Abstract
The sea cucumber Apostichopus japonicus is exploited as a commercial species owing to their high nutritive and medicinal value. Recent high summer temperatures have caused high mortality rates in A. japonicus. In this study, we applied the isobaric tag for relative and absolute quantitation (iTRAQ) technique to investigate the global protein expression profile under an acute short-term (48 h) heat stress. In total, 3432 proteins were identified, and 127 proteins showed significant heat stress responses, with 61 upregulated proteins and 66 downregulated proteins. Our results suggest that heat stress influenced the expression of proteins involved in various biological processes, such as tissue protection and detoxification, lipid and amino acid metabolism, energy production and usage, transcription and translation, cell apoptosis, and cell proliferation. These findings provide a better understanding about the response and thermo-tolerance mechanisms of A. japonicus under heat stress.
Keywords: Apostichopus japonicus, heat stress, iTRAQ, proteomics analysis
1. Introduction
The effects of global warming include rising mean annual temperatures and dramatic increase in the frequency and amplitude of severe temperature events [1]. These fluctuations constitute a major threat to aquatic organisms, as they are naturally exposed to changing water temperature. The sea cucumber Apostichopus japonicus, is an echinoderm distributed along the coast of northern China, southeastern Russia, Japan, the Republic of Korea, and the Democratic People’s Republic of Korea [2]. A. japonicus has been exploited as a commercial species owing to their high nutritive and medicinal value. Temperature is the pivotal environmental factor affecting the growth and physiology of A. japonicus [3]. Recent high summer temperatures have caused high mortality rates in cultured A. japonicus. Therefore, a better understanding of the mechanisms involved in the A. japonicus heat shock response would be significant and would lay the theoretical foundation for breeding traits for thermo-tolerance. Though specific heat response genes, such as genes from the heat shock protein (HSP) family, have been characterized, a lack of transcriptome and proteome data severely hampers revealing global gene changes and the key pathways that are active in heat stressed A. japonicus [4,5,6].
Proteomic approaches have been used to identify stress-responsive genes and proteins regulated by high temperatures. Two-dimensional electrophoresis (2DE) is the most frequently utilized approach for a proteomic analysis. However, not all proteins are amenable to gels, and proteins in low abundance are hard to be characterized in 2DE approach [7]. Besides, the quantification accuracy and ability of 2DE to identify proteins may be compromised by co-migration or partial co-migration of proteins [8]. A new technique called iTRAQ (isobaric tag for relative and absolute quantitation) has become popular in proteomic analysis in recent years, which provides more reliable quantitative measurements and comparisons among samples [9]. Additionally, the iTRAQ approach has largely improved proteomic analyses throughput and has been used in pathway studies.
In the current study, we applied the iTRAQ technique to assess the proteomic changes in A. japonicus intestinal tissues after heat shock. Our results suggest that heat stress influenced the expression of proteins involved in diverse biological processes, such as tissue protection and detoxification, lipid and amino acid metabolism, energy production and usage, transcription and translation, cell apoptosis, and cell proliferation. These findings provide a better understanding of the response and thermo-tolerance mechanisms in A. japonicus under heat stress.
2. Results
2.1. Overview of the Proteomics Data
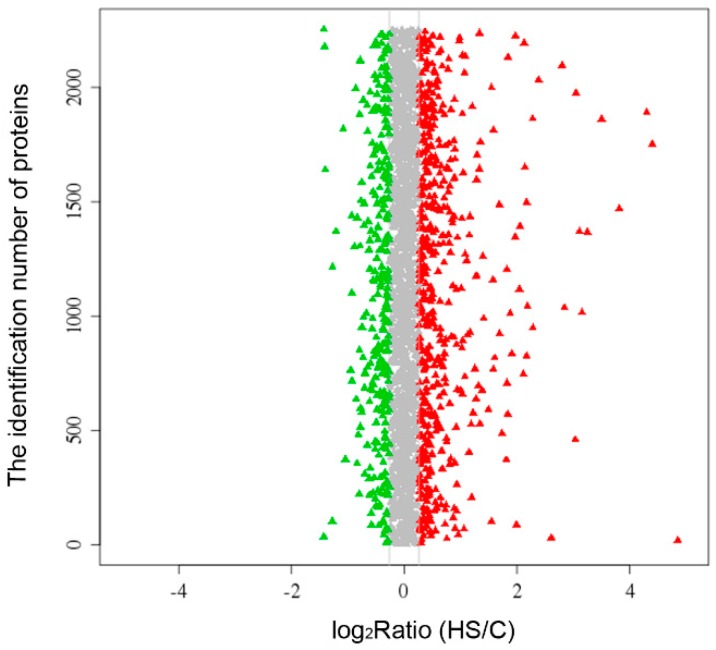
The proteomics data have been deposited to the ProteomeXchange via the PRIDE (Database ID: PXD002660) [10]. Totally 272,754 spectra were obtained, of which 38,588 unique spectra were detected (Table 1). A total 3423 proteins were identified at a global false discovery rate of 1% (Table S1). The global expression changes of these proteins under heat stress were shown in Figure 1. Finally, 127 proteins showed significant heat stress responses, with 61 upregulated proteins (Table 2) and 66 downregulated proteins (Table 3).
Table 1.
Overview of the proteomics sequencing results.
| Group Name | Number |
|---|---|
| Total spectra | 272,754 |
| Spectra | 41,330 |
| Unique spectra | 38,588 |
| Peptide | 10,908 |
| Unique peptide | 10,486 |
| Protein | 3432 |
| Upregulated protein | 61 |
| Downregulated protein | 66 |
Figure 1.
The change level of global proteins in the intestine in the heat shock group (HS) compared with the control group (C). Only the proteins with log2Ratio (HS/C) >0.26 or <−0.26 were colored (fold changes >1.20 as red and <0.83 as green).
Table 2.
Sixty-one upregulated proteins under heat stress in the intestine of the sea cucumber A. japonicas.
| Accession Number | Protein Description | HS vs. Control | |
|---|---|---|---|
| Mean | SD | ||
| HSPs and Related Proteins | |||
| Unigene28963 | heat shock protein 90 | 6.10 | 4.25 |
| CL6821.Contig1 | heat shock protein 70 | 1.56 | 0.62 |
| CL5625.Contig2 | heat shock protein 110 | 1.41 | 0.22 |
| Unigene15437 | heat shock protein 10 | 1.20 | 0.01 |
| CL12434.Contig1 | heat repeat-containing protein 7A | 1.36 | 0.19 |
| Detoxification and Tissue Protection | |||
| Unigene61290 | glutathione S-transferase | 1.36 | 0.07 |
| CL6008.Contig2 | glutathione S-transferase α-4-like, partial | 2.23 | 0.72 |
| Unigene25399 | sigma class glutathione S-transferase 2 | 1.29 | 0.26 |
| CL7884.Contig2 | phospholipid hydroperoxide glutathione peroxidase | 1.45 | 0.16 |
| Unigene29285 | prostaglandin D2 synthase, hematopoietic-like | 1.53 | 0.35 |
| Unigene25766 | cytochrome P450 4V2-like | 2.43 | 1.45 |
| Cell Apoptosis and Proliferation | |||
| Unigene29013 | apoptosis-inducing factor 1, mitochondrial-like, partial | 1.28 | 0.20 |
| CL4411.Contig2 | prohibitin-like | 1.19 | 0.12 |
| CL2387.Contig1 | autocrine proliferation repressor protein A-like | 3.57 | 3.08 |
| Unigene33274 | suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) | 1.27 | 0.09 |
| CL10790.Contig2 | erlin-1 | 1.29 | 0.04 |
| Unigene35102 | mesoderm-specific transcript protein (MEST) | 2.61 | 2.19 |
| Lipid Transport and Metabolism | |||
| Unigene64084 | long-chain specific acyl-CoA dehydrogenase | 1.41 | 0.24 |
| Unigene20467 | long-chain specific acyl-CoA dehydrogenase, mitochondrial-like | 2.22 | 0.72 |
| Unigene22338 | short chain dehydrogenase/reductase family 16C, member 5-like | 1.74 | 0.58 |
| Unigene4389 | 17-β-hydroxysteroid dehydrogenase type 4 | 1.94 | 1.11 |
| Unigene5795 | 11-β-hydroxysteroid dehydrogenase | 2.54 | 1.24 |
| Unigene6420 | enoyl-CoA Hydratase family member-like | 1.62 | 0.33 |
| CL12084.Contig1 | enoyl-CoA hydratase, mitochondrial-like | 1.30 | 0.16 |
| CL759.Contig2 | hydroxyacyl-Coenzyme A dehydrogenase | 1.34 | 0.31 |
| Unigene19362 | epidermal retinol dehydrogenase 2-like | 1.97 | 0.83 |
| Unigene15259 | carnitine O-palmitoyltransferase 2, mitochondrial-like | 1.21 | 0.06 |
| Unigene11008 | non-specific lipid-transfer protein-like | 2.99 | 1.98 |
| CL4289.Contig1 | nuclear progesterone receptor | 1.46 | 0.23 |
| CL6901.Contig2 | 2′-deoxynucleoside 5′-phosphate N-hydrolase 1 | 1.43 | 0.27 |
| CL8136.Contig1 | acyl-CoA-binding protein like, ACBP2 | 2.33 | 1.46 |
| Unigene18754 | oxysterol-binding protein-related protein 9 | 1.20 | 0.18 |
| Carbohydrate Transport and Metabolism | |||
| Unigene2131 | lactase | 1.48 | 0.10 |
| Amino Acid Transport and Metabolism | |||
| CL4095.Contig2 | sphingosine-1-phosphate lyase 1 | 1.33 | 0.27 |
| Unigene23212 | branched-chain-amino-acid aminotransferase-like protein 1 | 1.39 | 0.32 |
| Energy Production and Conversion | |||
| CL10773.Contig1 | isocitrate dehydrogenase | 1.68 | 0.19 |
| Unigene18857 | electron transfer flavoprotein subunit α, mitochondrial-like | 1.27 | 0.25 |
| CL6007.Contig1 | aldehyde dehydrogenase, dimeric NADP-preferring isoform | 1.37 | 0.22 |
| Unigene22955 | α-methylacyl-CoA racemase-like | 1.65 | 0.66 |
| Unigene175 | d-glucosyl-N-acylsphingosine glucohydrolase | 1.85 | 0.31 |
| Unigene22578 | α-galactosidase | 1.91 | 1.26 |
| Unigene29260 | ATPase family AAA domain-containing protein 1 | 1.35 | 0.05 |
| CL5389.Contig1 | ATPase inhibitor, mitochondrial-like | 1.31 | 0.21 |
| Protein Synthesis | |||
| CL9215.Contig1 | aspartyl-tRNA synthetase | 1.17 | 0.11 |
| CL7807.Contig3 | RNA-binding motif protein, X chromosome | 1.25 | 0.19 |
| Unigene8195 | elongation factor Tu, mitochondrial-like | 1.34 | 0.20 |
| Others/Uncharacterized | |||
| Unigene49395 | toposome | 1.83 | 0.81 |
| Unigene28479 | natterin-3-like | 2.63 | 1.01 |
| CL6732.Contig2 | calpain-5 isoform 2 | 1.38 | 0.37 |
| Unigene22143 | phospholipase C delta isoform | 2.55 | 0.58 |
| CL1115.Contig1 | endophilin-B1-like isoform 1 | 1.32 | 0.19 |
| Unigene322 | myosin VIb-like | 1.32 | 0.24 |
| CL7807.Contig3 | atlastin-2 | 1.33 | 0.23 |
| CL9074.Contig2 | cysteine rich protein 1 | 1.59 | 0.18 |
| Unigene11767 | suppressor of G2 allele of SKP1 homolog | 1.53 | 0.14 |
| CL8638.Contig1 | development-specific protein LVN1.2 | 1.71 | 0.62 |
| Unigene22386 | uncharacterized | 1.39 | 0.35 |
| Unigene1947 | uncharacterized | 3.76 | 1.54 |
| Unigene5634 | uncharacterized | 1.66 | 0.38 |
| Unigene16247 | uncharacterized | 2.11 | 1.58 |
| Unigene62712 | uncharacterized | 11.32 | 9.13 |
Table 3.
Sixty-six downregulated proteins under heat stress in the intestine of the sea cucumber A. japonicas.
| Accession Number | Protein Description | HS vs. Control | |
|---|---|---|---|
| Mean | SD | ||
| Cytoskeletal Proteins | |||
| Unigene32477 | twitchin-like | 0.67 | 0.21 |
| Unigene32260 | laminin subunit α-like | 0.68 | 0.13 |
| CL221.Contig4 | α-actinin-like | 0.76 | 0.10 |
| Unigene3881 | galectin-9-like | 0.56 | 0.29 |
| Unigene27394 | fibrillin-1-like | 0.52 | 0.17 |
| CL5005.Contig4 | cohesin subunit SA-1-like | 0.69 | 0.25 |
| Unigene9716 | titin isoform 3 | 0.72 | 0.17 |
| CL3832.Contig7 | filamin-C isoform 1 | 0.83 | 0.01 |
| Unigene30625 | muscle M-line assembly protein unc-89-like | 0.71 | 0.10 |
| Transcription and Translation | |||
| Unigene29879 | 60S ribosomal protein L8-like | 0.79 | 0.11 |
| Unigene26472 | 60S ribosomal protein L6, partial | 0.82 | 0.15 |
| Unigene8941 | ribosomal protein L4, partial | 0.82 | 0.09 |
| CL1672.Contig2 | 60S ribosomal protein L10-like | 0.73 | 0.14 |
| CL4437.Contig3 | splicing factor, proline- and glutamine-rich | 0.62 | 0.05 |
| CL5572.Contig1 | THO complex subunit 4 | 0.64 | 0.25 |
| Unigene22920 | small nuclear ribonucleoprotein-associated proteins B and B′ | 0.81 | 0.12 |
| Unigene15894 | malectin | 0.75 | 0.08 |
| DNA Replication and Repair | |||
| CL1035.Contig9 | histone H3.3 | 0.66 | 0.29 |
| Unigene5846 | histone H1-β, late embryonic | 0.61 | 0.05 |
| CL5357.Contig1 | legumain-like | 0.67 | 0.20 |
| CL64.Contig2 | poly(ADP-ribose) polymerase pme-5-like | 0.77 | 0.08 |
| Unigene9968 | ATP-binding cassette, sub-family C, member 9-like | 0.69 | 0.01 |
| Amino Acid Transport and Mechanism | |||
| CL4631.Contig1 | choline dehydrogenase, mitochondrial-like | 0.65 | 0.17 |
| Unigene11760 | branched-chain-amino-acid aminotransferase, cytosolic | 0.71 | 0.08 |
| CL3226.Contig1 | tyrosine aminotransferase-like | 0.66 | 0.28 |
| Unigene25781 | aminopeptidase N-like | 0.72 | 0.13 |
| CL12737.Contig1 | glutamyl aminopeptidase | 0.79 | 0.16 |
| CL2682.Contig1 | cytosolic serine hydroxymethyltransferase | 0.73 | 0.08 |
| CL9582.Contig2 | xaa-Pro aminopeptidase 1 isoform X3 | 0.76 | 0.09 |
| Unigene36365 | betaine homocysteine S-methyltransferase 1 | 0.58 | 0.22 |
| Unigene3911 | betaine homocysteine S-methyltransferase 1-like | 0.38 | 0.10 |
| Lipid Transport and Mechanism | |||
| Unigene27722 | peroxisomal bifunctional enzyme-like | 0.78 | 0.14 |
| Unigene10407 | peroxisomal bifunctional enzyme | 0.81 | 0.07 |
| CL8765.Contig2 | dihydropteridine reductase | 0.74 | 0.09 |
| CL1598.Contig1 | γ-butyrobetaine dioxygenase-like | 0.60 | 0.22 |
| Carbohydrate Transport and Metabolism | |||
| Unigene27857 | α-mannosidase 2C1-like | 0.47 | 0.24 |
| Unigene18547 | pyruvate carboxylase, mitochondrial | 0.73 | 0.15 |
| Hormonal and Nerve Regulation | |||
| Unigene25501 | thyroid hormone-induced protein B-like | 0.41 | 0.28 |
| CL9528.Contig2 | proactivator polypeptide | 0.72 | 0.23 |
| Unigene19435 | angiotensin-converting enzyme | 0.38 | 0.18 |
| CL7652.Contig1 | potassium channel tetramerization domain containing 6-like | 0.37 | 0.29 |
| CL5238.Contig2 | glutamate receptor 1-like | 0.68 | 0.08 |
| Others/Uncharacterized | |||
| Unigene1888b1 | sterigmatocystin biosynthesis dehydrogenase stcV | 0.65 | 0.26 |
| CL5732.Contig1 | N-acetylated-α-linked acidic dipeptidase 2-like isoform 1 | 0.58 | 0.11 |
| CL9717.Contig2 | peroxiredoxin-4-like | 0.53 | 0.07 |
| Unigene25753 | homogentisate 1,2-dioxygenase | 0.56 | 0.32 |
| CL2303.Contig2 | cathepsin L | 0.70 | 0.26 |
| Unigene8231 | α-parvin-like | 0.77 | 0.11 |
| CL7154.Contig1 | sorcin | 0.82 | 0.10 |
| Unigene32921 | thiopurine S-methyltransferase isoform X2 | 0.75 | 0.07 |
| CL2660.Contig13 | phosphatidylinositol-binding clathrin assembly protein unc-11-like isoform 6 | 0.80 | 0.09 |
| Unigene15838 | cytochrome P450 2N2 | 0.70 | 0.10 |
| CL2540.Contig2 | cytochrome P450 2U1-like | 0.58 | 0.05 |
| Unigene23131 | oocyst wall protein 4 precursor | 0.58 | 0.10 |
| CL4607.Contig1 | uterine-ovary specific-44 protein | 0.79 | 0.14 |
| CL10965.Contig2 | MAM and LDL-receptor class A domain-containing protein 2-like | 0.73 | 0.13 |
| Unigene11852 | uncharacterized | 0.74 | 0.17 |
| Unigene8201 | uncharacterized | 0.62 | 0.21 |
| Unigene63234 | uncharacterized | 0.55 | 0.34 |
| Unigene11761 | uncharacterized | 0.77 | 0.08 |
| CL11132.Contig3 | uncharacterized | 0.59 | 0.23 |
| Unigene23211 | uncharacterized | 0.77 | 0.20 |
| CL12015.Contig1 | uncharacterized | 0.58 | 0.19 |
| CL709.Contig5 | uncharacterized | 0.37 | 0.27 |
| Unigene8605 | uncharacterized | 0.78 | 0.03 |
| CL3869.Contig4 | uncharacterized | 0.58 | 0.12 |
2.2. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analyses
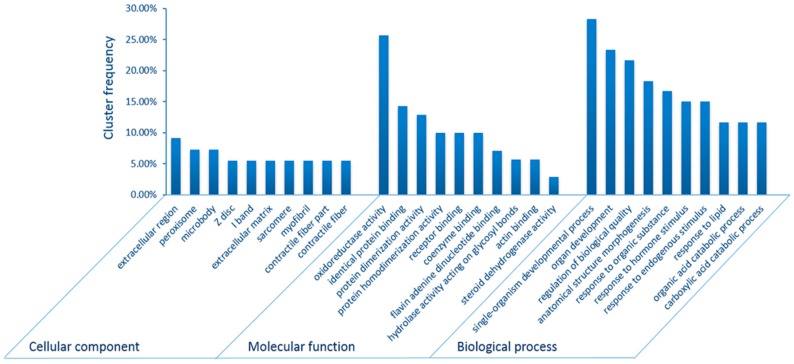
A GO analysis was performed to evaluate the functions of the differentially expressed proteins. Totals of 15, 12, and 59 categories were enriched in cellular component (CC), molecular function (MF) and biological process (BP) categories, respectively (Table S2). Fiber components (sarcomere, myofibril, contractile fiber part, and contractile fiber), binding functions (identical protein binding, receptor binding, coenzyme binding, and flavin adenine dinucleotide binding, and actin binding) and response processes (response to organic substance, response to hormone stimulus, response to endogenous stimulus, and response to lipid) were of the top 10 enriched GO-terms by cluster frequency in CC, MF and BP, respectively (Figure 2).
Figure 2.
Enriched Gene Ontology (GO) analysis of differentially expressed proteins in the intestine under heat stress. The top 10 enriched GO-terms by cluster frequency in “Cellular component”, “Molecular function” and “Biological process” were shown, respectively.
A KEGG pathway enrichment analysis revealed seven upregulated pathways, of which xenobiotics metabolism and fatty acid related metabolisms were included (Table 4). Systemic lupus erythematosus and the renin-angiotensin system were identified as downregulated pathways.
Table 4.
Enriched pathways of different expressed proteins (heat stress (HS) vs. Control).
| Pathway ID | Pathway Term | All Proteins with Pathway Annotation (2664) | Differential Proteins with Pathway Annotation (100) | p-Value |
|---|---|---|---|---|
| Upregulated Pathways | ||||
| ko00480 | Glutathione metabolism | 30 (1.13%) | 6 (6%) | 1.37 × 10−3 |
| ko00980 | Metabolism of xenobiotics by cytochrome P450 | 46 (1.73%) | 7 (7%) | 1.51 × 10−3 |
| ko00071 | Fatty acid metabolism | 60 (2.25%) | 8 (8%) | 2.31 × 10−3 |
| ko03320 | Peroxisome proliferator-activated receptors (PPAR) signaling pathway | 64 (2.4%) | 8 (8%) | 4.35 × 10−3 |
| ko00120 | Primary bile acid biosynthesis | 56 (2.1%) | 7 (7%) | 8.82 × 10−3 |
| ko00982 | Drug metabolism-cytochrome P450 | 51 (1.91%) | 6 (6%) | 1.10 × 10−2 |
| ko05215 | Prostate cancer | 37 (1.39%) | 5 (5%) | 1.13 × 10−2 |
| Downregulated Pathways | ||||
| ko05322 | Systemic lupus erythematosus | 23 (0.86%) | 4 (4%) | 9.55 × 10−3 |
| ko04614 | Renin-angiotensin system | 51 (1.91%) | 6 (6%) | 1.10 × 10−2 |
3. Discussion
The sea cucumber A. japonicus in the northern China experienced the highest temperature between 26 and 30 °C in the field [11]. Besides, A. japonicus enters a state of aestivation when the ambient temperature is maintained at 26 °C [3]. Previous reports showed that catalase (CAT) and superoxide dimutase (SOD) activities and HSPs levels varied significantly at 26 °C, indicating that this temperature is beyond the normal temperature limit for A. japonicus [3,4,5]. Therefore, we investigate the global protein expression profile under 26 °C heat stress.
3.1. Tissue Protection and Detoxification
HSP families play crucial roles protecting organisms against stress by re-establishing normal protein conformation and cellular homeostasis [12]. In our study, heat shock protein 90 (HSP90), HSP70, HSP100, and HSP10 were upregulated 6.10-, 1.56-, 1.41-, and 1.20-fold, respectively (Table 2). The protein family HSP90 helps in the processes of protein folding, degradation and transport, and is involved in cell-signal and cell-cycle control [13,14]. Our previous A. japonicus study showed that HSP90 also responds to HS at the mRNA level [5]. HSP70 helps prevent protein aggregation, assists in re-folding of abnormal proteins, and is essential for protein import and translocation processes [15,16]. HSP70 expression increased under heat stress in this study, which agreed with our previous western blot research of HSP70 [17]. HSP100 expression was also upregulated under heat stress. It is now clear that HSP100 plays a major role in thermo-tolerance, particularly in plants [18]. Recent HSP100 studies have focused on its cooperation with HSP70 during protein disaggregation [19]. HSP10 participates in various processes with HSP60, including the stress response and tumorigenesis [20,21]. In our study, HSP10 was more abundantly expressed under heat stress, which also agreed with our previous HSP10 mRNA study [4]. Taken together, our proteomics data show that four HSPs responded significantly to heat stress, indicating that these HSPs played crucial roles in alleviating heat stress in the sea cucumber A. japonicus.
Glutathione (GSH) is involved in many biological processes either as a co-factor of enzymatic reactions or as the major thiol-disulfide redox buffer [22]. Furthermore, GSH and GSH-associated metabolism provide important defense from many forms of stress [23]. In our study, phospholipid hydroperoxide peroxidase (GPx4) and three glutathione transferases (GSTs) were upregulated after heat stress in A. japonicus (Table 2). GPx4, a 20–22 kDa monomer, reduces hydroperoxides of complex lipids by transferring GSH to glutathione disulfide [22]. This process is crucial for scavenging or reducing excess quantities of reactive oxygen species (ROS), thereby maintaining cell redox homeostasis [23]. GSTs are essential enzymes in GSH metabolism, as GSH forms conjugates with a variety of electrophilic compounds, including various xenobiotic compounds, through the actions of GSTs [24]. The GSH conjugates are then exported out of the cell, which is an important component of detoxification [23]. Therefore, upregulation of GSH metabolic enzymes under heat stress is widely regarded as an essential way that cells protect against toxic damage [22,25,26].
3.2. Lipid, Amino Acid and Carbohydrate Metabolism
Fifteen proteins involved in lipid transport and mechanisms were upregulated under heat stress in A. japonicus, such as long-chain specific acyl-CoA dehydrogenases (ACADs) and enoyl-CoA hydratases (ECHs). ACADs are a class of enzymes that function to catalyze the initial step of fatty acid β-oxidation in the mitochondria [27]. Long-chain specific ACADs catalyze the breaking of long chain fatty acids into acetyl-CoA molecules. Two long-chain specific ACADs were highly expressed under heat stress in A. japonicus, reflecting the increasing demand for fatty acid metabolism. ECHs catalyze the second step of β-oxidation to breakdown fatty acids to produce acetyl-CoA and energy in the form of NADH [28]. These enzymes are highly efficient, allowing cells to metabolize fatty acids into energy very quickly. In our study, 17-β-ECH type 4 and 11-β-ECH were upregulated in the intestinal tissues of A. japonicus exposed to heat stress, suggesting a shift to lipid metabolism during energy production. These findings correlate well with previous reports suggesting that upregulation of fatty acid metabolism is an important energy budget strategy in a disadvantageous environment [26,29,30,31].
Nine proteins with roles in amino acid metabolism were less abundant in the HS group (Table 3). Notably, two types of betaine homocysteine S-methyltransferase (BHMT), BHMT1 and BHMT1-like, were downregulated 0.58- and 0.38-fold, respectively. BHMTs use betaine to catalyze the conversion of homocysteine (Hcy) to methionine (Met), which helps regulate Hcy levels and Met biosynthesis [32,33]. In our study, the expression of BHMTs decreased significantly under heat stress, which would result in changes in the concentrations of many metabolites and enzymes activities involved in Met, Hcy, and one-carbon metabolism [34]. Together with the reduction in other enzymes involved with amino acid metabolism, we suggest that heat stress may disrupt amino acid homeostasis. Hence, more attention should be paid to the nutrient requirements and amino acid deficiency diseases in sea cucumbers under stress.
In contrast, fewer proteins of carbohydrate transport and metabolism were involved under stress, as only one protein (lactase) was upregulated and two (α-mannosidase 2C1-like and pyruvate carboxylase) were downregulated. Expression of the key enzymes in the glycolytic pathway did not change, such as 6-phosphofructo-2-kinase and pyruvate kinase, indicating that short-term heat stress has no significant influence on this process [26] (Table S1). Pyruvate carboxylase, which synthesizes phosphoenolpyruvate from pyruvate during gluconeogenesis, was downregulated [35]. Thus, gluconeogenesis was likely depressed under heat stress.
3.3. Energy Production and Usage
Eight proteins involved in energy production and conversion were upregulated. Electron transfer flavoprotein (ETF) is located on the matrix face of the inner mitochondrial membrane and is a specific electron acceptor [36]. This protein is an important part of the electron transport chain, as it creates an electrochemical proton gradient that drives ATP synthesis. ETF was more abundantly expressed in A. japonicus under heat stress, suggesting that regulating HS demands additional energy [25]. However, the electron transport chain is the major site of ROS production, which may be the reason why high temperature increases the ROS levels in the cell [37,38].
Isocitrate dehydrogenase, a critical tricarboxylic acid (TCA) cycle enzyme, catalyzes the oxidative decarboxylation of isocitrate to produce α-ketoglutarate, using NAD+ or NADP+ as a co-factor [39]. Upregulation of isocitrate dehydrogenase accelerates the TCA cycle, suggesting an urgent need for energy. Additionally, isocitrate dehydrogenase is involved in controlling the mitochondrial redox balance and cellular defense against ROS, and overexpression of this enzyme results in protection from ROS-induced damage in mouse cells [40]. Isocitrate dehydrogenase increased in abundance in blue mussel Mytilus trossulus under heat stress while decreased in sea urchin Strongylocentrotus purpuratus exposed to stressful ultraviolet radiation [41,42]. Hence species may have different approaches to sense and deal with ROS [42].
These findings suggest that high temperature induces increase in the ROS production, which functions as a signal for activating a shift in metabolic pathways to enhance ROS-scavenging in A. japonicus.
3.4. Transcription and Translation
Our proteomics data show that the majority of proteins involved in transcription and translation were downregulated, including splicing factor, THO complex subunit 4, small nuclear ribonucleoprotein-associated proteins B and B′ and 60S ribosomal proteins. Global transcription and translation decrease in response to most types of cellular stress [29,43,44]. It is estimated that up to 50% of cellular energy, depending on the organism, is consumed in the translation process [45,46]. Hence, this decrease allows for a notable cellular energy savings. Furthermore, reducing protein synthesis avoids exposing nascent polypeptides to denaturing conditions that could further intensify the cellular stress response [44].
Only a few specific proteins participating in protein synthesis were more abundantly expressed in the HS group, such as elongation factor thermo unstable (EF-Tu). Actually, the functions of EF-Tu are not limited to a translation elongation factor but include chaperoning [47]. EF-Tu is an important HS response protein in many species, and high EF-Tu expression is correlated with thermo-tolerance [48,49,50].
3.5. Cell Apoptosis and Proliferation
Cell apoptosis signal occurs under heat stress. Apoptosis-inducing factor 1, a ubiquitous mitochondrial flavoprotein that participates in the degradation phase of apoptosis, rose 1.28-fold under heat stress in A. japonicus [51]. This result indicate that apoptosis is more predominant under heat stress, which agrees with the apoptotic signals detected in our previous ultrastructural observations, such as condensed chromatin and disappearing cytoplasm [17].
The evidence for decreased cell proliferation is quite clear. For example, two key antiproliferative proteins (prohibitin and autocrine proliferation repressor protein) were upregulated 1.19- and 3.57-fold, respectively [52,53]. Moreover, two types of histone proteins (histone H1 and histone H3.3) are less abundant under heat stress. Histone proteins are responsible for regulating DNA-templating processes, including DNA replication and repair [54]. Therefore, downregulation of histone proteins reflects decreased cell proliferation under stress.
3.6. Other Processes
Many other proteins were involved in the A. japonicus heat stress response. For example, we identified nine cytoskeletal proteins with decreased expression, suggesting the induction of apoptosis and depressed cell proliferation under heat stress [55,56]. Furthermore, cytoskeletal elements are composed of sarcomeres and reducing their expression decreases muscle contraction under heat stress [57,58]. The levels of hormonal and nerve regulation change under heat stress, which influence metabolism, signal transport, and other physiological functions [59,60,61]. Additionally, many uncharacterized proteins and proteins whose roles in the HS response remain unknown were detected. These results show the complexity of the HS response.
4. Materials and Methods
4.1. Animals and Samples
A. japonicus (mean weight, 99 ± 13 g) were supplied by a commercial farm in Qingdao (Shandong, China) in April 2015. Seawater temperature of the farm was about 13 °C. The sea cucumbers were transported to our laboratory and maintained in seawater tanks (30‰ salinity, 15 °C) for 2 weeks. The sea cucumbers were fed with a formulated diet (5.04% ± 0.19% (w/w) crude protein, 0.26% ± 0.05% (w/w) fat, and 72.20% ± 0.19% (w/w) ash) during the acclimation and experimental periods, and remaining feed was removed daily.
A rapid temperature-change regime was carried out in the treatment tank, using a 2-kW heating rod. The rate of heating was about 2 °C/h. The moment when water temperature rose to 26 °C was regarded as the initial time, and water temperature maintained at 26 °C in the subsequent experiment. Intestinal tissues of A. japonicus after a 48 h exposure were sampled as the heat stress (HS) group while those from an untreated tank were sampled as the control (C) group. No sea cucumbers died during the experiment. The intestinal tissues were frozen in liquid nitrogen and stored at −80 °C.
4.2. Protein Extraction, Digestion, and iTRAQ Labeling
Three biological replicates of the frozen intestinal tissues were prepared for the iTRAQ analysis. The tissue was ground to powder in liquid nitrogen and dissolved in lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, and 40 mM Tris-HCl, pH 8.5) containing 1 mM PMSF and 2 mM EDTA. 10 mM DTT was added to the lysis buffer after 5 min. An ultrasound on ice for 15 min was carried out to mix the suspension, which was then centrifuged at 25,000× g for 20 min at 4°C. The supernatant was transferred to chilled acetone and precipitated at −20 °C overnight. The supernatant was discarded after centrifugation at 25,000× g for 30 min at 4 °C, and the precipitate was washed three times with chilled acetone for 30 min each at 4 °C. The pellets were air-dried and dissolved in lysis buffer using ultrasound. The supernatant was reduced with 10 mM DTT at 56 °C for 1 h after centrifugation at 25,000× g for 30 min at 4 °C and alkylated immediately with 55 mM iodoacetamide in the dark at room temperature for 1 h. The treated proteins were precipitated in acetone at −20 °C for 3 h. The proteins were dissolved in buffer containing 1 mM PMSF and 2 mM EDTA using ultrasound after centrifugation at 25,000× g for 20 min at 4 °C and air-drying. The proteins were recovered after centrifugation at 25,000× g for 20 min at 4 °C and quantified using the Bradford method.
The protein samples were digested with Trypsin Gold (Promega, Madison, WI, USA) at 37 °C for 16 h, and the peptides were dried by vacuum centrifugation. An isobaric tag was labeled to the control (113, 114 and 116 Da) and HS samples (118, 119 and 121 Da), following the manufacturer’s instructions for the iTRAQ 8-plex reagents (Applied Biosystems, Foster City, CA, USA).
4.3. Fractionation by Strong Cation Exchange Chromatography (SCX) and Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
The labeled samples were fractionated on a SCX column using the LC-20AB high performance liquid chromatography (HPLC) pump system (Shimadzu, Kyoto, Japan). The peptides were eluted with a gradient of buffer A (25 mM NaH2PO4 in 25% ACN, pH 2.7) and buffer B (25 mM NaH2PO4 and 1 M KCl in 25% ACN, pH 2.7). The specific fractionating procedures were as follows: 100% buffer A for the first 10 min, 5%–60% buffer B for 27 min, 60%–100% buffer B for 1 min, and 100% buffer B for 1 min. Absorbance of the eluate was measured at 214 nm, and fractions were collected every min. The eluted peptides were desalted with a Strata X C18 column (Phenomenex, Torrance, CA, USA) and vacuum-dried.
A LC-20AD nanoHPLC (Shimadzu, Kyoto, Japan) and a 10 cm eluting C18 column were used to analyze the peptide fractions. Mass spectrometry data were acquired with the Triple TOF 5600 system (AB SCIEX, Concord, ON, Canada) fitted with the Nanospray III source (AB SCIEX) and a pulled quartz tip emitter (New Objectives, Woburn, MA, USA).
4.4. Protein Identification and Quantification
The raw LC-MS/MS data were converted to MGF files using Proteome Discovery 1.2 (Thermo, Pittsburgh, PA, USA). The proteins were identified using Mascot search engine 2.3.02 (Matrix Science, London, UK) with the A. japonicus transcriptomics database containing 30,622 sequences. Proteins containing at least two unique spectra were used for the follow-up quantification analysis. The quantitative protein ratios were weighted and normalized in Mascot. We only identified proteins with p-values <0.05 and fold changes >1.20 or <0.83 as being differentially expressed [62].
4.5. GO and KEGG Pathway Enrichment Analyses
The GO and KEGG databases were used to classify and group the identified proteins [63,64]. The hypergeometric test was used to identify significantly enriched GO terms and pathways of differentially expressed proteins. A p-value <0.05 was considered as significant.
5. Conclusions
This study provides a global view of the proteins differentially expressed in the intestinal tissues of A. japonicus under heat stress using the iTRAQ technique. Heat stress influences the expression of proteins involved in various biological processes, such as tissue protection and detoxification, lipid and amino acid metabolism, energy production and usage, transcription and translation, cell apoptosis, and cell proliferation. These results reveal possible molecular events in A. japonicus under heat stress.
Acknowledgments
This research was supported by the NSFC-Shandong Joint Fund for Marine Science Research Centers (U1406403), the National Natural Science Foundation of China (41406168), and Agricultural Seed Project of Shandong Province, Ecological Security and Environmental Guarantee of Marine Ranching (XDA11020703) and Chinese National 863 Project (2012AA10A412).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/2/150/s1.
Author Contributions
Dongxue Xu, Lina Sun and Hongsheng Yang conceived and supervised the study; Dongxue Xu and Hongsheng Yang designed the experiments; Dongxue Xu and Lina Sun performed the experiment and analyzed data; Dongxue Xu wrote the manuscript; Shilin Liu and Libin Zhang made manuscript revision.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Stocker T.F., Qin D., Plattner G.-K., Tignor M., Allen S.K., Boschung J., Nauels A., Xia Y., Bex V., Midgley P.M. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press; Cambridge, UK: 2013. Climate change 2013: The physical science basis. [Google Scholar]
- 2.Liao Y. Class Holothuroidea (in Chinese) Science Press; Beijing, China: 1997. Fauna sinica: Phylum echinodermata. [Google Scholar]
- 3.Ji T., Dong Y., Dong S. Growth and physiological responses in the sea cucumber, Apostichopus japonicus selenka: Aestivation and temperature. Aquaculture. 2008;283:180–187. doi: 10.1016/j.aquaculture.2008.07.006. [DOI] [Google Scholar]
- 4.Xu D., Sun L., Liu S., Zhang L., Ru X., Zhao Y., Yang H. Molecular cloning of heat shock protein 10 (Hsp10) and 60 (Hsp60) cDNAs and their expression analysis under thermal stress in the sea cucumber Apostichopus japonicus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2014;171:49–57. doi: 10.1016/j.cbpb.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H., Yang H., Zhao H., Chen M., Wang T. The molecular characterization and expression of heat shock protein 90 (Hsp90) and 26 (Hsp26) cDNAs in sea cucumber (Apostichopus japonicus) Cell Stress Chaperones. 2011;16:481–493. doi: 10.1007/s12192-011-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang P., Lu Y., Li C., Su X., Wang Z., Jin C., Li Y., Li T. Identification of differential expressed proteins and characterization their mRNA expression in thermally stressed Apostichopus japonicus. Comp. Biochem. Physiol. D Genom. Proteom. 2013;8:194–200. doi: 10.1016/j.cbd.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Zieske L.R. A perspective on the use of iTRAQ™ reagent technology for protein complex and profiling studies. J. Exp. Bot. 2006;57:1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 8.Wu W.W., Wang G., Baek S.J., Shen R.-F. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel-or LC-MALDI TOF/TOF. J. Proteome Res. 2006;5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 9.Karp N.A., Huber W., Sadowski P.G., Charles P.D., Hester S.V., Lilley K.S. Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell. Proteom. 2010;9:1885–1897. doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vizcaíno J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J.A., Sun Z., Farrah T., Bandeira N. Proteomexchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q., Yu S., Dong Y. Parental effect of long acclimatization on thermal tolerance of juvenile sea cucumber Apostichopus japonicus. PLoS ONE. 2015;10:150. doi: 10.1371/journal.pone.0143372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsell D., Lindquist S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 13.Young J.C., Moarefi I., Hartl F.U. Hsp90: A specialized but essential protein-folding tool. J. Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter K., Buchner J. Hsp90: Chaperoning signal transduction. J. Cell. Physiol. 2001;188:281–290. doi: 10.1002/jcp.1131. [DOI] [PubMed] [Google Scholar]
- 15.Daugaard M., Rohde M., Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 16.Clerico E.M., Tilitsky J.M., Meng W., Gierasch L.M. How Hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 2015;427:1575–1588. doi: 10.1016/j.jmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu D., Sun L., Liu S., Zhang L., Yang H. Histological, ultrastructural and heat shock protein 70 (HSP70) responses to heat stress in the sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 2015;45:321–326. doi: 10.1016/j.fsi.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Queitsch C., Hong S.-W., Vierling E., Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogk A., Kummer E., Bukau B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2015;2:22. doi: 10.3389/fmolb.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calderwood S.K., Khaleque M.A., Sawyer D.B., Ciocca D.R. Heat shock proteins in cancer: Chaperones of tumorigenesis. Trends Biochem. Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Höhfeld J., Hartl F.U. Role of the chaperonin cofactor Hsp10 in protein folding and sorting in yeast mitochondria. J. Cell Biol. 1994;126:305–315. doi: 10.1083/jcb.126.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maher P. The effects of stress and aging on glutathione metabolism. Ageing Res. Rev. 2005;4:288–314. doi: 10.1016/j.arr.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Dickinson D.A., Forman H.J. Glutathione in defense and signaling: Lessons from a small thiol. Ann. N. Y. Acad. Sci. 2002;973:488–504. doi: 10.1111/j.1749-6632.2002.tb04690.x. [DOI] [PubMed] [Google Scholar]
- 24.Eaton D.L., Bammler T.K. Concise review of the glutathione S-transferases and their significance to toxicology. Toxicol. Sci. 1999;49:156–164. doi: 10.1093/toxsci/49.2.156. [DOI] [PubMed] [Google Scholar]
- 25.Wei D., Jia F.-X., Tian C.-B., Tian Y., Smagghe G., Dou W., Wang J.-J. Comparative proteomic analysis of Bactrocera dorsalis (Hendel) in response to thermal stress. J. Insect Physiol. 2015;74:16–24. doi: 10.1016/j.jinsphys.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Liu G.-T., Ma L., Duan W., Wang B.-C., Li J.-H., Xu H.-G., Yan X.-Q., Yan B.-F., Li S.-H., Wang L.-J. Differential proteomic analysis of grapevine leaves by iTRAQ reveals responses to heat stress and subsequent recovery. BMC Plant Biol. 2014;14:110–126. doi: 10.1186/1471-2229-14-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorpe C., Kim J.J.P. Flavoprotein structure and mechanism of action of the acyl-coa dehydrogenases. FASEB J. 1995;9:718–725. doi: 10.1096/fasebj.9.9.7601336. [DOI] [PubMed] [Google Scholar]
- 28.Bahnson B.J., Anderson V.E., Petsko G.A. Structural mechanism of enoyl-CoA hydratase: Three atoms from a single water are added in either an E1cb stepwise or concerted fashion. Biochemistry. 2002;41:2621–2629. doi: 10.1021/bi015844p. [DOI] [PubMed] [Google Scholar]
- 29.Heunis T., Deane S., Smit S., Dicks L.M. Proteomic profiling of the acid stress response in Lactobacillus plantarum 423. J. Proteome Res. 2014;13:4028–4039. doi: 10.1021/pr500353x. [DOI] [PubMed] [Google Scholar]
- 30.Thorne M.A.S., Burns G., Fraser K.P.P., Hillyard G., Clark M.S. Transcription profiling of acute temperature stress in the Antarctic plunderfish Harpagifer antarcticus. Mar. Genom. 2010;3:35–44. doi: 10.1016/j.margen.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Sun J., Mu H., Zhang H., Chandramouli K.H., Qian P.-Y., Wong C.K.C., Qiu J.-W. Understanding the regulation of estivation in a freshwater snail through iTRAQ-based comparative proteomics. J. Proteome Res. 2013;12:5271–5280. doi: 10.1021/pr400570a. [DOI] [PubMed] [Google Scholar]
- 32.Skiba W.E., Taylor M., Wells M., Mangum J.H., Awad W. Human hepatic methionine biosynthesis. Purification and characterization of betaine homocysteine S-methyltransferase. J. Biol. Chem. 1982;257:14944–14948. [PubMed] [Google Scholar]
- 33.Pajares M.A., Pérez-Sala D. Betaine homocysteine S-methyltransferase: Just a regulator of homocysteine metabolism? Cell. Mol. Life Sci. 2006;63:2792–2803. doi: 10.1007/s00018-006-6249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teng Y.-W., Mehedint M.G., Garrow T.A., Zeisel S.H. Deletion of betaine-homocysteine S-methyltransferase in mice perturbs choline and L-carbon metabolism, resulting in fatty liver and hepatocellular carcinomas. J. Biol. Chem. 2011;286:36258–36267. doi: 10.1074/jbc.M111.265348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bahl J.J., Matsuda M., DeFronzo R.A., Bressler R. In vitro and in vivo suppression of gluconeogenesis by inhibition of pyruvate carboxylase. Biochem. Pharmacol. 1997;53:67–74. doi: 10.1016/S0006-2952(96)00660-0. [DOI] [PubMed] [Google Scholar]
- 36.Weidenhaupt M., Rossi P., Beck C., Fischer H.M., Hennecke H. Bradyrhizobium japonicum possesses two discrete sets of electron transfer flavoprotein genes: fixA, fixB and etfS, etfL. Arch. Microbiol. 1996;165:169–178. doi: 10.1007/s002030050312. [DOI] [PubMed] [Google Scholar]
- 37.Hasanuzzaman M., Nahar K., Alam M.M., Roychowdhury R., Fujita M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013;14:9643–9684. doi: 10.3390/ijms14059643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomanek L. Proteomics to study adaptations in marine organisms to environmental stress. J. Proteom. 2014;105:92–106. doi: 10.1016/j.jprot.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Alp P.R., Newsholme E.A., Zammit V.A. Activities of citrate synthase and NAD+-linked and NADP+-linked isocitrate dehydrogenase in muscle from vertebrates and invertebrates. Biochem. J. 1976;154:689–700. doi: 10.1042/bj1540689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jo S.-H., Son M.-K., Koh H.-J., Lee S.-M., Song I.-H., Kim Y.-O., Lee Y.-S., Jeong K.-S., Kim W.B., Park J.-W. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 2001;276:16168–16176. doi: 10.1074/jbc.M010120200. [DOI] [PubMed] [Google Scholar]
- 41.Fields P.A., Zuzow M.J., Tomanek L. Proteomic responses of blue mussel (Mytilus) congeners to temperature acclimation. J. Exp. Biol. 2012;215:1106–1116. doi: 10.1242/jeb.062273. [DOI] [PubMed] [Google Scholar]
- 42.Adams N., Campanale J., Foltz K. Proteomic responses of sea urchin embryos to stressful ultraviolet radiation. Integr. Comp. Biol. 2012;52:665–680. doi: 10.1093/icb/ics058. [DOI] [PubMed] [Google Scholar]
- 43.Ji C., Wu H., Wei L., Zhao J. ITRAQ-based quantitative proteomic analyses on the gender-specific responses in mussel Mytilus galloprovincialis to tetrabromobisphenol A. Aquat. Toxicol. 2014;157:30–40. doi: 10.1016/j.aquatox.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 45.Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/S0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 46.Rudra D., Warner J.R. What better measure than ribosome synthesis? Genes Dev. 2004;18:2431–2436. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki H., Ueda T., Taguchi H., Takeuchi N. Chaperone properties of mammalian mitochondrial translation elongation factor Tu. J. Biol. Chem. 2007;282:4076–4084. doi: 10.1074/jbc.M608187200. [DOI] [PubMed] [Google Scholar]
- 48.Bhadula S.K., Elthon T.E., Habben J.E., Helentjaris T.G., Jiao S., Ristic Z. Heat-stress induced synthesis of chloroplast protein synthesis elongation factor (EF-Tu) in a heat-tolerant maize line. Planta. 2001;212:359–366. doi: 10.1007/s004250000416. [DOI] [PubMed] [Google Scholar]
- 49.Ristic Z., Wilson K., Nelsen C., Momcilovic I., Kobayashi S., Meeley R., Muszynski M., Habben J. A maize mutant with decreased capacity to accumulate chloroplast protein synthesis elongation factor (EF-Tu) displays reduced tolerance to heat stress. Plant Sci. 2004;167:1367–1374. doi: 10.1016/j.plantsci.2004.07.016. [DOI] [Google Scholar]
- 50.Buckley B.A., Gracey A.Y., Somero G.N. The cellular response to heat stress in the goby Gillichthys mirabilis: A cDNA microarray and protein-level analysis. J. Exp. Biol. 2006;209:2660–2677. doi: 10.1242/jeb.02292. [DOI] [PubMed] [Google Scholar]
- 51.Susin S.A., Lorenzo H.K., Zamzami N., Marzo I., Snow B.E., Brothers G.M., Mangion J., Jacotot E., Costantini P., Loeffler M. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 52.Ikonen E., Fiedler K., Parton R.G., Simons K. Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS Lett. 1995;358:273–277. doi: 10.1016/0014-5793(94)01444-6. [DOI] [PubMed] [Google Scholar]
- 53.Brock D.A., Gomer R.H. A secreted factor represses cell proliferation in dictyostelium. Development. 2005;132:4553–4562. doi: 10.1242/dev.02032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 55.Marushige Y., Marushige K. Alterations in focal adhesion and cytoskeletal proteins during apoptosis. Anticancer Res. 1997;18:301–307. [PubMed] [Google Scholar]
- 56.Cordeiro O.D., Silva T.S., Alves R.N., Costas B., Wulff T., Richard N., de Vareilles M., Conceição L.E., Rodrigues P.M. Changes in liver proteome expression of senegalese sole (Solea senegalensis) in response to repeated handling stress. Mar. Biotechnol. 2012;14:714–729. doi: 10.1007/s10126-012-9437-4. [DOI] [PubMed] [Google Scholar]
- 57.Gunst S.J., Zhang W. Actin cytoskeletal dynamics in smooth muscle: A new paradigm for the regulation of smooth muscle contraction. Am. J. Physiol. Cell Physiol. 2008;295:C576–C587. doi: 10.1152/ajpcell.00253.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell K.P. Three muscular dystrophies: Loss of cytoskeleton-extracellular matrix linkage. Cell. 1995;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 59.Soffer R.L. Angiotensin-converting enzyme and the regulation of vasoactive peptides. Annu. Rev. Biochem. 1976;45:73–94. doi: 10.1146/annurev.bi.45.070176.000445. [DOI] [PubMed] [Google Scholar]
- 60.Dingledine R., Borges K., Bowie D., Traynelis S.F. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–62. [PubMed] [Google Scholar]
- 61.Miller C. An overview of the potassium channel family. Genome Biol. 2000;1:1–5. doi: 10.1186/gb-2000-1-4-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Briolant S., Almeras L., Belghazi M., Boucomont-Chapeaublanc E., Wurtz N., Fontaine A., Granjeaud S., Fusaï T., Rogier C., Pradines B. Research Plasmodium falciparum proteome changes in response to doxycycline treatment. Malar. J. 2010;9:141–154. doi: 10.1186/1475-2875-9-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gene Ontology Consortium. [(accessed on 20 May 2015)]. Available online: http://www.geneontology.org/
- 64.KEGG: Kyoto Encyclopedia of Genes and Genomes. [(accessed on 20 May 2015)]. Available online: http://www.genome.jp/kegg/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.