pathologic stress1 and is associated with an increased demand for oxygen, owing to changes in coronary resistance, cardiac vascularisation and perfusion2. The ensuing state of hypoxia activates hypoxia-inducible factor (HIF)1α that mediates adaptive transcriptional responses to low-oxygen tension3 to support cardiomyocyte growth, myocardial angiogenesis and reprogramming of metabolism4. Unremitted pathologic stress leads ultimately to functional deterioration of the heart with the clinical signs and symptoms of heart failure5.
Given that both HIF1α transcription and alternative pre-mRNA splicing are central to the control of metabolism, we embarked on a systematic and functional analysis of HIF1α-dependent splice factor expression and pre-mRNA splicing in pathologic cardiac hypertrophy. We identified splice factors as HIF1α targets, which prompted the analysis of their RNA targets, unveiling mechanistic and functional linkages between HIF1α, splice factor 3b subunit 1 (SF3B1), KHK-C splice isoform production and fructose metabolism in cardiac hypertrophy.
The HIF1α–SF3B1–KHK-C axis
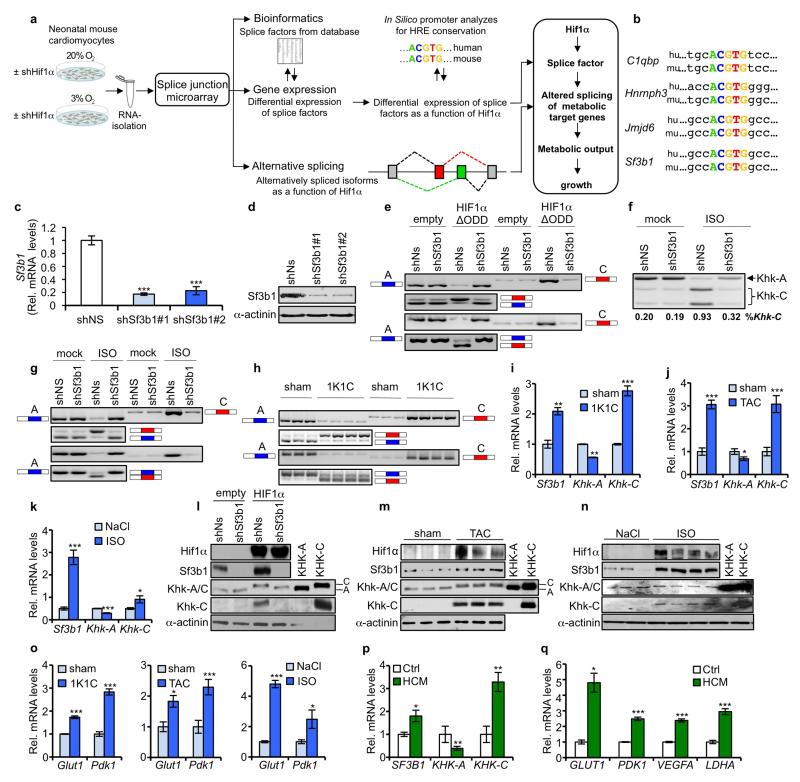
To deduce changes in both gene expression and alternative splicing as a function of Hif1α, we followed the experimental workflow illustrated in Extended Data Fig. 1a. RNA isolated from normoxic and hypoxic neonatal mouse cardiomyocytes (NMCs), depleted (or not) of Hif1α by specific short hairpin RNAs (shHif1α), was subjected to a splice-junction microarray analysis. Genes exhibiting a Hif1α-dependent expression profile were manually checked for an involvement in pre-mRNA splicing and, in parallel, matched to a list of 329 unique genes comprised of validated and in silico-predicted splice factors6–8 (Supplementary Fig. 1 and Supplementary Table 1). Twenty genes with characteristics of splice factors/regulators displayed a Hif1α-dependent expression pattern (Fig. 1a), of which four, C1qbp, Hnrnph3, Jmjd6 and Sf3b1, contained conserved hypoxia-responsive elements (HREs) in their promoters (Extended Data Fig. 1b).
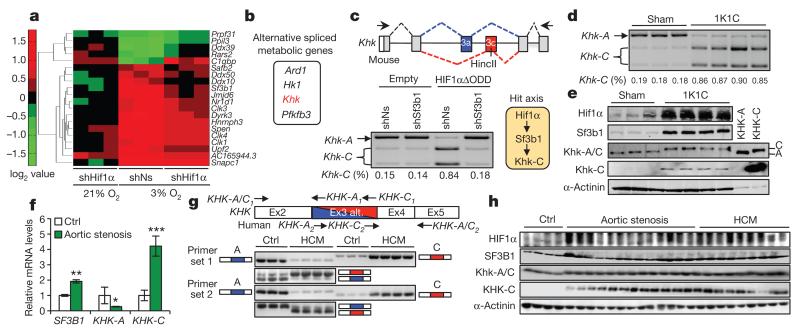
Figure 1. Identification of Hif1α-dependent splicing factors and splicing events.
a, Expression of splice factors in NMCs transduced and cultured as indicated versus NMCs treated with non-silencing shRNA (shNs) and cultured at 21% O2. b, Hit list of metabolic genes. c, d, NMCs transduced as indicated (c) and ventricular samples from sham- and 1K1C-treated mice (d) were analysed for Khk-A and Khk-C expression by RT–PCR and HincII digestion. Primers to amplify the KHK variable region are indicated (c, upper). The hypothetical HIF1α–SF3B1–KHK-C axis (c, right). e, Ventricular samples from sham- and 1K1C-treated mice and lysates of NMCs overexpressing KHK-A or KHK-C were processed for immunoblotting with antibodies against denoted proteins. A and C indicate the differential migration of KHK-C and KHK-A on SDS gels. f, Biopsies of left ventricles from aortic stenosis patients and healthy controls were assessed for SF3B1, KHK-A and KHK-C expression by qPCR (n = 16 for patient samples and n = 6 for controls; error bars are s.e.m.; **P < 0.01; ***P < 0.001; two-tailed unpaired t-test). g, Top, schematic representation of exon 2 to 5 of human KHK. Alternative usage of exon 3 (Ex3 alt.) is displayed in blue (exon 3A) and red (exon 3C). Primer sets location and orientation used for RT–PCR analysis, are indicated with arrows. Bottom, assessment of KHK-A and KHK-C expression by RT–PCR in ventricles of controls and HCM patients using primer set 1 and 2 as described above. h, Lysates of left-ventricular samples from healthy controls and aortic stenosis or HCM patients were processed for immunoblotting with antibodies against denoted proteins.
Data derived from the splice-junction microarray analysis were also interrogated for changes in gene splicing as a function of Hif1α and further filtered for their potential association to metabolism, leading to Ard1 (also known as Naa10), Hk1, Khk, and Pfkfb3, which exhibited the most consistent and significant changes (Fig. 1b). The Hif1α dependency of Khk alternative splicing implied a link between hypoxia and fructose metabolism, especially since the dominant Khk isoform induced by hypoxia, Khk-C, has a superior activity over Khk-A9 (Supplementary Fig. 2).
Analysis of Khk alternative splicing in NMCs by real-time PCR (RT–PCR) revealed Hif1α-mediated, Sf3b1-dependent induction of Khk-C mRNAs, while mRNA levels of Khk-A declined (Fig. 1c and Extended Data Fig. 1c–e). Isoproterenol treatment yielded similar results (Extended Data Fig. 1f, g). Also, in mouse models of cardiac hypertrophy, specifically one-kidney one-clip (1K1C), trans-aortic constriction (TAC) and isoproterenol infusion, Khk-C mRNA and protein production increased over sham-treated control mice (Fig. 1d, e and Extended Data Fig. 1h–n). Hif1α target and hypertrophic marker gene expression were dramatically increased in all three cardiomyopathy models (Extended Data Fig. 1o and Supplementary Fig. 3a–c).
Analysis of HIF1α, SF3B1 and KHK-C in ventricular biopsies of aortic stenosis or hypertrophic cardiomyopathy (HCM) patients corroborated the results obtained in mice both at the level of mRNA and protein expression (Fig. 1f–h and Extended Data Fig. 1p). mRNAs of HIF1α targets and hypertrophic markers were also induced (Extended Data Fig. 1q and Supplementary Fig. 3d). Thus, HIF1α, SF3B1 and KHK-C are coordinately upregulated in different mouse models of cardiac hypertrophy and biopsies of human cardiomyopathy patients.
The HIF1α target SF3B1 mediates splicing of KHK
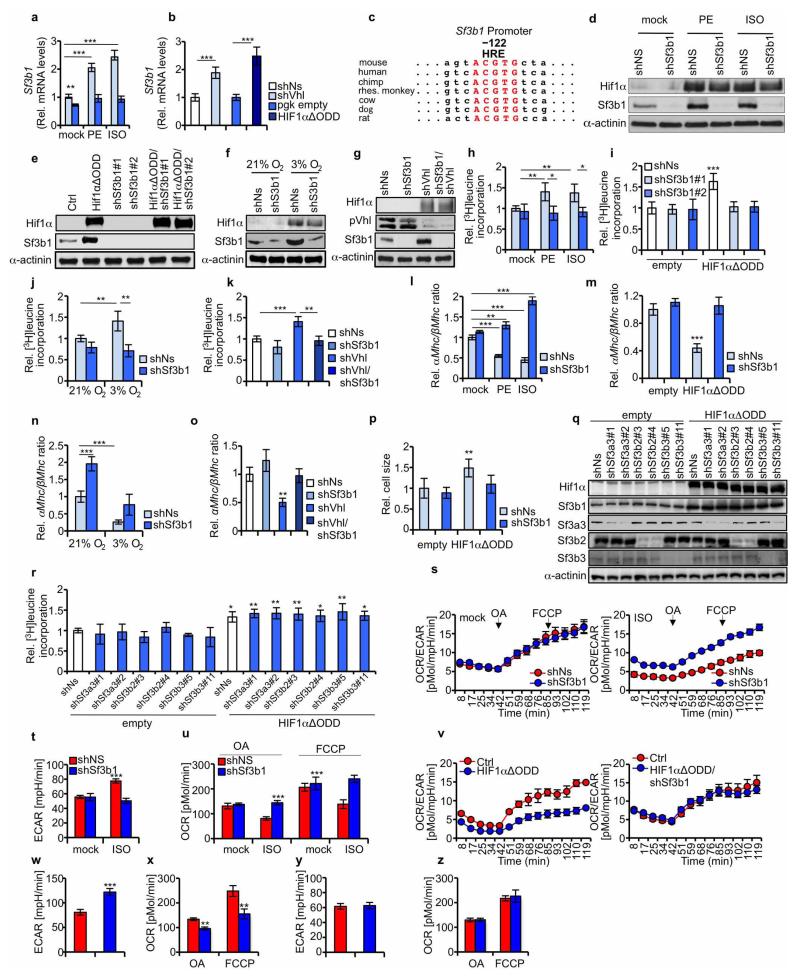
Sf3b1 appears to represent a direct Hif1α target. First, treatment of NMCs with phenylephrine or isoproterenol, known inducers of Hif1α and Hif1α-target genes10, increased Sf3b1 mRNA expression in a Hif1α-dependent manner (Extended Data Fig. 2a). Second, Sf3b1 mRNA expression also increased upon depletion of the Von Hippel–Lindau protein (pVhl) or ectopic expression of HIFlα lacking the oxygen-dependent degradation domain (HIF1αΔODD) (Extended Data Fig. 2b). Third, the promoter of SF3B1 contains a highly conserved HRE (Extended Data Fig. 2c). Fourth, expression of HIF1αΔODD activated a mouse wild-type Sf3b1-promoter-luciferase reporter but not the corresponding reporter harbouring a mutant HRE (Fig. 2a). Fifth, chromatin immunoprecipitation (ChIP) assays demonstrate an association of Hif1α with the HRE in the Sf3b1 promoter in native chromatin (Fig. 2b).
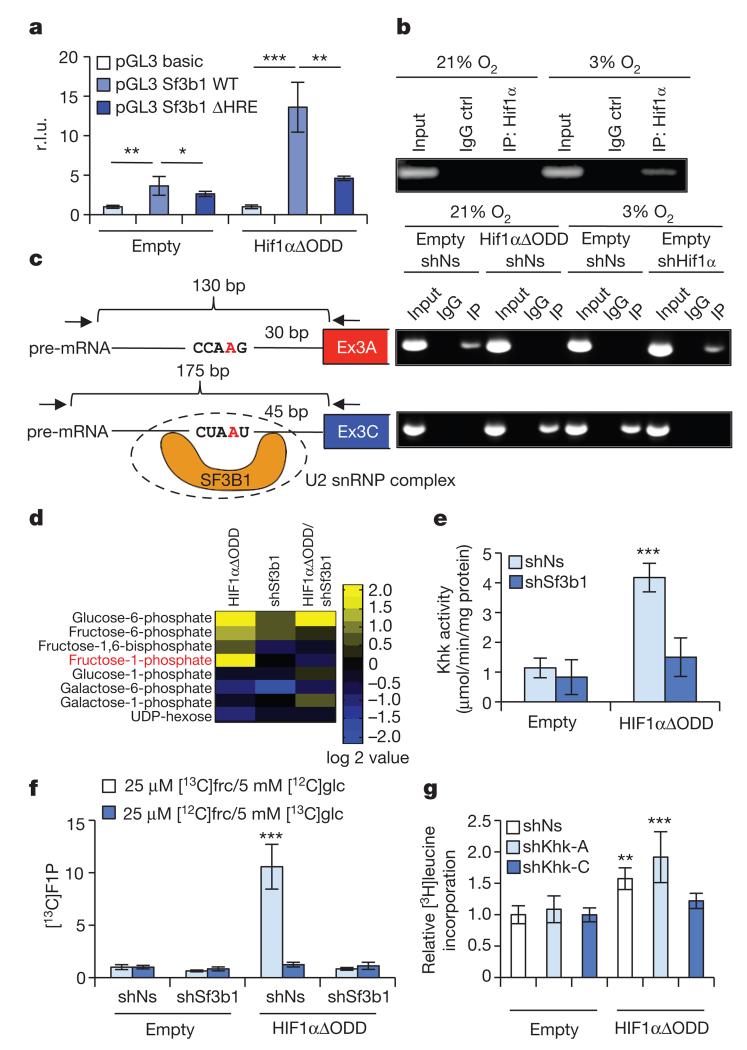
Figure 2. SF3B1 is a HIF1α-target gene that mediates splicing of KHK.
a, Co-transfection of wild type (WT) or HRE-mutated SF3B1 promoters fused to luciferase with either an empty vector control or HIF1αΔODD (values are relative light units (r.l.u.); n = 4 biological replicates per group). b, NMCs were cultured as indicated and processed for chromatin immunoprecipitation with a HIF1α-specific antibody (IP: HIF1α) or with a control isotype-matched antibody (IgG control). c, Schematic representing branch-point sequences located in front of exon 3A (upper left) or exon 3C (lower left). Branch point adenine is labelled in red. Location and orientation of primers (arrows), expected length of fragments to be amplified by RT–PCR and potential Sf3b1 binding sites are depicted. NMCs treated as indicated were processed for RNA-immunoprecipitation assay with antibodies specific for Sf3b1 (IP) or control-isotype matched antibody (IgG). Recovered RNA was purified and analysed by RT–PCR (upper right and lower right). d, Mass spectrometry-based metabolomic profiling of NMCs transduced with lentiviruses as denoted focused on hexose-phosphates. Metabolite values are compared to amounts in control transduced NMCs (n = 4 biological replicates per group). e, KHK activity in lysates of NMCs transduced with lentiviruses as indicated (n = 7 biological replicates per group). f, Relative amount of [13C]fructose-1-phosphate derived from [13C]fructose ([13C]frc) or [13C]glucose ([13C]glc) in NMCs transduced as denoted (n = 4 biological replicates per group). g, [3H]leucine incorporation in NMCs infected as indicated (n = 3 biological replicates per group, data show 1 of 3 representative experiments). For panels a, e–g, *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed unpaired t-test (a) one-way ANOVA followed by Dunnett’s multiple comparison post-test (e–g). Error bars are s.d. (a, g) or s.e.m. (e, f).
Next we measured the incorporation of radioactively labelled [3H]leucine in NMCs depleted of Sf3b1 and exposed to various treatments including phenylephrine or isoproterenol (Extended Data Fig. 2d), expression of HIF1αΔODD (Extended Data Fig. 2e), hypoxia (Extended Data Fig. 2f) or simultaneous depletion of pVhl (Extended Data Fig. 2g). We observed that [3H]leucine incorporation (Extended Data Fig. 2i–k), myosin heavy chain α/β mRNA ratio (Extended Data Fig. 2l–o) and cell size (Extended Data Fig. 2p) changed as a function of Sf3b1. Other components of the U2 small nuclear ribonucleoprotein (U2 snRNP) complex, namely Sf3b2, Sf3b3 and Sf3a3, neither increased in abundance in response to Hif1α expression nor did they affect, when downregulated by different shRNAs, to a measurable extent, Hif1α-stimulated cell growth (Extended Data Fig. 2q, r and Supplementary Fig. 4).
Also, known parameters of Hif1α-driven metabolism including extracellular acidification rate (ECAR; an indicator lactate production) and oxygen consumption rate (OCR; an indicator of oxidative phosphorylation) changed in a Sf3b1-dependent manner in NMCs. At basal levels, ECAR was not affected by depletion of Sf3b1 but increased upon addition of isoproterenol (Extended Data Fig. 2s, t). OCR was repressed, as expected, in isoproterenol-treated cells upon addition of either oleic acid or carbonyl cyanide-4-(trifluoromethoxy)-phenylhydrazone, an effect diminished by depletion of Sf3b1 (Extended Data Fig. 2u). Ectopic expression of HIF1αΔODD produced similar effects on ECAR and OCR as isoproterenol (Extended Data Fig. 2v–z), while depletion of Sf3b1 alone had no effect (Supplementary Fig. 5). In sum, these results suggest a key contributory role for Sf3b1 in Hif1α-mediated growth and metabolic programs.
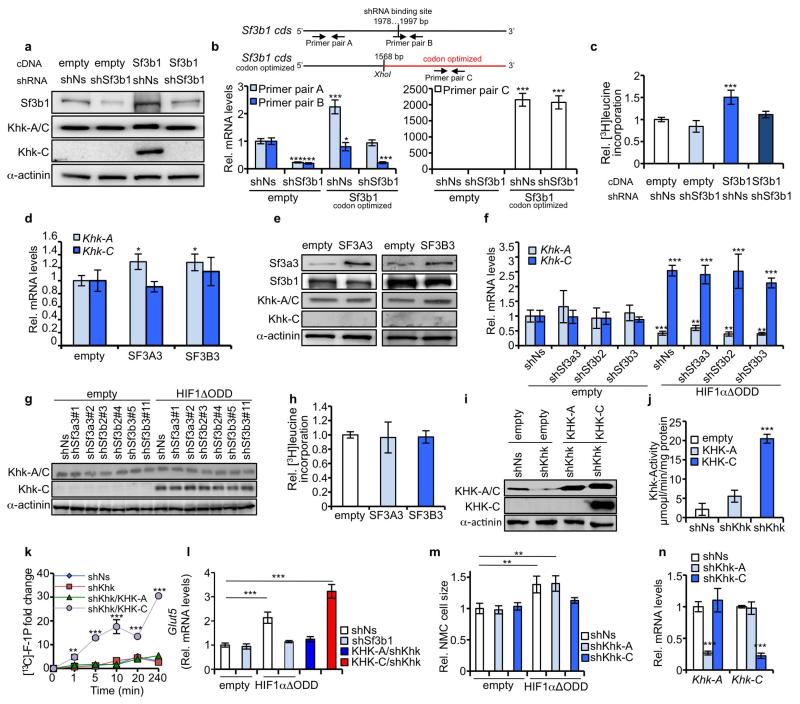
Sf3b1 participates in the catalysis of splicing11, can be crosslinked to the 5′ and 3′ of the branch point adenosine12, and displays differential effects on specific splice targets under conditions of reduced function13,14. Indeed, splice-junction analysis revealed widespread effects on both alternative splicing and gene expression that included changes in Khk isoform expression when Sf3b1 was depleted in hypoxic NMCs (Supplementary Tables 2 and 3). RNA-immunoprecipiations of Sf3b1 from extracts of NMCs exposed to hypoxia or HIF1αΔODD overexpression, demonstrate that Sf3b1 occupies RNA sequences upstream of exon 3C (and not of exon 3A) of Khk pre-mRNA in hypoxia but not normoxia (Fig. 2c). Upon depletion of Hif1α, Sf3b1 was redirected towards branch-point sequences of exon 3A (Fig. 2c). This suggests that Sf3b1 association with RNA sequences upstream of exon 3C of Khk requires Hif1α-signalling.
Sf3b1 acts as a direct mediator of Khk splicing as its overexpression caused a shift from Khk-A to Khk-C (Extended Data Fig. 3a). Selective depletion of endogenously (but not exogenously) produced Sf3b1 prevented this shift (Extended Data Fig. 3a). Quantitative PCR (qPCR) analysis confirmed the overexpression and/or reduction of Sf3b1 mRNAs where expected (Extended Data Fig. 3b). Concomitant with the shift in Khk isoform expression towards Khk-C, cell growth was induced (Extended Data Fig. 3c). Neither the overproduction of SF3A3 or SF3B3 (Extended Data Fig. 3d, e), nor the depletion of Sf3a3, Sf3b3 or Sf3b2 by corresponding shRNAs, generated a similar robust splice shift towards Khk-C despite efficient depletion of the respective proteins (Extended Data Fig. 3f, g and Supplementary Fig. 4). Cell growth was also not affected by SF3A3 or SF3B3 expression (Extended Data Fig. 3h).
Next we proceeded with mass-spectrometry-based metabolomic profiling of hexose-phosphates in NMCs expressing HIF1αΔODD in the absence or presence of shSf3b1. We observed profoundly increased levels of fructose-1-phosphate (F1P) and glucose-6-phosphate in response to HIF1α activation (Fig. 2d), but only the level of the former was Sf3b1-dependent (Fig. 2d). In accord with increased levels of F1P, HIF1αΔODD also increased total cellular Khk activity by 3.6-fold compared to control NMCs (Fig. 2e). In a comparative analysis, overexpressed human KHK-C displayed, in NMCs depleted for endogenous Khk, a 3.7-fold higher kinase activity than KHK-A (Extended Data Fig. 3i, j). Thus, activation of the Hif1α-Sf3b1 axis activates Khk-C to stimulate F1P production.
To assess whether increased levels of F1P are derived from external fructose or from conversion of glucose via the polyol pathway15, we supplied NMC culture media with [13C]fructose and unlabelled glucose or [13C]glucose and unlabelled fructose and measured [13C]F1P production. [13C]F1P levels increased upon HIF1αΔODD expression selectively in NMCs cultured with [13C]fructose and unlabelled glucose but not [13C]glucose and unlabelled fructose in a manner dependent on Sf3b1 (Fig. 2f). Moreover, KHK-C, but not KHK-A, expression induced accumulation of [13C]F1P levels in a time-dependent fashion in NMCs cultured with [13C]fructose (Extended Data Fig. 3k). Glut5 fructose transporter mRNA expression was likewise induced by KHK-C and HIF1αΔODD (Extended Data Fig. 3l).
Khk-C is also a critical downstream mediator of Hif1α-induced cell growth as selective depletion of Khk-C, but not Khk-A, rescued HIF1αΔODD-induced cardiomyocyte growth (Fig. 2g and Extended Data Fig. 3m). Knockdown efficiency and isoform specificity of the corresponding shRNAs targeting Khk isoforms was confirmed by qPCR (Extended Data Fig. 3n). Taken together, Sf3b1 appears to be a direct mediator of Khk pre-mRNA splicing, resulting in Khk-C expression, increased Khk activity, fructose uptake and cell growth.
Expression of KHK-C promotes anabolic metabolism
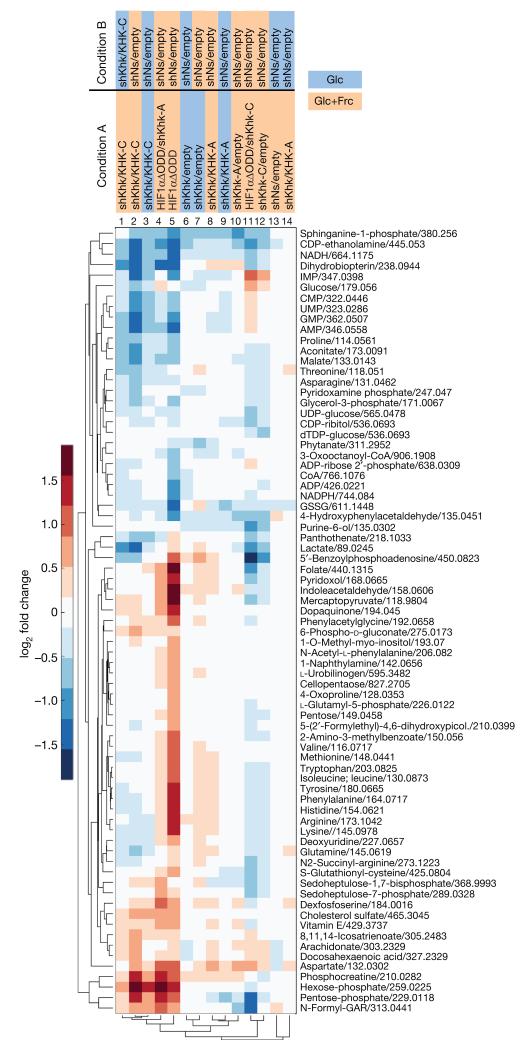
To characterize metabolic phenotypes in NMCs associated with activation of the HIF1α–SF3B1–KHK-C axis and as a function of fructose availability, we performed metabolomic profiling. These efforts established the following experimental points. First, in the presence of fructose and glucose, HIF1αΔODD expression increased glycolysis (for example, hexose-phosphates), amino acid metabolism and biosynthesis (for example, dexfosfoserine, aspartate, arginine), the non-oxidative pentose-phosphate pathway (PPP; for example, sedoheptulose-7-phosphate, pentose phosphates) and to a lesser degree also the oxidative PPP (for example, 6-phospho-d-gluconate, pentose phosphates) and intermediates of DNA/RNA biosynthesis (for example, N-formyl-GAR, folate, deoxyuridine) (Fig. 3). Changes in redox balance are exemplified by lower levels of NADPH, NADH and of the reduced state of glutathione (GSSG) (Fig. 3, lanes 4 and 5). Reduced levels of these metabolites are probably the result of extensive utilization of reducing equivalents for macromolecular biosynthesis16. Second, depletion of Khk-C (but not Khk-A) reverted HIF1αΔODD-induced metabolic changes (Fig. 3, compare lane 11 with 5 and 4), while its overexpression mimicked them (Fig. 3, compare lane 1–3 and 4–5). Third, when KHK-C was overexpressed under fructose-lacking conditions, levels of glycolytic and PPP metabolites decreased and the reduction of NADPH and NADH was less pronounced (Fig. 3, compare lane 2 and 3; Extended Data Fig. 4a). Hence, exogenous fructose contributes significantly to the metabolic effects mediated by Khk-C.
Figure 3. Anabolic metabolism and growth as a function of KHK-C.
Untargeted analysis of metabolites in NMCs treated as denoted. Depicted are metabolites with log2(fold change) > 0.5 and adjusted P value < 0.01 in at least one treatment group compared to corresponding control (n = 4 biological replicates per group).
In accord with the above, we detected [3H] radiolabel derived from [3H]fructose in the RNA, DNA and protein fractions of NMCs in a HIF1αΔODD- and KHK-C-dependent manner (Extended Data Fig. 4b). Expression of KHK-C stimulated also glucose uptake in the presence of fructose (Extended Data Fig. 4c). Moreover, [3H]leucine incorporation into NMCs increased as a function of fructose availability in a KHK-C-dependent fashion (Extended Data Fig. 4d). Increasing glucose availability in the presence of fructose had similar effects (Extended Data Fig. 4d). Immunoblotting confirmed the expected expression pattern of KHK-A and KHK-C (Supplementary Fig. 6a). These results suggest a critical role for KHK-C in HIF1α-dependent metabolic reprogramming and growth.
Since phosphorylation of fructose by Khk-C is unrestrained, it could contribute to ATP-depletion as shown previously in liver17, thereby limiting ATP-driven allosteric inactivation of glycolysis at the level of phosphofructokinase (PFK)18,19. When HIF1αΔODD was overexpressed in NMCs, the ADP/ATP ratios increased (Extended Data Fig. 4e). Simultaneous depletion of either Sf3b1 or Khk-C (but not Khk-A) prevented this (Extended Data Fig. 4e). Khk-C but not Khk-A, when overexpressed, enhanced ADP/ATP ratios (Extended Data Fig. 4e). Thus, activation of the HIF1α–SF3B1–KHK-C axis increases ADP/ATP ratios.
Hypoxia or activation of HIF1α drives an accumulation of lipids in heart hypertrophy10,20. Since the F1P product DHAP is important for formation of the glycerol backbone in triglyceride synthesis, we tested the Khk-C-dependency of this phenomenon by Oil Red O staining of lipid droplets in oleic-acid-treated NMCs. Overexpression of HIF1αΔODD or exposure to hypoxia induced lipid droplet formation, which was prevented upon depletion of Khk-C but not Khk-A (Extended Data Fig. 4f–h). Similar results were obtained when lipid levels were quantified with Nile Red (Supplementary Fig. 6b, c). Khk-C, but not Khk-A, was also required for phenylephrine-induced lipid accumulation (Supplementary Fig. 6d). Fructose carbon contributes to triglyceride synthesis in NMCs as measured by the amount of fructose-derived [3H] in the lipid fraction of NMCs expressing HIF1αΔODD (Supplementary Fig. 6e). Downregulation of Khk-A had no effect, but depletion of either Khk-C, Glut5 or Sf3b1 prevented HIF1α-driven conversion of fructose carbon into lipids (Supplementary Fig. 6e, f). In a gain-of-function experiment, over-expression of KHK-C promoted fructose carbon to lipid conversion while KHK-A expression did not (Supplementary Fig. 6e).
We also investigated the contribution of the two different Khk-isoforms to Hif1α-driven changes in OCR and ECAR in NMCs (Extended Data Fig. 4i–l). Khk-A depletion affected neither the ECAR nor the OCR of HIF1αΔODD-expressing cells (Extended Data Fig. 4i), while depletion of Khk-C did (Extended Data Fig. 4j). In the absence of HIF1αΔODD, neither Khk-A nor Khk-C depletion alone demonstrated any measurable effects on ECAR or OCR (Supplementary Fig. 6g, h). In a converse experiment, overexpression of KHK-A alone had little, if any, effect on these parameters while KHK-C overexpression increased ECAR and decreased OCR (Extended Data Fig. 4k, l). Thus, by stimulating fructose metabolism in cardiomyocytes, Khk-C expression promotes conversion of fructose carbon to lipids, suppresses mitochondrial oxidative phosphorylation and increases glycolysis.
Cardiac growth and dysfunction depends on Sf3b1
Next we assessed whether myocardial fructose and glucose uptake is induced in an in vivo model of cardiac hypertrophy (Extended Data Fig. 5a). When TAC-treated and sham-operated control mice were simultaneously exposed to [3H]fructose and [14C]deoxyglucose, they accumulated to higher levels in diseased hearts (Extended Data Fig. 5b, c). Hearts of mice after 1K1C surgery (Extended Data Fig. 5d), demonstrated a 2.5-fold increase of fructose levels (Extended Data Fig. 5e). [14C]fructose or [14C](deoxy)glucose uptake was also stimulated under conditions of hypertrophy in ex vivo ventricular tissue (Extended Data Fig. 5f, g). In line with this, Glut5 expression was induced in these hypertrophy models (Extended Data Fig. 5h, i) in hearts of isoproterenol-treated mice and in left-ventricular biopsies of aortic stenosis and HCM-patients (Extended Data Fig. 5j–l). In contrast, expression of aldose reductase (AKR1B1), which converts glucose to sorbitol in the polyol pathway, was not induced in the latter samples (Extended Data Fig. 5k, l). Thus, cardiac hypertrophy is associated with increased fructose levels and uptake.
To study the Sf3b1-dependency of these phenomena in response to pathologic stress, we designed a viral-based vector system enabling ventricular-specific knockdown of Sf3b1 that is based on an AAV9 backbone containing a U6 promoter followed by a TATA-lox site, a CMV promoter driving eGFP, another TATA-lox site and shRNAs targeting Sf3b1 (Fig. 4a). We constructed two viruses each expressing different shRNAs targeting Sf3b1 (AAV9-fl/fl-shSf3b1#1 and AAV9-fl/fl-shSf3b1#2). Validation of these recombinant viruses was performed both in vitro in NMCs and in vivo in Mlc2v-cre+ and Mlc2v-cre− mice (Extended Data Fig. 5m, n and Supplementary Fig. 7a–d).
Figure 4. SF3B1 is required for pathologic cardiac growth and function.
a, Schematic representation of the AAV9-fl/fl-shSf3b1 virus before and after Cre-mediated recombination (left panel) and of the experimental timeline (right panel). b–d, Left-ventricular weight normalized to tibia length (LV/TL) (b), isovolumic relaxation time (IVRT) (c) and ventricular fructose levels (d) of sham- or 1K1C-treated Mlc2v-cre− and Mlc2v-cre+ mice injected with AAV9-fl/fl-shSf3b1#1 viruses. e, Immunoblots of heart lysates from Mlc2v-cre− and Mlc2v-cre+ mice operated and transduced as in b–d using indicated antibodies. f, g, LV/TL (f) and ejection fraction (EF) (g) of sham- or TAC-treated Mlc2v-cre− and Mlc2v-cre+ mice injected as indicated. h, Lysates of hearts from mice transduced and operated as in f were processed for immunoblotting with antibodies against denoted proteins. For a–d, f, g, number of mice per group is given in Extended Data Table 1. Error bars are s.e.m. *P < 0.05; **P < 0.01; two-tailed unpaired t-test (b–d, f, g).
While injection of AAV9-fl/fl-shSf3b1#1 into Mlc2v-cre+ or Mlc2v-cre− mice failed to cause any effects on ventricular size in sham-operated mice, AAV9-fl/fl-shSf3b1#1 injection protected Mlc2v-cre+ but not Mlc2v-cre− littermates from 1K1C-induced cardiac hypertrophy (Fig. 4b) and diastolic dysfunction (Fig. 4c and Extended Data Table 1). Like hypertrophic growth and function, the increase in fructose levels was also dependent on Sf3b1 (Fig. 4d). Immunoblotting of lysates from these sham- or 1K1C-operated Mlc2v-cre− and Mlc2v-cre+ mice confirmed that Sf3b1 was depleted where expected, preventing the generation of Khk-C isoform in ventricles of 1K1C-treated Mlc2v-cre+ mice (Fig. 4e).
We also analysed the role of Sf3b1 in the TAC model. Application of both AAV9-fl/fl-shSf3b1#1 and AAV9-fl/fl-shSf3b1#2 viruses protected Mlc2v-cre+ but not Mlc2v-cre− littermates from cardiac hypertrophy (Fig. 4f; Extended Data Fig. 5o and Extended Data Table 1) and systolic dysfunction (Fig. 4g; Extended Data Fig. 5p and Extended Data Table 1). Immunoblotting confirmed that Sf3b1 was downregulated and prevented the switch to the Khk-C isoform in response to TAC-induced pathologic stress in hearts of Mlc2v-cre+ mice (Fig. 4h and Extended Data Fig. 5q). These results suggest that Sf3b1 is required for the development of pathologic cardiac hypertrophy.
Deletion of Khk protects from cardiac growth
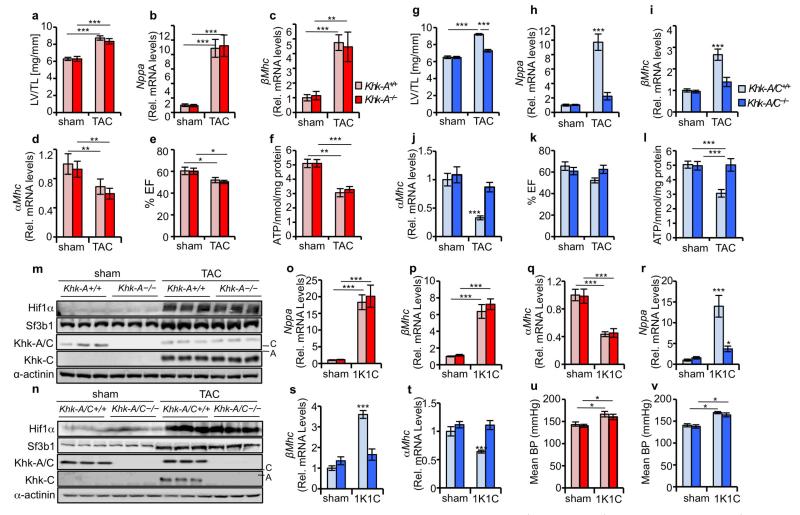
To assess the importance of Khk-C in cardiac hypertrophy and function in the context of pathologic stress, we investigated two genetic mouse models, which were subjected to TAC and 1K1C stresses. Khk-A−/− mice only express Khk-C, whereas Khk-A/C−/− mice are deleted for both isoforms21. Exposure of wild-type and Khk-A−/− mice to TAC treatment induced cardiac hypertrophy and systolic dysfunction in both mouse strains (Extended Data Fig. 6a–e and Extended Data Table 2). ATP measurements in left ventricles of sham- or TAC-treated wild-type and Khk-A−/− mice revealed a reduction of ATP levels in hearts after TAC compared to control mice (Extended Data Fig. 6f).
Hearts of Khk-A/C−/− mice, when exposed to TAC, did not display signs of hypertrophy or systolic dysfunction (Extended Data Fig. 6g–k and Extended Data Table 2). Moreover, ATP levels remained unchanged in Khk-A/C−/− mice exposed to TAC (Extended Data Fig. 6l). All TAC-treated hearts accumulated Hif1α and Sf3b1 in the left ventricle (Extended Data Fig. 6m, n). Khk-C expression was only detectable in TAC-treated hypertrophied ventricles, but not in the corresponding hearts from Khk-A/C−/− mice (Extended Data Fig. 6m, n).
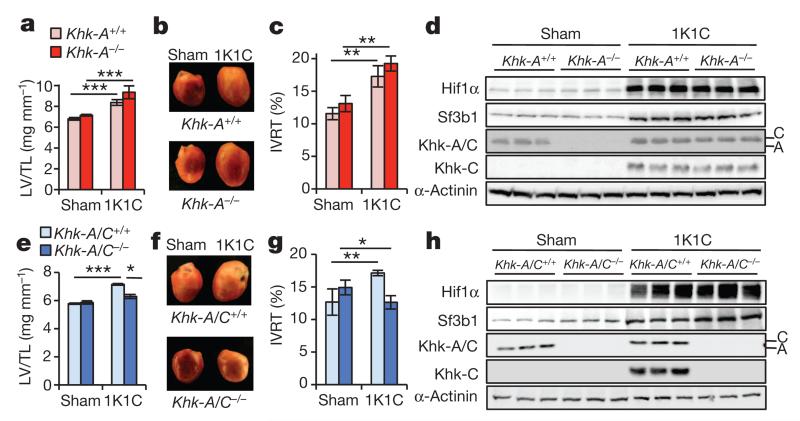
1K1C treatment of Khk-A−/− and Khk-A+/+ mice caused significant left-ventricular growth (Fig. 5a, b; Extended Data Fig. 6o–q and Extended Data Table 3) and diastolic dysfunction (Fig. 5c and Extended Data Table 3). Hif1α and Sf3b1 levels were induced in left ventricles of both 1K1C-treated strains, whereas Khk-A expression in sham-treated control mice was depleted as expected (Fig. 5d). Additional deletion of Khk-C protected hearts of these pathologies (Fig. 5e–g; Extended Data Fig. 6r–t and Extended Data Table 3). Immunoblotting confirmed activation of Hif1α and Sf3b1 upon pathologic stress, and that Khk-C was only expressed in wild-type and Khk-A−/− but not in Khk-A/C−/− ventricles (Fig. 5h). Arterial blood pressure was similar in these mouse strains following 1K1C treatment (Extended Data Fig. 6u, v). In sum, only deletion of Khk-A in combination with Khk-C, but not Khk-A alone, prevents the development of pathologic growth and cardiac dysfunction upon different stresses.
Figure 5. Khk-A/C−/− mice are protected from pathologic cardiac growth and contractile dysfunction.
a–c, Left-ventricular weight normalized to tibia length (LV/TL) (a), representative images of left ventricles (b) and isovolumic relaxation time (IVRT) (c) of sham- or 1K1C-operated Khk-A+/+ and Khk-A−/− mice. d, Lysates from ventricles of sham- or 1K1C-operated Khk-A+/+ and Khk-A−/− mice were processed for immunoblotting with antibodies as indicated. e–g, Left-ventricular weight normalized to tibia length (e), representative images of left ventricles (f) and isovolumic relaxation time (g) of sham- or 1K1C-operated Khk-A/C+/+ and Khk-A/C−/− mice. h, Lysates from ventricles of sham- or 1K1C-operated Khk-A/C+/+ and Khk-A/C−/− mice processed for immunoblotting with antibodies as indicated. For a, c, e, g, *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed unpaired t-test; number of mice per group is given in Extended Data Table 2. Error bars are s.e.m.
Discussion
Our results demonstrate that fructose metabolism can be provoked in the heart by pathologic stimuli through HIF1α activation of SF3B1-mediated splicing of KHK pre-mRNA (Extended Data Fig. 7). The potential general importance of these findings draw on the central role occupied by HIF1α in human disease, highlighting the HIF1α–SF3B1–KHK-C axis as a promising therapeutic target.
Splicing output of the HIF1α target SF3B1, which contributes to intron removal by anchoring pre-mRNA onto the spliceosome12, is sensitive to changes in the abundance of SF3B113,22. This feature of SF3B1 to expand and/or change splice target spectrum as a function of abundance may be exploited by HIF1α to affect KHK-C production. That the latter is less sensitive to changes in the levels of other factors of the U2snRNP complex investigated here implies that SF3B1 is either limiting for U2snRNP assembly/function or occupies a specific role in this context. Irrespective, this process is likely influenced by additional positive- and/or negative-acting factors, as has been demonstrated for PKM223.
The criticality of KHK-C function in pathologic cardiac growth may manifest at multiple levels. Unrestrained KHK-C-driven phosphorylation of fructose may limit ATP levels and thus restrict ATP-mediated negative feedback inhibition of PFK, facilitating high glycolytic flux24. The latter may be further enhanced by an increased availability of F1P, as this metabolite has been shown to induce glucokinase and pyruvate kinase, at least in the liver25. Apart from positively affecting glucose metabolism, KHK-C and fructose availability also increased nucleic acid, lipid and protein biosynthesis, which may support growth. Although our work points to a fructose-metabolism-related function of KHK-C in cardiac hypertrophy, the source of fructose could, under certain circumstances, vary and originate alternatively from glucose15. In addition, yet-to-be-discovered functions of KHK-C unrelated to fructose metabolism (that may, for example, relate to gene regulatory activities) could contribute to cell growth control26.
Biopsies of heart disease patients demonstrate increased expression of HIF1α, SF3B1 and KHK-C. Since aortic stenosis and diabetes are frequent comorbidities in senescent humans27 and diabetic patients tend to have also higher fructose levels in serum28, the impact of the HIF1α-SF3B1-KHK-C axis in these patients might be particularly important. With respect to other disease states, SF3B1 is found recurrently mutated in human cancers12. It is conceivable that some of these cancer-associated mutations affect alternative splicing of KHK and fructose metabolism in cancer cells independent of hypoxia, thereby contributing to tumour development.
METHODS
Human ventricular biopsies
Left-ventricular tissues were obtained from patients with hypertrophic cardiomyopathy and aortic stenosis (Supplementary Table 4) and healthy controls. Additional left-ventricular biopsies were provided by W. Linke (University of Münster). Clinical and demographical data from the corresponding patients were previously published29. All procedures were in compliance with local ethical committees. An informed consent was obtained from all patients included in the study.
Mouse strains
Khk-A−/− and Khk-A/C−/− mice were obtained from A. Asipu (University of Leeds) and R. Johnson (University of Colorado). MLC2v-cre line was from J. Chen (University of California). All mice were maintained in a conventional animal facility at RCHCI and EPIC, ETH Zürich. Maintenance and animal experimentation were in accordance with the Swiss Federal Veterinary Office (BVET) guidelines. Data presented in this manuscript represent studies with male mice. After baseline echocardiography mice were randomly distributed to sham or treatment groups. Animal surgeries and echocardiography was performed blinded, that is the involved veterinarian was not aware of the genotype of the mice before or during operation. Animal numbers for experiments were chosen based on expected mortality rates and phenotypical or functional changes of hearts in wild-type mice due to treatment/surgery. Animals were excluded from the study in case of death before the experimental endpoint or based on the evaluation of pain using a standardized score sheet, approved by BVET.
One-kidney, one-clip
Six- to seven-week-old Khk-A−/−, Khk-A/C−/− and corresponding Khk-A+/+ and Khk-A/C+/+ male mice with a pure C57/BL6J background as well as Mlc2v-cre+ and Mlc2v-cre− male mice with a C57/BL6J 129S mixed background were anaesthetized and surgery was performed as previously described10 by inhalation of isoflurane (4% in oxygen, 1 l min−1). A sham procedure, which included the entire surgery with the exception of artery clipping, was applied to control mice.
Trans-aortic constriction
10–12-week-old Khk-A−/−, Khk-A/C−/− and corresponding Khk-A+/+ and Khk-A/C+/+ male mice as well as Mlc2v-cre+ and Mlc2v-cre− male mice were subjected to pressure overload by trans-aortic constriction (TAC) through constriction of the descending aorta as described30. The mice were monitored for 14–18 days after surgery, and their heart functions determined by echocardiography.
In vivo isoproterenol delivery
Alzet model 2002 mini-osmotic pumps (Alzet) were filled with isoproterenol, and were set to deliver at 30 mg kg−1 per day for 14 days in C57/BL6J wild-type mice. Pumps were then implanted into 9–10-week-old male mice under isoflurane anaesthesia as previously described10.
Echocardiography
Transthoracic echocardiography was performed using a 30 MHz probe and the Vevo 770 Ultrasound machine (Visualsonics) under mild anaesthesia with 1.5% isoflurane as previously described31.
Direct arterial blood pressure measurements in mice
The arterial blood pressure measurements in mice were performed as described previously32. Blood pressure was monitored for about 1 h in awake animals that were unconstrained.
Isolation and maintenance of primary neonatal mouse cardiomyocytes
Isolation of neonatal mouse cardiomyocytes was performed as previously described10. Experiments involving HIF1α, SF3B1 and KHK-C were performed with maintenance medium supplemented with physiologic concentrations of glucose (5 mM) and fructose (25 μM). The medium was changed daily. Pharmacological treatment was done with phenylepinephrine at a concentration of 100 μM and with isoproterenol at 10 μM for 48 h before preparation of SDS samples or processing for RNA isolation.
Lentivirus production and transduction
Lentiviral particles were produced in HEK-293T cells as previously described33. HEK-293T cells were purchased from American Type Culture Collection (ATCC) and regularly checked for mycoplasma contamination using a PCR-based myocoplasma detection kit (Sciencell). Cells were transduced with lentivirus 3 h after isolation by adding virus-containing supernatants to the cell culture medium and incubating cells at 37 °C/5% CO2 overnight.
Transient transfection
The pcDNA3 Sf3b1 construct was transiently transfected by using the Lipofectamin 2000 (Invitrogen) in 2-day-old NMCs according to the manufacturer’s protocol. Cells were harvested and analysed 36 h after transfection.
AAV9-fl/fl-shSf3b1
For the generation of the viral constructs, we cloned two shRNAs targeting Sf3b1 in a pSico vector where the U6 promoter driven shRNA expression is Cre–lox regulated34. The shRNA sequences were the following: AAV9-fl/fl-shSf3b1#1, sense (5′–3′):TGCATCGTCCTCCAAAGATTTTCAA GAGAAATCTTTGGAGGACGATGCTTTTTTC, antisense (5′–3′): TCGAG AAAAAAGCATCGTCCTCCAAAGATTTCTC TT GAAAATCTTTGGAGG ACGATGCA; AAV9-fl/fl-shSf3b1#2, sense (5′–3′): TG CTGCCTTACGTC AGATTATTCAAGAGATAATCTGACGTAAGGCAGCTTTTTTC, antisense (5′–3′): TCGAGAAAAAAGCTGCCTTACGTCAGATTATCTCTTGAATAA TCTGACGTAAGGCAGCA.
After in vitro validation of shRNA efficiency using AdenoCre expression to induce Cre-mediated recombination and activation of shSf3b1 expression, the region bearing the U6 promoter, the shRNA and the TATA-lox sites of the pSico construct was amplified and ligated into NheI/XhoI linearized AAV-bGH(+) vector. Viral amplification was done by Vectorbiolabs. AAV9 viruses were injected at a concentration of 1.2 × 1013 genome copies (GC) per kg body weight into tail veins of Mlc2v-cre+ or Mlc2v-cre− mice.
Plasmid constructions
The pLenti-pgk-HIF1αΔODD puro lentiviral expression vector was generated by subcloning the HIF1αΔODD fragment from a pcDNA3-HA-HIF1αΔODD(401–603) plasmid35 as described previously36. The KHK-A and KHK-C overexpression constructs were generated by subcloning KHK-A and KHK-C from the pET11a-KHK-A and pET11a-KHK-C expression constructs (provided by A. Asipu) into the Gateway Entry vector pENTR1a and by LR-reaction in the pLenti-pgk-puro lentiviral expression vector. As a corresponding control the pLenti-pgk puro empty vector was used. SF3A3 and SF3B3 over-expression constructs were generated by subcloning SF3A3 (from human ORFeome 8.1 collection, Thermo Scientific) or SF3B3 (from Addgene, plasmid #11241) in the pLKO.1 CMV puro construct provided by A. Ittner (ETH Zürich), by using EcoRI and SalI (SF3A3) or EcoRI and MluI (SF3B3) restriction sites. As a control vector the empty pLKO.1 CMV puro construct was used. Since the wild-type coding sequence of Sf3b1 is toxic to bacteria, we used a codon optimized Sf3b1 to perform overexpression experiments (the DNA and amino acid sequence can be found in Supplementary Data 1). First, we amplified Sf3b1 from mouse cDNA until the internal XhoI restriction site (1–1,568 bp) and subcloned it into a pcDNA3 vector by using EcoRI/XhoI restriction sites. The synthesis and codon optimisation of the second part (1,578–3,915 bp) was performed by Genscript. Finally we subcloned the two parts of Sf3b1 by double ligation in a pcDNA3 vector by using EcoRI, XhoI and XbaI restriction sites.
Sf3b1 promoter-luciferase construct
1.5 kilobases (kb)of the Sf3b1 promoter was amplified from mouse BAC genomic DNA and cloned into the pGL3 luciferase reporter vector (Stratagene). HRE mutants of the respective promoters were generated by recombinant PCR37,38. Sense and antisense primers were generated bearing HRE mutations, which were used to amplify the mutant 5′ and 3′ regions of the promoter, respectively. The 5′ and 3′ products generated from the respective PCR reactions were pooled afterwards at a 1:1 ratio and re-amplified using primers targeting the 5′ and 3′ ends of the respective promoters. To check for the correctness of the HRE mutation in the right direction, HRE mutants of the respective promoters were verified by sequencing (Microsynth).
shRNA knockdown of Khk-A and Khk-C
Khk-A- or Khk-C-specific shRNAs were designed using the Life Technologies BLOCK-iT RNAi Designer online tool (Life Technologies), searching for 21mer targets of either Khk-A- or Khk-C-specific transcripts. shRNAs were automatically compared to the mouse database using BLAST. As a loop sequence, the TRC-library loop sequence TCTGAG was used. Per isoform, three different shRNAs were chosen as promising, according to the program’s algorithm and cloned into the pLKO.1 lentiviral vector. The presence of the shRNA was confirmed by sequencing (Microsynth). shRNAs were then tested in NMCs for knockdown efficiency and specificity for either Khk-A or Khk-C. After validation, shRNAs shKhk-A#3, referred as shKhk-A and shKhk-C#2, referred as shKhk-C were chosen for all experiments described. The sequences are as follows: shKhk-A, sense (5′–3′): CCGGGATGACCTCCGCC AACAT TCTCTCGAGAGAATGTTGGCGGAGGTCATCTTTTTG; shKhk-C, sense (5′–3′): CCGGGCTCTTGCTGCATCGTCAACACTCGAGTGTTGAC GATGCAGCAAGAGC TTTTTG.
TRC lentiviral shRNA constructs
For lentiviral shRNA mediated knockdown the following shRNAs from the TRC library were used in a pLKO.1 construct: Sf3b1 (shSf3b1#1, TRCN0000295293; shSf3b1#2, TRCN0000295294), Sf3a3 (shSf3a3#1, TRCN0000285834; shSf3a3#2, TRCN0000277028), Sf3b2 (shSf3b2#3, TRCN0000123811; shSf3b2#4, TRCN0000123812), Sf3b3 (shSf3b3#5, TRCN0000311577; shSf3b3#11 TRCN0000109223), Glut5 (shGlut5#4, TRCN0000079392; shGlut5#, TRCN0000079388), Hif1α (shHif1α, TRCN0000232220), Khk (shKhk, TRCN0000416511). As a non-targeting shRNA construct the pLKO.1 vector SHC002 (Sigma) was used.
RNA-isolation and reverse transcription
RNA was isolated with the RNeasy Mini Kit (Qiagen) and reverse-transcribed using a EcoDry Premix (OligoDT) (Clontech) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT–PCR) reactions were prepared with Roche LightCycler 480 SYBR Green I Master as recommended by the manufacturer and run on a LightCycler 480 (Roche). Ct values were normalized to the housekeeping gene Hprt1. Primer sequence of qRT–PCR primers used in this study are as follows. Mouse: Hprt1 5′-GGGGCTGTACTGCTTAACCAG-3′ and 5′-TGC TCATCAGTTGCCACTTC-3′; Sf3b1 (primer pair A in Extended Data Fig. 3a) 5′-GCATCGTCCTCCAAAGATTGC-3′ and 5′-GCATTCATTTTAGGATCG GGAGT-3′; Sf3b1 (primer pair B in Extended Data Fig. 3a) 5′-CTCATGG GCTGTGCTATCTTGC-3′ and 5′-GTTGCTGCCTCAGCCAAAGC-3′; Sf3b1 (codon optimized, primer pair C in Extended Data Fig. 3a) 5′-CGTTAAA GGCCATCGTGAACG-3′ and 5′-CAATTTTCCTGCACTTTCTCATGC-3′; Sf3a3 5′-GAACGGCTTATGGATGTTATGGC-3′ and 5′-GGTGATCGGAA TTGATCTGGTC-3′; Sf3b2 5′-CATCCCGAACCTCCCAAAGG-3′ and 5′-GC TCCT ATCTCTGCCAATCTGG-3′; Sf3b3 5′-ATTTTGGAGTTACTACGCCC AG-3′ and 5′-GCCATAAGTGACCGGATAACAC-3′; Khk-A 5′-CCAACATT CTGTGGACTTACG-3′ and 5′-CCTTCTCAAAGTCCTTAGCAG-3′; Khk-C 5′-GCTGACTTCAGGCAGAGG-3′ and 5′-CCTTCTCAAAGTCC TTAGC AG-3′; Glut-1 5′-CAGTTCGGCTATAACACTGGTG-3′ and 5′-GCCCCCGAC AGAGAAGAT-3′; Pdk1 5′-GGACTTCGGGTCAGTGAATGC-3′ and 5′-CGC AGAAACATAAACGAGGTC-3′; Pkm2 5′-CGCCTGGACATTGACTCTG-3′ and 5′-GAAATTCAGCCGAGCCACATT-3′; Glut5 5′-CCAATATGGGTACA ACGTAGCTG-3′ and 5′-CCAATATGGGTACAACGTAGCTG-3′; sk.α-Actin 5′-TATGTGGCTATCCAGGCGGTG-3′ and 5′-CCCAGAATCCAACACGAT GC-3′; Nppa 5′-GTGCGGTGTCCAA CACAGAT-3′ and 5′-TCCAATCCTGTC AATCCTACCC-3′; Nppb 5′-AGTCCTTCG GTCTCAAGGCA-3′ and 5′-CCG ATCCGGTCTATCTTGTGC-3′; αMhc 5′-GCCCAGTACCTCCGAAAGTC-3′ and 5′-GCCTTAACATACTCCTTGTC-3′; βMhc 5′-GCATTCTCCTGCTGTT TCCTT-3′ and 5′-TGGATTCTCAAACGTGTCTAGTGA-3′; human: HPRT1 5′-CCTGGCGTCGTGATTAGTGAT-3′ and 5′-AGACGTTCAGTCCTGTCC ATAA-3′; SF3B1 5′-GTGGGCCTCGATTCTACAGG-3′ and 5′-GATGTCAC GTATCCAGCAAATCT-3′; KHK-A 5′-GCTATTCTGTGGACCTACGCT-3′ and 5′-CAATGTGGATCCACTTGAACTG-3′ and KHK-C 5′-CTCCTGCT GCATCATCAAC-3′ and 5′-CAATGTGGATCCACTTGAACTG-3′; GLUT1 5′-GGCCAAGAGTGTGCTAAAGAA-3′ and 5′-ACAGCGTTGATGCCAGA CAG-3′; PDK1 5′-CTGTGATACGGATCAGAAACCG-3′ and 5′-TCCACCAA ACAATAAAGAGTGCT-3′; GLUT5 5′-TCCAGTATGGGTACAACGTGG-3′ and 5′-CATGGACACGGTTACAGACCA-3′; AKR1B1 5′-TTTTCCCATTGG ATGAGTCGG-3′ and 5′-CCTGGAGATGGTTGAAGTTGG-3′; NPPA 5′-CA ACGCAGACCTGATGGATTT-3′ and 5′-AGCCCCCGCTTCTTCATTC-3′; NPPB 5′-TGGAAACGTCCGGGTTACAG-3′ and 5′-CTGATCCGGTCCAT CTTCCT-3′; αMHC 5′-GCCCTTTGACATTCGCACTG-3′ and 5′-CGGGA CAAAATCTTGGCTTTGA-3′; βMHC 5′-ACTGCCGAGACCGAGTATG-3′ and 5′-GCGATCCTTGAGGTTGTAGAGC-3′.
Splice-junction microarrays
Total RNA was isolated from NMCs infected with shNs and shHif1α lentiviruses and cultured in either 21% or 3% O2.. In another experiment NMCs were transduced with shNs and shSf3b1 lentiviruses and cultured at 3% O2 before RNA was isolated. Complementary DNA samples were prepared using the GeneChip WT cDNA Synthesis and Amplification Kit (Affymetrix). cDNA was hybridized to AltSplice splice-junction microarrays and resulting data were analysed with version 3 of ASPIRE software as previously described39. The unprocessed cel files with microarray data are available from ArrayExpress with accession numbers E-MTAB-2652 and E-MTAB-3203, respectively.
Luciferase assays
The luciferase assay was performed from material of NMCs using a luciferase kit (Promega) as recommended by the manufacturer and analysed on the MicroLumatPlus LB 96V (Berthold Technologies).
Chromatin and RNA immunoprecipitations
ChIP assays were performed with material from NMCs and the assays carried out using the ChIP-IT Kit (Active Motif) according to the manufacturer’s instructions. ChIP was performed with a ChIP-grade antibody from Abcam. In silico promoter analyses were performed using MatInspector and DiAlignTF (Genomatix). Sequences of primers used are 5′-TTTCGCTGGCTTTTTAAGGA-3′ and 5′-ATCTTCGCCATTTTGTCCAC-3′.
RNA immunoprecipitation assays were performed with material from NMCs using a RNA-ChIP-IT kit from Active Motif as recommended by the manufacturer. RNA immunoprecipitation was performed using a Sf3b1 antibody from MBL International. Sequences of primers used to detect potential binding of Sf3b1 on the branch points of Khk-A and Khk-C are the following: branch point primer for Khk-A, 5′- CTACTTCCCTCTGCTCAGCC-3′ and 5′-GGTCATCC AGGACAAAATGC-3′; branch point primer for Khk-C, 5′-GGACCCTTC TTCTCTGAGGC-3′ and 5′-CACTTGAGACACATCCACGC-3′.
Immunoblotting
Dissected hearts and NMCs lysates were homogenized by freeze slamming and solubilized in a modified SDS sample buffer sonicated and boiled for 5 min40). Protein lysates were resolved on 8%, 10% or 15% polyacrylamide minigels (BioRad) and transferred onto nitrocellulose membrane (GE Healthcare) by wet or semi-dry transfer. The following antibodies were used for immunoblotting rabbit HIF1α (catalogue no. NB-100-47; Novus Biologicals), a rabbit SF3B1 (catalogue no. ab172634; Abcam), a rabbit-KHK (catalogue no. HPA007040; Sigma), a rabbit KHK-C (custom made by Eurogentec) a rabbit Cre (catalogue no. 69050; Novagen), a rabbit SF3A3 (catalogue no. ab140605), a mouse SF3B2 (catalogue no. ab56800), a rabbit SF3B3 (catalogue no. NBP2-20258) mouse α-actinin (catalogue no. A7811; Sigma) and a chicken-GFP (catalogue no. GFP-1020; Aves Labs). Immunodetection and visualization of signals by chemiluminescence was carried out as described40, or by using the ImageQuant LAS 4000 mini (GE Healthcare). Immunoblots for Khk-A/C and Khk-C were visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Ketohexokinase splicing assay I
This Khk splicing assay including amplification of part of the Khk cDNA and the identified alternative spliced region was adapted from a splice assay for PKM1/M2 isoforms23. RNA isolated from NMCs transduced with shNs or shSf3b1 lentiviruses after treatment with isoproterenol or HIF1αΔODD and RNA isolated from sham- or 1K1C-treated wild-type left ventricles was reverse-transcribed then amplified using a forward primer (5′-GAAGCAGATCCTGTGCGTG-3′) that bound to shared Khk A/C exon 1 and a reverse primer (5′-ATTGTGCTCCTCTATCCGC-3′) that bound to shared Khk A/C exon 5 sequences. The 468-bp PCR products representing both Khk-A and Khk-C transcripts were then digested with HincII and run on a 1.5% agarose gel, after which the 468 bp (uncut Khk-A-specific amplicon), 294 bp and 174 bp (HincII-cleaved Khk-C-specific amplicon) products were quantitated using ImageJ software.
Ketohexokinase splicing assay II
To analyse KHK isoforms in human and mouse hearts or NMCs RT–PCR was performed. Total RNA was isolated from hearts or NMCs and reverse-transcribed into cDNA using the RNA to cDNA EcoDry Premix (OligoDT) kit (Clontech). Full length KHK-A/C cDNA was amplified by RT–PCR, loaded onto an agarose gel and purified by gel extraction. One hundred nanograms of full-length KHK-A/C cDNA was used as a template to amplify KHK-A and KHK-C specific fragments, respectively, using primer pairs consisting of a common KHK A/C forward and KHK-A- or KHK-C-specific reverse primer set. To be able to distinguish between KHK-A and KHK-C isoforms, the corresponding amplicons differed in length. The influence of fragment length on fragment amplification was minimised by designing two sets of primer pairs for each isoform (primer set 1 vs primer set 2): Khk-A1 < Khk-C1 and Khk-A2 > Khk-C2. The primer sequences are the following: primer pair 1 (human): KHK-A/C 5′-GAGAAGCAGATCCTGTGC-3′, KHK-A 5′-AGCGTAGGTCC ACAGAATAGC-3′ and KHK-C 5′-GGAGTTGTTGATGATGCAGC-3′; primer pair 2 (human): KHK-A 5′-GCTATTCTGTGGACCTACGCT-3′ and KHK-C 5′-CAGTTCAAGTGGATCCACATTG-3′; KHK-A/C 5′-GGTGTTGT GTGCGTCTATCC-3′; primer pair 1 (mouse); Khk-A/C 5′-GAAGCAGATC CTGTGCGTG-3′, Khk-A 5′-GGTCTGAAGGACCACATATCG-3′ and Khk-C 5′-CATTGGAGTTGTTGACGATGC-3′; primer pair 2 (mouse): Khk-A 5′-CAT TCTGTGGACTTACGATATGTG-3′ and Khk-C 5′-GTACCATTATACTCT ACGACACGAAC-3′; Khk-A/C 5′-GGTGTTGTGTGCGTCTATCC-3′.
Ketohexokinase activity assay
The ketohexokinase activity assay41 was modified by adding Roche Complete Protease Inhibitor Cocktail and Roche PhosSTOP to the reaction mixture, addition of 140 μg NMCs protein per reaction and incubating for 2 h at 37 °C. Spectrophotometric absorbance measurements of the remaining fructose were performed in 96-well plates at 515 nm using a Spectra MAX 190 plate reader (Molecular Devices). Wild-type liver protein was used as a positive control and reactions without protein were used as negative controls.
Metabolomics
4 × 105 NMCs were cultured per 3-cm dish. Cells were washed with PBS and whole-cell culture plates were snap frozen on dry ice. Metabolites were extracted twice from plates with hot (75 °C) 70% ethanol. Targeted analysis of hexose-phosphates levels and of their 13C-labelling pattern was performed by ion-pairing, reverse phase, ultra-performance liquid chromatography–tandem mass spectrometry on a Thermo Quantum Ultra instrument (Thermo Scientific)42. Technical quadruplicates were merged using their mean.
Untargeted analysis of metabolites was performed by flow injection–time-of-flight mass spectrometry on a Agilent 6550 Instrument in negative mode 4 GHz, high resolution in a m/z range of 50–1,000 as reported earlier43. Ions were annotated to ionic derivatives of the metabolites listed in the KEGG database (organism hsa) using 0.001 atomic mass units (a.m.u.) tolerance. Data processing and analysis was done with Matlab.
Immunofluorescent stainings
Immunofluorescent stainings were performed as described previously10. Cells were permeabilized and incubated with sarcomeric α-actinin antibody (Sigma) diluted 1:500 in 2% horse serum, 0.025% TritonX-100/PBS overnight at 4 °C. Cells were washed with PBS and incubated with 4′,6-diamidino-2-phenylindole (DAPI), Phalloidin and AlexaFluor488 anti-mouse secondary antibody for 1 h at room temperature. Afterwards, stainings were washed and mounted in Lisbeth medium.
[3H]leucine incorporation assay
Cardiomyocyte hypertrophy was evaluated by measuring [3H]leucine incorporation into newly synthesized proteins44. Cells were counted in a Neubauer chamber after trypsinization and scintillation counts were normalized to absolute cell number.
Ex vivo [3H]fructose/[14C](deoxy)-glucose uptake
Animals were euthanized 8 weeks after surgery and hearts were shortly placed into 1 M KCl solution. The left ventricle were cut into equal tissue pieces, dried on tissue, weighted and placed into 6-well plates with 2 ml medium (serum-free DMEM, 5 mM glucose and 8 μM fructose). Tissue explants were incubated in the same medium containing 1 mCi ml−1 [3H]fructose (specific activity 5 Ci mmol−1) and 0.1 mCi ml−1 [14C]deoxy-d-glucose (specific activity 58 mCi mmol−1) and harvested at respective time points. Harvested explants were washed two times with PBS and dissolved in 0.5 ml 1 M KOH (70 °C, 10 min, shaking). 5 ml scintillation liquid was added and radioactivity was measured in a Beckman LS6500 scintillation counter. The radioactivity counts were normalized to tissue weight.
In vivo [3H]fructose/[14C](deoxy)-glucose uptake and biodistribution
A mixture of 20 μl 1 mCi ml−1 [3H]fructose (specific activity 5 Ci mmol−1) and 200 μl 0.1 mCi ml−1 [14C]deoxy-d-glucose (specific activity 55 mCi mmol−1) was applied to sham- or TAC-treated C57/Bl6 J mice at 3 weeks post operation by oral gavage. After 45 min animals were weighed and euthanized. Denoted organs were collected and weighed. Tissue was digested with 4 M ethanolic KOH, radioactivity was measured in a Beckman LS6500 scintillation counter (Beckman) and calculated as disintegrations per minute (d.p.m.) per mg tissue weight.
Fructose content
Heart fructose content was measured using the fructose colorimetric/fluorometric assays kit according manufacturer’s instructions (Biovision) and as described previously45. Values were normalized to protein content.
[3H]fructose incorporation into RNA, DNA and protein
4 × 105 NMCs were seeded in 3-cm dishes and transduced in quadruplicates. On day 3, NMCs were incubated in maintenance medium containing 5 mM glucose, 8 μM unlabelled Fructose and 1 mCi ml−1 [3H]fructose (specific activity 5 Ci mmol−1) overnight. NMCs were washed three times with PBS before cell lysis was performed with 0.7 ml TRIzol (Invitrogen). Fractionation into RNA, DNA and protein was performed according to the manufacturer’s instructions with adaptations of the volumes according to the starting volume.
ADP/ATP Assay
5 × 104 NMCs were cultured in a 96-well format and treated as denoted. After 3 days in culture NMCs were analysed using a Bioassay Systems Enzylight ATP:ADP ratio kit (ELDT-100, Bioassay Systems) according to manufacturer’s instructions. Cellular ATP from 10 μl of lysate reacted with d-luciferin to generate a light signal that was proportional to the intracellular ATP concentration. In a second step, ADP was converted to ATP by an enzymatic reaction, and reacting with d-luciferin as in the first step. Luminometry was analysed using a MicroLumatPlus LB 96V (Berthold Technologies).
Myocardial ATP content
The level of myocardial ATP was measured by using a colorimetric/fluorometric assay kit (Abcam) according to the manufacturer’s instructions. Left-ventricular samples of about 10 mg weight were used as a starting material. The absorbance was read at 550 nm using a Tecan Infinite M1000 Pro plate reader and ATP content was calculated based on a standard curve generated from ATP-standards on the same 96-well plate. Values were normalized to protein concentrations obtained from Bradford assays.
[3H]fructose incorporation into lipids
4 × 105 cells were seeded in 3-cm dishes and transduced in quadruplicates. On day 3, cells were loaded with 0.25 mM oleic acid overnight and 1 μCi ml−1 [3H]fructose (specific activity 5.0 Ci mmol−1) was added to the medium. On day 4, cells were washed twice with PBS and lipids were extracted by hexane-isopropanol extraction46. Protein concentration was measured by BCA protein assay and radioactivity count was normalized to protein content.
Oil Red O staining
Oil Red O staining was combined with immunofluorescence according to a protocol in ref. 47. Spot-detection was performed with the Anchor image analysis platform (http://www.microscopyimages.org/). Nuclei are segmented by first thresholding the (Gaussian-blurred) DAPI channel with Otsu’s method, and then a watershed segmentation on the inverted Euclidian distance transform. Neighbouring segments are merged to increase an ellipticity score. A merge occurs if: ell(xy) > 0.5[ ell(x) + ell(y)], where xy is the merge of segments x and y. Segments with an area >50 μm2 are deemed nuclei. Cells are segmented by thresholding the histogram-stretched FITC channel, combining with the nuclei binary-image, and performing ultimate erosion using the segmented nuclei as seeds. Spots are segmented by first performing rolling-ball background correction, thresholding, and then a watershed segmentation on the inverted intensity after median filtering. Segments with an area >0.2 μm2 and a minimum gradient signal are deemed spots.
Nile red staining
4 × 105 NMCs were incubated with 0.25 mM oleic acid (Sigma-Aldrich) overnight and stained with 10 μg ml−1 Nile red (Sigma-Aldrich) for 10 min in PBS48. Values were normalized to cell number.
Cell size quantification
NMCs were stained after 3 days of culturing for sarcomeric α-actinin (1:500, Sigma), phalloidin and DAPI. Pictures of all channels were taken using a 20× magnification. Using the software Cell Profiler, sarcomeric α-actinin-positive cells were identified and cell area was quantified. Multi-nucleated cells were counted manually and average area per cell was corrected taking into account the number of multi-nucleated cells.
Measurements of extracellular acidification and oxygen consumption rate
1 × 105 NMCs per well were seeded in a 24-well Seahorse cell culture plate (SeaHorse Bioscience) and assayed at day 3. ECAR and OCR were measured in parallel using a SeaHorse XF24 Flux Analyzer (SeaHorse Bioscience) in NMCs incubated in low-buffered Krebs-Henseleit buffer, supplemented with glucose (5 mM) and fructose (25 μM). After five consecutive basal measurements of ECAR to measure glycolysis related lactate production, 0.25 mM oleic acid was added and oxygen consumption rates were measured again for five consecutive times. Finally, 1 μM carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone was injected to stimulate maximal OCR in NMCs, reflecting mitochondrial reserve capacity.
Statistical analysis
For statistical analyses, unpaired, two-tailed Student’s t-tests (Excel) or one-way ANOVA analyses followed by a Dunnet’s multiple comparison post-test were used as indicated in the respective figure legends. No statistical methods were used to predetermine sample size.
Extended Data
Extended Data Figure 1. HIF1α induces SF3B1 gene expression that mediates KHK-C isoform expression.
a, Schematic experimental workflow. b, Comparison of C1QBP, HNRNPH3, JMJD6 and SF3B1 promoter sequences of human and mouse illustrate putative HREs up to 1,000 bp upstream of the transcription start site (TSS). The core consensus HRE motif is shown in colour. c, d, qPCR (c) and immunoblotting (d) for Sf3b1 expression in NMCs transduced with scrambled shNs viruses or viruses bearing two distinct shRNAs against Sf3b1. All values obtained by qPCR are presented in relation to Sf3b1 expression in shNs (set as 1.0) (n = 3 biological replicates per group). e, RT–PCR analysis of Khk mRNA as in Fig. 1 g in NMCs transduced as indicated. f, RT–PCR analyses of Khk mRNA from NMCs infected as indicated and stimulated with mock or isoproterenol (ISO). PCR products were digested with HincII as in Fig. 1c . Splicing is quantified as percent of the Khk-C isoform (%Khk-C). g, RT–PCR based assessment of Khk-A and Khk-C isoform expression as done in Fig. 1 g. h, i, Ventricular samples of sham- or 1K1C-operated mice were analysed for Khk isoform expression (h) as done in Fig. 1 g or Sf3b1, Khk-A and Khk-C mRNA expression (i) by qPCR; n = 6 for each group. j, k, Ventricular samples of sham- or TAC-treated mice (j) and NaCl- or isoproterenol-treated mice (k) were analysed for expression of Sf3b1, Khk-A and Khk-C mRNA expression by qPCR (n = 6 for each individual group). l, NMCs transduced as specified were validated for protein expression of indicated proteins by using immunoblots. In addition lysates of NMCs overexpressing KHK-A or KHK-C were processed for immunoblotting Khk antibodies. m, n, Ventricular samples of sham- or TAC-treated (m) and sham- or isoproterenol-treated mice (n) were analysed for indicated proteins by immunoblotting. o, Probes from left ventricles of 1K1C-, TAC- or isoproterenol-treated mice and their corresponding controls were analysed for mRNA expression of Hif1α targets. All values were presented in relation to sham-operated controls set as 1.0 (n = 6 for each individual group). p, q, Left-ventricular biopsies of HCM patients and healthy controls were analysed for mRNA expression of SF3B1, KHK-A and KHK-C (p) and HIF1α targets (q). All values are presented in relation to healthy controls set as 1.0 (n = 6 for controls and n = 16 for patient samples). Error bars are s.d. (c) or s.e.m. (i–k, o–q). *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed unpaired t-test (c, i–k, o–q).
Extended Data Figure 2. SF3B1 is a HIF1α-target gene, whose product regulates growth and metabolic shift in vitro.
a, NMCs infected as denoted, were stimulated with either mock, phenylephrine and isoproterenol, respectively. Sf3b1 mRNA expression was examined by qPCR. (n = 3 biological replicates). b, NMCs were transduced as indicated and processed for qPCR analysis of Sf3b1 mRNA. (n = 3 biological replicates). c, Comparative analysis of a conserved HRE (shown in red) in the promoter of SF3B1 of diverse mammalian species located 122 bp downstream of the transcription start site (TSS). d–g, Immunoblot detection of indicated proteins of NMCs transduced with shNs, and shSf3b lentiviruses treated with phenylephrine or isoproterenol (d) co-infected with empty vector or HIF1αΔODD (e) cultured under normoxic (21% O2) or hypoxic (3% O2) conditions (f) and co-transduced with shVhl lentiviruses (g). h–o, Evaluation of [3H]leucine incorporation and the myosin heavy chain α/β mRNA expression ratio in NMCs treated with either phenylephrine or isoproterenol (h, l), HIF1αΔODD overexpression (i, m), hypoxia (j, n) or pVhl deletion (k, o), with or without shSf3b1 mediated Sf3b1 depletion. Obtained values are shown relative to incorporated radioactivity in control (shNS) NMCs (set as 1.0) (n = 4 per individual group). p, NMCs treated as in i were microscopically analysed for cell size using CellProfiler. Data represents 1 of 3 independent experiments with approximately 200 analysed cells per experiment. q, Lysates of NMCs treated as indicated, were assessed for protein expression of specified proteins by immunoblots. r, Evaluation of [3H]leucine incorporation of NMCs infected as indicated. Obtained values are shown relative to incorporated radioactivity in control (shNs/empty) NMCs (set as 1.0) (n = 3 per individual group). s–z, Assessment of extracellular acidification rate (ECAR), and oleic acid- and carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP)-induced oxygen consumption rate (OCR) in NMCs transduced and treated as indicated. Highlighted ECAR measurements were done at baseline. All data are compared to shNs-transduced NMCs. (n = 8 biological replicates in the group shNs + isoproterenol; n = 10 biological replicates in all other groups). Error bars are s.d. (a, b, h–o, r) or s.e.m. (p, s–z). *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed unpaired t-test (b, t, u, w–z). One-way ANOVA followed by Dunnett’s multiple comparison post-test (a, h–p, r).
Extended Data Figure 3. Influence of U2snRNP-complex members on KHK isoform expression and validation of KHK-C-mediated fructose uptake and metabolism.
a, NMCs were transduced with either shNs or shSf3b1 and transfected with codon optimized Sf3b1 or empty construct and expression of denoted proteins was assessed. b, NMCs were transduced with either shNs or shSf3b1 and transfected with codon-optimized Sf3b1 or empty construct, respectively. Exogenous Sf3b1-expression was evaluated by qRT–PCR. Three different primer pairs were used. Primer pair A detects both endogenous and exogenous codon-optimized Sf3b1 (top). Primer pair B recognizes Sf3b1 close to the shRNA binding site and is specific for endogenous Sf3b1 (left). Primer pair C is located in the codon-optimized region (right). Values were normalized to shNs/empty-vector-infected NMCs (n = 3 biological replicates per group). c, [3H]leucine incorporation normalized to cell number in NMCs treated as in b (n = 4 biological replicates per group). d, NMCs transduced with empty vector or human SF3A3 or SF3B3 were assessed for mRNA expression of Khk-A and Khk-C by qRT–PCR. Values were normalized to empty-vector-transduced NMCs (n = 3 biological replicates per group). e, NMCs transduced as in d were immunoblotted for indicated proteins. f, NMCs transduced as indicated were probed for mRNA expression of Khk-A or Khk-C by qRT–PCR. Values were normalized to shNs/empty-vector-transduced NMCs (n = 3 biological replicates). g, NMCs treated with either scrambled shRNA or two different shRNAs targeting either Sf3a3, Sf3b2 or Sf3b3 were co-transduced with empty vector or HIF1αΔODD. Protein expression of Khk-A/C or Khk-C specifically was evaluated by immunoblotting. h, Evaluation of [3H]leucine incorporation of NMCs infected as in d normalized to empty-vector-transduced NMCs (n = 4 biological replicates per group). i, Lysates of NMCs treated as indicated were immunoblotted for specified proteins. j, KHK-activity in lysates of NMCs transduced with a combination of lentiviruses expressing either shKhk targeting mouse Khk and human KHK-A or shKhk and human KHK-C or a scrambled shRNA control together with an empty vector. k, Evaluation of the kinetics of [13C]fructose-1-phosphate accumulation in NMCs treated as in i at the respective time points (n = 3 biological replicates). l, Glut5 mRNA expression in NMCs transduced as indicated. (n = 3 biological replicates per group). m, NMCs transduced as indicated were microscopically analysed for cell size. Data represents 1 of 3 experiments with approximately 50 analysed cells per experiment n, Khk-A- and Khk-C-specific shRNAs were evaluated for their potential to inhibit Khk-A and Khk-C isoform expression, respectively. NMCs were infected with shNs, shKhk-A or shKhk-C lentiviruses. Data are shown in relation to shNs NMCs (set to 1.0) (n = 3 biological replicates). Error bars are s.d. (b–d, f, h, k, l, n) or s.e.m. (m). *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA followed by Dunnett’s multiple comparison post-test (b–d, f, k, l, m, n).
Extended Data Figure 4. Downstream metabolic effects of fructose metabolism in NMCs.
a, log2 fold change of metabolites in NMCs overexpressing KHK-C and cultured with glucose or glucose and fructose compared to the corresponding control transduced NMCs (n = 4 biological replicates per group). b, Incorporation of [3H]fructose into RNA (top), DNA (middle) and protein (bottom) of NMCs treated as indicated (n = 4 biological replicates per group). c, Uptake of [14C]deoxyglucose in NMCs infected as in b at depicted time points (n = 4 biological replicates per group). d, [3H]leucine incorporation in NMCs transduced with shNs or shKhk and co-transduced with either empty overexpression vector or KHK-A or KHK-C respectively. NMCs were cultured in media with increasing fructose concentrations, under physiologic glucose amounts or with increasing glucose concentrations under physiologic fructose amounts. Data are presented relative to shNs/empty-vector-transduced NMCs at 5 mM glucose/0 μM fructose (n = 4 biological replicates). e, ADP/ATP ratios in NMCs transduced as indicated. (n = 4 biological replicates, data show 1 of 3 representative experiments). f, Immunofluorescence images of NMCs transduced with empty vector (upper panel) or HIF1αΔODD (middle panel) lentiviruses or exposed to hypoxia (lower panel) were additionally transduced as indicated. Prior to staining for sarcomeric α-actinin, Oil Red O and DAPI, NMCs were incubated with oleic acid. g, Quantification of lipid droplets/cell in NMCs of immunofluorescent images shown in f. h, Ratio of lipid droplets/cell area of NMCs shown in g. i–l, NMCs were infected as indicated and processed for oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measurements under basal conditions or after injection of OA or FCCP. Depicted are rates expressed as OCR to ECAR ratios (upper panels) or individual ECAR (for measurements at baseline) or OCR values (n = 8 biological replicates for HIF1αΔODD/shKhk-A; n = 10 biological replicates for all other groups). Error bars are s.d. (b–e) or s.e.m. (g–l). *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA followed by Dunnett’s multiple comparison post-test (b–e, g, h). Two-tailed unpaired t-test (i–l).
Extended Data Figure 5. Fructose and glucose uptake ex vivo and validation of AAV9-fl/fl-shSf3b1 viruses.
a, Ratio of left-ventricular weight to body weight of sham- or TAC-treated C57/Bl6J mice; (n = 7 per group). b, c, [3H]fructose (b) and [14C]deoxy-glucose (c) were co-applied by oral gavage and biodistribution was measured at 45 min post injection (n = 7 per group). d, Ratio of left-ventricular weight to tibia length (LV/TL) of sham- or 1K1C-treated C57/Bl6J wild-type mice (n = 6 for sham and n = 7 for 1K1C-treated mice). e, Fructose levels in probes from left ventricles of indicated mice as determined by a quantitative colorimetric assay, normalized to protein amount (n = 4 for sham and n = 5 for hearts of 1K1C-treated mice). f, g, Freshly isolated left-ventricular pieces of sham- or 1K1C-treated mice incubated in medium containing either [14C]fructose (f) or [14C]deoxyglucose (g) as indicated were processed for uptake measurements. Counts were normalized to tissue weight (n = 6 per time point and treatment). h–j, qPCR analyses of Glut5 mRNA expression in left ventricles of 1K1C- (h), TAC- (i) or isoproterenol-treated (j) mice. All values are presented in relation to Glut5 expression in shams (set as 1.0). n = 5 for sham-treated mice as controls for TAC-treated mice (n = 6), all other groups n = 7. k, l, Relative mRNA expression of GLUT5 and aldose reductase (AKR1B1) in left-ventricular biopsies of patients diagnosed with aortic stenosis (k) or HCM (l) in relation to healthy controls (set as 1.0) was analysed by qPCR (n = 6 for controls; n = 16 for HCM and n = 17 for patients with aortic stenosis). m, Integration of AAV9-fl/fl-shSf3b1#1 viruses in the myocardium of Mlc2-cre− or Mlc2v-cre+ mice was examined 12 weeks after intravenous injection by confocal microscopy (using 20× magnification) of left-ventricular cryoslices stained against GFP and sarcomeric α-actinin and counterstained for DAPI. n, Ventricular samples of Mlc2v-cre− or Mlc2-cre+ mice transduced with AAV9-fl/fl-shSf3b1#1 or AAV9-fl/fl-shSf3b1#2 viruses were assessed for expression of indicated proteins by immunoblotting. o, p, Ratio of left-ventricular weight to tibia length (o) and ejection fraction (%EF) (p) of sham- or TAC-treated Mlc2v-cre− and Mlc2v-cre+ mice injected with AAV9-fl/fl-shSf3b1#1 (number of mice per group is given in Extended Data Table 1). q, Lysates of hearts from sham- or TAC-operated Mlc2v-cre− and Mlc2v-cre+ mice transduced with AAV9-fl/fl-shSf3b1#1 viruses were processed for immunoblotting with antibodies as denoted. Error bars are s.e.m. (a–l, o, p). *P < 0.05; **P < 0.01; ***P < 0.001, two-tailed unpaired t-test (a–l, o, p).
Extended Data Figure 6. Pathologic growth is dependent on HIF1α-driven and SF3B1-mediated KHK-C expression.
a–f, Ratio of left-ventricular weight to tibia length (LV/TL) (a), mRNA expression of the hypertrophic markers Nppa (b), myosin heavy chain β (βMhc) (c) and myosin heavy chain α (αMhc) (d), ejection fraction (%EF) (e) and left-ventricular ATP content (f) of sham- or TAC-treated Khk-A+/+ and Khk-A−/− mice. g–l, Ratio of left-ventricular weight to tibia length (LV/TL) (g), mRNA expression of the hypertrophic markers Nppa (h), myosin heavy chain β (βMhc) (i) and myosin heavy chain α (αMhc) (j), ejection fraction (%EF) (k) and left-ventricular ATP content (l) of Khk-A/C+/+ and Khk-A/C−/− mice. m, n, Lysates from ventricles of sham- or TAC-operated Khk-A+/+ and Khk-A−/− mice (m) and Khk-A/C+/+ and Khk-A/C−/− mice (n) were processed for immunoblotting with antibodies as indicated. o–t, Assessment of hypertrophic marker gene expression (Nppa, βMhc and αMhc) in ventricles of TAC- vs sham-treated Khk-A−/− mice and their corresponding Khk-A+/+ controls (o–q) as well as Khk-A/C−/− mice and their corresponding Khk-A/C+/+ controls (r–t). u, v, Mean blood pressure (BP) in mmHg of Khk-A+/+ and Khk-A−/− (u) as well as Khk-A/C+/+ and Khk-A/C−/− mice (v) 8 weeks after sham or 1K1C treatment. For a–l, o–v, the number of mice per group is given in Extended Data Table 2. Error bars are s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed unpaired t-test.
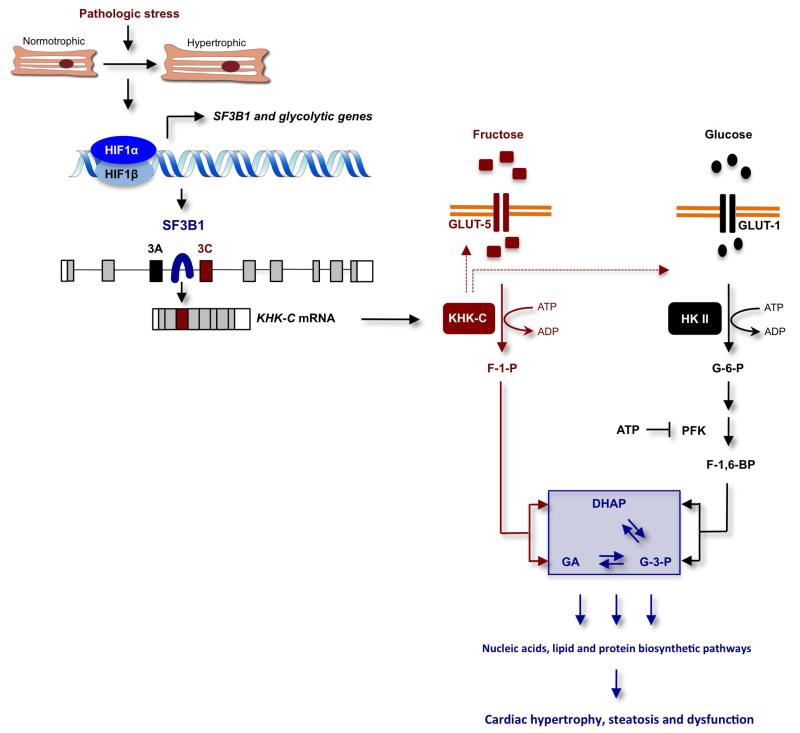
Extended Data Figure 7. Model explaining how HIF1α activation of SF3B1-dependent splicing of KHK regulates fructose and glucose metabolism to promote cardiac hypertrophy in response to pathologic stress.
In this model, pathologic stress leads to increased expression of HIF1α and HIF1α-dependent activation of genes encoding glycolytic enzymes and the splicing factor SF3B1. SF3B1, in turn, assembles at the branch-point sequence upstream of exon 3C of KHK pre-mRNA leading to an inclusion of this exon and KHK-C protein production. This shift in isoform expression from KHK-A to KHK-C in response to HIF1α–SF3B1 pathway activation drives KHK-C-dependent fructose uptake via stimulation of GLUT5 expression, the conversion of fructose to fructose-1-phosphate (F1P) and contributes simultaneously to the activation of glucose uptake and metabolism through a yet-to-be-determined mechanism. The model holds that the unrestrained conversion of fructose to F1P by KHK-C limits ATP levels, thereby alleviating potential allosteric inhibition of phosphofructokinase (PFK) by ATP to maintain a high glycolytic flux. F1P is further metabolized to dehydroxyacetone phosphate (DHAP) and glyceraldehyde (GA). While DHAP serves as a precursor for glycerol synthesis, GA can be further converted to glyceraldehyde-3-phosphate (G3P), a key glycolytic intermediate. G3P can be channelled into the non-oxidative pentose phosphate pathway (PPP) supporting nucleic and amino acid biosynthesis. This metabolic constellation, created by the activation of the HIF1α–SF3B1–KHK-C axis, increases macromolecular biosynthetic capacity essential for hypertrophic growth, steatosis and cardiac dysfunction. HKII and F-1,6-BP denotes hexokinase II and fructose-1,6-bisphosphate, respectively.
Extended Data Table 1. Echocardiographic analysis of sham-, 1K1C- or TAC-treated Mlc2v-cre−/-cre+ mice injected with AAV9-fl/fl-shSf3b1 viruses.
| Mlc2v-cre − | Mlc2v-cre + | |||
|---|---|---|---|---|
| AAV9 fl/fl shSf3b1#1 | AAV9 fl/fl shSf3b1#1 | |||
|
|
||||
| sham | 1K1C | sham | 1K1C | |
| n | 4 | 3 | 5 | 3 |
| BW (g) | 27.5 ± 0.6 | 27.3 ± 0.7 | 27.3 ± 0.6 | 27.8 ± 0.8 |
| IVS;d (mm) | 0.71 ± 0.03 | 1.00 ± 0.03** | 0.70 ± 0.02 | 0.80 ± 0.04 |
| IVS;s (mm) | 0.98 ± 0.02 | 1.30 ± 0.05** | 1.04 ± 0.09 | 1.08 ± 0.08 |
| LVID;d (mm) | 3.97 ± 0.08 | 4.13 ± 0.10 | 3.85 ± 0.11 | 4.09 ± 0.10 |
| LVID;s (mm) | 3.08 ± 0.10 | 3.30 ± 0.11 | 2.86 ± 0.15 | 2.98 ± 0.18 |
| LVPW;d (mm) | 0.69 ± 0.01 | 0.85 ± 0.03** | 0.67 ± 0.02 | 0.82 ± 0.01 |
| LVPW;s (mm) | 0.91 ± 0.35 | 1.08 ± 0.12 | 0.93 ± 0.09 | 0.97 ± 0.04 |
| %FS | 27.8 ± 1.9 | 25.3 ± 2.1 | 29.8 ± 1.6 | 27.1 ± 2.8 |
| E/A | 2.21 ± 0.26 | 0.80 ± 0.06*** | 2.17 ± 0.17 | 1.73 ± 0.11 |
|
| ||||
| Mlc2v-cre − | Mlc2v-cre + | |||
| AAV9 fl/fl shSf3b1#1 | AAV9 fl/fl shSf3b1#1 | |||
|
|
||||
| sham | TAC | sham | TAC | |
|
| ||||
| n | 5 | 4 | 5 | 5 |
| BW (g) | 27.4 ± 0.8 | 28.0 ± 0.9 | 26.5 ± 0.5 | 26.9 ± 0.3 |
| IVS;d (mm) | 0.70 ± 0.03 | 0.98 ± 0.03*** | 0.68 ± 0.02 | 0.84 ± 0.04 |
| IVS;s (mm) | 0.96 ± 0.06 | 1.37 ± 0.04** | 0.97 ± 0.05 | 1.18 ± 0.05 |
| LVID;d (mm) | 3.94 ± 0.06 | 4.04 ± 0.09 | 3.82 ± 0.06 | 3.68 ± 0.06 |
| LVID;s (mm) | 2.89 ± 0.13 | 2.80 ± 0.09 | 2.71 ± 0.13 | 2.54 ± 0.11 |
| LVPW;d (mm) | 0.68 ± 0.02 | 0.98 ± 0.04*** | 0.70 ± 0.02 | 0.82 ± 0.03 |
| LVPW;s (mm) | 0.97 ± 0.06 | 1.37 ± 0.10** | 0.99 ± 0.07 | 1.11 ± 0.03 |
| %FS | 30.4 ± 2.3 | 19.9 ± 3.9* | 30.04 ± 3.8 | 34.2 ± 1.3 |
| E/A | 1.62 ± 0.09 | 1.72 ± 0.23 | 1.68 ± 0.02 | 1.74 ± 0.12 |
| Aortic Vel. (mm/s) | 923 ± 58 | 3920± 305*** | 878 ±72 | 3670± 190*** |
|
| ||||
| Mlc2v-cre − | Mlc2v-cre + | |||
| AAV9 fl/fl shSf3b1#2 | AAV9 fl/fl shSf3b1#2 | |||
|
|
||||
| sham | TAC | sham | TAC | |
|
| ||||
| n | 5 | 4 | 5 | 5 |
| BW (g) | 27.4 ± 0.8 | 28.0 ± 0.9 | 26.5 ± 0.5 | 26.9 ± 0.3 |
| IVS;d (mm) | 0.70 ± 0.03 | 0.98 ± 0.03*** | 0.68 ± 0.02 | 0.84 ± 0.04 |
| IVS;s (mm) | 0.96 ± 0.06 | 1.37 ± 0.04** | 0.97 ± 0.05 | 1.18 ± 0.05 |
| LVID;d (mm) | 3.94 ± 0.06 | 4.04 ± 0.09 | 3.82 ± 0.06 | 3.68 ± 0.06 |
| LVID;s (mm) | 2.89 ± 0.13 | 2.80 ± 0.09 | 2.71 ± 0.13 | 2.54 ± 0.11 |
| LVPW;d (mm) | 0.68 ± 0.02 | 0.98 ± 0.04*** | 0.70 ± 0.02 | 0.82 ± 0.03 |
| LVPW;s (mm) | 0.97 ± 0.06 | 1.37 ± 0.10** | 0.99 ± 0.07 | 1.11 ± 0.03 |
| %FS | 30.4 ± 2.3 | 19.9 ± 3.9* | 30.04 ± 3.8 | 34.2 ± 1.3 |
| E/A | 1.62 ± 0.09 | 1.72 ± 0.23 | 1.68 ± 0.02 | 1.74 ± 0.12 |
| Aortic Vel. (mm/s) | 923 ± 58 | 3920± 305*** | 878 ±72 | 3670± 190*** |
BW, body weight; IVS;d, interventricular septum diameter in diastole, IVS;s, interventricular septum diameter in systole; LVID;d, left-ventricular inner diameter in diastole; LVID;s, left-ventricular inner diameter in systole; LVPW;d, left-ventricular posterior wall diameter in diastole, LVPW;s, left-ventricular posterior wall diameter in systole; %FS, fractional shortening; E/A, ratio of early (E) to late (A) ventricular filling velocities; Aortic Vel. = aortic velocity. Values shown are mean ± s.e.m.;
P < 0.05;
P < 0.01;
P < 0.001; two-tailed unpaired t-test.
Extended Data Table 2. Echocardiographic analysis of sham- or TAC-operated Khk-A+/+/Khk-A−/− and Khk-A/C+/+/Khk-A/C−/− mice.
| Khk-A+/+ | Khk-A−/− | |||
|---|---|---|---|---|
|
|
||||
| sham | TAC | sham | TAC | |
| n | 6 | 6 | 5 | 6 |
| BW (g) | 25.6 ± 0.7 | 27.3 ± 0.8 | 25.4 ± 0.7 | 25.6 ± 0.7 |
| IVS;d (mm) | 0.73 ± 0.01 | 0.98 ± 0.02*** | 0.72 ± 0.01 | 1.00 ± 0.03*** |
| IVS;s (mm) | 1.03 ± 0.07 | 1.34 ± 0.04** | 1.12 ± 0.01 | 1.36 ± 0.06** |
| LVID;d (mm) | 3.85 ± 0.08 | 3.80 ± 0.06 | 3.75 ± 0.10 | 3.73 ± 0.07 |
| LVID;s (mm) | 2.71 ± 0.16 | 2.65 ± 0.11 | 2.66 ± 0.20 | 2.70 ± 0.10 |
| LVPW;d (mm) | 0.72 ± 0.01 | 0.99 ± 0.04*** | 0.72 ± 0.01 | 0.99 ± 0.03*** |
| LVPW;s (mm) | 1.04 ± 0.03 | 1.35 ± 0.03*** | 1.05 ± 0.01 | 1.32 ± 0.05*** |
| %FS | 34.0 ±2.1 | 26.3 ±1.5* | 34.3 ± 1.7 | 26.0 ± 1.2* |
| E/A | 1.38 ± 0.11 | 2.20 ± 0.05** | 1.97 ± 0.4 | 1.90 ± 0.4 |
| Aortic Vel. (mm/s) | 938 ± 55 | 4250± 275*** | 880 ± 68 | 3700± 308*** |
|
| ||||
| Khk-A/C+/+ | Khk-A/C−/− | |||
|
|
||||
| sham | TAC | sham | TAC | |
|
| ||||
| n | 5 | 6 | 6 | 5 |
| BW (g) | 25.8 ± 0.6 | 24.9 ± 0.8 | 24.51 ± 0.6 | 24.4 ± 0.6 |
| IVS;d (mm) | 0.72 ± 0.01 | 1.00 ± 0.05** | 0.64 ± 0.04 | 0.83 ± 0.06 |
| IVS;s (mm) | 1.15 ± 0.01 | 1.42 ± 0.07* | 1.06 ± 0.07 | 1.19 ± 0.13 |
| LVID;d (mm) | 3.53 ± 0.13 | 3.70 ± 0.23 | 3.65 ± 0.13 | 3.67 ± 0.04 |
| LVID;s (mm) | 2.28 ± 0.27 | 2.61 ± 0.33 | 2.49 ± 0.17 | 2.45 ± 0.10 |
| LVPW;d (mm) | 0.69 ± 0.03 | 1.00 ± 0.05*** | 0.67 ± 0.02 | 0.81 ± 0.04* |
| LVPW;s (mm) | 1.12 ± 0.03 | 1.25 ± 0.03* | 1.00 ± 0.06 | 1.11 ± 0.07 |
| %FS | 35.5 ± 3.0 | 27.5 ±1.6* | 32.1 ± 2.30 | 33.4 ± 2.9 |
| E/A | 1.51 ± 0.09 | 1.56 ± 0.17 | 1.77 ± 0.10 | 1.64 ± 0.21 |
| Aortic Vel. (mm/s) | 928 ± 120 | 4020± 232*** | 937 ± 59 | 3600± 292*** |
BW, body weight; IVS;d, interventricular septum diameter in diastole, IVS;s, interventricular septum diameter in systole; LVID;d, left-ventricular inner diameter in diastole; LVID;s, left-ventricular inner diameter in systole; LVPW;d, left-ventricular posterior wall diameter in diastole, LVPW;s, left-ventricular posterior wall diameter in systole; %FS, fractional shortening; E/A, ratio of early (E) to late (A) ventricular filling velocities; Aortic Vel. = aortic velocity. Values shown are mean ± s.e.m.;
P < 0.05;
P < 0.01;
P < 0.001; two-tailed unpaired t-test.
Extended Data Table 3. Echocardiographic analysis of sham- or 1K1C-operated Khk-A+/+/Khk-A−/− and Khk-A/C+/+/Khk-A/C−/− mice.
| Khk-A+/+ | Khk-A−/− | |||
|---|---|---|---|---|
|
|
||||
| sham | 1K1C | sham | 1K1C | |
| n | 5 | 6 | 5 | 6 |
| BW (g) | 27.2 ± 0.7 | 27.1 ± 0.6 | 28.2 ± 2.0 | 29.7 ± 0.5 |
| IVS;d (mm) | 0.75 ± 0.01 | 0.94 ± 0.03*** | 0.72 ± 0.02 | 1.01 ± 0.06** |
| IVS;s (mm) | 1.08 ± 0.05 | 1.31 ± 0.05** | 1.00 ± 0.04 | 1.31 ± 0.09* |
| LVID;d (mm) | 4.02 ± 0.10 | 3.93 ± 0.12 | 3.91 ± 0.18 | 4.37 ± 0.32 |
| LVID;s (mm) | 3.02 ± 0.08 | 3.03 ± 0.14 | 2.90 ± 0.20 | 3.45 ± 0.40 |
| LVPW;d (mm) | 0.73 ± 0.02 | 0.99 ± 0.04*** | 0.74 ± 0.02 | 0.97 ± 0.04** |
| LVPW;s (mm) | 1.01 ± 0.05 | 1.30 ± 0.05** | 1.06 ± 0.05 | 1.29 ± 0.06* |
| %FS | 27.1 ± 1.5 | 22.6 ± 1.4* | 28.8 ± 3.4 | 19.2 ± 3.05* |
| E/A | 1.97 ± 0.17 | 1.47 ± 0.10* | 1.83 ± 0.55 | 1.27 ± 0.14 |
|
| ||||
| Khk-A/C+/+ | Khk-A/C−/− | |||
|
|
||||
| sham | 1K1C | sham | 1K1C | |
|
| ||||
| n | 5 | 6 | 6 | 5 |
| BW (g) | 25.8 ± 0.6 | 24.9 ± 0.8 | 24.51 ± 0.6 | 24.4 ± 0.6 |
| IVS;d (mm) | 0.72 ± 0.01 | 1.00 ± 0.05** | 0.64 ± 0.04 | 0.83 ± 0.06 |
| IVS;s (mm) | 1.15 ± 0.01 | 1.42 ± 0.07* | 1.06 ± 0.07 | 1.19 ± 0.13 |
| LVID;d (mm) | 3.53 ± 0.13 | 3.70 ± 0.23 | 3.65 ± 0.13 | 3.67 ± 0.04 |
| LVID;s (mm) | 2.28 ± 0.27 | 2.61 ± 0.33 | 2.49 ± 0.17 | 2.45 ± 0.10 |
| LVPW;d (mm) | 0.69 ± 0.03 | 1.00 ± 0.05*** | 0.67 ± 0.02 | 0.81 ± 0.04* |
| LVPW;s (mm) | 1.12 ± 0.03 | 1.25 ± 0.03* | 1.00 ± 0.06 | 1.11 ± 0.07 |
| %FS | 35.5 ± 3.0 | 27.5 ± 1.6* | 32.1 ± 2.30 | 33.4 ± 2.9 |
| E/A | 1.51 ± 0.09 | 1.56 ± 0.17 | 1.77 ± 0.10 | 1.64 ± 0.21 |
BW, body weight; IVS;d, interventricular septum diameter in diastole, IVS;s, interventricular septum diameter in systole; LVID;d, left-ventricular inner diameter in diastole; LVID;s, left-ventricular inner diameter in systole; LVPW;d, left-ventricular posterior wall diameter in diastole, LVPW;s, left-ventricular posterior wall diameter in systole; %FS, fractional shortening; E/A, ratio of early (E) to late (A) ventricular filling velocities; Aortic Vel. = aortic velocity. Values shown are mean ± s.e.m.;
P < 0.05;
P < 0.01;
P < 0.001; two-tailed unpaired t-test.
Supplementary Material
Acknowledgements
We thank S. Georgiev, T. Simka, S. Xu, C. Bischoff, J. M.Dominguez, W. Kovacs and M. Piontek and other members of the Krek laboratory for discussions, help and technical assistance. We are grateful to M. Stoffel for performing tail vein injections. K. Chien, A. Asipu and R. J. Johnson provided mouse lines. This work was supported by grants from Sinergia (Swiss National Science Foundation) to W.K., T.P. and J. U. and the Swiss Heart Foundation to W.K.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Yip GW, Fung JWH, Tan Y-T, Sanderson JE. Hypertension and heart failure: a dysfunction of systole, diastole or both? J. Hum. Hypertens. 2009;23:295–306. doi: 10.1038/jhh.2008.141. [DOI] [PubMed] [Google Scholar]
- 2.Shiojima I, et al. Disruption of coordinated cardiac hypertrophy and angiogenesis contributes to the transition to heart failure. J. Clin. Invest. 2005;115:2108–2118. doi: 10.1172/JCI24682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu. Rev. Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 5.Hill JA, Olson EN. Cardiac plasticity. N. Engl. J. Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 6.Grosso AR, et al. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res. 2008;36:4823–4832. doi: 10.1093/nar/gkn463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbosa-Morais NL, Carmo-Fonseca M, Aparício S. Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res. 2006;16:66–77. doi: 10.1101/gr.3936206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diggle CP, et al. Ketohexokinase: expression and localization of the principal fructose-metabolizing enzyme. J. Histochem. Cytochem. 2009;57:763–774. doi: 10.1369/jhc.2009.953190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnan J, et al. Activation of a HIF1α-PPARγ axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab. 2009;9:512–524. doi: 10.1016/j.cmet.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, et al. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonnal S, Vigevani L, Valcárcel J. The spliceosome as a target of novel antitumour drugs. Nature Rev. Drug Discov. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 13.Corrionero A, Minana B, Valcarcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolatshad H, et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia. 2015;29:1092–1103. doi: 10.1038/leu.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramasamy R, Goldberg IJ. Aldose reductase and cardiovascular diseases, creating human-like diabetic complications in an experimental model. Circ. Res. 2010;106:1449–1458. doi: 10.1161/CIRCRESAHA.109.213447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis CA, et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirillo P, et al. Ketohexokinase-dependent metabolism of fructose induces proinflammatory mediators in proximal tubular cells. J. Am. Soc. Nephrol. 2009;20:545–553. doi: 10.1681/ASN.2008060576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang M, et al. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell. 2010;143:711–724. doi: 10.1016/j.cell.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Vander Heiden MG, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma S, et al. Intramyocardial lipid accumulation in the failing human heart resembles the lipotoxic rat heart. FASEB J. 2004;18:1692–1700. doi: 10.1096/fj.04-2263com. [DOI] [PubMed] [Google Scholar]
- 21.Diggle CP, et al. Both isoforms of ketohexokinase are dispensable for normal growth and development. Physiol. Genomics. 2010;42A:235–243. doi: 10.1152/physiolgenomics.00128.2010. [DOI] [PubMed] [Google Scholar]
- 22.Gao Y, Trivedi S, Ferris RL, Koide K. Regulation of HPV16 E6 and MCL1 by SF3B1 inhibitor in head and neck cancer cells. Sci. Rep. 2014;4:6098. doi: 10.1038/srep06098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiota M, Galassetti P, Monohan M, Neal DW, Cherrington AD. Small amounts of fructose markedly augment net hepatic glucose uptake in the conscious dog. Diabetes. 1998;47:867–873. doi: 10.2337/diabetes.47.6.867. [DOI] [PubMed] [Google Scholar]
- 26.Luo W, et al. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falcaõ-Pires I, et al. Diabetes mellitus worsens diastolicleft ventricular dysfunction in aortic stenosis through altered myocardial structure and cardiomyocyte stiffness. Circulation. 2011;124:1151–1159. doi: 10.1161/CIRCULATIONAHA.111.025270. [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki T, Akanuma H, Yamanouchi T. Increased fructose concentrations in blood and urine in patients with diabetes. Diabetes Care. 2002;25:353–357. doi: 10.2337/diacare.25.2.353. [DOI] [PubMed] [Google Scholar]
- 29.van Heerebeek L, et al. Myocardial structure and function differ in systolic and diastolic heart failure. Circulation. 2006;113:1966–1973. doi: 10.1161/CIRCULATIONAHA.105.587519. [DOI] [PubMed] [Google Scholar]
- 30.Kassiri Z, et al. Combination of tumor necrosis factor-α ablation and matrix metalloproteinase inhibition prevents heart failure after pressure overload in tissue inhibitor of metalloproteinase-3 knock-out mice. Circ. Res. 2005;97:380–390. doi: 10.1161/01.RES.0000178789.16929.cf. [DOI] [PubMed] [Google Scholar]
- 31.Nemir M, et al. The Notch pathway controls fibrotic and regenerative repair in the adult heart. Eur. Heart J. 2014;21:2174–2185. doi: 10.1093/eurheartj/ehs269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haefliger JA, et al. Connexin43-dependent mechanism modulates renin secretion and hypertension. J. Clin. Invest. 2006;116:405–413. doi: 10.1172/JCI23327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuroda H, Kutner RH, Bazan NG, Reiser J. Simplified lentivirus vector production in protein-free media using polyethylenimine-mediated transfection. J. Virol. Methods. 2009;157:113–121. doi: 10.1016/j.jviromet.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 34.Ventura A, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl Acad. Sci. USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl Acad. Sci. USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Troilo A, et al. HIF1α deubiquitination by USP8 is essential for ciliogenesis in normoxia. EMBO Rep. 2014;15:77–85. doi: 10.1002/embr.201337688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casonato A, et al. A new L1446P mutation is responsible for impaired von Willebrand factor synthesis, structure, and function. J. Lab. Clin. Med. 2004;144:254–259. doi: 10.1016/j.lab.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Krishnan J, et al. Dietary obesity-associated Hif1 activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 2012;26:259–270. doi: 10.1101/gad.180406.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.König J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature Struct. Mol. Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirschy A, Schatzmann F, Ehlers E, Perriard JC. Establishment of cardiac cytoarchitecture in the developing mouse heart. Dev. Biol. 2006;289:430–441. doi: 10.1016/j.ydbio.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 41.Ishimoto T, et al. Opposing effects of fructokinase C and A isoforms on fructose-induced metabolic syndrome in mice. Proc. Natl Acad. Sci. USA. 2012;109:4320–4325. doi: 10.1073/pnas.1119908109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buescher JM, Moco S, Sauer U, Zamboni N. Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal. Chem. 2010;82:4403–4412. doi: 10.1021/ac100101d. [DOI] [PubMed] [Google Scholar]
- 43.Fuhrer T, Heer D, Begemann B, Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 2011;83:7074–7080. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- 44.Fukuzawa J, et al. Cardiotrophin-1 increases angiotensinogen mRNA in rat cardiac myocytes through STAT3: an autocrine loop for hypertrophy. Hypertension. 2000;35:1191–1196. doi: 10.1161/01.hyp.35.6.1191. [DOI] [PubMed] [Google Scholar]
- 45.Son N-H, et al. Cardiomyocyte aldose reductase causes heart failure and impairs recovery from ischemia. PLoS ONE. 2012;7:e46549. doi: 10.1371/journal.pone.0046549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radin NS. Extraction of tissue lipids with a solvent of low toxicity. Methods Enzymol. 1981;72:5–7. doi: 10.1016/s0076-6879(81)72003-2. [DOI] [PubMed] [Google Scholar]
- 47.Koopman R, Schaart G, Hesselink MKC. Optimisation of oil red O staining permits combination with immunofluorescence and automated quantification of lipids. Histochem. Cell Biol. 2001;116:63–68. doi: 10.1007/s004180100297. [DOI] [PubMed] [Google Scholar]
- 48.Kim K-Y, et al. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J. Clin. Invest. 2011;121:3701–3712. doi: 10.1172/JCI44736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.