Abstract
The actual state of intestinal long-term colonization by extended-spectrum β-lactamase (ESBL)-producing Escherichia coli in healthy Japanese people remains unclear. Therefore, a total of 4,314 fecal samples were collected from 2,563 food handlers from January 2010 to December 2011. Approximately 0.1 g of each fecal sample was inoculated onto a MacConkey agar plate containing cefotaxime (1 μg/ml). The bacterial colonies that grew on each plate were checked for ESBL production by the double-disk synergy test, as recommended by the Clinical and Laboratory Standards Institute. The bacterial serotype, antimicrobial susceptibility, pulsotype, sequence type (ST), and ESBL genotype were checked, and the replicon types of plasmids harboring the ESBL gene were also determined after conjugation experiments. ESBL producers were recovered from 70 (3.1%) of 2,230 participants who were checked only once. On the other hand, ESBL producers were isolated at least once from 52 (15.6%) of 333 participants who were checked more than twice, and 13 of the 52 participants carried ESBL producers for from more than 3 months to up to 2 years. Fluoroquinolone (FQ)-resistant E. coli strains harboring blaCTX-M were repeatedly recovered from 11 of the 13 carriers of blaCTX-M-harboring E. coli. A genetically related FQ-resistant E. coli O25b:H4-ST131 isolate harboring blaCTX-M-27 was recovered from 4 of the 13 carriers for more than 6 months. Three FQ-resistant E. coli O1:H6-ST648 isolates that harbored blaCTX-M-15 or blaCTX-M-14 were recovered from 3 carriers. Moreover, multiple CTX-M-14- or CTX-M-15-producing E. coli isolates with different serotypes were recovered from 2 respective carriers. These findings predict a provable further spread of ESBL producers in both community and clinical settings.
INTRODUCTION
The production of extended-spectrum β-lactamases (ESBLs) is one of the most common cephalosporin resistance mechanisms acquired by bacteria belonging to the members of the family Enterobacteriaceae (1). In particular, the incidence of CTX-M-type-ESBL-producing Escherichia coli isolates among clinical isolates recovered from community-acquired infections, such as urinary tract infections and bacteremia, has been increasing (2). The increasing rates of isolation of these microbes alert us to the probable growing risk to public health in the future (3). The exact reason for the rapid spread of ESBL-producing E. coli isolates among healthy individuals remains unclear, although alterations of the bacterial phenotypic and genetic properties that permit easier colonization in the human intestines, as well as the considerable contamination of foods with ESBL producers, have been speculated to underlie the phenomenon (4).
The prevalence of intestinal carriage of ESBL-producing bacteria belonging to the family Enterobacteriaceae, in particular, the CTX-M-15-producing E. coli O25b-sequence type 131 (ST131) clone, has recently been regarded to be a growing public health concern worldwide (5), because CTX-M-type-ESBL-producing E. coli O25b ST131 clones usually show coresistance to fluoroquinolones (FQs) and sometimes cause community-acquired infections, such as urinary tract infections, as well as nosocomial infections. Some investigators have reported that the rate of enteric carriage of ESBL producers among healthy individuals, including travelers and soldiers, is considerable (6–10). However, although most studies have so far focused on hospitalized patients, the augmented prevalence of cephalosporin-resistant Enterobacteriaceae in clinical settings at present would also be greatly affected by the ESBL producers increasingly colonizing the intestines of healthy people in the community. Woerther et al. have recently reported on the trends in the fecal carriage of ESBL producers in the community, as well as their regional specificities, their dissemination routes, and the way to control the bacterial populations in members of the community colonized with ESBL producers (5). Moreover, some studies on the duration of fecal carriage of ESBL-producing bacteria in patients have been conducted in Thailand and Sweden (11–13). Titelman et al. have noted that the fecal carriage of ESBL-producing Enterobacteriaceae in patients was common for 12 months after the time of initial infection (14). Information on the duration of fecal carriage of ESBL producers is important to determine how to deal with healthy carriers of ESBL producers at the time of their admission to hospitals.
The aim of our study, therefore, was to elucidate the states of colonization by ESBL-producing E. coli in the intestines of 2,563 Japanese food handlers.
MATERIALS AND METHODS
Sample collection and bacterial isolation.
The investigation was performed with 2,563 different participants at the Okazaki City Public Health Center in Japan from January 2010 to December 2011. Among the 2,563 participants, 2,230 were checked for colonization with ESBL producers only once and 333 participants were checked more than twice during the period of investigation (see Fig. S1 in the supplemental material). In accordance with the requirements of the Ministry of Health, Labour and Welfare of Japan, workers who handle food are expected to be checked every month and, at a minimum, at least twice a year at regional health centers or appointed private microbiology laboratories to determine whether they carry any pathogenic bacteria, including enterohemorrhagic E. coli, Shigella spp., or Salmonella spp., in their intestinal tracts, although the periodic checking of the feces of food handlers is not strictly enforced in Japan. Therefore, we considered that it would be suitable to follow up healthy people with ESBL-producing microbes in their intestines for a long period through the use of this evaluation system. The study was conducted with the approval of the ethics committee for epidemiological studies of the Nagoya University Graduate School of Medicine. All people were informed of the ethical points, including the purpose of the study and the study protocol, prior to the investigation, and those who consented to join the study on their own were enrolled as participants.
A total of 4,314 stool specimens were collected from 2,563 different participants (1,050 males and 1,513 females; age range, 18 to 74 years for males and 18 to 79 years for female; average age ± standard deviation, 48.9 ± 14.4 years for males and 48.8 ± 14.0 years for females). The stool specimen (approximately 0.1 g) from each participant was directly inoculated by use of a sterilized cotton swab onto MacConkey agar plates (Eiken Chemical Co., Ltd., Tokyo, Japan) supplemented with 1 μg of cefotaxime (CTX) per ml (CTX-MacConkey). When the colonies that uniformly grew on an agar plate showed very similar features, three colonies were picked from the plate and the bacterial species was identified by using conventional biochemical tests and an API 20E system (Sysmex bioMérieux Co., Ltd., Tokyo, Japan). Four to 10 colonies were picked and subjected to further testing when multiple colonies with different features appeared on an agar plate.
Screening for ESBL producers and genetic identification.
Screening of probable ESBL producers was performed by the double-disk synergy test using commercially available ceftazidime, cefotaxime, and amoxicillin-clavulanic acid disks purchased from Eiken Chemical Co., Ltd. (15). PCR analyses were performed using 6 sets of PCR primers for the detection of genes for CTX-M group 1, CTX-M group 2, CTX-M group 8, CTX-M group 9, TEM-type, and SHV-type ESBLs, and their genotypes were further determined by nucleotide sequencing analyses of the PCR amplicons (16, 17).
Serotyping of E. coli by antisera and detection of serotype O25b by PCR.
The serotype was determined using E. coli antisera, Seiken set 1 (Denka Seiken Co., Ltd., Tokyo, Japan) for the O antigen and Seiken set 2 (Denka Seiken) for the H antigen, according to the manufacturer's instructions. Serotypes that could not be determined by this method were designated O antigen untypeable (OUT) or H antigen untypeable (HUT). In addition, genetic O serotyping by PCR was performed as previously described only for the E. coli isolates determined to be serogroup O25 by the E. coli antisera (18). This is because ESBL-producing and FQ-resistant E. coli isolates are usually identified to be serotype O25b:H4, a serological variant of serotype O25:H4, but serotype O25b:H4 cannot be discriminated from serotype O25:H4 by commercially available antisera.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed using a control strain, E. coli ATCC 25922, and the agar dilution method, which was done as recommended by the Clinical and Laboratory Standards Institute (CLSI) (19). Interpretation of the MIC results was done in accordance with the CLSI criteria in document M7-A9 (19). The MICs of the following antimicrobials were purchased from the indicated sources: piperacillin, cefotaxime, ceftazidime, imipenem, aztreonam, gentamicin, minocycline, fosfomycin, ciprofloxacin, and levofloxacin were from Wako Pure Chemical Co., Inc., Tokyo, Japan; cefmetazole, amikacin, and chloramphenicol were from Sigma-Aldrich Japan, Tokyo, Japan; and flomoxef was provided by Shionogi & Co., Ltd., Tokyo, Japan.
PFGE and MLST.
Pulsed-field gel electrophoresis (PFGE) analysis of each ESBL-producing isolate was performed as described elsewhere (20). In brief, a plug containing whole-genomic DNA was digested with XbaI (TaKaRa Bio. Inc., Tokyo, Japan), and electrophoresis was performed using a CHEF-DR III system (Bio-Rad Laboratories, Hercules, CA, USA) with pulse times ranging from 2.2 to 54.2 s and at a voltage of 6 V/cm at 14°C for 19 h. Strain H9812 of Salmonella enterica serotype Braenderup was used as the control strain. A dendrogram showing the genetic relatedness among the isolates was prepared with Fingerprinting II software (Bio-Rad Laboratories). The isolates obtained from each participant were regarded to have the same genetic background when they possessed a pulsotype with ≥85% similarity, and one representative isolate of each pulsotype was selected and used for further study. When a genetic similarity of less than 85% was observed between two isolates, they were considered genetically different, and both isolates were separately characterized in the present study, as performed previously (21).
Multilocus sequence typing (MLST) of the blaCTX-M-harboring E. coli isolates repeatedly recovered from the same participant was performed by analysis of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) according to a protocol provided by a website for MLST of E. coli (http://mlst.ucc.ie/mlst/dbs/Ecoli). The sequence types (STs) were compared with the population structure of the species using the eBURST program (http://eburst.mlst.net/).
Conjugation study, plasmid replicon typing, and PCR detection of blaCTX-M genes.
All the blaCTX-M-harboring donor E. coli isolates that colonized the participants for a long period were uniformly susceptible to rifampin. Thus, rifampin-resistant E. coli CSH-2 (metB F−, resistant to nalidixic acid and rifampin) was used as the recipient in the conjugation experiments, performed by the broth mating method (22). Transconjugants were selected on Luria-Bertani (LB) agar plates supplemented with CTX (20 μg/ml) and rifampin (100 μg/ml) (Wako Pure Chemical Co., Inc.). The pulsotype of each transconjugant was compared to that of the recipient strain to avoid the acquisition of rifampin resistance by the donor strain. In addition, the acquisition of blaCTX-M by the transconjugant was also confirmed by PCR using positive-control strains for each ESBL gene (16, 17). For the resultant blaCTX-M-harboring transconjugants, as well as their parent isolates repeatedly recovered from the same participant, the replicon types of the plasmids were checked by PCR-based replicon typing (PBRT) using 18 pairs of primers as described previously (23).
Statistical analysis.
Comparisons of the proportions of participants by age and gender were made by a continuity-adjusted χ2 test with SPSS software (version 20.0 for Windows; SPSS Inc., Chicago, IL, USA). A P value of <0.05 was considered to denote a statistically significant difference.
RESULTS
Isolation of ESBL producers from fecal samples and their characteristics.
ESBL producers were recovered from 197 of the 4,314 fecal specimens evaluated in the present study. More than 2 ESBL producers were recovered at the same time from each of 10 of the 197 fecal specimens. Two ESBL producers were recovered from 9 of these 10 fecal specimens, and 3 ESBL producers were from 1 of the 10 fecal specimens. When the shapes of the colonies that grew on a plate were apparently the same, three colonies were picked up and subjected to typing of the ESBL by PCR. If the shapes of the colonies that grew on a plate were different in size and/or color, 4 to 10 colonies with a distinct appearance were fished out and tested. As a result, from 1 to 3 ESBL producers with different genotypes were isolated from each participant, and 208 ESBL producers were finally obtained from 122 participants. If the ESBL producers recovered from the same participant in each test showed very similar PFGE profiles and the same ESBL type, only 1 isolate was selected and kept for further analyses. Thus, 59 of the 208 ESBL producers were excluded. Consequently, 149 ESBL producers possessing different genetic backgrounds were recovered from 122 (4.8%; 62 males and 60 females) of the 2,563 participants (1,050 males and 1,513 females) and evaluated in this study (see Fig. S1 in the supplemental material). Neither age nor gender bias in the rate of detection of the 149 ESBL producers from the 122 participants was observed (P ≥ 0.05). Of these 149 isolates, 145 were identified to be E. coli, 3 were identified to be Klebsiella pneumoniae, and 1 was identified to be Aeromonas hydrophila (Table 1). Two ESBL-producing isolates, one E. coli isolate and one K. pneumoniae isolate, were coisolated from 2 participants, and an ESBL-producing K. pneumoniae isolate alone was recovered from 1 participant.
TABLE 1.
Characteristics of nonduplicate 149 ESBL producers obtained from 122 participants
| Species | No. of isolates in each of the following CTX-M groups: |
|||||
|---|---|---|---|---|---|---|
| CTX-M-1 | CTX-M-2 | CTX-M-8 | CTX-M-9 | Non-CTX-Ma | Total | |
| E. coli | 30 | 19 | 7b | 82 | 7 | 145 |
| K. pneumoniae | 2c | 1 | 3 | |||
| A. hydrophila | 1 | 1 | ||||
| Total | 30 | 21 | 7 | 83 | 8 | 149 |
Isolates harboring the blaSHV gene.
One of seven isolates harbored both the blaCTX-M-8 and blaTEM-52 genes.
These isolates harbored both the blaCTX-M-2 and blaSHV-1 genes.
One hundred thirty-eight (95.2%) of the 145 ESBL-producing E. coli isolates from 120 participants harbored a blaCTX-M gene, and the remaining 7 isolates harbored blaSHV-5-group genes, such as blaSHV-12, as shown in Table 2. The types of blaCTX-M genes were mainly blaCTX-M-14 (43.5%), followed by blaCTX-M-15 (17.9%), blaCTX-M-27 (13.1%), and blaCTX-M-2 (13.1%). The most frequent O serogroup of the blaCTX-M-harboring E. coli isolates was O25 (27 isolates, 18.6%), followed by O1 (11 isolates, 7.6%) and O153 (8 isolates, 5.5%). Evaluation of the relationship between blaCTX-M genes and O serogroups showed that the serogroups of the isolates harboring blaCTX-M-14, blaCTX-M-2, or blaCTX-M-15 were diverse, i.e., O1, O74, and O153; however, 16 (84.2%) of the 19 isolates harboring blaCTX-M-27 were O25b.
TABLE 2.
O serogroups and CTX-M types of nonduplicate 145 ESBL-producing E. coli isolates
| Serogroup | No. (%) of isolates of each CTX-M type |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CTX-M-1 group |
CTX-M-2 group, CTX-M-2 | CTX-M-8 group, CTX-M-8 | CTX-M-9 group |
Non-CTX-Ma | Total | |||||
| CTX-M-1 | CTX-M-15 | CTX-M-3 | CTX-M-55 | CTX-M-14 | CTX-M-27 | |||||
| O1 | 3 | 2b | 5 | 1 | 11 (7.6) | |||||
| O8 | 3 | 1 | 1 | 5 (3.4) | ||||||
| O15 | 2 | 1 | 3 (2.1) | |||||||
| O18 | 1 | 1 (0.7) | ||||||||
| O25 | 1 | 1 | 1 | 1 | 7 | 16 | 27 (18.6) | |||
| O27 | 1 | 1 (0.7) | ||||||||
| O29 | 1 | 1 | 2 (1.4) | |||||||
| O74 | 1 | 4 | 5 (3.4) | |||||||
| O78 | 3 | 1 | 4 (2.8) | |||||||
| O86a | 1 | 1 (0.7) | ||||||||
| O125 | 1 | 2 | 1 | 4 (2.8) | ||||||
| O128 | 1 | 1 | 2 (1.4) | |||||||
| O142 | 3 | 3 (2.1) | ||||||||
| O148 | 1 | 1 (0.7) | ||||||||
| O153 | 1 | 1 | 5 | 1 | 8 (5.5) | |||||
| O157 | 1c | 1 (0.7) | ||||||||
| O166 | 1 | 1 (0.7) | ||||||||
| O169 | 1 | 1 | 2 (1.4) | |||||||
| OUT | 2 | 11 | 1 | 11 | 3 | 30 | 1 | 4 | 63 (43.4) | |
| Total | 2 (1.4) | 26 (17.9) | 1 (0.7) | 1 (0.7) | 19 (13.1) | 7 (4.8) | 63 (43.5) | 19 (13.1) | 7 (4.8) | 145 (100) |
Isolates harboring a blaSHV-5-group gene, such as blaSHV-12.
One of the two isolates harbored both the blaCTX-M-8 and blaTEM-52 genes.
This isolate did not harbor genes for Shiga-like toxins.
As shown in Table 3, most ESBL-producing E. coli isolates were susceptible to cefmetazole, flomoxef, imipenem, amikacin, and fosfomycin. However, these isolates tended to show a phenotype of multidrug resistance to aztreonam, minocycline, and FQs. The isolates harboring the blaCTX-M-27 gene, especially the serotype O25b isolates, usually showed resistance to FQs, such as ciprofloxacin and levofloxacin, but no isolate showing resistance to gentamicin and amikacin was found among the 19 isolates harboring the blaCTX-M-27 gene, although 23 of 63 isolates harboring the blaCTX-M-14 gene showed resistance to gentamicin and/or amikacin. The FQ MICs for serotype O25b:H4 isolates harboring the blaCTX-M-27 gene were significantly higher than those for the isolates of the other O serotypes harboring any of the other blaCTX-M genes (P < 0.05) (see Fig. S2 in the supplemental material). The MIC values of aztreonam and ceftazidime for the isolates harboring the blaCTX-M-15 gene were significantly higher than those for the isolates harboring any of the other blaCTX-M genes regardless of their O serotypes (P < 0.05) (see Fig. S3 in the supplemental material). On the other hand, no significant differences in the MIC values of minocycline, amikacin, and fosfomycin (P ≥ 0.05) were found among the groups harboring different blaCTX-M genes.
TABLE 3.
Antimicrobial resistance profiles of 145 nonduplicate ESBL-producing E. coli isolates
| Antimicrobial agentb | No. of resistant isolates of each CTX-M typea |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CTX-M-1 group |
CTX-M-2 group, CTX-M-2 (n = 19) | CTX-M-8 group, CTX-M-8 (n = 7) | CTX-M-9 group |
Non- CTX-Mc (n = 7) | Total (n = 145) | |||||
| CTX-M-1 (n = 2) | CTX-M-15 (n = 26) | CTX-M-3 (n = 1) | CTX-M-55 (n = 1) | CTX-M-14 (n = 63) | CTX-M-27 (n = 19) | |||||
| Ceftazidime | 0 | 17 | 0 | 1 | 0 | 0 | 1 | 1 | 5 | 25 |
| Aztreonam | 2 | 25 | 0 | 1 | 10 | 0 | 11 | 3 | 7 | 59 |
| Gentamicin | 0 | 6 | 1 | 0 | 4 | 0 | 23 | 0 | 0 | 34 |
| Amikacin | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Minocycline | 2 | 17 | 1 | 1 | 15 | 3 | 37 | 15 | 5 | 96 |
| Chloramphenicol | 0 | 7 | 0 | 0 | 1 | 0 | 14 | 0 | 1 | 23 |
| Fosfomycin | 0 | 1 | 1 | 0 | 1 | 0 | 5 | 1 | 0 | 9 |
| Ciprofloxacin | 0 | 16 | 1 | 1 | 1 | 0 | 28 | 18d | 2 | 67 |
| Levofloxacin | 0 | 16 | 1 | 1 | 0 | 0 | 28 | 18d | 2 | 66 |
Interpretation of MIC results was done in accordance with the CLSI criteria in document M7-A9 (19).
All ESBL-producing E. coli isolates were resistant to piperacillin and cefotaxime but were susceptible to cefmetazole, flomoxef, and imipenem.
Isolates harboring a blaSHV-5-group gene, such as blaSHV-12.
Sixteen of 18 E. coli isolates harboring the blaCTX-M-27 gene were serogroup O25.
PFGE analysis.
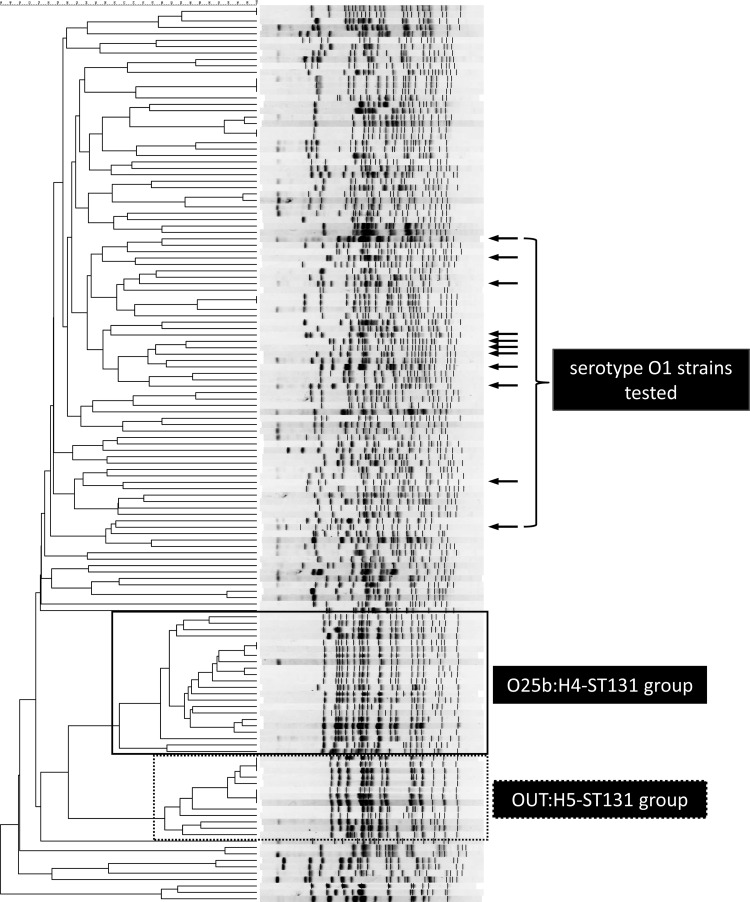
As shown in Fig. 1, the O25b:H4-ST131 group (including 19 isolates of O25b:H4-ST131, 2 isolates of O25b:HNM-ST131 [where NM indicates nonmotile], and 1 isolate of O25b:HUT-ST131) and the OUT:H5-ST131 group (including 12 isolates of OUT:H5-ST131 and 1 isolate of O25:H5-ST131) constituted 2 large clusters which were defined at the 71% and the 80% similarity levels, respectively, but the isolates belonging to the O1 serogroup showed a very wide genetic diversity.
FIG 1.
Dendrogram of PFGE types among the ESBL-producing E. coli isolates. The results of PFGE of XbaI-digested whole-genome DNA from 142 of the 145 ESBL-producing E. coli isolates recovered from 120 participants are shown. DNA from the remaining three ESBL-producing E. coli isolates (including one E. coli serogroup O15 isolate and two E. coli serogroup OUT isolates) became smeared during digestion with XbaI. The dendrogram was produced by the unweighted pair group method using arithmetic averages (UPGMA) algorithm based on the Dice similarity coefficient with a 1.0% band position tolerance. The O25b:H4-ST131 group and the OUT:H5-ST131 group are surrounded by squares with solid lines and dotted lines, respectively. Arrows, the rows for the serogroup O1 isolates tested.
Long-term colonization by blaCTX-M-harboring E. coli.
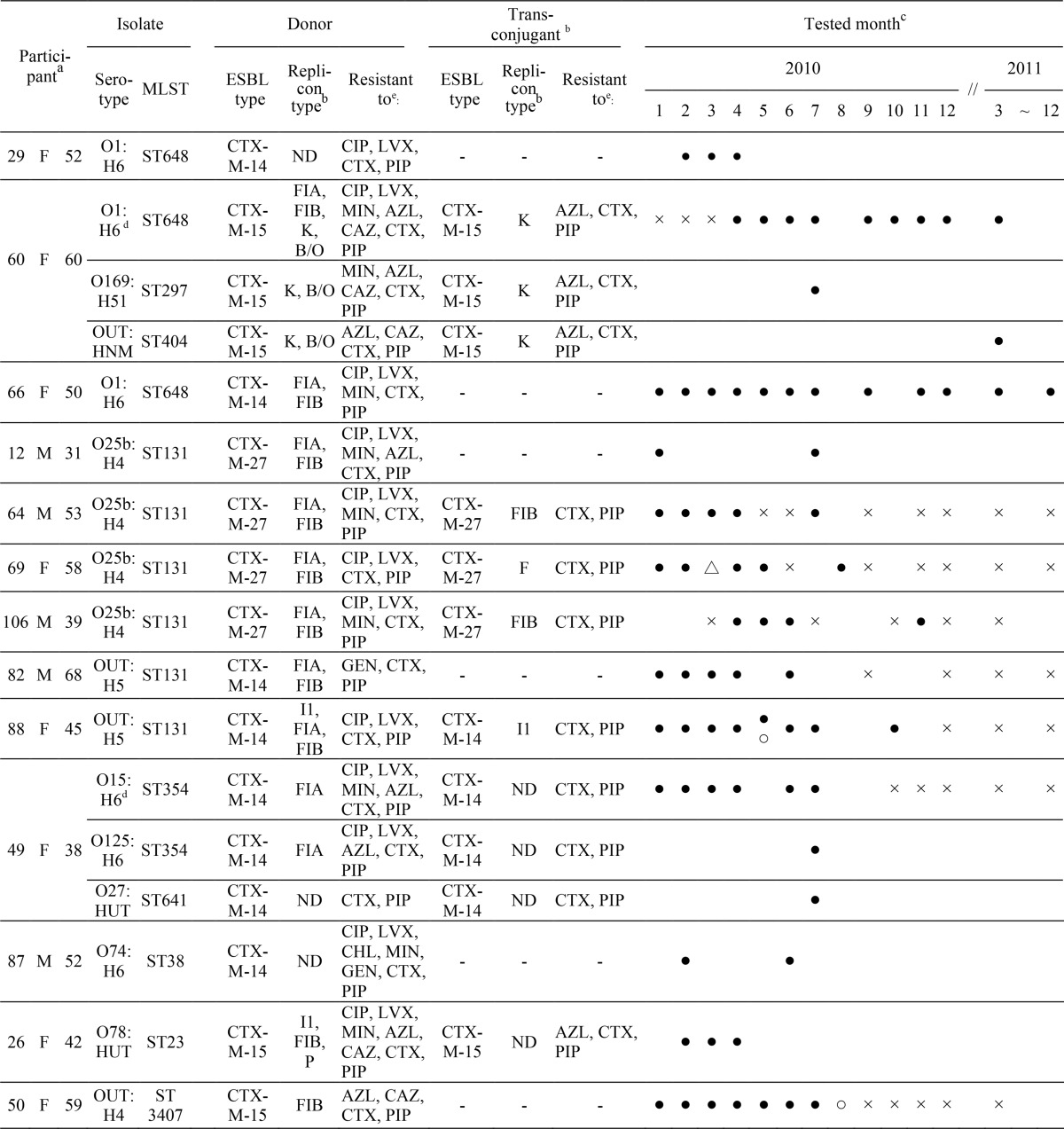
Among the 122 carriers of ESBL producers, 52 carriers could be checked at least 2 times, and blaCTX-M-harboring E. coli isolates were recovered more than twice from 13 (25%) of the 52 carriers during the study period (Table 4; see also Table S1 and Fig. S1 in the supplemental material). Since some of the blaCTX-M-harboring E. coli isolates from each of the 13 participants showed the same PFGE profile, we defined the 13 participants to be long-term carriers. The periods of detection of the blaCTX-M-harboring E. coli isolates in each participant ranged from 3 months to up to 2 years. Neither age nor gender bias was observed (P ≥ 0.05) among the 13 long-term carriers. In two of the long-term carriers (participants 60 and 66), blaCTX-M-harboring E. coli O1:H6-ST648 isolates were repeatedly recovered for more than 1 year. Interestingly, blaCTX-M-14-harboring E. coli O1:H6-ST648 isolates were repeatedly recovered for up to 2 years in a long-term carrier (participant 66). In three long-term carriers (participants 64, 69, and 106), blaCTX-M-harboring E. coli isolates were intermittently recovered but the participants were negative over different periods. In four cases (participants 12, 26, 29, and 87), the durations of colonization of blaCTX-M-harboring E. coli in 2011 were uncertain because they were not checked in 2011. It was confirmed from the questionnaires submitted by all the participants at the time of submission of each fecal sample that all but one carrier (participant 69) received any obvious antimicrobial treatment during the period of investigation. Participant 69 received antimicrobial treatment for pneumonia in March 2010, but the name of the antimicrobial and its period of administration were unidentified. An ESBL-producing E. coli O25b:H4-ST131 isolate harboring blaCTX-M-27 was first isolated from participant 69 in January 2010, prior to antimicrobial administration in March 2010, and the same ESBL producers were repeatedly recovered from this participant for 5 months even after antimicrobial treatments were stopped, as shown in Table 4. Two carriers (participants 50 and 88) traveled abroad during the investigation period, and ESBL-producing E. coli OUT:H4-ST3407 harboring blaCTX-M-15 disappeared from one carrier (participant 50) just after overseas travel, while E. coli OUT:H5-ST131 harboring blaCTX-M-14 was repeatedly recovered from participant 88 for 5 months after the foreign travel.
TABLE 4.
Characteristics of blaCTX-M-harboring E. coli isolates repeatedly recovered from 13 long-term carriers and blaCTX-M-harboring E. coli isolates simultaneously recovered from 2 participants
The data in the first three columns represent the personal identification number, gender (F, female; M, male), and age (in years), respectively.
-, not transferred; ND, the replicon type was not determined by PBRT.
•, detection of blaCTX-M-harboring E. coli; ×, no detection of blaCTX-M-harboring E. coli, even though fecal screening was performed; △, prescription of antimicrobial treatment; ○, overseas travel.
Isolates detected for a long period.
CIP, ciprofloxacin; LVX, levofloxacin; CTX, cefotaxime; PIP, piperacillin; MIN, minocycline; AZL, aztreonam; CAZ, ceftazidime; GEN, gentamicin; CHL, chloramphenicol.
Serotypes and multilocus sequence types of blaCTX-M-harboring E. coli isolates from long-term carriers.
As shown in Table 4, the most frequent serotype of blaCTX-M-harboring E. coli isolates that colonized any of the participants for long periods was O25b:H4 (4 carriers), followed by O1:H6 (3 carriers) and OUT:H5 (2 carriers). The serotypes of the remaining isolates were diverse, i.e., O15, O74, and O78.
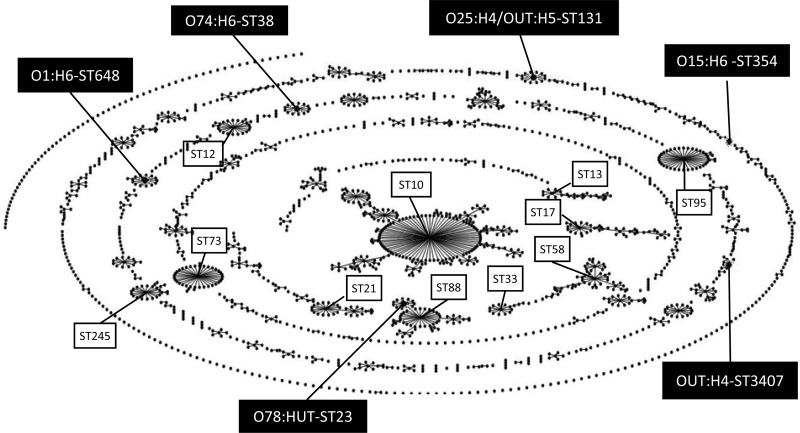
MLST analysis identified 6 different STs, and the most predominant one was ST131 (6 isolates), followed by ST648 (3 isolates). The remaining STs were ST354, ST38, ST23, and ST3407, and the 6 STs were scattered evenly among the ST population structure of E. coli, as illustrated by eBURST analysis (Fig. 2). No apparent genetic relationship between ST648 and ST131 according to the nucleotide sequences of the seven housekeeping genes for which they were tested was found.
FIG 2.
Result of eBURST analysis. A snapshot of the population created by eBURST analysis (http://eburst.mlst.net/) shows clusters of linked and unlinked sequence types in the E. coli MLST database (2,000 sequence types; http://mlst.ucc.ie/mlst/dbs/Ecoli). Sequence type labels were removed, but the serotypes and sequence types found for the ESBL-producing E. coli isolates repeatedly recovered from healthy people in the present study are shown.
Characteristics of blaCTX-M-harboring E. coli isolates from long-term carriers.
As shown in Table 4, 6 (46%), 4 (31%), and 3 (23%) individuals were long-term carriers of E. coli isolates harboring blaCTX-M-14, blaCTX-M-27, and blaCTX-M-15, respectively. Most blaCTX-M-harboring E. coli isolates colonizing participants for long periods tended to show multidrug resistance profiles, and the blaCTX-M-harboring E. coli isolates recovered from 11 of 13 long-term carriers showed coresistance to FQs. Interestingly, the ceftazidime MICs for blaCTX-M-27-harboring E. coli isolates recovered from four long-term carriers (participants 12, 64, 69, and 106) were about 2 or 4 μg/ml, although CTX-M-27 generally hydrolyzes ceftazidime as well as cefotaxime.
Replicon types of blaCTX-M-carrying plasmids.
The conjugation experiment and replicon typing of plasmids were performed with the 17 blaCTX-M-harboring E. coli isolates recovered from the 13 long-term carriers. As shown in Table 4, the conjugal transfer of blaCTX-M-carrying plasmids to recipient cells was successful for 11 isolates, and the replicon types of the plasmids transferred were IncF (1 isolate), IncFIB (2 isolates), IncK (3 isolate), and IncI1 (1 isolate), as determined by PBRT. The Inc types of the plasmids from the remaining 4 isolates could not be determined. The blaCTX-M-27 genes in the E. coli O25b:H4-ST131 isolates were carried by plasmids belonging to IncF or IncFIB.
Detection of multiple blaCTX-M-harboring E. coli isolates with diverse serotypes.
In 2 of the 13 long-term carriers (participants 49 and 60), blaCTX-M-14- and blaCTX-M-15-harboring E. coli isolates, respectively, were continuously recovered during the period of investigation, but the serotypes of the E. coli isolates were different in each participant (Table 4). In the case of participant 49, a blaCTX-M-14-harboring E. coli O15:H6-ST354 isolate was repeatedly recovered for 7 months, and blaCTX-M-14-harboring E. coli O125:H6 isolates belonging to ST354 were also recovered together with a blaCTX-M-14-harboring E. coli O27:HUT-ST641 isolate in July 2010. Conjugal transfer of a blaCTX-M-14-carrying plasmid to recipient cells was successful in the conjugation experiment, but the replicon types of the plasmids carrying the blaCTX-M-14 gene could not be determined by PBRT. blaCTX-M-15-harboring E. coli O169:H51 and OUT:HNM isolates, together with E. coli O1:H6-ST648, were recovered from the feces of participant 60, a long-term carrier, on different occasions. However, the replicon types of all 3 plasmids carrying the blaCTX-M-15 gene were IncK, suggesting the probable conjugal transfer of the blaCTX-M-15-carrying plasmids among genetically different commensal E. coli lineages in the bowel of participant 60.
DISCUSSION
We first elucidated that blaCTX-M-harboring E. coli isolates have colonized the intestinal tracts of healthy Japanese people, even in the absence of antimicrobial pressure, for from 3 months to up to 2 years. At one point in the study of ESBL producers from 2,230 participants, 70 (3.1%) participants were positive for ESBL producers. This proportion is comparable to that (6.4%) determined in other investigations involving 218 participants in Japan (24). On the other hand, 52 (15.6%) of the 333 participants whose feces were checked more than twice were found to be positive for ESBL producers. This proportion is far greater than that for the 2,230 participants evaluated once, but we assume that this value may well more accurately reflect the state of fecal carriage of ESBL producers among healthy Japanese people. To our knowledge, our study, which involved more than 2,500 participants, is the first one to report the state of colonization by blaCTX-M-harboring E. coli isolates in the intestinal tracts of people living in the community who have not apparently been exposed to antimicrobials.
There are some reports concerning the duration of fecal carriage of ESBL-producing E. coli isolates in humans who had been checked in clinical settings. An investigation enrolling 24 outpatients in Thailand demonstrated that the average period of carriage of ESBL producers in feces was 98 days and that 8 (33%) out of 24 patients carried ESBL-producing E. coli for 6 months (11). In a French study, 180 (40%) of 448 patients were persistent carriers of ESBL-producing Enterobacteriaceae for a median period of 6.6 months (25). After a nosocomial outbreak in southern Sweden, the long-term carriage of ESBL-producing E. coli (41 to 59 months) was observed in 5 (13%) of 39 patients (12). In a Netherlands study with 521 participants, 19 (16.8%) of 113 participants who acquired ESBL producers during foreign travel kept the ESBL producers for 6 months (26). A Swedish study also showed that 10 (24%) of 41 patients with traveler's diarrhea who carried ESBL-producing E. coli still carried the ESBL producers after 3 to 8 months (13). These findings are somewhat similar to our observation that 13 (25%) of the 52 carriers kept the ESBL producers for long periods. The present study may well be unprecedented, because more than 2,500 healthy participants were involved and the investigation was continued for more than 6 months with 10 carriers of ESBL producers and for up to 2 years in one case. Therefore, the results of our investigation suggest the actual state of the fecal carriage of ESBL producers among healthy people to whom no obvious antimicrobial was administered.
The affinity of particular E. coli serotypes to human intestines or the fitness of E. coli in human intestines would contribute to the long-term colonization of ESBL-producing E. coli in the intestinal tract. As for the serogroups of E. coli often recovered from human intestines, O1 (4.81%), both O1 (7.1%) and O25 (7.1%), and O25 (9.1%) were reportedly predominant in healthy subjects (27), patients suffering from sporadic diarrhea (28), and outpatients (29), respectively. These findings suggest that E. coli serogroups O1 and O25 are the ones that are most able to adapt to the human bowel. Serogroups O1 and O25 were predominant in the enteric flora of both American and Romanian people (30, 31), indicating that E. coli serogroups O1 and O25 generally accommodate themselves to the human intestines regardless of the human race. Among the people who participated in the present study, blaCTX-M-harboring E. coli serogroup O1 or O25 was indeed continuously recovered from 7 of 13 carriers. Moreover, 1 case was found to carry blaCTX-M-14-harboring E. coli O1:H6 for 2 years in the absence of antimicrobial treatment. These findings are consistent with those of several previous studies conducted in Japan and other countries. In the present study, the rates of detection of ESBL-producing E. coli serogroups O1 and O25 from the 120 people positive for ESBL producers were 7.6% (11/145) and 18.6% (27/145), respectively (Table 5). On the other hand, the rates of recovery of ESBL-producing E. coli isolates of serogroups O1 and O25 from patients who carried the ESBL producers for long periods were 23.1% (3/13) and 30.8% (4/13), respectively, and these values are much higher than those found for the 120 carriers of ESBL producers (Table 5). The acquisition of blaCTX-M genes by E. coli serogroups O1 and O25 might well contribute to the long-term carriage of ESBL-producing E. coli in the intestines of healthy humans receiving no antimicrobial treatment. In addition, according to the results of PFGE analyses, it was suggested that the E. coli isolates belonging to serogroup O25 formed a clonal lineage, while the E. coli isolates belonging to serogroup O1 were genetically diverse (Fig. 1). It has been considered that bacterial strains harboring plasmids carrying antimicrobial resistance genes tend to disappear sooner or later from human intestines and also that the plasmids carrying antimicrobial resistance genes are apt to be deleted from bacterial cells before long in the absence of antimicrobial treatment because the acquisition of antimicrobial resistance mechanisms usually impedes bacterial growth. However, ESBL-producing E. coli serogroups O1 and O25, which have a probable affinity for blaCTX-M-bearing plasmids, could persistently inhabit the intestines of healthy people. This finding may suggest the possible acquisition of an additional ability for intraintestinal long-term colonization of E. coli isolates belonging to serogroups O1 and O25 that harbor IncF-group plasmids, which often carry various blaCTX-M genes.
TABLE 5.
Characteristics of the ESBL-producing E. coli isolates from the 120 overall carriers of ESBL producers and 13 long-term carriers of ESBL producers
| Characteristic | No. (%) of isolates among: |
|
|---|---|---|
| 145 isolates from 120 carriers | 13 isolates from 13 long-term carriers | |
| O serogroup | ||
| O1 | 11 (7.5) | 3 (23.1) |
| O25 | 27 (18.6) | 4 (30.8) |
| O15 | 3 (2.1) | 1 (7.7) |
| O74 | 5 (3.4) | 1 (7.7) |
| O78 | 4 (2.8) | 1 (7.7) |
| Other O serogroups | 32 (22.2) | 0 (0) |
| OUT | 63 (43.4) | 3 (23.1) |
| Molecular type of ESBL | ||
| CTX-M-1 group | 30 (20.7) | 3 (23.1) |
| CTX-M-2 group | 19 (13.1) | 0 (0) |
| CTX-M-8 group | 7 (4.8) | 0 (0) |
| CTX-M-9 group | 82 (56.6) | 10 (76.9) |
| Non-CTX-M | 7 (4.8) | 0 (0) |
| Resistance to: | ||
| Ceftazidime | 25 (17.2) | 3 (23.1) |
| Aztreonam | 59 (40.7) | 5 (38.5) |
| Gentamicin | 34 (23.4) | 2 (15.4) |
| Amikacin | 1 (0.7) | 0 (0) |
| Minocycline | 96 (66.2) | 8 (61.5) |
| Chloramphenicol | 23 (15.9) | 1 (7.7) |
| Fosfomycin | 9 (6.2) | 0 (0) |
| Ciprofloxacin | 67 (46.2) | 11 (84.6) |
| Levofloxacin | 66 (45.5) | 11 (84.6) |
In the present study, we detected plasmids belonging to the IncF group, especially FIA and FIB, from 11 of 13 blaCTX-M-harboring E. coli isolates repeatedly recovered, as reported previously (14). We also found that the blaCTX-M-27 gene in E. coli O25b:H4-ST131 isolates was carried by plasmids belonging to the IncF or IncFIB group. The plasmid should have affinity for human-associated E. coli if the blaCTX-M-carrying plasmid is kept for a long period in an E. coli isolate that colonizes human bowels even in the absence of antimicrobials. In fact, the plasmids bearing blaCTX-M-15, blaCTX-M-14, or blaCTX-M-27 often belong to the IncF type (32–34). These IncF-type plasmids were also frequently found in human-associated E. coli strains that can usually accept various types of plasmids carrying multiple drug resistance genes (35, 36). The IncF group of plasmids has frequently been detected in blaCTX-M gene-harboring E. coli isolates repeatedly recovered from the feces of healthy people and could contribute to long-term carriage in the human bowel and to the widespread nature of blaCTX-M genes among various Gram-negative bacilli belonging to the family Enterobacteriaceae.
The CTX-M-15-producing E. coli O25b:H4-ST131 isolates distributed worldwide are usually resistant to FQs. In the present study, FQ-resistant and blaCTX-M-harboring E. coli isolates were continuously recovered from 11 (85%) of 13 long-term carriers, and FQ-resistant and blaCTX-M-27-harboring E. coli O25b:H4-ST131 isolates were repeatedly isolated from 4 participants. The FQ resistance rates of ESBL-producing and ESBL-nonproducing E. coli isolates recovered from Japanese hospitals were about 63.3% and 30%, respectively (37, 38). Interestingly, the FQ resistance rate (85%) of the blaCTX-M-harboring E. coli isolates colonizing Japanese food handlers for long periods found in the present study was considerably higher than the average FQ resistance rates of ESBL-producing and ESBL-nonproducing E. coli clinical isolates. This might predict the future endemicity of FQ-resistant E. coli O25b:H4-ST131 isolates harboring blaCTX-M genes, especially blaCTX-M-27, in Japan and surrounding Asian countries, as well as the global spread of FQ-resistant and CTX-M-15-producing E. coli O25b:H4-ST131 isolates.
In conclusion, our study elucidated that 70 (3.1%) of 2,230 healthy people were positive for ESBL producers when fecal specimens from these individuals were checked once and 52 (15.6%) of 333 people were positive for ESBL producers when fecal specimens were rechecked. This volunteer-based investigation could not systematically check all 2,563 participants through the investigation period, but 13 of the 52 carriers of ESBL producers who were checked more than twice (see Table S1 in the supplemental material) were found to carry ESBL producers for from more than 3 months to up to 2 years even in the absence of obvious antimicrobial treatment. Increasing human intestinal colonization of E. coli O25b:H4-ST131 and O1:H6-ST648 isolates that harbor blaCTX-M-carrying plasmids belonging to the IncF, IncFIB, or IncK group would contribute to the augmented rates of long-term carriage of ESBL-producing E. coli isolates in the bowels of ordinary people receiving no antimicrobial treatment, as well as in patients admitted to hospital settings. FQ-resistant E. coli O25b:H4-ST131 isolates harboring blaCTX-M-27 were repeatedly isolated from 4 of the 13 long-term carriers for several months. These observations might have implications for the choice of antimicrobial agents used for the treatment of community-acquired infections in the future, such as urinary tract infections and bacteremia caused by E. coli.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Ministry of Health, Labour and Welfare of Japan (grant no. H21-Shinkou-Ippan-008 and H24-Shinkou-Ippan-010) and in part by a grant from the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development, AMED.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02929-15.
REFERENCES
- 1.Paterson DL, Bonomo RA. 2005. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantón R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, Coque TM. 2008. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect 14(Suppl 1):S144–S153. [DOI] [PubMed] [Google Scholar]
- 3.Pitout JD, Laupland KB. 2008. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 4.Chong Y, Shimoda S, Yakushiji H, Ito Y, Miyamoto T, Kamimura T, Shimono N, Akashi K. 2013. Community spread of extended-spectrum β-lactamase-producing Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis: a long-term study in Japan. J Med Microbiol 62:1038–1043. doi: 10.1099/jmm.0.059279-0. [DOI] [PubMed] [Google Scholar]
- 5.Woerther PL, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janvier F, Delacour H, Tessé S, Larréché S, Sanmartin N, Ollat D, Rapp C, Mérens A. 2014. Faecal carriage of extended-spectrum β-lactamase-producing enterobacteria among soldiers at admission in a French military hospital after aeromedical evacuation from overseas. Eur J Clin Microbiol Infect Dis 33:1719–1723. doi: 10.1007/s10096-014-2141-8. [DOI] [PubMed] [Google Scholar]
- 7.Arcilla MS, van Hattem JM, Bootsma MC, van Genderen PJ, Goorhuis A, Schultsz C, Stobberingh EE, Verbrugh HA, de Jong MD, Melles DC, Penders J. 2014. The Carriage of Multiresistant Bacteria after Travel (COMBAT) prospective cohort study: methodology and design. BMC Public Health 14:410. doi: 10.1186/1471-2458-14-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George EA, Sankar S, Jesudasan MV, Sudandiradoss C, Nandagopal B. 2014. Incidence of extended spectrum beta lactamase producing Escherichia coli among patients, healthy individuals and in the environment. Indian J Med Microbiol 32:172–174. doi: 10.4103/0255-0857.129810. [DOI] [PubMed] [Google Scholar]
- 9.Solé M, Pitart C, Oliveira I, Fàbrega A, Muñoz L, Campo I, Salvador P, Alvarez-Martínez MJ, Gascón J, Marco F, Vila J. 2014. Extended spectrum β-lactamase-producing Escherichia coli faecal carriage in Spanish travellers returning from tropical and subtropical countries. Clin Microbiol Infect 20:O636–O639. doi: 10.1111/1469-0691.12592. [DOI] [PubMed] [Google Scholar]
- 10.Villar HE, Baserni MN, Jugo MB. 2013. Faecal carriage of ESBL-producing Enterobacteriaceae and carbapenem-resistant Gram-negative bacilli in community settings. J Infect Dev Ctries 7:630–634. doi: 10.3855/jidc.2900. [DOI] [PubMed] [Google Scholar]
- 11.Apisarnthanarak A, Bailey TC, Fraser VJ. 2008. Duration of stool colonization in patients infected with extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Clin Infect Dis 46:1322–1323. doi: 10.1086/533475. [DOI] [PubMed] [Google Scholar]
- 12.Alsterlund R, Axelsson C, Olsson-Liljequist B. 2012. Long-term carriage of extended-spectrum β-lactamase-producing Escherichia coli. Scand J Infect Dis 44:51–54. doi: 10.3109/00365548.2011.592987. [DOI] [PubMed] [Google Scholar]
- 13.Tham J, Walder M, Melander E, Odenholt I. 2012. Duration of colonization with extended-spectrum β-lactamase-producing Escherichia coli in patients with travellers' diarrhoea. Scand J Infect Dis 44:573–577. doi: 10.3109/00365548.2011.653582. [DOI] [PubMed] [Google Scholar]
- 14.Titelman E, Hasan CM, Iversen A, Nauclér P, Kais M, Kalin M, Giske CG. 2014. Faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae is common 12 months after infection and is related to strain factors. Clin Microbiol Infect 20:O508–O515. doi: 10.1111/1469-0691.12559. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. 2009. Change in the prevalence of extended-spectrum-β-lactamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother 63:72–79. doi: 10.1093/jac/dkn463. [DOI] [PubMed] [Google Scholar]
- 16.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 17.Yagi T, Kurokawa H, Shibata N, Shibayama K, Arakawa Y. 2000. A preliminary survey of extended-spectrum β-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS Microbiol Lett 184:53–56. doi: 10.1111/j.1574-6968.2000.tb08989.x. [DOI] [PubMed] [Google Scholar]
- 18.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J Antimicrob Chemother 61:1024–1028. doi: 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2011. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 8th ed Approved standard M7-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.Barrett TJ, Lior H, Green JH, Khakhria R, Wells JG, Bell BP, Greene KD, Lewis J, Griffin PM. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol 32:3013–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrico JA, Pinto FR, Simas C, Nunes S, Sousa NG, Frazão N, de Lencastre H, Almeida JS. 2005. Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J Clin Microbiol 43:5483–5490. doi: 10.1128/JCM.43.11.5483-5490.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray JL, Nielsen KM. 2005. Experimental methods for assaying natural transformation and inferring horizontal gene transfer. Methods Enzymol 395:491–520. doi: 10.1016/S0076-6879(05)95026-X. [DOI] [PubMed] [Google Scholar]
- 23.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 24.Luvsansharav UO, Hirai I, Niki M, Nakata A, Yoshinaga A, Moriyama T, Yamamoto Y. 2011. Prevalence of fecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae among healthy adult people in Japan. J Infect Chemother 17:722–725. doi: 10.1007/s10156-011-0225-2. [DOI] [PubMed] [Google Scholar]
- 25.Birgand G, Armand-Lefevre L, Lolom I, Ruppe E, Andremont A, Lucet JC. 2013. Duration of colonization by extended-spectrum β-lactamase-producing Enterobacteriaceae after hospital discharge. Am J Infect Control 41:443–447. doi: 10.1016/j.ajic.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 26.Paltansing S, Vlot JA, Kraakman ME, Mesman R, Bruijning ML, Bernards AT, Visser LG, Veldkamp KE. 2013. Extended-spectrum β-lactamase-producing Enterobacteriaceae among travelers from the Netherlands. Emerg Infect Dis 19:1206–1213. doi: 10.3201/eid1908.130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura N, Kozaki A, Sasaki T, Komatsubara A. 1999. Comparison of O-serotype distribution of Escherichia coli isolated from faecal specimens of patients with sporadic diarrhea and healthy persons and regional difference in Japan. Kansenshogaku Zasshi 73:53–61. (In Japanese.) doi: 10.11150/kansenshogakuzasshi1970.73.53. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi I, Osawa H, Harada T, Saika T, Kanayama A, Muraoka H, Uchino U, Hasegawa M, Sato Y, Numata K, Hashiguchi N, Nishida M. 1997. Study of diarrhea-inducing strains of Escherichia coli mainly isolated in Kanto area between June and September 1996. Kansenshogaku Zasshi 71:495–500. (In Japanese.) doi: 10.11150/kansenshogakuzasshi1970.71.495. [DOI] [PubMed] [Google Scholar]
- 29.Hibi H, Ishikawa K, Izumitani M, Tanaka T, Naide Y. 1991. Studies on the virulent factor of Escherichia coli isolated from urogenital infection—pilus type and adherence to human exfoliated uroepithelial cells etc. Hinyokika Kiyo 37:1525–1529. (In Japanese.). [PubMed] [Google Scholar]
- 30.Kennedy RP, Plorde JJ, Ptersdorf RG. 1965. Studies on the epidemiology of Escherichia coli infections. IV. Evidence for a nosocomial flora. J Clin Invest 44:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popovici M, Georgescu C, Dumitrescu M, Florescu D, Racoviţă DC. 1976. Evaluation of the etiopathogenic significance of E. coli serotypes O1–O25. Rev Ig Bacteriol Virusol Parazitol Epidemiol Pneumoftiziol Bacteriol Virusol Parazitol Epidemiol 21:85–94. (In Romanian.) [PubMed] [Google Scholar]
- 32.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu FY, Yao D, Pan JY, Chen C, Qin ZQ, Parsons C, Yang LH, Li QQ, Zhang XQ, Qu D, Wang LX. 2010. High prevalence of plasmid-mediated 16S rRNA methylase gene rmtB among Escherichia coli clinical isolates from a Chinese teaching hospital. BMC Infect Dis 10:184. doi: 10.1186/1471-2334-10-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumura Y, Yamamoto M, Higuchi T, Komori T, Tsuboi F, Hayashi A, Sugimoto Y, Hotta G, Matsushima A, Nagao M, Takakura S, Ichiyama S. 2012. Prevalence of plasmid-mediated AmpC β-lactamase-producing Escherichia coli and spread of the ST131 clone among extended-spectrum β-lactamase-producing E. coli in Japan. Int J Antimicrob Agents 40:158–162. doi: 10.1016/j.ijantimicag.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Naseer U, Sundsfjord A. 2011. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist 17:83–97. doi: 10.1089/mdr.2010.0132. [DOI] [PubMed] [Google Scholar]
- 36.Peirano G, Pitout JD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents 35:316–321. doi: 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Matsumura Y, Yamamoto M, Nagao M, Hotta G, Matsushima A, Ito Y, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group . 2012. Emergence and spread of B2-ST131-O25b, B2-ST131-O16 and D-ST405 clonal groups among extended-spectrum-β-lactamase-producing Escherichia coli in Japan. J Antimicrob Chemother 67:2612–2620. doi: 10.1093/jac/dks278. [DOI] [PubMed] [Google Scholar]
- 38.Ministry of Health, Labour and Welfare. 2011. Open report of The Japan Nosocomial Infections Surveillance (JANIS) database, January to December 2010. Ministry of Health, Labour and Welfare, Tokyo, Japan: (In Japanese.) http://www.nih-janis.jp/report/open_report/2010/3/1/ken_Open_Report_201000.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.