Abstract
Methanobactin, a small modified polypeptide synthesized by methanotrophs for copper uptake, has been found to be chromosomally encoded. The gene encoding the polypeptide precursor of methanobactin, mbnA, is part of a gene cluster that also includes several genes encoding proteins of unknown function (but speculated to be involved in methanobactin formation) as well as mbnT, which encodes a TonB-dependent transporter hypothesized to be responsible for methanobactin uptake. To determine if mbnT is truly responsible for methanobactin uptake, a knockout was constructed in Methylosinus trichosporium OB3b using marker exchange mutagenesis. The resulting M. trichosporium mbnT::Gmr mutant was found to be able to produce methanobactin but was unable to internalize it. Further, if this mutant was grown in the presence of copper and exogenous methanobactin, copper uptake was significantly reduced. Expression of mmoX and pmoA, encoding polypeptides of the soluble methane monooxygenase (sMMO) and particulate methane monooxygenase (pMMO), respectively, also changed significantly when methanobactin was added, which indicates that the mutant was unable to collect copper under these conditions. Copper uptake and gene expression, however, were not affected in wild-type M. trichosporium OB3b, indicating that the TonB-dependent transporter encoded by mbnT is responsible for methanobactin uptake and that methanobactin is a key mechanism used by methanotrophs for copper uptake. When the mbnT::Gmr mutant was grown under a range of copper concentrations in the absence of methanobactin, however, the phenotype of the mutant was indistinguishable from that of wild-type M. trichosporium OB3b, indicating that this methanotroph has multiple mechanisms for copper uptake.
INTRODUCTION
Methanotrophs, or methane-oxidizing bacteria, are a group of microbes with great environmental and industrial importance. For example, methanotrophs are well known to play a key role in controlling the net emission of methane from soils, a potent greenhouse gas with a global warming potential of ∼34 times that of carbon dioxide over a 100-year time frame (1). In fact, it is estimated that as much as 90% of methane generated in anaerobic soils via methanogenesis may be removed via methanotrophy (2). Further, methanotrophs oxidize methane under ambient temperatures and pressures and, thus, are attractive platforms for the valorization of methane to products such as single-cell protein, bioplastics, biofuels, and osmoprotectants (3–5).
Methanotrophs are fairly ubiquitous and are found in many different environments, including forest soils, landfill cover soils, agricultural soils, freshwater and marine sediments, and many other locations (4, 6, 7). Although methane oxidation is commonly associated with oxygen reduction, in the past 15 years, methane oxidation has also been shown to be coupled with sulfate, nitrite, and nitrate reduction (4, 8–10). Methanotrophs also show remarkable phylogenetic diversity, with aerobic methanotrophs grouping in the Gammaproteobacteria and Alphaproteobacteria as well as in the NC10 and Verrucomicrobia phyla (4, 6, 8).
A key issue affecting aerobic methanotrophic activity, particularly the activities in the Gammaproteobacteria and Alphaproteobacteria, is the availability of copper. It was first discovered >30 years ago that some methanotrophs exhibited a unique “copper switch,” where the form and activity of the methane monooxygenase (MMO) dramatically changes with changing copper availability. Specifically, it was found that under copper-limiting conditions, some methanotrophs synthesized a cytoplasmic or soluble methane monooxygenase (sMMO). As copper levels increased, expression of sMMO decreased, while expression and activity of a membrane-bound or particulate methane monooxygenase (pMMO) increased (4, 11, 12). The sMMO has a broad substrate range and, as a result, has great versatility for use in biocatalysis and bioremediation, but it also has a relatively poor affinity for methane (4, 13–16). pMMO, conversely, has a relatively narrow substrate range and a greater specificity for methane, suggesting that strategies to utilize methanotrophs to reduce methane emissions and/or remove methane from the atmosphere should target pMMO-expressing methanotrophs (14, 17, 18).
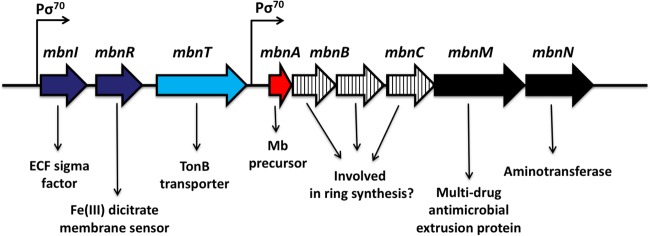
The mechanism underlying this copper switch was recently found to involve a novel copper-binding compound or chalkophore called methanobactin. Methanobactin is a small, modified polypeptide (<1,200 Da) with two heterocyclic rings, either an imidazole, an oxazolone, or a pyrazinedione ring, each with an associated enethiol group, that together are responsible for copper binding (19–22). Biochemical analyses indicated that methanobactin may be formed from a polypeptide precursor with the heterocyclic rings derived from an -X-Cys dipeptide sequence (22). Interrogation of available methanotrophic genomes found one possible candidate gene, mbnA. Deletion of mbnA in Methylosinus trichosporium OB3b showed that it is indeed the precursor of methanobactin and that it is part of a gene cluster (Fig. 1) with many genes of unknown function (possibly involved in methanobactin formation) as well as an aminotransferase (also possibly involved in methanobactin formation) and an extrusion protein (that may serve to secrete methanobactin). Upstream of mbnA is a gene encoding a TonB-dependent transporter (mbnT) that has been suggested, but not shown, to be involved in methanobactin uptake (23).
FIG 1.
Methanobactin gene cluster in Methylosinus trichosporium OB3b. ECF, extracytoplasmic function; Mb, methanobactin.
To elucidate the role of mbnT in methanobactin uptake, we created mutants of M. trichosporium OB3b in which mbnT has been selectively knocked out via marker exchange mutagenesis.
MATERIALS AND METHODS
Growth conditions.
Wild-type Methylosinus trichosporium OB3b and the mbnT::Gmr mutant (constructed as described below) were grown on nitrate mineral salt (NMS) medium (24) at 30°C with CH4 added at a methane-to-air ratio of 1:2. Liquid cultures were grown in 250-ml sidearm Erlenmeyer flasks with 30 to 50 ml of medium shaken at 200 rpm. Copper (as CuCl2) and methanobactin from M. trichosporium OB3b were filter sterilized and were added to NMS medium as described earlier (25). Growth was monitored by measuring the optical density at 600 nm (OD600) with a Genesys 20 visible spectrophotometer (Spectronic Unicam, Waltham, MA) at 3- to 12-hour intervals. Cultures were grown in at least duplicate biological replicates and were harvested at late exponential phase for analysis of specific gene expression and metal distribution.
Knockout of mbnT.
Marker exchange mutagenesis was applied to create a knockout of mbnT, which encodes a TonB-dependent transporter using the protocol described in the work of Semrau et al. (23). Briefly, 3′ and 5′ DNA regions of mbnT (arms A and B, respectively) were selectively amplified by PCR using the primers listed in Table 1. These PCR products were then digested with BamHI, separated by gel electrophoresis, and purified using the QIAquick gel extraction kit (Qiagen) by following the manufacturer's instructions. Arms A and B were ligated and were again PCR amplified. The amplified product was digested with EcoRI and HindIII and was inserted into pK18mobsacB, yielding the construct pWG01. The gentamicin resistance gene (Gmr) was then excised from plasmid p34S-Gm using BamHI. This was then inserted into the BamHI site between arms A and B to give the construct pWG011. This was then used to transform Escherichia coli S17.1 (26). E. coli S17.1 was then conjugated with M. trichosporium OB3b as described by Martin and Murrell (27). Transconjugants were identified by plating cells onto NMS plates with 2.5 μg · ml−1 gentamicin. Residual contamination by E. coli S17.1 was then removed by subsequently growing the resulting mbnT::Gmr mutant of M. trichosporium OB3b in NMS medium with 2.5 μg · ml−1 gentamicin and 10 μg · ml−1 nalidixic acid. Successful knockout of mbnT via double homologous recombination was confirmed by screening the kanamycin-sensitive and sucrose-resistant phenotype, by PCR, and by sequencing.
TABLE 1.
Primers used in this study
| Primer | Targeted gene | Sequencea (5′–3′) | Reference |
|---|---|---|---|
| Arm A forward | mbnT | ATTTTTgaattcCCAGAAATATGAGATTCCGCb | This study |
| Arm A reverse | ATTTTTggatccCACGACCAGATCGATGATACb | ||
| Arm B forward | mbnT | ATTTTTggatccTTCGGTTCGATCAACGAGGb | This study |
| Arm B reverse | ATTTTTaagcttGCCAATCAGCGTGGAGAACCb | ||
| qpmoA_FO | pmoA | TTCTGGGGCTGGACCTAYTTC | 48 |
| qpmoA_RO | CCGACAGCAGCAGGATGATG | ||
| qmmoX_FO | mmoX | TCAACACCGATCTSAACAACG | 48 |
| qmmoX_RO | TCCAGATTCCRCCCCAATCC | ||
| q16S rRNA_FO | 16S rRNA | GCAGAACCTTACCAGCTTTTGAC | 48 |
| q16S rRNA_RO | CCCTTGCGGGAAGGAAGTC | ||
| qmbnA_FO | mbnA | TGGAAACTCCCTTAGGAGGAA | 23 |
| qmbnA_RO | CTGCACGGATAGCACGAAC |
Y, S, and R are the IUPAC DNA codes for the C/T, C/G, and A/G nucleobases, respectively.
Lowercase letters indicate EcoRI, BamHI, or HindIII restriction site sequences included in these primers.
RNA extraction and RT.
RNA was isolated using a method described previously (23). Briefly, 2.5 ml of stop solution (5% buffer equilibrated phenol [pH 7.3] in ethanol) was first added to cultures (22.5 ml) to stop synthesis of new mRNA. Cell pellets were then collected by centrifugation at 4,300 × g for 15 min at 4°C. The cells were resuspended in 0.75 ml of extraction buffer (100 mM Tris-HCl [pH 8.0], 1.5 M NaCl, and 1% [wt/vol] hexadecyltrimethylammonium bromide [CTAB]) before lysis using 20% SDS, 20% lauryl sarcosine, and bead beating. Subsequent steps of RNA extraction were then performed as described previously (23–25). Total RNA was then subjected to RNase-Free DNase treatment until free of DNA contamination as proven via PCR amplification of the 16S rRNA gene. The purified RNA was quantified spectrophotometrically using NanoDrop (NanoDrop ND-1000; NanoDrop Technologies, Inc., Wilmington, DE). RNA samples were stored at −80°C and were used for cDNA synthesis within 2 days of extraction. DNA-free total RNA (500 ng) was treated with SuperScript III reverse transcriptase for reverse transcription (RT) of mRNA to cDNA (Invitrogen, Carlsbad, CA) by following the manufacturer's instructions.
RT-qPCR.
Reverse transcription-quantitative PCR (RT-qPCR) analyses were performed to determine the relative expression of the pmoA, mmoX, and mbnA genes in M. trichosporium OB3b and in the mbnT::Gmr mutant strains grown at various concentrations of copper and methanobactin. Gene-specific primers (Table 1) were used for the RT-qPCR analyses, and their specificity was verified by sequencing and gel electrophoresis. Measurements were performed in 96-well PCR plates using the CFX Connect real-time PCR detection system (Bio-Rad, Hercules, CA). In each well, quantitative PCRs (qPCRs) (20 μl) consisted of 0.8 μl cDNA, 1× iTaq universal SYBR green supermix (Bio-Rad, Hercules, CA), 0.5 μM each of the forward and reverse primers, and nuclease-free sterile water (Ambion/Life Technologies, Grand Island, NY). A three-step thermal cycler program, with an initial denaturation at 95°C for 3 min and 40 cycles of denaturation (94°C for 20 s), annealing (58°C for 20 s), and extension (68°C for 30 s), was performed. The specificity of qPCR products was again confirmed by melting curve analysis with temperatures ranging from 55°C to 95°C after the completion of amplification cycles. The threshold amplification cycle (CT) values were then imported from CFX Manager software (Bio-Rad) into Microsoft Excel to quantify the relative levels of expression of different genes. The comparative CT method (2−ΔΔCT) (28) was used to calculate relative gene expression levels using 16S rRNA as the housekeeping gene.
Metal analysis.
Copper associated with the biomass of wild-type M. trichosporium OB3b and the mbnT::Gmr mutant was determined as described previously (29). Briefly, cultures were harvested by centrifugation at 4,300 × g for 15 min. The cell pellets were resuspended in 1 ml morpholinepropanesulfonic acid (MOPS) buffer before being stored at −80°C. Before metal measurement, 1 ml of 70% nitric acid (vol/vol) was added to the cell suspension and was incubated for 2 h at 95°C with inversion every 20 min. Copper associated with biomass was subsequently analyzed using an inductively coupled plasma mass spectrometer (Agilent Technologies, Santa Clara, CA). At least duplicate biological samples for every condition were analyzed.
Methanobactin in spent medium and in cell extracts.
For characterization of the location of methanobactin from wild-type M. trichosporium OB3b and the mbnT::Gmr mutant, cells were cultured in 12 liters of NMS medium amended with 0.2 μM CuCl2 in a 15-liter New Brunswick fermentor at 30°C for 48 h. Following the incubation period, 10 liters of the culture was removed and 10 liters of fresh NMS medium was added to the fermentor, and the copper concentration increased to 5 μM. This sequence was then repeated with increasing copper concentrations to 10 and 20 μM in subsequent fermentor turnovers.
The extracellular fraction and cells from each 10-liter sample were separated via tangential-flow filtration using a 10,000-Da molecular mass filter as previously described (30). The cells from the retentate were then harvested by centrifugation at 13,200 × g at 4°C. The pellet was resuspended in 10 mM phosphate buffer, pH 7.3, and was centrifuged at 13,200 × g at 4°C. This cell pellet was then resuspended in a minimal volume of 10 mM phosphate buffer at a pH of 6.8 plus 1 μg DNase · ml−1 and was lysed by three passes through an EmulsiFlex-C3 high-pressure homogenizer at 15,000 lb/in2 (Avestin Inc., Ottawa, ON, Canada) at 4°C. The cell extract was then centrifuged at 13,000 × g for 20 min to remove unlysed cells followed by filtration though 0.2-μm Millipore filters (Billerica, MA).
Methanobactin antibody generation.
Antibodies to methanobactin (Amb) from M. trichosporium OB3b were produced in LOU/c rats, which were immunized subcutaneously and intraperitoneally with a methanobactin-ovalbumin fusion protein (50 μg), 5 nmol CpG oligonucleotide (Tib Molbiol, Berlin, Germany), 500 μl phosphate-buffered saline, and 500 μl incomplete Freund's adjuvant. A boost without adjuvant was given 6 weeks after the primary injection. Tissue culture supernatants (TCS) were tested in a solid-phase immunoassay with methanobactin coupled to bovine serum albumin (BSA) or an irrelevant peptide coupled to BSA-coated enzyme-linked immunosorbent assay (ELISA) plates at a concentration of 4 μg · ml−1. Monoclonal antibodies (MAbs) from TCS bound to methanobactin were detected with horseradish peroxidase (HRP)-conjugated MAbs against the rat IgG isotypes (TIB173 IgG2a, TIB174 IgG2b, and TIB170 IgG1 [all from the ATCC] and R-2c IgG2c [homemade]), thus avoiding MAbs of the IgM class. HRP was visualized with ready-to-use 3,3′,5,5′-tetramethylbenzidine (TMB) (1-step Ultra TMB ELISA; Thermo Fisher, Waltham, MA). Hybridomas that reacted specifically with methanobactin were frozen, and the antibody containing TCS was used in subsequent blots.
Derivation of polyvinylidene difluoride membranes.
The N terminus of methanobactin from M. trichosporium OB3b is lost during ring formation (19, 21), preventing methanobactin from binding to polyvinylidene difluoride (PVDF) membranes (31). Poly(allylamine) was therefore attached to PVDF membranes, which enables methanobactin binding via its C terminus. Poly(allylamine) was attached to PVDF membranes by the derivatization procedure described by Rodrigues et al. (32). Briefly, PVDF sheets were etched in alcoholic KOH and then reacted with poly(allylamine) under alkaline conditions. Next, the amino groups were reacted with 1,4-phenylene diisothiocyanate (DITC), converting the amino-modified PVDF to DITC-functionalized membranes (DITC-phosphonoacetic acid [PAA]-PVDF membranes).
Chemiluminescence Western dot blots.
DITC-PAA-PVDF transfer membranes were sized to fit an 8-by-12 well Bio-Dot dot blot (Bio-Rad Inc., Hercules, CA); they were washed with 20 mM Tris-HCl plus 0.5 M NaCl (Tris-buffered saline [TBS]) at pH 7.5 and were loaded with filter paper onto this dot blotter. Samples (spent medium and cell extracts) were then loaded under vacuum and were dried for 30 min under vacuum. The membrane was then wetted with 50% CH3OH-50% H2O and was washed twice in TBS at room temperature. The membrane was then incubated overnight in 0.2% nonfat dry milk in TBS at 4°C with the TBS subsequently decanted. The membrane was then resuspended in TBS plus 0.1% Tween 20 (TTBS) at pH 7.5 and incubated for 10 min at room temperature. The membrane was then suspended in fresh TTBS and incubated at room temperature for 1 h. The TTBS was decanted, and the membrane was resuspended in the primary antibody buffer (TTBS plus 0.2% nonfat dry milk [antibody buffer] plus 5.7 μg Amb · ml−1 [primary antibody solution]) and incubated overnight at 4°C. Following incubation, the primary antibody solution was decanted, and the membrane was resuspended in TTBS and incubated for 10 min with agitation, followed by one change in TTBS with incubation for an additional 10 min. The TTBS was decanted, and the membrane was resuspended in secondary antibody solution consisting of 33 μl of goat anti-rat Ig (H/L)-alkaline phosphatase from AbD Serotec (Atlanta, GA) to 100 ml of antibody buffer and incubated for 2 h. Following incubation, the secondary antibody solution was decanted, and the membrane was washed three times with TTBS as described above. Visualization of the blot was done via the Bio-Rad Immun-Star AP substrate for chemiluminescence (Hercules, CA, USA) by following the manufacturer's suggested procedure.
RESULTS
Using marker exchange protocols, a transconjugant colony with a double homologous recombination event in which mbnT was successfully knocked out was identified (Fig. 2). This was confirmed by sequencing as well as by verifying that the mutant was gentamicin and sucrose resistant but sensitive to kanamycin (data not shown).
FIG 2.
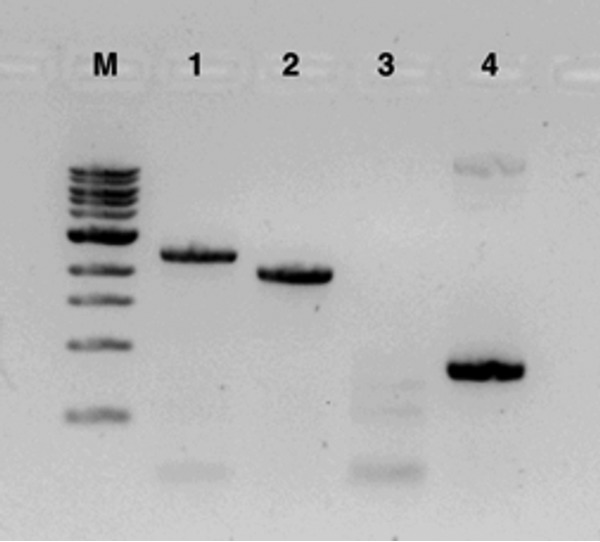
Verification of knockout of mbnT in M. trichosporium by PCR. M, molecular weight markers; lane 1, PCR of mbnT from the M. trichosporium OB3b mbnT::Gmr mutant; lane 2, PCR of mbnT from wild-type M. trichosporium OB3b; lane 3, PCR of pK18mobsacB backbone in M. trichosporium OB3b mbnT::Gmr; lane 4, PCR of pK18mobsacB backbone in pWG011.
The phenotype of the mbnT::Gmr mutant was then further examined and compared to that of wild-type M. trichosporium. When grown in various copper concentrations, both the mbnT::Gmr mutant and the wild type had increasing amounts of copper associated with biomass (Fig. 3A). Further, gene expression in both the mutant and the wild type showed clear evidence of the copper switch; i.e., as copper increased, expression of mmoX decreased by several orders of magnitude, while pmoA expression increased over an order of magnitude (Fig. 3B and C). Finally, expression of mbnA, encoding the precursor polypeptide of methanobactin, decreased substantially in wild-type M. trichosporium OB3b and in the mbnT::Gmr mutant as copper increased, indicating that the knocking out of mbnT did not affect methanobactin expression (Fig. 3D).
FIG 3.
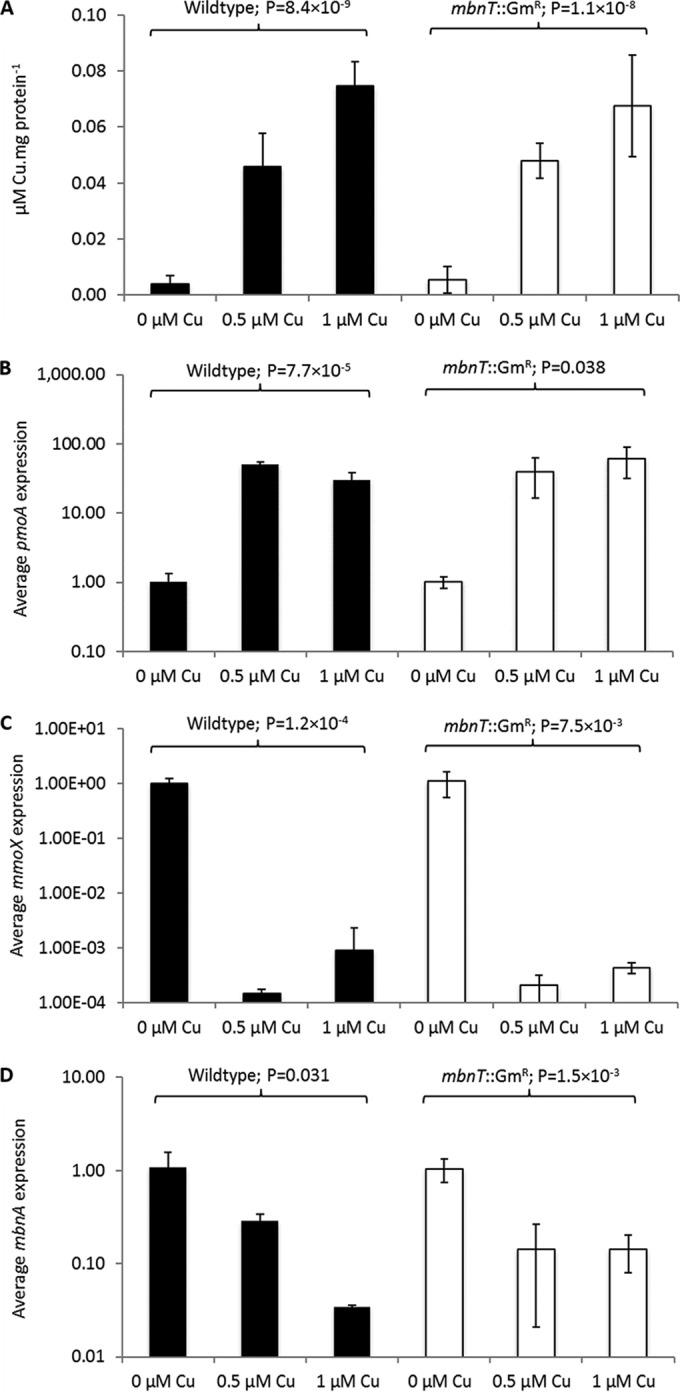
Characterization of wild-type M. trichosporium OB3b (black bars) and the mbnT::Gmr mutant (white bars) grown in the presence of various amounts of copper. (A) Copper associated with biomass; (B) RT-qPCR of pmoA; (C) RT-qPCR of mmoX; (D) RT-qPCR of mbnA. Error bars indicate standard deviations from at least duplicate biological replicates. Indicated P values are from one-way analysis of variance (ANOVA).
These findings suggest either that mbnT is not involved in copper uptake (i.e., binding of copper-methanobactin complexes) or that there are multiple mechanisms for copper uptake in M. trichosporium OB3b, such that the copper switch is still operative. To differentiate between these possibilities, methanobactin in the spent medium and cell extracts of the mbnT::Gmr mutant and wild-type strain of M. trichosporium OB3b was assayed for a wide range of copper concentrations using immunoblotting assays. As shown in Fig. 4, as the growth concentration of copper increased, the amount of methanobactin in the spent medium decreased in wild-type M. trichosporium OB3b but was readily apparent in the spent medium of the mbnT::Gmr mutant at all tested copper concentrations. Conversely, methanobactin was found in the cell extract of M. trichosporium OB3b under all conditions, indicating that methanobactin was taken up after secretion. No methanobactin was ever observed in the cell extract of the mbnT::Gmr mutant, indicating that the mutant produced and secreted methanobactin but was unable to subsequently take it up.
FIG 4.
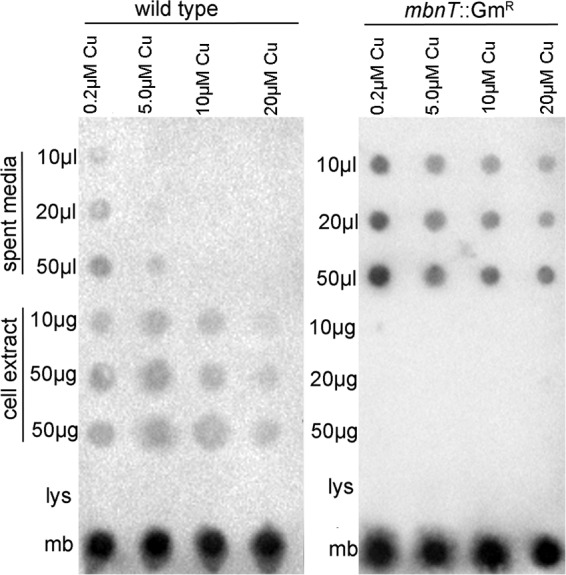
Immuno-blotting assays for location of methanobactin in wild-type M. trichosporium OB3b and in the mbnT::Gmr mutant as a function of the concentration of copper in the growth medium (0.2, 5, 10, or 20 μM copper). Fifty nanomoles lysozyme (lys) and 50 nmol methanobactin (mb) were used as negative and positive controls, respectively.
The mbnT::Gmr mutant and wild-type strain of M. trichosporium OB3b were then grown in the presence of 1 μM copper and various amounts of copper-free methanobactin. As shown in Fig. 5A, in the presence of either 5 or 50 μM methanobactin, copper associated with the biomass of the mbnT::Gmr mutant decreased >3-fold, while no significant change in the copper levels of wild-type M. trichosporium OB3b was observed. Further, expression of mmoX increased >3 orders of magnitude in the mbnT::Gmr mutant, while pmoA expression dropped by approximately 8-fold. No significant change in the expression of either mmoX or pmoA was observed, however, in wild-type M. trichosporium OB3b (Fig. 5B and C). Collectively, these data show that in the presence of a molar excess of methanobactin, copper was still bioavailable to wild-type M. trichosporium OB3b but was not for the mbnT::Gmr mutant. Additionally, it was assayed whether the addition of exogenous methanobactin affected mbnA expression in wild-type M. trichosporium OB3b and in the mbnT::Gmr mutant. As shown in Fig. 5D, as increasing amounts of methanobactin were added, mbnA expression increased in both the wild-type and mutant strains.
FIG 5.
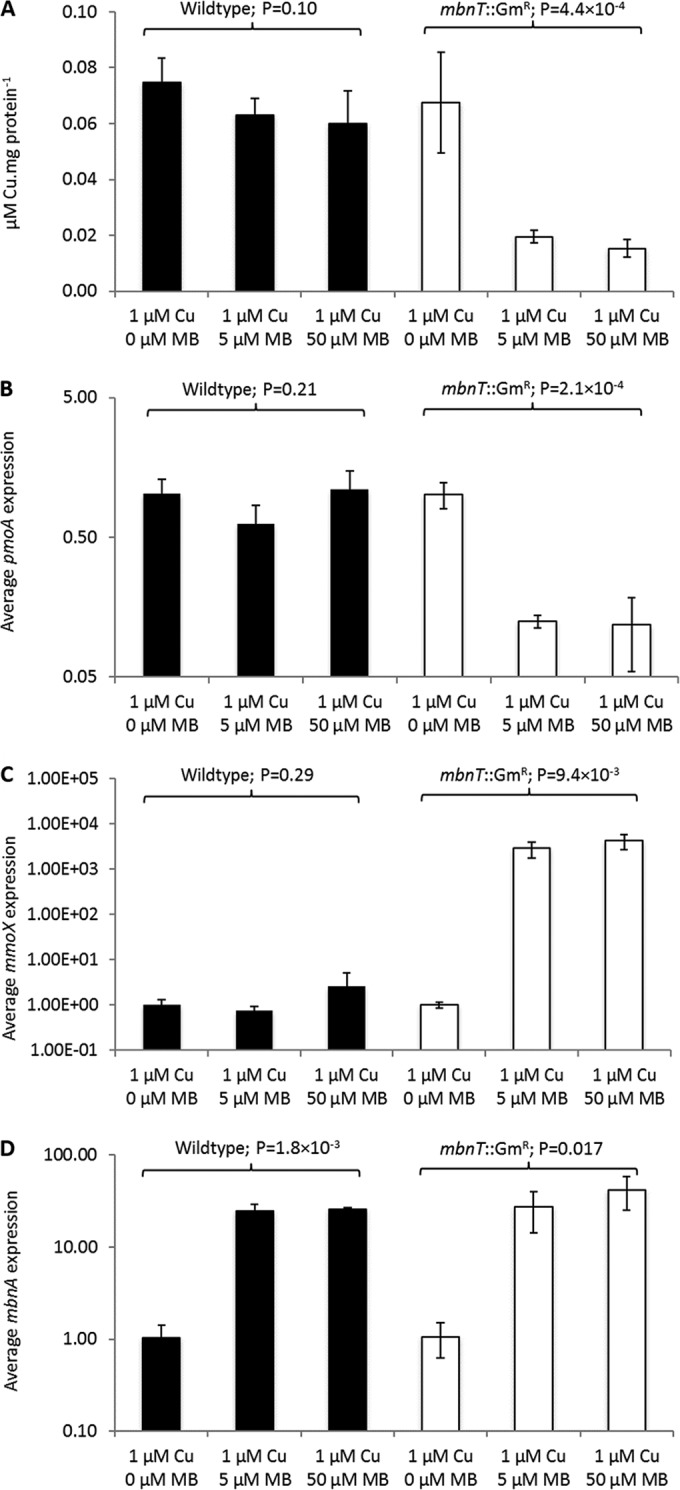
Characterization of wild-type M. trichosporium OB3b (black bars) and the mbnT::Gmr mutant (white bars) grown in the presence of 1 μM copper and various amounts of methanobactin (MB). (A) Copper associated with biomass; (B) RT-qPCR of pmoA; (C) RT-qPCR of mmoX; (D) RT-qPCR of mbnA. Error bars indicate standard deviations from at least duplicate biological replicates. Indicated P values are from one-way analysis of variance (ANOVA).
DISCUSSION
Since the discovery of the methanobactin gene cluster, it has been speculated that a TonB-dependent transporter encoded by mbnT is responsible for methanobactin uptake (23). Here, we show that methanobactin uptake is indeed mediated by mbnT, as (i) methanobactin was taken up by wild-type M. trichosporium OB3b but not by the mbnT::Gmr mutant and (ii) the mbnT::Gmr mutant of M. trichosporium OB3b was unable to take up copper if methanobactin was exogenously added to bind copper, but wild-type M. trichosporium OB3b was able to take up copper.
The data also show, however, that M. trichosporium OB3b has an alternative mechanism(s) for copper uptake; i.e., in the absence of any exogenous methanobactin, the amounts of copper in the wild-type and mbnT::Gmr strains of M. trichosporium OB3b were indistinguishable. The conclusion of multiple copper uptake systems, however, is not novel, as it was reported earlier that at least two pathways for copper uptake exist in M. trichosporium OB3b (33). Such redundancy in copper uptake systems in methanotrophs, although unusual compared to those of other microbes, can be explained when one considers the importance of copper in methanotrophic metabolism. That is, methanotrophs expressing pMMO have a strong need for copper, as it occupies at least two of three metal centers found in purified pMMO (4, 33–35).
An interesting issue is that, as found earlier in a mutant of M. trichosporium OB3b where mbnA, encoding the precursor polypeptide of methanobactin, was knocked out, the copper switch still existed in the mbnT::Gmr mutant. Genomic analyses have found that mbnT is part of a FecIRA-like gene cluster; i.e., mbnT is preceded by mbnR and mbnI, encoding a putative membrane sensor and an extracytoplasmic function sigma factor, respectively (36). Such a system is frequently found in siderophore synthesis where an outer membrane transporter binds a ferrisiderophore, transmitting a signal to a membrane sensor that then activates an extracytoplasmic function sigma factor. This ultimately induces the expression of genes required for siderophore synthesis, as well as, in some cases, genes unrelated to siderophore production or uptake, e.g., genes encoding exotoxins and proteases (37–43). Given this similarity, it has been speculated that after MbnT binds copper-methanobactin, a signal cascade results whereby methanobactin synthesis and possibly expression of mmo and pmo operons are controlled (36).
The findings presented here, however, suggest that although such a signal cascade may exist after MbnT binds copper-methanobactin, such a regulatory scheme does not include the copper switch between sMMO and pMMO. It is also difficult to conclude from our data that this signal cascade affects the expression of mbnA. That is, mbnA expression in wild-type M. trichosporium OB3b and in the mbnT::Gmr mutant decreased significantly with increasing copper, but the magnitude of the drop in expression was greater in the wild-type strain (Fig. 3D). Further, in the presence of 1 μM copper and various amounts of exogenous methanobactin, mbnA expression in wild-type M. trichosporium OB3b and in the mbnT::Gmr mutant responded with the same pattern (Fig. 5D). It appears that another regulatory circuit is involved in controlling the expression of mbnA, but the possibility that such expression is also controlled to some extent by mbnI, which is indirectly activated by MbnT binding copper-methanobactin, cannot be excluded at this time.
In conclusion, here we report the successful knockout of mbnT and show that this is responsible for methanobactin uptake. The phenotype of the mbnT::Gmr mutant, however, indicates that M. trichosporium OB3b has multiple systems for copper uptake. It is tempting to speculate that methanobactin may serve as a high-affinity system to collect copper, but when copper is not limiting, an alternative lower-affinity system is used. Such a hypothesis is supported by the finding that expression of mbnA decreases with increasing copper in wild-type M. trichosporium OB3b and in the mbnT::Gmr mutant.
The nature of this imputed low-affinity copper uptake mechanism is still elusive, but clues from other methanotrophs, e.g., Methylomicrobium album BG8 and Methylococcus capsulatus Bath, may provide some suggestions. That is, it has been shown that in M. album BG8, there exists an outer membrane protein, CorA, that is copper repressible and may serve to bind copper (44). Further, it has been found that M. capsulatus Bath synthesizes a similar outer membrane protein, MopE, as well as a secreted truncated form, MopE*, both of which bind Cu(II) (45–47). A gene encoding a protein similar to CorA and MopE, mbnP, is adjacent to the methanobactin gene cluster in M. trichosporium OB3b (36), and it may be that this serves as an alternative copper uptake mechanism in M. trichosporium OB3b. To determine if this is indeed the case, it is recommended that the protein and lipid composition of the outer membrane of M. trichosporium OB3b be characterized under various copper concentrations to see if any significant changes in MbnP occur. It may also be informative to create double knockouts, e.g., knockouts of both mbnP and mbnT or mbnP and mbnA, to determine if the resulting double mutants of M. trichosporium OB3b are severely inhibited in their ability to collect copper.
Funding Statement
This research was supported by the Office of Science (Biological and Environmental Research), U.S. Department of Energy, grant DE-SC0006630, to J.D.S. and A.A.D. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Myhre G, Shindell D, Breon F-M, Collins W, Fuglestvedt J, Huang J, Koch D, Lamarque JF, Lee D, Mendoza B, Nakajima T, Robock A, Stephens G, Takamura T, Zhang H. 2013. Anthropogenic and natural radiative forcing, p 659–740. In Stocker TF, Qin D, Plattner GK, Tignor MMB, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM (ed), Climate change 2013: the physical science basis. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 2.Chowdury TR, Dick RP. 2013. Ecology of aerobic methanotrophs in controlling methane fluxes from wetlands. Appl Soil Ecol 65:8–22. doi: 10.1016/j.apsoil.2012.12.014. [DOI] [Google Scholar]
- 3.Khmelenina VN, Rozova ON, But SY, Mustakhimov II, Reshetnikov AS, Beschastnyl AP, Trotsenko YA. 2015. Biosynthesis of secondary metabolites in methanotrophs: biochemical and genetic aspects (review). Appl Biochem Microbiol 51:150–158. doi: 10.1134/S0003683815020088. [DOI] [PubMed] [Google Scholar]
- 4.Semrau JD, DiSpirito AA, Yoon S. 2010. Methanotrophs and copper. FEMS Microbiol Rev 34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 5.Strong PJ, Xie S, Clarke WP. 2015. Methane as a resource: can the methanotrophs add value? Environ Sci Technol 49:4001–4018. doi: 10.1021/es504242n. [DOI] [PubMed] [Google Scholar]
- 6.Op den Camp H, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland N-K, Pol A, Dunfield PF. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep 1:293–306. doi: 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- 7.Hanson RS, Hanson TE. 1996. Methanotrophic bacteria. Microbiol Rev 60:439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJ, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MS, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 9.Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 10.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 11.Choi D-W, Kunz R, Boyd ES, Semrau JD, Antholine WE, Han J-I, Zahn JA, Boyd JM, de la Mora A, DiSpirito AA. 2003. The membrane-associated methane monooxygenase (pMMO) and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J Bacteriol 185:5755–5764. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanley SH, Prior SD, Leak DJ, Dalton H. 1983. Copper stress underlies the fundamental change in intracellular location of methane monooxygenase in methane-oxidising organisms: studies in batch and continuous cultures. Biotechnol Lett 5:487–492. doi: 10.1007/BF00132233. [DOI] [Google Scholar]
- 13.Kalyuzhnaya MG, Puri AW, Lidstrom ME. 2015. Metabolic engineering in methanotrophic bacteria. Metab Eng 29:142–152. doi: 10.1016/j.ymben.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Lee SW, Keeney DR, Lim DH, DiSpirito AA, Semrau JD. 2006. Mixed pollutant degradation by Methylosinus trichosporium OB3b expressing either soluble or particulate methane monooxygenase: can the tortoise beat the hare? Appl Environ Microbiol 72:7503–7509. doi: 10.1128/AEM.01604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semrau JD. 2011. Bioremediation via methanotrophy: overview of recent findings and suggestions for future research. Front Microbiol 2:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trotsenko YA, Murrell JC. 2008. Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol 63:183–229. doi: 10.1016/S0065-2164(07)00005-6. [DOI] [PubMed] [Google Scholar]
- 17.Lontoh S, Semrau JD. 1998. Methane and trichloroethylene degradation by Methylosinus trichosporium expressing particulate methane monooxygenase. Appl Environ Microbiol 64:1106–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon S, Carey JN, Semrau JD. 2009. Feasibility of atmospheric methane removal using methanotrophic biotrickling filters. Appl Microbiol Biotechnol 83:949–956. doi: 10.1007/s00253-009-1977-9. [DOI] [PubMed] [Google Scholar]
- 19.Behling LA, Hartsel SC, Lewis DE, DiSpirito AA, Choi DW, Masterson LR, Veglia G, Gallagher WH. 2008. NMR, mass spectrometry and chemical evidence reveal a different chemical structure for methanobactin that contains oxazolone rings. J Am Chem Soc 130:12604–12605. doi: 10.1021/ja804747d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Ghazouani A, Baslé A, Gray J, Graham DW, Firbank SJ, Dennison C. 2012. Variations in methanobactin structure influences copper utilization by methane-oxidizing bacteria. Proc Natl Acad Sci U S A 109:8400–8404. doi: 10.1073/pnas.1112921109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, Asunskis D, Sherwood PMA. 2004. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science 305:1612–1615. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 22.Krentz BD, Mulheron HJ, Semrau JD, DiSpirito AA, Bandow N, Haft DH, Vuilleumier S, Murrell JC, McEllistrem MT, Hartsel SC, Gallagher W. 2010. A comparison of methanobactins from Methylosinus trichosporium OB3b and Methylocystis strain SB2 predicts methanobactins are synthesized from diverse peptide precursors modified to create a common core for binding and reducing copper ions. Biochemistry 49:10117–10130. doi: 10.1021/bi1014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semrau JD, Jagadevan S, DiSpirito AA, Khalifa A, Scanlan J, Bergman BH, Freemeier BC, Baral BS, Bandow NS, Vorobev A, Haft DH, Vuilleumier S, Murrell JC. 2013. Methanobactin and MmoD work in concert to act as the ‘copper-switch’ in methanotrophs. Environ Microbiol 15:3077–3086. [DOI] [PubMed] [Google Scholar]
- 24.Whittenbury R, Phillips KC, Wilkinson JF. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol 61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 25.Vorobev A, Jagadevan S, Baral BS, DiSpirito AA, Freemeier BC, Bergman BH, Bandow NL, Semrau JD. 2013. Detoxification of mercury by methanobactin from Methylosinus trichosporium OB3b. Appl Environ Microbiol 79:5918–5926. doi: 10.1128/AEM.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol Gen Genet 196:413–420. doi: 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- 27.Martin H, Murrell JC. 1995. Methane monooxygenase mutants of Methylosinus trichosporium constructed by marker-exchange mutagenesis. FEMS Microbiol Lett 127:243–248. doi: 10.1111/j.1574-6968.1995.tb07480.x. [DOI] [Google Scholar]
- 28.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 29.Kalidass B, Ul-Haque MF, Baral BS, DiSpirito AA, Semrau JD. 2015. Competition between metals for binding to methanobactin enables expression of soluble methane monooxygenase in the presence of copper. Appl Environ Microbiol 81:1024–2031. doi: 10.1128/AEM.03151-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bandow NL, Gallagher WH, Behling L, Choi DW, Semrau JD, Hartsel SC, Gilles VS, Dispirito AA. 2011. Isolation of methanobactin from the spent media of methane-oxidizing bacteria. Methods Enzymol 495:259–269. doi: 10.1016/B978-0-12-386905-0.00017-6. [DOI] [PubMed] [Google Scholar]
- 31.Zahn JA, DiSpirito AA. 1996. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J Bacteriol 178:1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodrigues JD, Combrink J, Brandt WF. 1994. Derivatization of polyvinylidene diflluoride membranes for solid-phase sequencing analysis of a phosphorylated sea urchin embryo histone H1 peptide. Anal Biochem 216:365–372. doi: 10.1006/abio.1994.1054. [DOI] [PubMed] [Google Scholar]
- 33.Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. 2010. Oxidation of methane by a biological dicopper centre. Nature 465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hakemian AS, Kondapalli KC, Tesler J, Hoffman BM, Stemmler TL, Rosenzweig AC. 2008. The metal centers of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry 47:6793–6801. doi: 10.1021/bi800598h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinho M, Choi DW, DiSpirito AA, Antholine WE, Semrau JD, Münck E. 2007. Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus Bath: evidence for a dinuclear iron center. J Am Chem Soc 129:15783–15785. doi: 10.1021/ja077682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenney GE, Rosenzweig AC. 2013. Genome mining for methanobactins. BMC Biol 11:17. doi: 10.1186/1741-7007-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braun V, Mahren S, Sauter A. 2006. Gene regulation by transmembrane signaling. Biometals 19:103–113. doi: 10.1007/s10534-005-8253-y. [DOI] [PubMed] [Google Scholar]
- 38.Brooks BE, Buchanan SK. 2008. Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochim Biophys Acta 1778:1930–1945. doi: 10.1016/j.bbamem.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crosa JH. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev 61:319–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grosse C, Friedrich S, Nies DH. 2007. Contribution of extracytoplasmic function sigma factors to transition metal homeostasis in Cupriavidus metallidurans strain CH34. J Mol Microbiol Biotechnol 12:227–240. doi: 10.1159/000099644. [DOI] [PubMed] [Google Scholar]
- 41.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 99:7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahren S, Braun V. 2003. The FecI extracytoplasmic-function sigma factor of Escherichia coli interacts with the β′ subunit of RNA polymerase. J Bacteriol 185:1796–1802. doi: 10.1128/JB.185.6.1796-1802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visca P, Leoni L, Wilson MJ, Lamont IL. 2002. Iron transport and regulation, cell signaling and genomics: lessons from Escherichia coli and Pseudomonas. Mol Microbiol 45:1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x. [DOI] [PubMed] [Google Scholar]
- 44.Berson O, Lidstrom ME. 1997. Cloning and characterization of corA, a gene encoding for a copper-repressible polypeptide in the type I methanotroph, Methylomicrobium albus BG8. FEMS Microbiol Lett 148:169–174. doi: 10.1111/j.1574-6968.1997.tb10284.x. [DOI] [PubMed] [Google Scholar]
- 45.Karlsen OA, Berven FS, Stafford GP, Larsen Ø, Murrell JC, Jensen HB, Fjellbirkeland A. 2003. The surface-associated and secreted MopE protein of Methylococcus capsulatus (Bath) corresponds to changes in the concentration of copper in the growth medium. Appl Environ Microbiol 69:2386–2388. doi: 10.1128/AEM.69.4.2386-2388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helland R, Fjellbirkeland A, Karlsen OA, Ve T, Lillehaug JR, Jensen HB. 2008. An oxidized tryptophan facilitates copper binding in Methylococcus capsulatus-secreted protein MopE. J Biol Chem 283:13897–13904. doi: 10.1074/jbc.M800340200. [DOI] [PubMed] [Google Scholar]
- 47.Ve T, Mathisen K, Helland R, Karlsen OA, Fjellbirkeland A, Røhr Å Andersson KK, Pedersen R-B, Lillehaug JR, Jensen HB. 2012. The Methylococcus capsulatus (Bath) secreted protein, MopE*, binds both reduced and oxidized copper. PLoS One 7(8):e43146. doi: 10.1371/journal.pone.0043146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knapp CW, Fowle DA, Kulczycki E, Roberts JA, Graham DW. 2007. Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc Natl Acad Sci U S A 104:12040–12045. doi: 10.1073/pnas.0702879104. [DOI] [PMC free article] [PubMed] [Google Scholar]