Abstract
The rapid rise in DNA sequencing has led to an expansion in the number of glycoside hydrolase (GH) families. The GH43 family currently contains α-l-arabinofuranosidase, β-d-xylosidase, α-l-arabinanase, and β-d-galactosidase enzymes for the debranching and degradation of hemicellulose and pectin polymers. Many studies have revealed finer details about members of GH43 that necessitate the division of GH43 into subfamilies, as was done previously for the GH5 and GH13 families. The work presented here is a robust subfamily classification that assigns over 91% of all complete GH43 domains into 37 subfamilies that correlate with conserved sequence residues and results of biochemical assays and structural studies. Furthermore, cooccurrence analysis of these subfamilies and other functional modules revealed strong associations between some GH43 subfamilies and CBM6 and CBM13 domains. Cooccurrence analysis also revealed the presence of proteins containing up to three GH43 domains and belonging to different subfamilies, suggesting significant functional differences for each subfamily. Overall, the subfamily analysis suggests that the GH43 enzymes probably display a hitherto underestimated variety of subtle specificity features that are not apparent when the enzymes are assayed with simple synthetic substrates, such as pNP-glycosides.
INTRODUCTION
Carbohydrates serve a range of functional purposes in biological systems, including energy storage, signal transduction, and intracellular trafficking, among others (1). Importantly, carbohydrates are the main end product of plant primary production, representing a large of majority of carbon fixation by plants (2). As a photosynthetically renewable form of fixed carbon, plant biomass represents a prime target for the replacement of petroleum-derived fuels for future sustainability efforts. The enzymatic degradation and modification of carbohydrates have thus been cast to the forefront of biofuel production research (3).
As functional efforts to discover plant cell wall polysaccharide (PCWP)-degrading enzymes identify novel activities and mechanisms (4, 5), it is important to derive and maintain a concise classification system for these enzymes. A sequence-based classification of carbohydrate-active enzymes (CAZymes) began in 1991 (6), with the classification of 35 families of glycoside hydrolases (GHs). Today the CAZy database (7) comprises 5 separate enzyme classes, namely, the aforementioned GHs, glycosyltransferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), and auxiliary activities (AAs), as well as associated carbohydrate binding modules (CBMs), that together correspond to over 530,000 individual sequences (at the time of submission of this article). The largest of these classes are the glycoside hydrolases, currently represented by over 241,000 sequences classified into 133 families based on amino acid sequence similarity.
The rapid advancements in DNA sequencing over the past decade have exponentially increased the number of sequences assigned to each family. Hence, some of the larger and multifunctional GH families, including GH5 (8), GH13 (9), and GH30 (10), as well as PL families (11), have been divided into subfamilies. The previous studies show a much stronger correlation between subfamily designations and substrate specificity than family-level assignments, allowing for improved accuracy of the prediction of activity for individual proteins during genomic or metagenomic analyses.
At present, the CAZy database reports 4,555 GH43 family members, making GH43 one of the largest GH families. The family is dominated by bacterial sequences (4,197), with eukaryotic (312) and archaeal (17) sequences also contained in the group. The functional characteristics of this family have been studied comparatively well, with 146 members that have been characterized biochemically against a natural or synthetic substrate, but this analysis still lags far behind the deposition of new sequences. The major reported activities are β-d-xylosidase (EC 3.2.1.37), α-l-arabinofuranosidase (EC 3.2.1.55), endo-α-l-arabinanase (EC 3.2.1.99), and 1,3-β-galactosidase (EC 3.2.1.145) activities. Taken together, this family comprises a range of debranching enzymes for aiding in the degradation of hemicellulose, particularly arabinoxylans, and pectin. The first crystal structure and catalytic residues of a GH43 protein were determined in 2002 by Nurizzo et al. (12), who reported a 5-bladed β-propeller structure. Subsequent studies have shown different binding mechanisms (13) and catalytic residues (14) within the family, suggesting the existence of different clades within the GH43 family.
The GH43 family has emerged as an important family for biomass deconstruction efforts, as studies have found it to be expanded in a number of plant cell wall-degrading microorganisms (15). Additionally, studies of the human gut microbiome have identified GH43 enzymes to be among the most abundant CAZymes present (16, 17). Such abundance in both cases suggests an important role in accessing a wide range of complex substrates and highlights the need for accurate functional predictions for GH43 enzymes in genomic and metagenomic studies.
Here we present a subfamily classification scheme for the GH43 family, based on phylogenetic analysis and backed, where possible, by functional and structural studies of individual enzymes. The introduction of these subfamilies will allow for more accurate functional annotation of discovered sequences and will guide structure and function analysis toward the characterization of enzymes significantly divergent from those previously studied. It is hoped that such analysis may enable a greater understanding of this abundant GH family.
MATERIALS AND METHODS
All complete GH43 domain sequences were taken from the CAZy database on 25 May 2015 and supplemented with sequences from other sources, including the Joint Genome Institute (JGI), the Broad Institute, the Human Microbiome Project (HMP), and GenBank, resulting in 7,664 sequences. To reduce redundancy and improve the processing time, sequences were clustered at 95% similarity by using CD-Hit (18), resulting in 4,189 remaining sequences. Multiple-sequence alignment was performed by MUSCLE (19). In order to generate high-quality and relevant alignments, MAFFT (20) was used to iteratively remove highly dissimilar sequences. Such sequences were defined as those having a gap of >3 residues or an insertion of >1 residue that was not also seen in at least 2 other sequences. Following these quality control measures, 3,337 sequences remained. With these sequences, FASTTree (21) was used to generate a phylogenetic tree based on the midpoint root method (Fig. 1).
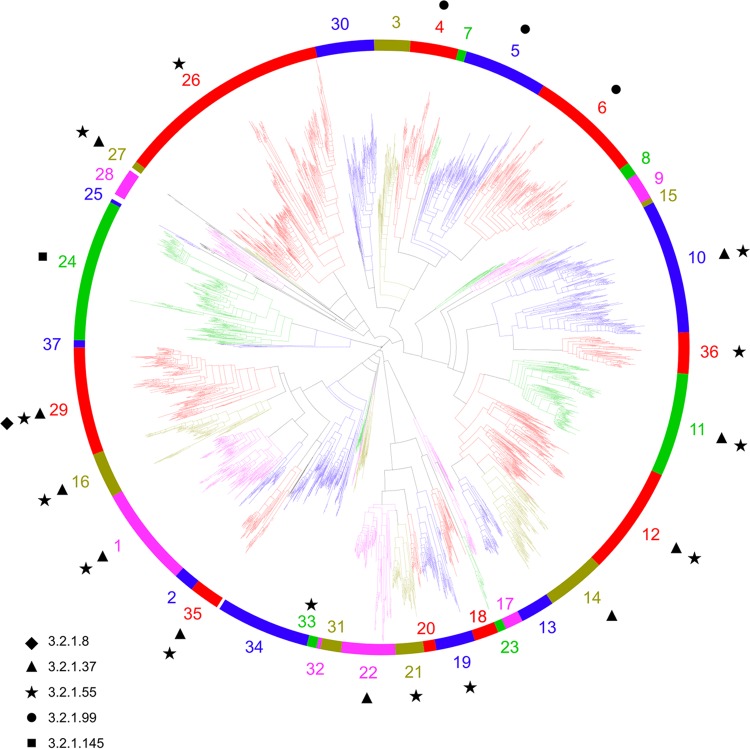
FIG 1.
Phylogenetic tree of the GH43 family, showing the 37 subfamilies as colored branches. Black branches represent sequences that could not be assigned to a subfamily. Activities found in the various subfamilies are shown as follows: ◆, xylanase (EC 3.2.1.8); ▲, xylosidase (EC 3.2.1.37); ★, arabinofuranosidase (EC 3.2.1.55); ●, arabinanase (EC 3.2.1.99); and ■, galactosidase (EC 3.2.1.145).
Manual separation of subfamilies was decided based on phylogenetic distances in this reduced tree. Subfamilies were required to contain at least 5 sequences found in this reduced tree in order to generate a proper multiple-sequence alignment. Additionally, each family was required to show taxonomic diversity above the class level to ensure that a subfamily was not comprised solely of taxonomically recent gene homologues due to genome sequencing bias for a particular class, order, or family. These criteria resulted in 37 putative subfamilies.
Hidden Markov models (HMMs) were created for each subfamily, as well as for the complete GH43 subfamily, by using HMMer3 (22). All GH43 sequences were compared to these HMMs by use of HMMer3 to assign a subfamily to each sequence. Each sequence was compared to all other GH43 sequences by using BLASTP (23), and the top 100 BLAST hits were retained. The top 100 BLAST hits for each sequence were grouped by subfamily, and the sum total bit score for each subfamily was calculated.
Criteria for assignment of a domain into a subfamily were 2-fold: (i) HMM comparison must have provided an E value that was e−20 lower for the specific subfamily than for any other subfamily HMM, including the HMM generated from all GH43 sequences; and (ii) the subfamily with the highest bit score ratio (resulting from the sum of the top 100 BLAST hits divided by the number of sequences in the subfamily) must have agreed with the HMM designation.
After assignment of all GH43 sequences meeting the above criteria, individual alignments, trees, and HMMs were built for each putative subfamily, using all sequences and excluding the CD-Hit and MAFFT procedures. Subfamilies containing proteins with known structures and catalytic residues were inspected manually to ensure the conservation of catalytic residues. This inspection identified an important distinction within one putative subfamily, resulting in the creation of subfamilies GH43_24, GH43_25, and GH43_37. The HMM and BLAST analysis was repeated with all complete subfamilies to assign subfamily membership to each sequence. The resulting 37 subfamilies collectively contained 7,040 sequences (91.5% of all GH43 domains analyzed), ranging from 10 to 870 sequences each.
In order to assess the functional capabilities of each subfamily, all GH43 proteins that have been characterized biochemically against natural or synthetic substrates were mapped onto subfamily trees (see the supplemental material) displaying either the EC number or the substrate against which these proteins have shown activity. In total, 21 of 37 subfamilies contained at least one characterized member.
Because CAZymes are often very modular proteins, i.e., containing modules corresponding to different families, we searched for other modules frequently found in the proteins of a given GH43 subfamily, producing a matrix of cooccurrence counts. These counts were normalized against the total number of domains in a given subfamily to generate a matrix showing frequencies of cooccurrence (Fig. 2).
FIG 2.
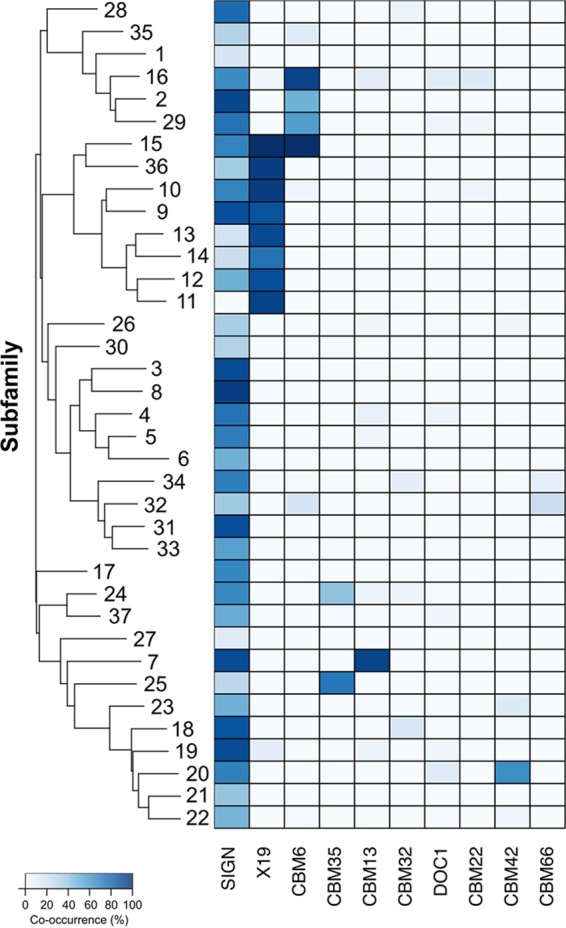
Heat map showing frequencies of cooccurrence of GH43 subfamily domains and major noncatalytic modules. Subfamilies are clustered according to the similarities of their respective HMM profiles. CBM, carbohydrate binding module; DOC, cellulosomal dockerin domain; X19, conserved noncatalytic module.
RESULTS AND DISCUSSION
Our classification procedure resulted in the assignment of 4,455 GH43 protein domains to 37 individual subfamilies. Specific information about each subfamily is presented in Table 1. Here we discuss the activities identified in these subfamilies as well as the main functional modules found appended to GH43 members.
TABLE 1.
Characteristics of each GH43 subfamily
| Subfamily | Size of family (no. of sequences) | Taxonomic distribution | EC no. (no. of sequences)a | No. of characterized enzymes | Ligand(s)b | PDB code for 3D structure (1 per enzyme) |
|---|---|---|---|---|---|---|
| GH43c | 122 | 3.2.1.55 | 2 | |||
| GH43_1 | 254 | 3.2.1.37 (11), (3.2.1.37 + 3.2.1.55) (2) | 13 | BWX, pNP-aLAraf, pNP-BXYL | 4MLG | |
| GH43_2 | 31 | Neocallimastigomycota | 0 | |||
| GH43_3 | 87 | 0 | ||||
| GH43_4 | 264 | Neocallimastigomycota | 3.2.1.99 | 12 | SBAR, LAR | 2X8F, 3KMV, 3LV4, 4KC7, 4KCA |
| GH43_5 | 211 | 3.2.1.99 | 9 | SBAR, LAR | 1GYD, 1UV4, 1WL7, 3CU9, 4KCB | |
| GH43_6 | 38 | Fungi | 3.2.1.99 | 9 | SBAR, LAR | |
| GH43_7 | 13 | Bacteria | 0 | |||
| GH43_8 | 26 | Bacteria | 0 | |||
| GH43_9 | 137 | Chytridiomycota | 0 | |||
| GH43_10 | 376 | 3.2.1.37 (1), (3.2.1.37 + 3.2.1.55) (1), 3.2.1.55 (7) | 9 | AAX, WAX, SBAR, MU-xylose, pNP-aLAraf, pNP-BXYL | ||
| GH43_11 | 583 | Ascomycota | 3.2.1.37 (14), (3.2.1.37 + 3.2.1.55) (4) | 18 | OSX, xylobiose, linear 1,4-β-xylan, pNP-aLAraf, pNP-BXYL | 1YI7, 1YIF, 1YRZ, 2EXH, 3C2U |
| GH43_12 | 395 | Mollusca | 3.2.1.37 (1), (3.2.1.37 + 3.2.1.55) (3), 3.2.1.55 (5) | 9 | WAX, MU-xylose, pNP-aLAraf | |
| GH43_13 | 15 | Fungi | 0 | |||
| GH43_14 | 34 | Fungi | 3.2.1.37 | 1 | WAX, pNP-BXYL | |
| GH43_15 | 9 | Bacteria | 0 | |||
| GH43_16 | 184 | Neocallimastigomycota | (3.2.1.37 + 3.2.1.55) (1), 3.2.1.55 (6) | 7 | WAX, linear arabinoxylan, OSX, RAX, MU-aLAraf | 1W0N, 3C7E |
| GH43_17 | 53 | 0 | 5C0P | |||
| GH43_18 | 61 | 0 | ||||
| GH43_19 | 53 | Neocallimastigomycota | 3.2.1.55 | 1 | ||
| GH43_20 | 19 | Basidiomycota | 0 | |||
| GH43_21 | 19 | Fungi | 3.2.1.55 | 1 | SBAR, pNP-aLAraf | |
| GH43_22 | 118 | 3.2.1.37 | 1 | Xylan oligosaccharide (unspecified source) | ||
| GH43_23 | 36 | 0 | ||||
| GH43_24 | 228 | 3.2.1.145 | 8 | LWG, 1,3-β-galactan | 3NQH, 3VSF | |
| GH43_25 | 9 | Fungi | 0 | |||
| GH43_26 | 851 | 3.2.1.55 | 3 | pNP-aLAraf | 3AKF, 3CPN | |
| GH43_27 | 32 | Bacteria | 3.2.1.37 (1), 3.2.1.55 (1) | 2 | OSX, pNP-BXYL, pNP-aLAraf | |
| GH43_28 | 75 | 0 | ||||
| GH43_29 | 218 | (3.2.1.37 + 3.2.1.55) (2), 3.2.1.55 (1), 3.2.1.8 (2) | 5 | pNP-BXYL, pNP-aLAraf | 3QED, 4NOV | |
| GH43_30 | 111 | 0 | ||||
| GH43_31 | 54 | Bacteroidetes | 0 | 3KST | ||
| GH43_32 | 32 | Rotifera | 0 | |||
| GH43_33 | 25 | 3.2.1.55 | 1 | pNP-aLAraf | 4QQS | |
| GH43_34 | 185 | 0 | 3QZ4 | |||
| GH43_35 | 63 | Neocallimastigomycota | (3.2.1.37 + 3.2.1.55) | 3 | pNP-BXYL, pNP-aLAraf | |
| GH43_36 | 47 | 3.2.1.55 | 2 | No substrates specified | 3ZXJ | |
| GH43_37 | 13 | 0 |
EC number for characterized enzymes (multiple EC numbers in parentheses belong to the same enzyme).
BWX, birchwood xylan; AAX, AZCL-arabinoxylan; MU, methylumbelliferyl; BXYL, beta-xylan (unspecified source); AAR, alpha-arabinan; WAX, wheat arabinoxylan; SBAR, sugar beet arabinan; OSX, oat spelt xylan; RAX, rye arabinoxylan; LWG, larchwood galactan; LAR, linear arabinan. Arabinan (sugar beet) corresponds to any of the following: linear 1,5-α-l-arabinan, debranched arabinan, linear arabinan, or red debranched arabinan.
Sequences that could not be assigned to a subfamily.
β-d-Xylosidases (EC 3.2.1.37) and α-l-arabinofuranosidases (EC 3.2.1.55).
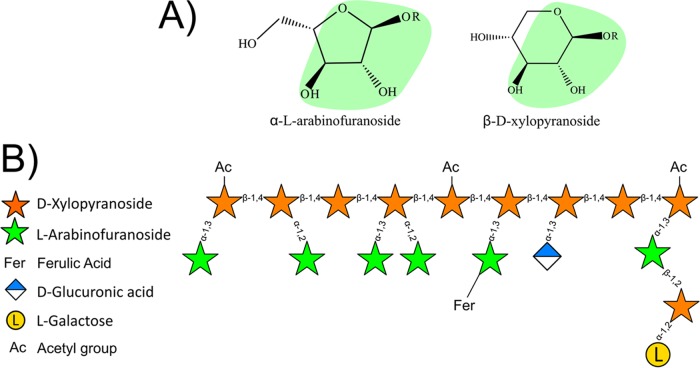
β-d-Xylosidases and α-l-arabinofuranosidases constitute the majority of characterized GH43 enzymes. To date, all subfamilies that have been characterized and shown to be polyspecific harbor both β-d-xylosidase (EC 3.2.1.37) and α-l-arabinofuranosidase (EC 3.2.1.55) activities. The overlap of these activities within a subfamily, and in some cases within a single protein, is not altogether unsurprising, as the α-l-arabinofuranose and β-d-xylose conformations are sterically similar near the glycosidic bond (Fig. 3), and indeed, this cooccurrence has been reported many times previously (24, 25).
FIG 3.
(A) Stereochemical representation of α-l-arabinofuranoside and β-d-xylopyranoside. The green areas indicate similar stereochemistries around the glycosidic bond. (B) Schematic representation of arabinoxylan, showing common linkages found in complex arabinoxylan polysaccharides.
Of particular note is subfamily 36, which has demonstrated activity against disubstituted (1,2- and 1,3-arabinofuranoside) xylopyranose residues (26, 27). These residues are typically recalcitrant to enzymatic attack, and as such, this subfamily has significant biotechnological interest.
The abundance of characterized enzymes with these activities highlights the reliance of functional screening efforts on easily available synthetic pNP-sugars, in particular pNP-β-d-xyloside and pNP-α-l-arabinofuranoside, for identification of activity.
Endo-α-l-arabinanases (EC 3.2.1.99).
Subfamilies GH43_4, GH43_5, and GH43_6 are the only ones showing α-l-arabinanase activity. These three closely related but distinct subfamilies are among the most well characterized, with each showing endo-α-l-arabinanase activity. Subfamilies GH43_4 and GH43_5 both have multiple crystal structures available, with subfamily 5 having the first obtained crystal structure for a GH43 enzyme (12).
β-1,3-Galactosidases (EC 3.2.1.145).
Subfamily GH43_24 is well characterized, with 8 members having been characterized biochemically. This is the only subfamily shown to have β-1,3-d-galactosidase activity. Members of GH43_24 have been characterized and their structures obtained (14), and these show a shift of the catalytic base residue from Asp38 (in the CjArb43A reference sequence of Nurizzo et al. [12]) to Glu112 (in the Ct1,3Gal43A reference sequence of Jiang et al. [14]).
With subfamily GH43_24, the uncharacterized subfamily GH43_37 shares a similar motif at the catalytic base, except that a glycine replaces the glutamic acid residue. The effect of such a shift is unknown, but it may result in a loss of functional activity or a repurposing of this domain for other functions. This potential repurposing of domains has been addressed by Aspeborg et al. (8) and includes inactivated chitinases repurposed as xylanase inhibitors in GH18 (28) and amino acid transporters arising from ancestral α-amylases of GH13 (9). It is also impossible to rule out a change in protein structure that may bring a different residue into the active site to serve as the catalytic base or the requirement of a cofactor for deglycosylation, as seen for ascorbate in the myrosinases of the GH1 family (29).
Uncharacterized subfamilies.
Of the 37 subfamilies defined here, only 19 subfamilies contain at least one protein that has been characterized through biochemical assay. Furthermore, only 10 subfamilies have at least 5 characterized members, which may explain why some subfamilies harboring only EC 3.2.1.37 or EC 3.2.1.55 activity are identified as monospecific. This lack of biochemical characterization stretches throughout the GH43 tree, with some poorly explored regions of the tree being related only distantly to subfamilies with characterized members. The analysis presented here brings these clades into focus as targets for further functional and structural exploration.
Functional modules found appended to GH43 domains.
To identify CAZy domains frequently found together, we examined the modularity of all proteins containing a GH43 domain, which revealed a number of significant associations between individual GH43 subfamilies and other CAZy modules (Fig. 2). Such associations have the potential to inform the functions of subfamilies that have no characterized members as well as to inform us about the evolutionary trajectory followed by each subfamily.
(i) CBM6.
The most frequently cooccurring module was found to be CBM6, a module with a demonstrated function of binding to amorphous cellulose and β-1,4-xylan, with 6.0% of all GH43 domain-containing proteins also harboring a CBM6 domain. This module was found to be associated with 10 different subfamilies but was most striking in subfamilies GH43_15 and GH43_16, with 100% and 64% cooccurrence rates, respectively. These CBM6 modules demonstrated increased catalytic improvement of associated enzymes on nonsoluble substrates (30). That previous study of CBM6 domains identified four clades of CBM6 family domains. The CBM6 modules found in the present study either are not assigned to any of the four subfamilies defined by Abbott et al. (77 of 237 domains) (30) or belong to subfamily CBM6b (160 domains), a clade with demonstrated xylan-binding capabilities, which matches the activity of characterized members of this subfamily.
(ii) CBM35.
CBM35 modules are found in GH43_24 and GH43_25 and are known to bind xylan and manno-oligosaccharides but also bind 1,3-β-d-galactose (31). Such binding is expected, as the GH43_24 subfamily is the only one containing a characterized 1,3-β-d-galactosidase activity.
(iii) CBM13.
Subfamily GH43_7 has yet to have a member characterized, but we observed that all GH43_7 proteins also harbor a CBM13 module. A CBM13-containing α-l-arabinofuranosidase (AbfB) has been characterized for the soil actinomycete Streptomyces lividans (32) and has demonstrated xylan-binding functionality. This cooccurrence hints toward β-d-xylosidase (EC 3.2.1.37) and α-l-arabinofuranosidase (EC 3.2.1.55) activities, as seen in the subfamilies above associated with CBM6 xylan-binding domains.
(iv) CBM42.
CBM42 modules are seen in 59% of subfamily GH43_20 proteins. Assays of a CBM42-containing α-l-arabinofuranosidase (AkAbfB) from Aspergillus kawachii (33) revealed an arabinose-binding capacity for CBM42, which differs from the xylan-binding CBMs found associated with other GH43 subfamilies. This correlation suggests that these proteins recognize the arabinofuranoside component of arabinoxylans rather than the xylan backbone. Such a strategy could avoid competition for limited space of binding to the xylan backbone or may represent an organism's utilization of the arabinose side chains rather than the more abundant xylan backbone. Subfamily GH43_20 contains only bacterial sequences, none of which have been characterized biochemically; additional study would be necessary to further either of these hypotheses.
The cooccurrence of these CBM modules with separate GH43 subfamilies is congruent with the findings of previous studies and supports the notion that the GH43 family is a pectin and arabinoxylan debranching and degradation family.
(v) X19.
The three-dimensional structures of several GH43 members show a C-terminal extension, here termed X19, which folds independently of the β-propeller but has no apparent function. The systematic C-terminal position of X19 relative to GH43, as well as the absence of any linking peptide or domain between them, suggests that it is a C-terminal cap that may aid in protein stability. This X19 domain is found only in a subset of GH43 subfamilies (GH43_9 through GH43_14 and GH43_36), and its presence originates at a single point in the GH43 subfamily tree. It is possible that this domain is an evolutionary remnant that is unnecessary for catalytic function. An additional note is that outside the GH43 family, the X19 domain is not seen to occur with any other GH families.
(vi) Proteins containing multiple GH43 modules.
In addition to searching for other CAZy modules found with GH43 subfamily domains, we searched for proteins containing multiple GH43 domains from different subfamilies. We identified 297 proteins that contained two GH43 domains and 20 proteins that contained three GH43 domains (Table 2). The most common GH43 domain to be found with another GH43 domain was from subfamily GH43_34, with 156 of 228 total entries (approximately 70%) found with another GH43 domain. Furthermore, the GH43_34 domain was found to be the C-terminal domain in 143 of these entries, which may allude to a functional characteristic. This subfamily is currently uncharacterized, which opens the door for many questions related to both its membership and its position within multimodular proteins. Such a frequent level of cooccurrence may suggest a potential synergistic interaction, an auxiliary role for this subfamily in identifying or binding substrates, or a potential loss of function that would need to be confirmed through functional biochemical analysis of individual protein domains.
TABLE 2.
Subfamily membership and relative positions of catalytic domains in proteins with multiple GH43 modules
| First domain | Other domain(s) |
No. of proteinsa | |
|---|---|---|---|
| Second position | Third position | ||
| GH43b | GH43_18 | None | 1 |
| GH43_22 | GH43_29 | 1 | |
| GH43_26 | None | 4 | |
| GH43_31 | None | 2 | |
| GH43_3 | GH43_31 | None | 2 |
| GH43_9 | GH43_19 | GH43_34 | 11 |
| GH43_19 | None | 37 | |
| GH43_10 | GH43_16 | None | 11 |
| GH43_16 | GH43_22 | None | 2 |
| GH43_17 | GH43_19 | None | 2 |
| GH43_18 | GH43_34 | None | 52 |
| GH43_19 | GH43_9 | GH43_34 | 1 |
| GH43_24 | None | 5 | |
| GH43_26 | None | 7 | |
| GH43_29 | GH43_34 | 1 | |
| GH43_29 | None | 4 | |
| GH43_34 | None | 59 | |
| GH43_22 | GH43_24 | None | 1 |
| GH43_26 | GH43_34 | 5 | |
| GH43_27 | None | 1 | |
| GH43_34 | None | 82 | |
| GH43_23 | GH43_34 | None | 2 |
| GH43_24 | GH43_22 | None | 1 |
| GH43_26 | GH43_17 | None | 1 |
| GH43_23 | None | 5 | |
| GH43_31 | None | 2 | |
| GH43_34 | None | 1 | |
| GH43_27 | GH43_22 | GH43 | 1 |
| GH43 | None | 1 | |
| GH43_29 | GH43_10 | None | 2 |
| GH43_31 | GH43_26 | None | 1 |
| GH43_34 | GH43_3 | None | 13 |
Number of observed cases among 4,455 GH43 proteins.
Domains that could not be assigned to any of the 37 subfamilies.
(vii) Signal peptides.
To identify the cellular locations of GH43-containing proteins, we identified secretion signal peptides found to cooccur with GH43 domains. Across all GH43-containing proteins, 69% also contain a signal peptide directing the translated protein outside the cytoplasm. One exception is subfamily GH43_11, which does not have any of its 600 members cooccurring with a signal peptide. This subfamily is limited to the Ascomycota, and this lack of a signal peptide suggests that this subfamily is involved in intracellular processes, such as the degradation of imported disaccharides or cell wall remodeling. In contrast, proteins containing multiple GH43 domains are found to contain a signal peptide in over 92% of cases, suggesting that they play a role in degradation of extracellular substrates.
Conclusions.
The recent interest in biomass-degrading enzymes and gut microbiome studies have led to a rapid expansion in glycoside hydrolase sequences, including those of the GH43 family. This abundance of data allows for a finer-detailed analysis of the family but also exposes limitations arising from the existence of such a large GH family. Functional and structural characteristics assigned to the family are not shared among all members but are partitioned at a level below that of the current GH family designation. The aim of our work was to partition sequences into more homogeneous, finer subgroups in order to improve protein sequence annotation and functional prediction for future postgenomic studies.
We have presented here a subfamily classification system for the GH43 family of enzymes. The robustness of these subfamilies is necessary to ensure their survival with the steady increase in the number of sequences. These subfamilies encompass over 91% of all completely sequenced modules and show both phylogenetic and functional characteristics to support their assignments. Of these 37 subfamilies, just 21 have characterized members, representing four distinct EC numbers (EC 3.2.1.37, EC 3.2.1.55, EC 3.2.1.99, and EC 3.2.1.145), showing that progress in the field of functional characterization has lagged significantly behind sequencing progress. Nonetheless, for subfamilies with multiple characterized members, we see strong agreement within a subfamily toward a particular enzymatic activity.
The expansion of GH43 modules in several plant cell wall-degrading organisms, their prevalence in the gut microbiome, and their overall abundance in nature drive interest in this GH family. Continued genomic and metagenomic studies will contribute additional GH43 domains to the existing subfamilies and may result in the formation of new subfamilies. As the GH43 family is expanded and potentially further divided, the subfamily classification will be updated and released to the public on the CAZy website.
Compared to the previous subfamily classification of the GH5 family, we see a similar level of sequence diversity in the GH43 family, but the apparent functional diversity is much lower. This may be due to the overuse of synthetic pNP-monosaccharides as substrates for activity-based screening experiments. The natural substrates of GH43 enzymes are extremely diverse in terms of structure, with structures including pectin side chains, a range of arabinoxylans, and intracellular polysaccharides. It is likely that the diverse subfamilies introduced here reflect the diversity of substrate structures and that the current characterizations of these subfamilies are often distorted through the convenient use of synthetic pNP-sugars. The use of naturally sourced sugars for characterization may allow finer-resolution details of the activity found in each subfamily. Thus, bioinformatic studies not only benefit from experimental science to improve predictions but also can guide and inform experimentalists beyond the obvious choice of targets for subfamilies with no characterized members.
Supplementary Material
Funding Statement
Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (NSERC) provided funding to Keith Mewis in the form of a Michael Smith Foreign Study Supplement.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03453-15.
REFERENCES
- 1.Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J. 1999. Essentials of glycobiology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 2.Lichtenthaler FW. 2007. Carbohydrates as renewable raw materials: a major challenge of green chemistry, p 23–63. In Tundo P, Perosa A, Zecchini F (ed), Methods and reagents for green chemistry: an introduction. Wiley, Hoboken, NJ. [Google Scholar]
- 3.Armstrong Z, Mewis K, Strachan C, Hallam SJ. 2015. Biocatalysts for biomass deconstruction from environmental genomics. Curr Opin Chem Biol 29:18–25. doi: 10.1016/j.cbpa.2015.06.032. [DOI] [PubMed] [Google Scholar]
- 4.Quinlan RJ, Sweeney MD, Leggio LL, Otten H, Poulsen J-CN, Johansen KS, Krogh KB, Jørgensen CI, Tovborg M, Anthonsen A. 2011. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci U S A 108:15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. 2014. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol 10:122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henrissat B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aspeborg H, Coutinho PM, Wang Y, Brumer H, Henrissat B. 2012. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol 12:186. doi: 10.1186/1471-2148-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B. 2006. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel 19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- 10.St John FJ, González JM, Pozharski E. 2010. Consolidation of glycosyl hydrolase family 30: a dual domain 4/7 hydrolase family consisting of two structurally distinct groups. FEBS Lett 584:4435–4441. doi: 10.1016/j.febslet.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 11.Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho P, Henrissat B. 2010. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J 432:437–444. doi: 10.1042/BJ20101185. [DOI] [PubMed] [Google Scholar]
- 12.Nurizzo D, Turkenburg JP, Charnock SJ, Roberts SM, Dodson EJ, McKie VA, Taylor EJ, Gilbert HJ, Davies GJ. 2002. Cellvibrio japonicus α-l-arabinanase 43A has a novel five-blade β-propeller fold. Nat Struct Mol Biol 9:665–668. doi: 10.1038/nsb835. [DOI] [PubMed] [Google Scholar]
- 13.Vandermarliere E, Bourgois T, Winn M, Van Campenhout S, Volckaert G, Delcour J, Strelkov S, Rabijns A, Courtin C. 2009. Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family. Biochem J 418:39–47. doi: 10.1042/BJ20081256. [DOI] [PubMed] [Google Scholar]
- 14.Jiang D, Fan J, Wang X, Zhao Y, Huang B, Liu J, Zhang XC. 2012. Crystal structure of 1,3Gal43A, an exo-β-1,3-galactanase from Clostridium thermocellum. J Struct Biol 180:447–457. doi: 10.1016/j.jsb.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canbäck B, Choi C, Cichocki N, Clum A. 2015. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet 47:410–415. doi: 10.1038/ng.3223. [DOI] [PubMed] [Google Scholar]
- 16.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 17.Wu M, McNulty NP, Rodionov DA, Khoroshkin MS, Griffin NW, Cheng J, Latreille P, Kerstetter RA, Terrapon N, Henrissat B, Osterman AL, Gordon JI. 2015. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science 350:aac5992. doi: 10.1126/science.aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res 41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huy ND, Thayumanavan P, Kwon T-H, Park S-M. 2013. Characterization of a recombinant bifunctional xylosidase/arabinofuranosidase from Phanerochaete chrysosporium. J Biosci Bioeng 116:152–159. doi: 10.1016/j.jbiosc.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Lagaert S, Pollet A, Courtin CM, Volckaert G. 2014. β-Xylosidases and α-l-arabinofuranosidases: accessory enzymes for arabinoxylan degradation. Biotechnol Adv 32:316–332. doi: 10.1016/j.biotechadv.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 26.McKee LS, Peña MJ, Rogowski A, Jackson A, Lewis RJ, York WS, Krogh KBRM, Viksø-Nielsen A, Skjøt M, Gilbert HJ, Marles-Wright J. 2012. Introducing endo-xylanase activity into an exo-acting arabinofuranosidase that targets side chains. Proc Natl Acad Sci U S A 109:6537–6542. doi: 10.1073/pnas.1117686109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Broek LM, Lloyd R, Beldman G, Verdoes J, McCleary B, Voragen AJ. 2005. Cloning and characterization of arabinoxylan arabinofuranohydrolase-D3 (AXHd3) from Bifidobacterium adolescentis DSM20083. Appl Microbiol Biotechnol 67:641–647. doi: 10.1007/s00253-004-1850-9. [DOI] [PubMed] [Google Scholar]
- 28.Durand A, Hughes R, Roussel A, Flatman R, Henrissat B, Juge N. 2005. Emergence of a subfamily of xylanase inhibitors within glycoside hydrolase family 18. FEBS J 272:1745–1755. doi: 10.1111/j.1742-4658.2005.04606.x. [DOI] [PubMed] [Google Scholar]
- 29.Burmeister WP, Cottaz S, Rollin P, Vasella A, Henrissat B. 2000. High resolution X-ray crystallography shows that ascorbate is a cofactor for myrosinase and substitutes for the function of the catalytic base. J Biol Chem 275:39385–39393. doi: 10.1074/jbc.M006796200. [DOI] [PubMed] [Google Scholar]
- 30.Abbott DW, Ficko-Blean E, van Bueren AL, Rogowski A, Cartmell A, Coutinho PM, Henrissat B, Gilbert HJ, Boraston AB. 2009. Analysis of the structural and functional diversity of plant cell wall specific family 6 carbohydrate binding modules. Biochemistry (Mosc) 48:10395–10404. doi: 10.1021/bi9013424. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh A, Luís AS, Brás JLA, Pathaw N, Chrungoo NK, Fontes CMGA, Goyal A. 2013. Deciphering ligand specificity of a Clostridium thermocellum family 35 carbohydrate binding module (CtCBM35) for gluco- and galacto-substituted mannans and its calcium induced stability. PLoS One 8:e80415. doi: 10.1371/journal.pone.0080415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent P, Shareck F, Dupont C, Morosoli R, Kluepfel D. 1997. New α-l-arabinofuranosidase produced by Streptomyces lividans: cloning and DNA sequence of the abfB gene and characterization of the enzyme. Biochem J 322:845–852. doi: 10.1042/bj3220845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyanaga A, Koseki T, Matsuzawa H, Wakagi T, Shoun H, Fushinobu S. 2004. Crystal structure of a family 54 α-l-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose. J Biol Chem 279:44907–44914. doi: 10.1074/jbc.M405390200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.