Abstract
Ectopic pregnancy is a very dangerous complication of pregnancy, affecting 1%–2% of all reported pregnancies. Due to ethical constraints on human biopsies and the lack of suitable animal models, there has been little success in identifying functionally important genes in the pathogenesis of ectopic pregnancy. In the present study, we developed a random walk–based computational method named TM-rank to prioritize ectopic pregnancy–related genes based on text mining data and gene network information. Using a defined threshold value, we identified five top-ranked genes: VEGFA (vascular endothelial growth factor A), IL8 (interleukin 8), IL6 (interleukin 6), ESR1 (estrogen receptor 1) and EGFR (epidermal growth factor receptor). These genes are promising candidate genes that can serve as useful diagnostic biomarkers and therapeutic targets. Our approach represents a novel strategy for prioritizing disease susceptibility genes.
Keywords: ectopic pregnancy, pathogenesis, text mining, gene prioritization
1. Introduction
Ectopic pregnancy is defined as embryo implantation outside of the uterine cavity, with the most common site being the Fallopian tube [1]. Its incidence is 1%–2% of all reported pregnancies [2]. Ectopic pregnancy is a very dangerous complication of pregnancy, which accounts for 10% of all maternal deaths during the early stage of pregnancy [2]. Notably, unlike humans, the major type of ectopic pregnancy in animals is abdominal pregnancy, and tubal pregnancy is rather rare [3]. The pathogenesis of ectopic pregnancy in humans is not fully understood.
It has been suggested that both impaired embryo transport in the Fallopian tube and abnormal tubal environment may contribute to the pathogenesis of ectopic pregnancy. The major risk factors for ectopic pregnancy include tubal infection and smoking [4,5]. These two conditions initiate inflammation within the Fallopian tube, leading to embryo retention by disrupting smooth muscle contractility and ciliary beat frequency [6]. The inflammatory environment may also provide necessary pro-implantation factors with roles in the establishment of Fallopian tube receptivity for the retained embryo [7]. A number of genes have been implicated in ectopic pregnancy [8,9,10]. However, due to ethical constraints on human biopsies and the lack of suitable animal models, most studies have focused on altered tubal gene or protein expression by comparing ectopic pregnancy with normal controls; thus, these studies are largely descriptive and speculative. There has been little success in identifying functionally important genes in the pathogenesis of ectopic pregnancy.
Recently, text mining methodology has been implemented [11], making it possible to retrieve all ectopic pregnancy candidate genes from published experimental results in an automated way. Because functional validation of these candidates using experimental methods is not feasible for now, computational methods may provide a useful option for prioritization of the most likely susceptibility genes. Among all computational methods, the random walk algorithm, which incorporates topological properties of the gene network, has attracted more and more attention in recent years [12,13]. It is a variant of the PageRank algorithm used by the Google search engine to rank search results on the world-wide web [14]. In the present study, we developed a random walk–based strategy named TM-rank to prioritize ectopic pregnancy–related genes by combining text mining data and gene network information. Our approach represents a novel strategy for prioritization of disease susceptibility genes.
2. Results
2.1. Identification of Susceptibility Genes for Ectopic Pregnancy by Using Text Mining
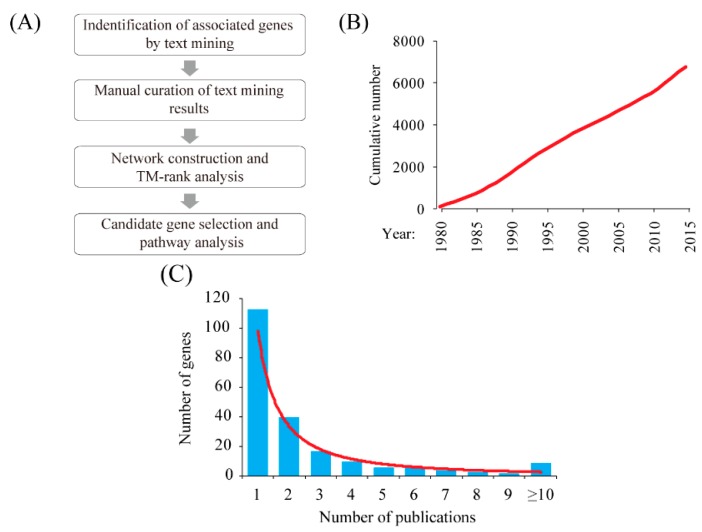
We carried out a key word search in the PubMed database and obtained 7363 publications related to ectopic pregnancy as a result (from January 1980 to October 2015). The titles and abstracts of these publications were fetched and analyzed through a computational pipeline (Figure 1A). Statistical analysis shows that the number of publications on ectopic pregnancy has been growing linearly in recent years (Figure 1B). From these publications, a total of 264 ectopic pregnancy–related genes were extracted (Figure 1C; Table S1).
Figure 1.
Systematic identification of susceptibility genes for ectopic pregnancy. (A) Overview of the experimental design; (B) Cumulative number of publications related to ectopic pregnancy by year (from 1980 to 2015); (C) Distribution of the number of publications per gene.
2.2. Construction of Gene Network
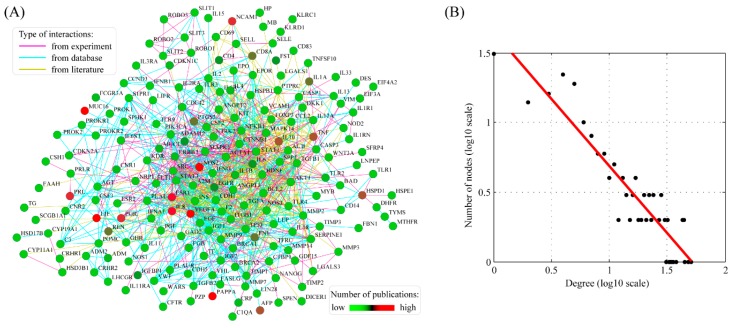
The gene network was generated by using the STRING (Search Tool for the Retrieval of INteracting Genes/proteins) database. With the combined score being set to 0.9, we obtained a network consisting of 198 nodes connected via 1007 edges (Figure 2A). Topological analysis showed that the network follows a power-law distribution (Figure 2B). In this network, we identified nine hub genes: IL6 (interleukin 6), SRC (v-src sarcoma viral oncogene homolog), STAT3 (signal transducer and activator of transcription 3), TP53 (tumor protein p53), VEGFA (vascular endothelial growth factor A), AKT1 (v-akt murine thymoma viral oncogene homolog 1), PIK3CA (phosphoinositide-3-kinase catalytic α polypeptide), NFKB1 (nuclear factor of kappa light polypeptide gene enhancer in B-cells 1), IGF1 (insulin-like growth factor 1), CTNNB1 (catenin β 1), TGFB1 (transforming growth factor β 1), EGFR (epidermal growth factor receptor), INS (insulin) and MMP9 (matrix metallopeptidase 9).
Figure 2.
The gene network underlying ectopic pregnancy susceptibility genes. (A) The structure of the gene network generated by the STRING software. The number of publications for each gene was represented in color gradient with a scale bar; (B) Degree distribution of the gene network. The degree distribution follows a power law distribution. With 95% confidence interval, the scaling exponent is estimated to be 0.9614 ± 0.1346.
2.3. Gene Prioritization by TM-Rank
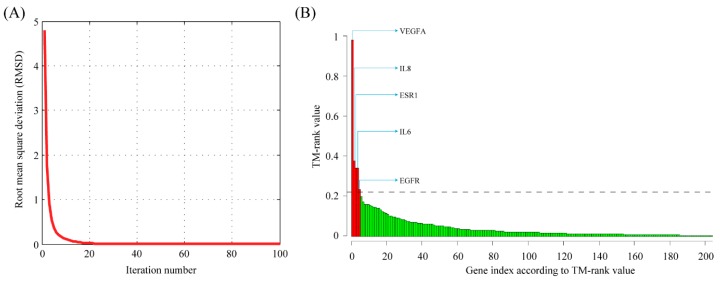
Initially, the gene rank (the importance of a gene to ectopic pregnancy) was determined by the number of publications for each gene. Considering the network underlying all ectopic pregnancy–related genes, we would like a gene to be highly ranked if it is linked to other highly ranked genes. Therefore, we developed the TM-rank algorithm, which extends the PageRank algorithm [14], for analyzing our text mining results. We demonstrated that the importance of a gene as determined by the number of publications could propagate fast through the entire network by iterative random walk (Figure 3A). Finally, genes were ranked according to TM-rank values in descending order. As a result, the top five genes exceeded a defined TM-rank cut-off (mean + 2 × SD = 0.1919). These five genes were: VEGFA (vascular endothelial growth factor A), IL8 (interleukin 8), IL6 (interleukin 6), ESR1 (estrogen receptor 1) and EGFR (epidermal growth factor receptor) (Figure 3B).
Figure 3.
Prioritization of susceptibility genes for ectopic pregnancy by the TM-rank algorithm. (A) Convergence of the TM-rank algorithm. Convergence was confirmed by observing an exponential decrease in the RMSD (root mean square deviation) values during the sequential iteration; (B) Gene ranking by the TM-rank algorithm. Genes were sorted according to rank values in descending order. Top-ranked genes, which exceeded the mean plus two standard deviations, were labeled.
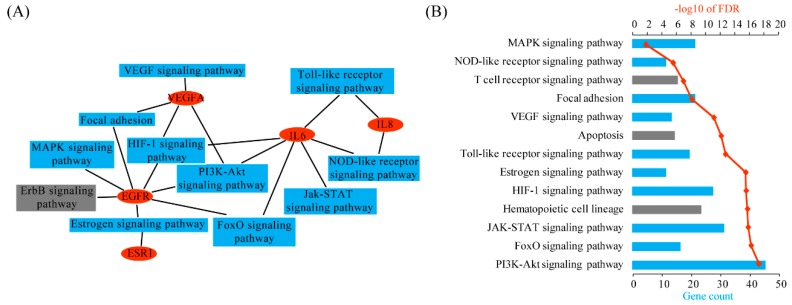
By searching the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database [15], these five genes are linked to 11 pathways (Figure 4A). EGFR is involved in seven pathways, namely the estrogen signaling pathway, focal adhesion pathway, ErbB signaling pathway, MAPK signaling pathway, PI3K-Akt signaling pathway, FoxO signaling pathway and HIF-1 signaling pathway. IL6 is associated with six pathways: the Jak-STAT signaling pathway, PI3K-Akt signaling pathway, HIF-1 signaling pathway, FoxO signaling pathway, NOD-like receptor signaling pathway and Toll-like receptor signaling pathway. Beyond the VEGF signaling pathway, VEGFA also plays a role in the PI3K-Akt signaling pathway, HIF-1 signaling pathway and focal adhesion pathway. IL8 participates in the NOD-like receptor signaling pathway and Toll-like receptor signaling pathway. ESR1 is the key component of the estrogen signaling pathway.
Figure 4.
Pathway analysis. (A) Pathway assignment for the top five genes recommended by the TM-rank algorithm. This graph was generated by using the Cytoscape software. Ellipse nodes in red represent genes and rectangle nodes in blue represent pathways; (B) Pathway enrichment analysis for all the 264 genes identified by text mining. This analysis was performed by using the DAVID tool. The significance cutoff for FDR was set at 0.01.
In addition, we performed pathway enrichment analysis on the complete list of 264 susceptibility genes by using the DAVID (Database for Annotation, Visualization and Integrated Discovery) tools. As a result, a total of 13 pathways, namely the PI3K-Akt signaling pathway, FoxO signaling pathway, Jak-STAT signaling pathway, hematopoietic cell lineage, HIF-1 signaling pathway, estrogen signaling pathway, Toll-like receptor signaling pathway, apoptosis, VEGF signaling pathway, focal adhesion, T cell receptor signaling pathway, NOD-like receptor signaling pathway, and MAPK signaling pathway, are significantly enriched (false discovery rate FDR < 0.05) (Figure 4B). Interestingly, all 11 pathways linked to top-ranked genes are overlapped with these 13 pathways enriched among all genes identified by text mining, indicating that our TM-rank algorithm is able to prioritize functionally important genes for ectopic pregnancy.
3. Discussion
Ectopic pregnancy is a very dangerous complication of pregnancy, the pathogenesis of which is not fully understood. There is a wealth of information that is hidden within published experimental results. In the present study we performed a text mining analysis of ectopic pregnancy–associated genes. A total of 264 genes were indentified from 7363 publications. Since a large number of publications were analyzed, our result may provide reasonably good coverage of the entire set of ectopic pregnancy–associated genes.
Interpretation of the text mining results is usually based on the number of publications for each gene. That is, the more publications a gene is cited in, the more important the gene is to ectopic pregnancy. The disadvantage is that genes are treated as separated entities and the interactions between genes are ignored. It has been shown that gene products or proteins usually form protein complexes to execute their functions instead of acting alone [16]. Hence, a gene is indicative of functional importance for its interacting partners, opening the possibility to integrate interaction information for gene prioritization. In the present study, a genome-wide gene network was constructed by using up-to-date interaction data available in the STRING database [17]. Using a threshold value of 0.9 for the combined score, we obtained a gene network consisting of 198 nodes connected via 1007 edges. So far, several studies have been conducted to incorporate the topology of the gene network in prioritization of disease candidate genes [18,19,20]. The main concern for these studies is that the incompleteness and noisiness of interaction data may affect the accuracy of the prioritization results. By weighting and integrating interaction data from high-throughput experiments and from the mining of databases and literature, the STRING database provides a comprehensive gene network at a reasonable false positive rate [17]. We expected that the use of the STRING database may alleviate this problem to a certain extent.
Using text mining data and gene network information as input, we developed a random walk–based algorithm named TM-rank to prioritize ectopic pregnancy–related genes. This algorithm is a variant of the PageRank algorithm used by the Google search engine to rank search results on the world-wide web [14]. Compared to previously described algorithms, our method is unique in that: (1) both text mining data and gene interaction information are used; (2) no training set of functionally confirmed genes is needed; and (3) valuable insights of genes with high susceptibility to ectopic pregnancy are recommended.
An arbitrary cutoff value for TM-rank is needed to select top-ranked genes. If an arbitrary cutoff of 0.1 is used, the number of selected genes will be 23. On the other hand, if the top 10 genes are selected, the TM-rank cutoff will be 0.15. In the present study, we used mean + 2 × SD, which is 0.1919, as the cutoff value. However, because the distribution of TM-rank is not a normal distribution (Kolmogorov-Smirnov goodness-of-fit test, p = 3.3 × 10−27), this cutoff is actually no better than any other one. Using this cutoff value, we identified a total of five top-ranked genes: VEGFA (vascular endothelial growth factor A, count = 21, degree = 44, TM-rank = 0.98909), IL8 (interleukin 8, count = 9, degree = 31, TM-rank = 0.38012), IL6 (interleukin 6, count = 5, degree = 51, TM-rank = 0.33978), ESR1 (estrogen receptor 1, count = 9, degree = 24, TM-rank = 0.33905) and EGFR (epidermal growth factor receptor, count = 5, degree = 33, TM-rank = 0.23342). In contrast, the top five genes ranked by count of publications are (six genes actually, count ≥ 10): MUC16 (mucin 16, count = 22, degree = 1, TM-rank = 0.13124), VEGFA (count = 21, degree = 44, TM-rank = 0.98909), PSG1 (pregnancy specific β1-glycoprotein 1, count = 13, degree = 0, TM-rank = 0.031113), NOS2 (nitric oxide synthase 2, count = 10, degree = 6, TM-rank = 0.083989), LIF (leukemia inhibitory factor, count = 10, degree = 4, TM-rank = 0.059718), and PAPPA (pregnancy-associated plasma protein A, count = 10, degree = 2, TM-rank = 0.034855). Of these genes, only VEGF is among the top five genes recommended by TM-rank. Therefore, our TM-rank algorithm provides a balance between the count of publication and the degree in the network.
These top five genes recommended by TM-rank can serve as useful diagnostic biomarkers and therapeutic targets. Serum VEGF is reported to be higher in ectopic pregnancy compared with normal pregnancy, indicating that VEGF may be a useful diagnostic biomarker for ectopic pregnancy [21]. Moreover, VEGF has been shown to be up-regulated at the implantation site compared to the non-implantation site of the Fallopian tube in ectopic pregnancy [22]. It is thought that elevated expression of VEGF may contribute to ectopic pregnancy by enhancing the supply of oxygen to the embryo via angiogenesis [23]. IL-6 and IL-8 are inflammatory cytokines and both of them are highly induced in the Fallopian tube upon chlamydial infection [24] as well as ectopic implantation [25]. Embryo implantation into the uterus is known to induce a local inflammatory response in a similar way as leukocytes accumulate at inflammatory sites [26]. It is likely that IL-6 and IL-8, induced by inflammation in the Fallopian tube, may provide signals to the embryo and promote tubal implantation. As the predominant estrogen receptor subtype in the Fallopian tube [27], ESR1 has been shown to serve as a crucial regulator in Fallopian tube development [28]. Exogenous estrogen treatment enhances embryo transport in the mouse Fallopian tube [29,30]. Interestingly, ESR1 protein has been shown to be absent in the Fallopian tube in human ectopic pregnancy [31], suggesting a role of ESR1 in the pathogenesis of ectopic pregnancy. EGFR is highly expressed at the ectopic implantation site [32]. It has been demonstrated that EGFR is a promising target for the treatment of ectopic pregnancy [33].
Unlike the supervised method with a positive training set, our un-supervised prioritization method is very difficult to provide validation data for. Wet experiments are often used; however, due to ethical constraints on human biopsies and the lack of suitable animal models, experimental validation is not a feasible option in this case. Alternatively, we performed pathway enrichment analysis on the complete list of 264 susceptibility genes by using the DAVID tools. A total of 13 enriched pathways were identified. In contrast, the top five genes selected by TM-rank are linked to 11 pathways. Interestingly, all 11 pathways are overlapped with the 13 enriched pathways, except for the ErbB signaling pathway. This result might provide validity of our methodology.
A limitation for text mining–based strategies is that there is no chance to discover new genes. In order to solve this problem, we enlarged the network by including new genes that are not reported to be involved in ectopic pregnancy. According to the rule of “guilty-by-association”, these new genes may be potential susceptibility genes. Finally, we made a list of 10 new genes, all of which have more than two connections with known genes (Table S2).
In conclusion, we developed a random walk–based algorithm named TM-rank to prioritize ectopic pregnancy–related genes based on text mining data and gene network information. Application of the TM-rank methodology yielded promising candidate genes that can serve as useful diagnostic biomarkers and therapeutic targets. Our study on ectopic pregnancy can act as an illustrative example for broader use in various diseases.
4. Materials and Methods
4.1. Text Mining
The PubMed database [34] was used as a source of publications for text mining. We conducted a search with the following combinations of query key words: “ectopic pregnancy” OR “tubal pregnancy” OR “eccyesis”. The search tag “(Title/Abstract)” was added after each key word. In order to make information extraction more precise, the relevant publications were retrieved in XML format with content enclosed within XML tag pairs. For each article, titles and abstract texts were fetched and transformed into the PubTator format [35] using in-house PERL scripts.
Text mining was performed by using the GNormPlus pipeline [36]. GNormPlus includes two modules: gene mention recognition and gene name normalization, respectively. Gene mentions were detected by using the CRF++ library [37]. Gene name normalization was performed by GenNorm [38]. The GNormPlus pipeline has achieved a precision of 87.1% and a recall of 86.4% [36]. To ensure accuracy, we manually checked each entry from the output of GNormPlus. A total of 1864 false positive errors out of a total of 6771 entries were identified. Thus, the observed precision was 4907/6771 = 72.47%. Finally, all the curated entries were summarized and a full gene list associated with ectopic pregnancy was compiled.
4.2. Construction of Gene Network
The STRING database v10.0 [17] was used to create gene network. The minimum combined score was set to 0.900 (highest confidence). The Cytoscape software [39] was applied for visualization and analysis of the gene network. The degree for each gene was analyzed by NetworkAnalyzer [40]. The threshold value for hub genes was defined as the mean plus two standard deviations.
4.3. The TM-Rank Algorithm
In order to prioritize susceptibility genes for ectopic pregnancy, TM-rank adopted a random walk in the gene network to infer which genes are most important. We followed a general implementation of a random walk in an iterative form:
| (1) |
where rt+1 and rt are the TM-rank vectors in which the i-th element represents the TM-rank for gene i at time steps t and t+1, respectively. The parameter d is the restart probability which is set to 0.15 in this study. The vector r° is the initial vector. Taking the number of publications into consideration, we biased the initial rank vector to the normalized number of publications for each gene. Assuming the total number of genes in the gene network is n, then the column-normalized network adjacency matrix can be written as:
| (2) |
For a given gene i, let Ni be the node set of immediate interacting neighbors of node i and ci be the number of publications for gene i. Then wij is defined as:
| (3) |
The TM-rank algorithm was implemented in MATLAB language (MathWorks, MA, USA).
Acknowledgments
This work was supported by National Natural Science Foundation of China (grant number 31271602 to Ji-Long Liu).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/2/191/s1.
Author Contributions
Ji-Long Liu designed/performed the experiments and wrote the paper; Miao Zhao helped in data analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Walker J.J. Ectopic pregnancy. Clin. Obstet. Gynecol. 2007;50:89–99. doi: 10.1097/GRF.0b013e31802f4f79. [DOI] [PubMed] [Google Scholar]
- 2.Farquhar C.M. Ectopic pregnancy. Lancet. 2005;366:583–591. doi: 10.1016/S0140-6736(05)67103-6. [DOI] [PubMed] [Google Scholar]
- 3.Corpa J.M. Ectopic pregnancy in animals and humans. Reproduction. 2006;131:631–640. doi: 10.1530/rep.1.00606. [DOI] [PubMed] [Google Scholar]
- 4.Tay J.I., Moore J., Walker J.J. Ectopic pregnancy. BMJ. 2000;320:916–919. doi: 10.1136/bmj.320.7239.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouyer J., Coste J., Shojaei T., Pouly J.L., Fernandez H., Gerbaud L., Job-Spira N. Risk factors for ectopic pregnancy: A comprehensive analysis based on a large case-control, population-based study in france. Am. J. Epidemiol. 2003;157:185–194. doi: 10.1093/aje/kwf190. [DOI] [PubMed] [Google Scholar]
- 6.Shaw J.L., Horne A.W. The paracrinology of tubal ectopic pregnancy. Mol. Cell. Endocrinol. 2012;358:216–222. doi: 10.1016/j.mce.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw J.L., Dey S.K., Critchley H.O., Horne A.W. Current knowledge of the aetiology of human tubal ectopic pregnancy. Hum. Reprod. Update. 2010;16:432–444. doi: 10.1093/humupd/dmp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji Y.F., Chen L.Y., Xu K.H., Yao J.F., Shi Y.F. Locally elevated leukemia inhibitory factor in the inflamed fallopian tube resembles that found in tubal pregnancy. Fertil. Steril. 2009;91:2308–2314. doi: 10.1016/j.fertnstert.2008.01.110. [DOI] [PubMed] [Google Scholar]
- 9.Salih S.M., Taylor H.S. Hoxa10 gene expression in human fallopian tube and ectopic pregnancy. Am. J. Obstet. Gynecol. 2004;190:1404–1406. doi: 10.1016/j.ajog.2004.01.066. [DOI] [PubMed] [Google Scholar]
- 10.Savaris R.F., da Silva L.C., Moraes Gda S., Edelweiss M.I. Expression of muc1 in tubal pregnancy. Fertil. Steril. 2008;89:1015–1017. doi: 10.1016/j.fertnstert.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 11.Rebholz-Schuhmann D., Oellrich A., Hoehndorf R. Text-mining solutions for biomedical research: Enabling integrative biology. Nat. Rev. Genet. 2012;13:829–839. doi: 10.1038/nrg3337. [DOI] [PubMed] [Google Scholar]
- 12.Chen L.C., Yeh H.Y., Yeh C.Y., Arias C.R., Soo V.W. Identifying co-targets to fight drug resistance based on a random walk model. BMC Syst. Biol. 2012;6:5. doi: 10.1186/1752-0509-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler S., Bauer S., Horn D., Robinson P.N. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brin S., Page L. The anatomy of a large-scale hypertextual web search engine. Comp. Net. ISDN Syst. 1998;30:107–117. doi: 10.1016/S0169-7552(98)00110-X. [DOI] [Google Scholar]
- 15.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. Kegg as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berggard T., Linse S., James P. Methods for the detection and analysis of protein-protein interactions. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 17.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J., Aronow B.J., Jegga A.G. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinform. 2009;10:73. doi: 10.1186/1471-2105-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo H., Dong J., Hu S., Cai X., Tang G., Dou J., Tian M., He F., Nie Y., Fan D. Biased random walk model for the prioritization of drug resistance associated proteins. Sci. Rep. 2015;5:10857. doi: 10.1038/srep10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison J.L., Breitling R., Higham D.J., Gilbert D.R. Generank: Using search engine technology for the analysis of microarray experiments. BMC Bioinform. 2005;6 doi: 10.1186/1471-2105-6-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felemban A., Sammour A., Tulandi T. Serum vascular endothelial growth factor as a possible marker for early ectopic pregnancy. Hum. Reprod. 2002;17:490–492. doi: 10.1093/humrep/17.2.490. [DOI] [PubMed] [Google Scholar]
- 22.Lam P.M., Briton-Jones C., Cheung C.K., Leung S.W., Cheung L.P., Haines C. Increased messenger RNA expression of vascular endothelial growth factor and its receptors in the implantation site of the human oviduct with ectopic gestation. Fertil. Steril. 2004;82:686–690. doi: 10.1016/j.fertnstert.2003.12.052. [DOI] [PubMed] [Google Scholar]
- 23.Daniel Y., Geva E., Lerner-Geva L., Eshed-Englender T., Gamzu R., Lessing J.B., Bar-Am A., Amit A. Levels of vascular endothelial growth factor are elevated in patients with ectopic pregnancy: Is this a novel marker? Fertil. Steril. 1999;72:1013–1017. doi: 10.1016/S0015-0282(99)00417-3. [DOI] [PubMed] [Google Scholar]
- 24.Mpiga P., Mansour S., Morisset R., Beaulieu R., Ravaoarinoro M. Sustained interleukin-6 and interleukin-8 expression following infection with chlamydia trachomatis serovar L2 in a HeLa/THP-1 cell co-culture model. Scand. J. Immunol. 2006;63:199–207. doi: 10.1111/j.1365-3083.2006.01734.x. [DOI] [PubMed] [Google Scholar]
- 25.Balasubramaniam E.S., Van Noorden S., El-Bahrawy M. The expression of interleukin (IL)-6, IL-8, and their receptors in fallopian tubes with ectopic tubal gestation. Fertil. Steril. 2012;98:898–904. doi: 10.1016/j.fertnstert.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Dominguez F., Yanez-Mo M., Sanchez-Madrid F., Simon C. Embryonic implantation and leukocyte transendothelial migration: Different processes with similar players? FASEB J. 2005;19:1056–1060. doi: 10.1096/fj.05-3781hyp. [DOI] [PubMed] [Google Scholar]
- 27.Shao R., Egecioglu E., Weijdegard B., Kopchick J.J., Fernandez-Rodriguez J., Andersson N., Billig H. Dynamic regulation of estrogen receptor-α isoform expression in the mouse fallopian tube: Mechanistic insight into estrogen-dependent production and secretion of insulin-like growth factors. Am. J. Physiol. Endocrinol. Metab. 2007;293:E1430–E1442. doi: 10.1152/ajpendo.00384.2007. [DOI] [PubMed] [Google Scholar]
- 28.Mowa C.N., Iwanaga T. Differential distribution of oestrogen receptor-α and -β mRNAs in the female reproductive organ of rats as revealed by in situ hybridization. J. Endocrinol. 2000;165:59–66. doi: 10.1677/joe.0.1650059. [DOI] [PubMed] [Google Scholar]
- 29.Parada-Bustamante A., Orihuela P.A., Rios M., Navarrete-Gomez P.A., Cuevas C.A., Velasquez L.A., Villalon M.J., Croxatto H.B. Catechol-o-methyltransferase and methoxyestradiols participate in the intraoviductal nongenomic pathway through which estradiol accelerates egg transport in cycling rats. Biol. Reprod. 2007;77:934–941. doi: 10.1095/biolreprod.107.061622. [DOI] [PubMed] [Google Scholar]
- 30.Shao R., Nutu M., Karlsson-Lindahl L., Benrick A., Weijdegard B., Lager S., Egecioglu E., Fernandez-Rodriguez J., Gemzell-Danielsson K., Ohlsson C., et al. Downregulation of cilia-localized IL-6R α by 17β-estradiol in mouse and human fallopian tubes. Am. J. Physiol Cell Physiol. 2009;297:C140–C151. doi: 10.1152/ajpcell.00047.2009. [DOI] [PubMed] [Google Scholar]
- 31.Horne A.W., King A.E., Shaw E., McDonald S.E., Williams A.R., Saunders P.T., Critchley H.O. Attenuated sex steroid receptor expression in fallopian tube of women with ectopic pregnancy. J. Clin. Endocrinol. Metab. 2009;94:5146–5154. doi: 10.1210/jc.2009-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsson U.W., Johns T.G., Wilmann T., Kaitu’u-Lino T., Whitehead C., Dimitriadis E., Menkhorst E., Saglam B., Gao Y., Greenall S.A., et al. Effects of gefitinib, an epidermal growth factor receptor inhibitor, on human placental cell growth. Obstet. Gynecol. 2013;122:737–744. doi: 10.1097/AOG.0b013e3182a1ba56. [DOI] [PubMed] [Google Scholar]
- 33.Horne A.W., Skubisz M.M., Doust A., Duncan W.C., Wallace E., Critchley H.O., Johns T.G., Norman J.E., Bhattacharya S., Mollison J., et al. Phase II single arm open label multicentre clinical trial to evaluate the efficacy and side effects of a combination of gefitinib and methotrexate to treat tubal ectopic pregnancies (GEM II): Study protocol. BMJ Open. 2013;3 doi: 10.1136/bmjopen-2013-002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The PubMed database. [(accessed on 15 October 2015)]; Available online: http://www.ncbi.nlm.nih.gov/pubmed.
- 35.Wei C.H., Kao H.Y., Lu Z. Pubtator: A web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013;41:W518–W522. doi: 10.1093/nar/gkt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei C.H., Kao H.Y., Lu Z. Gnormplus: An integrative approach for tagging genes, gene families, and protein domains. Biomed Res. Int. 2015;2015:918710. doi: 10.1155/2015/918710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The CRF++ library. [(accessed on 15 October 2015)]. Available online: http://crfpp.googlecode.com/svn/trunk/doc/index.html.
- 38.Wei C.H., Kao H.Y. Cross-species gene normalization by species inference. BMC Bioinform. 2011;12 doi: 10.1186/1471-2105-12-S8-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The Cytoscape software. [(accessed on 15 October 2015)]. Available online: http://cytoscape.org.
- 40.Assenov Y., Ramirez F., Schelhorn S.E., Lengauer T., Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.