Abstract
Extended-spectrum β-lactamase (ESBL)-producing Escherichia coli sequence type ST131 has emerged as the leading cause of community-acquired urinary tract infections and bacteremia worldwide. Whether environmental water is a potential reservoir of these strains remains unclear. River water samples were collected from 40 stations in southern Taiwan from February to August 2014. PCR assay and multilocus sequence typing (MLST) analysis were conducted to determine the CTX-M group and sequence type, respectively. In addition, we identified the seasonal frequency of ESBL-producing E. coli strains and their geographical relationship with runoffs from livestock and poultry farms between February and August 2014. ESBL-producing E. coli accounted for 30% of the 621 E. coli strains isolated from river water in southern Taiwan. ESBL-producing E. coli ST131 was not detected among the isolates. The most commonly detected strain was E. coli CTX-M group 9. Among the 92 isolates selected for MLST analysis, the most common ESBL-producing clonal complexes were ST10 and ST58. The proportion of ESBL-producing E. coli was significantly higher in areas with a lower river pollution index (P = 0.025) and regions with a large number of chickens being raised (P = 0.013). ESBL-producing E. coli strains were commonly isolated from river waters in southern Taiwan. The most commonly isolated ESBL-producing clonal complexes were ST10 and ST58, which were geographically related to chicken farms. ESBL-producing E. coli ST131, the major clone causing community-acquired infections in Taiwan and worldwide, was not detected in river waters.
INTRODUCTION
Escherichia coli sequence type ST131 (O25:H4), associated with CTX-M-15 extended-spectrum β-lactamase (ESBL), is the leading cause of community-acquired urinary tract infections (UTIs) and bacteremia worldwide (1–7). This clonal group is virulent and carries a broad range of resistance genes on transferable plasmids (1, 3–9). Studies conducted in southern Taiwan have reported that several clinical isolates of ESBL-producing E. coli belonged to the ST131-O25b lineage. The most prevalent ESBL-encoding gene is blaCTX-M-14 (10, 11). Most patients with bacteremia or UTIs were healthy previously and did not exhibit any apparent risk factors. Only 30% of infants with UTIs caused by this clone had identifiable potential risk factors (e.g., previous antimicrobial use, hospitalization, neonatal infection, or underlying disease), suggesting that these UTIs were mostly community acquired, not hospital acquired (10–12).
The established reservoirs of the ST131 E. coli clone are humans, companion animals, noncompanion animals, and foods (7). Adequate studies have not been conducted for evaluating whether environmental water is a potential reservoir of this multidrug-resistant clone (13–15). Outbreaks of enteric infections are caused by various bacteria, protozoa, fungi, and viruses present in contaminated drinking water (16, 17). In addition, E. coli in irrigation water contaminates fresh produce (18; http://www.pma.com/∼/media/pma-files/food-safety/cps/cps-research-reportag-water-200813version-11final.pdf). The contamination of rivers can occur through different sources, including human fecal contamination (sewer overflow during heavy rain or inadequate wastewater treatment), animals (particularly livestock), and runoff from pastures and sewage. In this study, we determined whether river water is a potential reservoir of ESBL-producing E. coli in southern Taiwan, focusing on the blaCTX-M clone found in human infections.
MATERIALS AND METHODS
Sampling of river water.
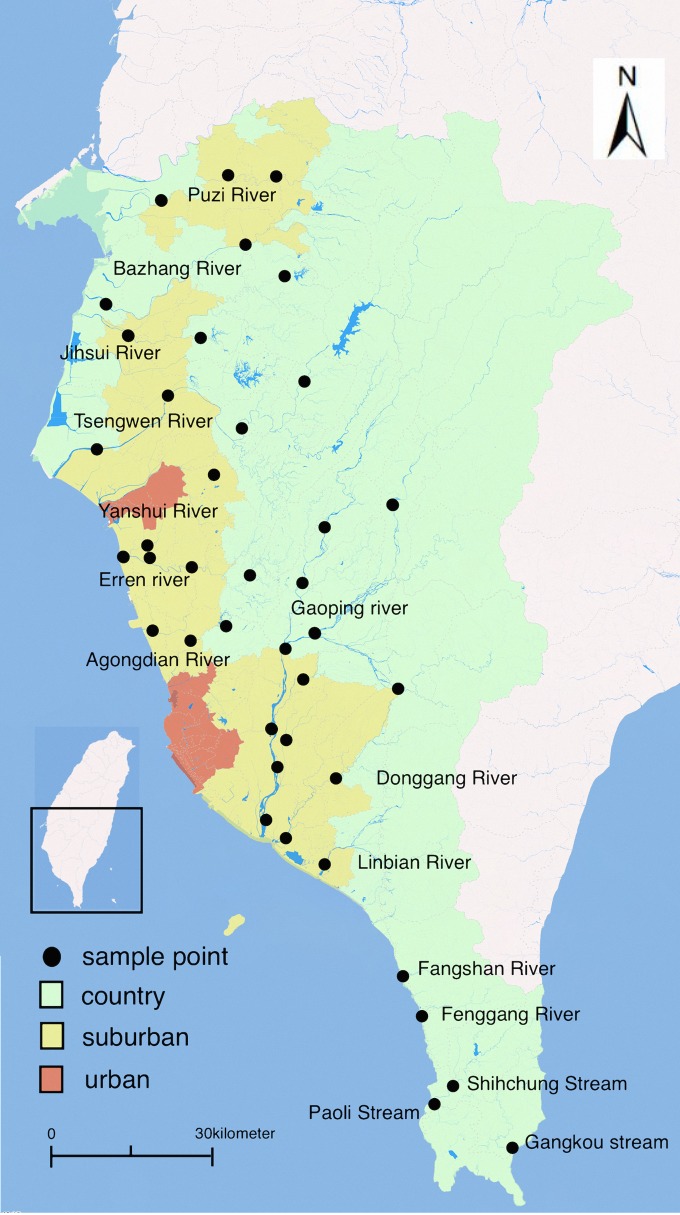
The Taiwan Environmental Protection Administration (TEPA) routinely examines river water in southern Taiwan for the presence of heavy metals and E. coli. Figure 1 displays 99 stations located in the upper and lower streams in southern Taiwan that are monitored every month, including the Puzi, Bazhang, Jihsui, Tsengwen, Yanshui, Linbian, Donggang, Fenggang, Fangshan, Gaoping, Erren, Agongdian, and Shihchung Rivers and the Paoli and Gangkou streams. One hundred milliliters of river water was sampled each time and analyzed in accordance with the standard procedures of the Environmental Analysis Laboratory of the TEPA (19). All of the sampling procedures followed the rules established by the Environmental Analysis Laboratory of the TEPA. TPEA method NIEA W104.51C was used as the standard basis for sampling (19). Water samples were analyzed immediately after sample collection; the time required from sample collection to laboratory work completion was <24 h. To perform cluster sampling for each river and county, we used a random-number generator in selecting 40 of the 99 stations. One water sample was collected at each sampled station per month. In total, 280 river water samples were collected between February and August of 2014 in southern Taiwan. The normal mean daily temperature in the study area varied between 20.3°C in February and 28.7°C in August. The monthly precipitation ranged between 20.5 mm in February and 416.7 mm in August (http://www.cwb.gov.tw/V7/climate/monthlyMean/Taiwan_tx.htm).
FIG 1.
Study area in southern Taiwan, including the Puzi, Bazhang, Jihsui, Tsengwen, Yanshui, Linbian, Donggang, Fenggang, Fangshan, Gaoping, Erren, and Agongdian Rivers and the Shihchung, Paoli, and Gangkou streams, with 40 river sampling points. Map created via Paint 2, version 5.3.1 (250) (TryBest Studio), using data from the Taiwan National Land Surveying and Mapping Center.
Parameters of sample collection.
Information on sampling sites, weather, human population grade, rainfall season, the river pollution index (RPI), distance to the river origin, and livestock and poultry densities around the sampling site were evaluated. In addition, the relationship between ESBL-producing E. coli isolates and these parameters was analyzed.
In accordance with the climate statistics provided by the Central Weather Bureau of Taiwan, the period from February to April was defined as the spring (dry) season, during which the mean temperature and precipitation of each month were <25°C and <100 mm, respectively. The period from May to August was defined as the summer (rainy) season, during which the mean temperature and precipitation of each month were >25°C and >100 mm, respectively. The human population around the sampling site and the population grade of each county or town were provided by the Ministry of the Interior of Taiwan. The population of a county or town was categorized as grade 1 for <49,000, grade 2 for between 49,000 and 93,000, grade 3 for between 93,000 and 137,000, and grade 4 for >137,000 people. The RPI was graded according to the definitions provided by the TEPA, which include the concentrations of 4 parameters in water: dissolved oxygen (DO), biochemical oxygen demand (BOD5), suspended solids (SS), and ammonia nitrogen (NH3-N). We defined the RPI as the average of these 4 parameters, wherein an RPI of <2, 2 to 3, 3 to 6, and >6 was determined as nonpolluted, lightly polluted, moderately polluted, and severely polluted, respectively (20). We defined the upper, middle, and lower streams of a river based on the distance from the sampling site to the river origin, wherein the upper, middle, and lower streams were the first third, middle third, and final third of the river length, respectively. We collected information on livestock and poultry distribution from the Council of Agriculture, Taiwan. We defined totals of <100,000, between 100,000 and 500,000, and >500,000 chickens being raised per month as low, middle, and high densities of chicken farming, respectively. In a similar manner, we defined totals of <10,000, between 10,000 and 50,000, and >50,000 pigs being farmed per month as low, middle, and high densities of pig farming, respectively. We included 128 districts or counties in our study region. After excluding the area without chicken farms, we selected the tertiles as the cut points, which are approximately 100,000 and 500,000 chickens per month, for categorization. For pig farms, we selected 10,000 and 50,000 pigs per month as the cut points, because the chicken-farming density scale is approximately 10 times greater than the pig-farming density scale in southern Taiwan.
Identification and purification of E. coli.
The river water samples were tested for E. coli by using the membrane filtration method. We used the decimal serial dilutions of river water filtered through 0.45-μm-pore-size membranes and followed the official method published by the TEPA (TEPA method NIEA E202.55B) (21). A series of 10-fold dilutions of the water sample were conducted using a sterile phosphate-buffered magnesium chloride solution as dilution blanks. The membranes were placed on an LES (Lawrence Experimental Station) endo agar plate (Bottle M Endo agar LES 500G; Becton Dickson and Company) and incubated at 35°C ± 1°C for 24 h. The E. coli was extracted from colonies that had a green metallic sheen and suspended in 1 ml of phosphate-buffered saline (PBS). We selected as many samples as possible and up to 10 colonies from each LES endo agar plate. One hundred microliters of the highest dilution displaying growth was spread on Luria-Bertani–ampicillin agar plates and incubated at 37°C overnight. Each of these colonies was subcultured 3 additional times. The purified isolates were inoculated on E. coli CHROMagar (ECC) plates (CHROMagar, Paris, France) (22) and further incubated overnight at 37°C to confirm the identification of E. coli.
CHROMagar ESBL plates (CHROMagar, Paris, France) were used to identify ESBL-producing E. coli according to the manufacturer's instructions.
Detection of O25b-ST131, multilocus sequence typing (MLST), and blaCTX-M gene groups.
For chromosomal DNA preparation, a single colony of E. coli was suspended in 200 μl of sterile double-distilled water and heated to 100°C for 10 min. After cooling, the samples were centrifuged, and 2 μl of the supernatant containing chromosomal DNA was used as a template for PCR amplification.
The O25b serotype was determined by employing the methods of Clermont et al. (23), and the following screening primers were used: rfb.1bis (5′-ATACCGACGACGCCGATCTG-3′) and rfbO25b.r (5′-TGCTATTCATTATGCGCAGC-3′) (24, 25).
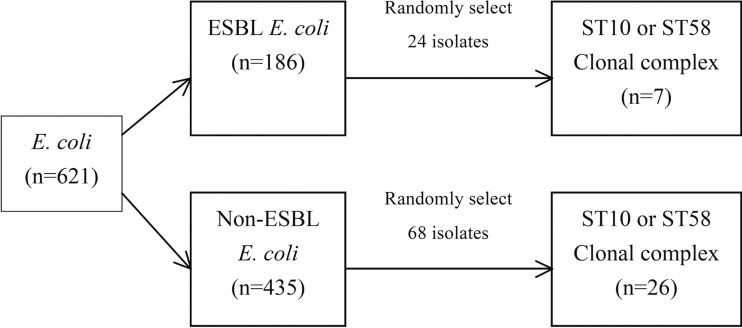
According to the stratification of the O25b status (Fig. 2), we used a random-number generator in selecting 92 isolates for MLST analysis. All isolates with the O25b serotype (12 ESBL-producing and 34 non-ESBL-producing) were evaluated through MLST analysis. Twelve O25-positive, ESBL-positive and 34 O25-positive, ESBL-negative E. coli strains were matched for randomly selecting O25-negative, ESBL-positive and O25-negative, ESBL-negative E. coli isolates.
FIG 2.
Sample selection for detecting the sequence type of E. coli from river waters in southern Taiwan.
The MLST grouping of E. coli was determined by analyzing 8 housekeeping gene sequences (adk, fumC, gyrB, icd, mdh, purA, and recA) on the MLST database website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) (26). The eBURST (http://eburst.mlst.net/) algorithm was used to determine the clonal complexes of sequence types (STs) and sequence type complexes (STCs), where STs sharing at least 5 loci in common with at least one other member of the group were assigned to defined STCs; STs with the highest number of single-locus variants (SLVs) were assigned as the founder. In addition, the blaCTX-M gene of all ESBL-producing E. coli isolates was investigated. The CTX-M groups 1, 2, 8, and 9, as well as SHV and TEM, were detected by performing multiplex PCR with specific primers, as shown in Table 1 (26, 27). Specific PCRs were performed to detect blaCTX-M-14 and blaCTX-M-15 by employing the methods of Sidjabat et al. and Chia et al. (25, 26).
TABLE 1.
O25b CTX-M group-specific primers used for multiplex PCR
| PCR and primer names | Sequencea (5′–3′) | Reference |
|---|---|---|
| Multiplex II, CTX-M group 1, group 2, and group 9 | ||
| MultiCTXMGp1_for | TTAGGAARTGTGCCGCTGYA | 27 |
| MultiCTXMGp1-2_rev | CGATATCGTTGGTGGTRCCAT | |
| MultiCTXMGp2_for | CGTTAACGGCACGATGAC | |
| MultiCTXMGp1-2_rev | CGATATCGTTGGTGGTRCCAT | |
| MultiCTXMGp9_for | TCAAGCCTGCCGATCTGGT | |
| MultiCTXMGp9_rev | TGATTCTCGCCGCTGAAG | |
| CTX-M group 8/25 | ||
| CTX-Mg8/25_for | AACRCRCAGACGCTCTAC | |
| CTX-Mg8/25_rev | TCGAGCCGGAASGTGTYAT | |
| CTX-M-14 | ||
| CTX-14F | TACCGCAGATAATACGCAGGTG | 26 |
| CTX-14R | CAGCGTAGGTTCAGTGCGATCC | |
| CTX-M-15 | ||
| CTX-M-15-SF | CACACGTGGAATTTAGGGACT | 25 |
| CTX-M-15-SR | GCCGTCTAAGGCGATAAACA | |
| Multiplex I, TEM and SHV | ||
| MultiTSO-T_for | CATTTCCGTGTCGCCCTTATTC | 27 |
| MultiTSO-T_rev | CGTTCATCCATAGTTGCCTGAC | |
| Multiplex PCR | ||
| SHV-F | AACGGAACTGAATGAGGCGCT | 26 |
| SHV-R | TCCACCATCCACTGCAGCAGCT |
Y = T or C; R = A or G; S = G or C; D = A, G, or T.
Statistical analysis.
All statistical analyses were performed using the Statistical Package for Social Sciences (version 22.0) software package for Windows (SPSS Inc., Chicago, IL). Data concerning patients with ESBL-producing and non-ESBL-producing E. coli were expressed as percentages of each subgroup of patients and were compared by conducting the χ2 test for categorical variables.
RESULTS
Identification of ESBL-producing E. coli in river water.
ESBL-producing E. coli accounted for 186 (30%) of the 621 E. coli strains isolated from 280 river samples in southern Taiwan over the 7-month study period. All 621 E. coli isolates were screened for the O25b serotype through PCR assay. Serotype O25b accounted for 46 (7.4%) of the 621 isolates (Table 2). Although this serotype was observed more frequently among the ESBL-producing E. coli than among the non-ESBL-producing E. coli (12/186 [6.5%] versus 34/435 [2.8%]), the difference was statistically nonsignificant (P = 0.55).
TABLE 2.
Numbers of O25b and non-O25b serotypes in ESBL-producing and non-ESBL-producing E. colia
| Serotype | No. of E. coli strains |
Total | |
|---|---|---|---|
| ESBL (n = 186) | Non-ESBL (n = 435) | ||
| O25b | 12 | 34 | 46 |
| Non-O25b | 174 | 401 | 575 |
| Total | 186 | 435 | 621 |
Numbers of O25b and non-O25b serotypes in ESBL-producing E. coli (n = 186) and non-ESBL-producing E. coli (n = 435) are from the 621 E. coli strains isolated from river waters in southern Taiwan.
As shown in Table 3, among the 12 ESBL-producing E. coli isolates belonging to the O25b serotype, 4 strains carried genes encoding CTX-M ESBLs, including 2 carrying the CTX-M group 2 gene, one carrying the CTX-M group 9 gene, and one carrying the CTX-M (3, 15) gene. Among the 186 ESBL-producing E. coli isolates, 18 carried the CTX-M group 1 gene, 3 carried the CTX-M group 2 gene, 6 carried the CTX-M group 8 gene, and 43 carried the CTX-M group 9 gene.
TABLE 3.
Number of CTX-M, SHV, and TEM genes detected in O25b ESBL-producing E. coli and non-O25b ESBL-producing E. colia
| Gene type | No. of genes in each E. coli serotype |
Total | |
|---|---|---|---|
| O25b ESBL (n = 12) | Non-O25b ESBL (n = 174) | ||
| CTX-M group 1 | 0 | 18 | 18 |
| CTX-M group 2 | 2 | 1 | 3 |
| CTX-M group 8 | 0 | 6 | 6 |
| CTX-M group 9 | 1 | 42 | 43 |
| SHV | 0 | 23 | 23 |
| TEM | 7 | 89 | 96 |
Shown are the numbers of genes of CTX-M group 1, group 2, group 8, and group 9 and SHV and TEM detected in O25b ESBL-producing E. coli (n = 12) and non-O25b ESBL-producing E. coli (n = 174) from 186 ESBL-producing E. coli strains isolated from river waters in southern Taiwan.
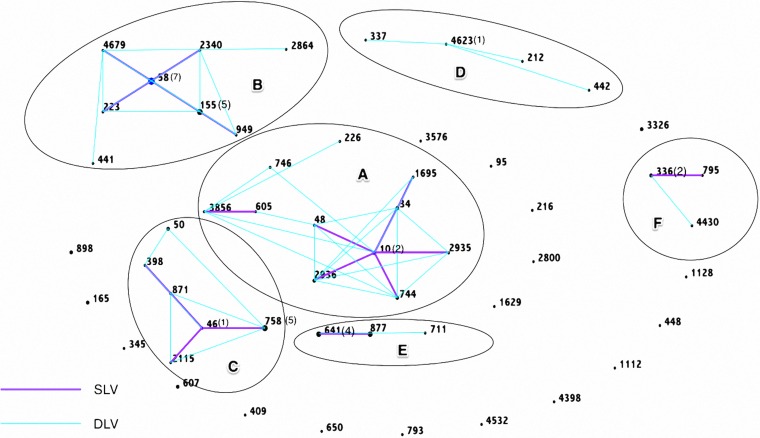
Among the 24 ESBL-producing E. coli isolates and 68 non-ESBL-producing E. coli isolates selected for MLST analysis, the sequence types were identified for 83 isolates. Genetic relatedness within each clonal complex is shown in Fig. 3 (based on data from the MLST database [http://mlst.warwick.ac.uk/mlst/]). Among the 83 E. coli isolates, the eBURST diagram revealed that the clustering of E. coli STs belonged to 6 main complexes (A, STC10; B, STC58; C, STC46; D, STC4623; E, STC641; and F, STC336); 15 isolates belonged to the ST10 clonal complex, and 18 belonged to the ST58 clonal complex (including 7 ST58 strains and 5 ST155 strains). No ST131 clonal complex was detected among all 92 E. coli isolates.
FIG 3.
eBURST diagram showing the clustering of E. coli STs belonging to the 6 main complexes isolated from river waters in southern Taiwan: A, clonal complex of sequence type 10 (STC10; n = 15); B, STC58 (n = 18); C, STC46 (n = 11); D, STC4623 (n = 4); E, STC641 (n = 9); F, STC336 (n = 4). E. coli strains from the same ancestor based on the entire MLST data set were grouped and are indicated with black circles. Each ST is represented as a node. Single-locus variants (SLV) are connected with a purple line, and double-locus variants (DLV) are connected with a blue line. Strains that shared 5 of the 7 alleles were grouped into a single clonal complex.
Among the identifiable 83 isolates detected from MLST analysis, 7 isolates belonged to the ST10 or ST18 clonal complex in the ESBL-producing group, and 26 isolates belonged to the ST10 or ST58 clonal complex in the non-ESBL-producing group.
Relationship between river water samples and evaluated parameters.
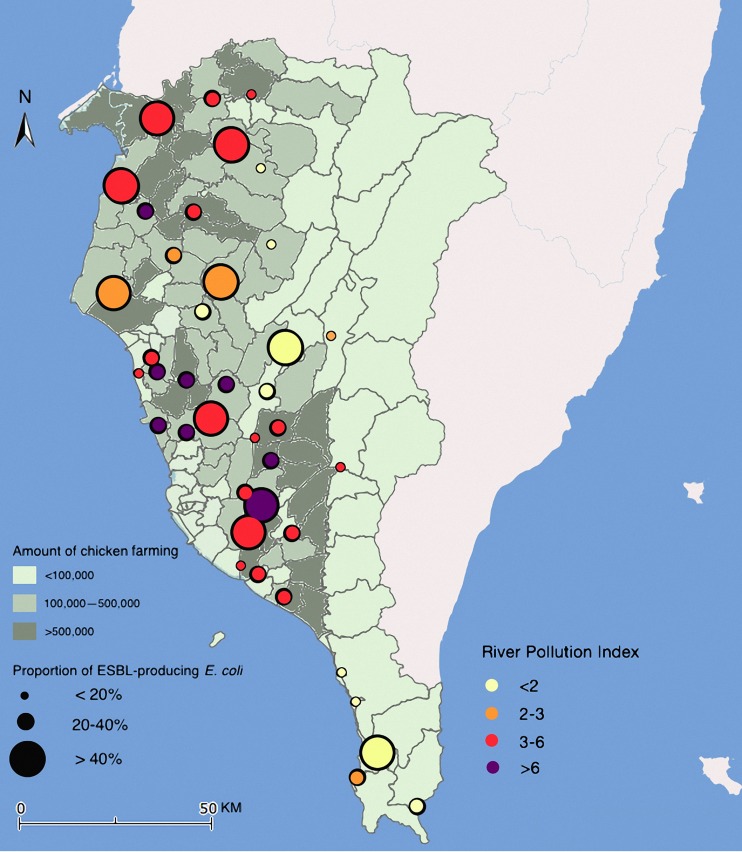
The geographic distribution of the proportion of ESBL-producing E. coli in relation to the number of chickens being raised per month and RPI in southern Taiwan is displayed in Fig. 4. As shown in Table 4, no significant differences in the proportion of ESBL-producing E. coli strains were observed among the seasons, stream sites, populations, and numbers of pigs. The proportion of ESBL-producing E. coli was significantly higher in the low-RPI group (P = 0.025). In addition, the proportion of ESBL-producing E. coli strains was significantly higher in regions with a large number of chickens being raised (P = 0.013). No significant difference was observed in the proportion of the ST10 or ST58 clonal complex between the high- and low-density chicken farming areas (P = 0.67).
FIG 4.
Geographic distribution of the proportion of ESBL-producing E. coli in relation to the number of chickens being raised and river pollution index (RPI) in southern Taiwan. The background of the map with the color gradient presents the number of chickens raised in each county; the dots with a size display the proportion of ESBL-producing E. coli in each sampling point, and the color of the dots represents the RPI. Map created via Paint 2, version 5.3.1 (250) (TryBest Studio), using data from the Taiwan National Land Surveying and Mapping Center.
TABLE 4.
Characteristics of ESBL-producing and non-ESBL-producing E. coli of 621 E. coli strains isolated from river waters in southern Taiwan
| Characteristic | No. (%) of isolates |
P value | |
|---|---|---|---|
| Non-ESBL (n = 435) | ESBL (n = 186) | ||
| Seasona | 0.80 | ||
| Dry (2–4 mo) | 133 (70.7) | 55 (29.3) | |
| Rain (5–8 mo) | 302 (69.7) | 131 (30.3) | |
| Regionb | 0.999 | ||
| Upper | 44 (69.8) | 19 (30.2) | |
| Middle | 166 (70.0) | 71 (30.0) | |
| Lower | 225 (70.1) | 96 (29.9) | |
| Populationc | 0.19 | ||
| ≤137,000 | 423 (70.5) | 177 (29.5) | |
| >137,000 | 12 (57.1) | 9 (42.9) | |
| No. of animals being raised | |||
| Chickend | 0.013 | ||
| <100,000 | 131 (78.9) | 35 (21.1) | |
| 100,000–500,000 | 192 (66.2) | 98 (33.8) | |
| >500,000 | 112 (67.9) | 53 (32.1) | |
| Pige | 0.55 | ||
| <1,000 | 205 (71.7) | 81 (28.3) | |
| 10,000–50,000 | 124 (67.0) | 61 (33.0) | |
| >50,000 | 106 (70.7) | 44 (29.3) | |
| RPIf | 0.025 | ||
| ≤6 | 327 (67.8) | 155 (32.2) | |
| >6 | 108 (77.7) | 31 (22.3) | |
Dry season is defined from February to April and wet season from May to August.
Water collected in the region is classified as upper, middle, and lower stream by the distance of the sampled station to the river origin.
Classified by the population density at the sampled station.
Classified by the number of chickens produced in the region of the sampled station in a month.
Classified by the number of pigs produced in the region of sampled station in a month.
Calculated by the average of the concentrations of four parameters in water: dissolved oxygen (DO), biochemical oxygen demand (BOD5), suspended solids (SS), and ammonia nitrogen (NH3-N).
DISCUSSION
Several studies have been conducted on the isolation of ESBL-producing E. coli from river water (13–15). The difference in the abundance of human and animal ESBL-producing E. coli clones in various river waters may be related to the nature of their drainage basins, the intensity of antibiotic use, and the relative amount of fecal contamination from humans and farm animals. Clonal groups similar to those found in human infections have been isolated from contaminated rivers adjacent to large cities (13–15). In contrast, nonhuman clonal groups more frequently are isolated from rural streams (28–30). The isolation rate of ESBL-producing E. coli was approximately 30% in our study, indicating their high numbers in environmental water. Zurfluh et al. reported an isolation rate of 36.2% for ESBL-producing E. coli from river and lake water in Switzerland (15). In a Chinese study, an isolation rate of 14.8% was reported for ESBL-producing E. coli from downstream water (31). However, research investigating river water as a potential reservoir of ESBL-producing E. coli is limited.
In our previous study, we observed that most ESBL-producing clinical E. coli strains isolated from infants belonged to CTX-M-14 in either ST131 or non-ST131 clones (10). In the current study, the most predominant clones observed were ST10 and ST58 clonal complexes. These ESBL-producing clones are associated with chickens and are unrelated to those found in human infections.
Colomer-Lluch et al. isolated quinolone-resistant E. coli belonging to the clonal groups O25b-H4 B2-ST131 and O25b:H4-D-ST69 from raw sewage and river water in Spain (13). In contrast, Dhanji et al. did not detect blaCTX-M-15, the most common ESBL-encoding gene in clinical E. coli strains in the United Kingdom, in the Thames in London (14, 32). Geser et al. (33) identified CTX-M-1 ESBL-encoding genes in Enterobacteriaceae isolates from food-producing animals in Switzerland. However, the predominant ESBL-producing clinical isolates in Switzerland belonged to CTX-M-15 (34).
As shown in Table 5, various ESBL-producing E. coli clones have been isolated from animals, although ST131 clones have not been detected in certain studies (28, 30, 35–40, 42–45). The ST10, ST58, and ST155 clones have been detected in animals, water, and human isolates in the Netherlands, Canada, Chile, and China (28, 30, 35, 38–45). We detected numerous ST10 or ST58 clonal complexes in high-density chicken-farming areas. Additional studies are warranted to elucidate the virulence potential of E. coli ST10 and ST58 clonal groups.
TABLE 5.
Summary of 13 major animal or human E. coli sequence type studies conducted after 2000
| Author | Yr | Country | Sample origin (no.) | Most common serotype (%) | Dominant ST | ST131 (no.) | ST10 (no.) | ST58/ST155 (no.) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Overdevest et al. | 2009 | Netherlands | Chicken meat (89), beef (85), pork (57), mixed meat (22), other meat (9) | Chicken (CTX-M-1; 58.1), all other meat (CTX-M-1; 62.5) | ST117, ST10 | Not found | Found | ST155 | 35 |
| Ma et al. | 2006 | China | Duck feces (230), water (15) | CTX-M-27/55/24/105/14 (27.7/22/21.1/16.2/12.2) | NA | NA | NA | NA | 36 |
| Dierikx et al. | 2009 | Denmark | Broiler feces (1,066), farmer's feces (18) | CTX-M-1, CMY-2, SHV-12 | ST93 | Not found | Not found | Not found | 37 |
| Rashid et al. | 2010 | Bangladesh | OBSa feces (170), water (8) | CTX-M-15 (100) | ST156 | Not found | ST10CC (1) | Not found | 28 |
| Hernandez et al. | 2009 | Chile | Gull feces (370), human feces (49) | CTX-M group 1 (94.6) | ST10 | Not found | ST10 (24) | ST58 (4), ST155 (1) | 30 |
| Bergeron et al. | 2005–2007 | Canada | Human urine (475), meat of chicken (253), beef (242), pork (242), animal feces (349) | NAb | ST117, ST10 | Not found | ST10(13) | Not found | 38 |
| Blaak et al. | 2011 | Netherlands | Flies (87), poultry manure in broiler farm (10), laying-hen farm (10) | SHV-12, CTX-M-1, TEM-52 | ST58, ST10 | Not found | ST10 (4) | ST58 (6), ST155 (2) | 39 |
| Cortes et al. | 2003 | Spain | Floor of poultry (59) and pig (27) farms | Poultry, CTX-M-9 group (64.9) and CTX-M-1 group (5.3); pig, CTX-M-1 group (69.0) | ST155, ST10 | ST131 (1) | ST10 (3) | ST155 (4) | 40 |
| Peirano et al. | 2000–2010 | Canada | Human blood (4,698) | CTX-M-15, 119 (60); CTX-M-14, 58 (29) | ST131 | ST131 (117) | ST10CC (13) | Not found | 41 |
| Leverstein-van Hall et al. | 2006–2010 | Netherlands | Chicken meat (98), floor of poultry farm (35), human (516) | Poultry, CTX-M-1 (49); chicken meat, CTX-M-1 (49); human, CTX-M-1 (24) | ST10, ST58 | Not found | Human (3), meat (4) | Human, ST58 (3) | 42 |
| Hu et al. | 2010–2013 | China | Swine feces (31), water (26), human urine, blood, sputum, and body secretion (82) | CTX-M-1 group (33), CTX-M-9 (99) | ST131, CC10 | ST131 (20) | CC10 (19) | NA | 43 |
| Ding et al. | 2006–2007 | China | Pig tissue (81) | NA | CC10 | Not found | CC10 (26) | CC58 (8) | 44 |
| Borjesson et al. | 2010 | Sweden | Broiler feces (10) | CTX-M-1 (10) | ST155 | Not found | Not found | ST155 (3) | 45 |
OBS, open bill stork (Anastomus oscitans).
NA, not applicable.
We observed a significant association of ESBL-producing E. coli clones isolated from river water with the presence of chicken farms but not with pig farms (P = 0.013). This finding may be explained by the shorter period and higher antibiotic usage per average body weight for poultry compared to those for pigs. In southern Taiwan, antibiotic usage is 10 times higher in chickens than in pigs (42), although dosing is higher in pigs (43). According to the Annual Report of Food and Drug Research of 2014, released by the Food and Drug Administration, Taiwan, antibiotic residues were detected with greater frequency in chicken meat than in pig meat (46). In addition, a significant difference was observed in the proportion of ESBL-producing E. coli among different RPIs in our series (P = 0.025); this difference may be due to chicken farms being located mostly in suburban areas, where the river water is less polluted. The relationship between resistant bacteria and the RPI remains controversial. Multidrug-resistant bacteria in rivers may have developed because of the deterioration of water quality resulting from the flow of untreated sewage from domestic wastewater or intensive agricultural and industrial activities (47). However, the detection of multidrug-resistant bacteria in chlorinated water also has been reported (48, 49).
Our study has several limitations. Data from southern Taiwan may be inapplicable to other countries because of the geographic diversity of E. coli strains and their variable virulence and resistance genes. For example, ESBL-producing E. coli strains expressing blaCTX-M-14 are prevalent in Taiwan, whereas those expressing blaCTX-M-15 are more common in other parts of the world (11). Although we selected sample stations based on the surveillance data from the TEPA, water samples may not be independent because certain stations are located on the same river; however, the distribution of sampling stations is essential for the analysis of regional characteristics. Furthermore, the replication of E. coli in a single water sample is possible. We did not isolate the DNA of all E. coli strains, as has been conducted in previous studies; therefore, it is difficult to evaluate the replication (13, 15). Moreover, we did not detect ESBL-associated genes, including blaCTX-M, blaSHV, and blaTEM, from certain ESBL-producing E. coli isolates in our study. We attribute this finding to the difference between the genes identified through PCR assay and the design of CHROMagar ESBL plates or other ESBL-associated genes, such as blaOXA, which we did not detect. In addition, we did not detect several resistant genes reported in other studies; for instance, blaCMY-2 may contribute to cefotaxime resistance. In an Asian surveillance study on ESBL-producing Enterobacteriaceae, isolates were reported to have a higher prevalence of AmpC blaCMY-type β-lactamases, particularly blaCMY-2, in Taiwan (50). Additional epidemiological studies are required to correlate river water, animal contact, poultry consumption, and other environmental exposures with the emergence of ESBL-producing E. coli.
In conclusion, this study investigated ESBL-producing E. coli in various river waters in southern Taiwan. We isolated several ESBL-producing E. coli clones found in poultry and other farm animals but not the O25b-ST131 clone, which causes human infections. These environmental ESBL-producing clones potentially can spread among humans, rendering treatment difficult. The continued surveillance of food and environment is essential to anticipate emerging infections.
ACKNOWLEDGMENTS
We thank Calvin M. Kunin for his review of the manuscript.
We have no conflicts of interest to disclose.
This work was supported by the Ministry of Science and Technology, Taiwan, R.O.C., under grant MOST 104-2314-B-075B-003-MY3.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Canton R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis 14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Chemother 53:2733–2739. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau SH, Kaufmann ME, Livermore DM, Woodford N, Willshaw GA, Cheasty T, Stamper K, Reddy S, Cheesbrough J, Bolton FJ, Fox AJ, Upton M. 2008. UK epidemic Escherichia coli strains A-E, with CTX-M-15 beta-lactamase, all belong to the international O25:H4-ST131 clone. J Antimicrob Chemother 62:1241–1244. doi: 10.1093/jac/dkn380. [DOI] [PubMed] [Google Scholar]
- 4.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. [DOI] [PubMed] [Google Scholar]
- 5.Peirano G, Pitout JD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents 35:316–321. doi: 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Platell JL, Cobbold RN, Johnson JR, Heisig A, Heisig P, Clabots C, Kuskowski MA, Trott DJ. 2011. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob Agents Chemother 55:3782–3787. doi: 10.1128/AAC.00306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 8.Oteo J, Perez-Vazquez M, Campos J. 2010. Extended-spectrum β-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr Opin Infect Dis 23:320–326. doi: 10.1097/QCO.0b013e3283398dc1. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 10.Cheng MF, Chen WL, Hung WY, Huang IF, Chiou YH, Chen YS, Lee SS, Hung CH, Wang JL. 2015. Emergence of extended spectrum-beta-lactamase-producing Escherichia coli O25b-ST131: a major community-acquired uropathogen in infants. Pediatr Infect Dis J 34:469–475. [DOI] [PubMed] [Google Scholar]
- 11.Chung HC, Lai CH, Lin JN, Huang CK, Liang SH, Chen WF, Shih YC, Lin HH, Wang JL. 2012. Bacteremia caused by extended-spectrum-beta-lactamase-producing Escherichia coli sequence type ST131 and non-ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob Agents Chemother 56:618–622. doi: 10.1128/AAC.05753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu YH, Cheng MF, Lai CH, Lin HH, Hung CH, Wang JL. 2014. The role of sequence type (ST) 131 in adult community-onset non-ESBL-producing Escherichia coli bacteraemia. BMC Infect Dis 14:579. doi: 10.1186/s12879-014-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colomer-Lluch M, Mora A, Lopez C, Mamani R, Dahbi G, Marzoa J, Herrera A, Viso S, Blanco JE, Blanco M, Alonso MP, Jofre J, Muniesa M, Blanco J. 2013. Detection of quinolone-resistant Escherichia coli isolates belonging to clonal groups O25b:H4-B2-ST131 and O25b:H4-D-ST69 in raw sewage and river water in Barcelona, Spain. J Antimicrob Chemother 68:758–765. doi: 10.1093/jac/dks477. [DOI] [PubMed] [Google Scholar]
- 14.Dhanji H, Murphy NM, Akhigbe C, Doumith M, Hope R, Livermore DM, Woodford N. 2011. Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum beta-lactamase from UK river water. J Antimicrob Chemother 66:512–516. doi: 10.1093/jac/dkq472. [DOI] [PubMed] [Google Scholar]
- 15.Zurfluh K, Hachler H, Nuesch-Inderbinen M, Stephan R. 2013. Characteristics of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl Environ Microbiol 79:3021–3026. doi: 10.1128/AEM.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anonymous. 2000. Waterborne outbreak of gastroenteritis associated with a contaminated municipal water supply, Walkerton, Ontario, May-June 2000. Canada Commun Dis Rep 26:170–173. [PubMed] [Google Scholar]
- 17.Hoxie NJ, Davis JP, Vergeront JM, Nashold RD, Blair KA. 1997. Cryptosporidiosis-associated mortality following a massive waterborne outbreak in Milwaukee, Wisconsin. Am J Public Health 87:2032–2035. doi: 10.2105/AJPH.87.12.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steele M, Odumeru J. 2004. Irrigation water as source of foodborne pathogens on fruit and vegetables. J Food Protect 67:2839–2849. [DOI] [PubMed] [Google Scholar]
- 19.Taiwan Environmental Protection Administration. 2004. Rivers, lakes, and reservoir water quality sampling general rule. TPEA method NIEA W104.51C Taiwan Environmental Protection Administration, Taipei, Taiwan: (In Chinese.) [Google Scholar]
- 20.Liou SM, Lo SL, Wang SH. 2004. A generalized water quality index for Taiwan. Environ Monit Assess 96:35–52. doi: 10.1023/B:EMAS.0000031715.83752.a1. [DOI] [PubMed] [Google Scholar]
- 21.Taiwan Environmental Protection Agency. 2012. Water coliform detection method–filter method. TPEA method NIEA E202.55B. Taiwan Environmental Protection Agency, Taipei, Taiwan: (In Chinese.) [Google Scholar]
- 22.Alonso JL, Soriano A, Carbajo O, Amoros I, Garelick H. 1999. Comparison and recovery of Escherichia coli and thermotolerant coliforms in water with a chromogenic medium incubated at 41 and 44.5°C. Appl Environ Microbiol 65:3746–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Mulvey MR, Nordmann P, Ruppe E, Sarthou JL, Frank T, Vimont S, Arlet G, Branger C, Woodford N, Denamur E. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother 64:274–277. doi: 10.1093/jac/dkp194. [DOI] [PubMed] [Google Scholar]
- 24.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J Antimicrob Chemother 61:1024–1028. doi: 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- 25.Sidjabat HE, Paterson DL, Adams-Haduch JM, Ewan L, Pasculle AW, Muto CA, Tian GB, Doi Y. 2009. Molecular epidemiology of CTX-M-producing Escherichia coli isolates at a tertiary medical center in western Pennsylvania. Antimicrob Agents Chemother 53:4733–4739. doi: 10.1128/AAC.00533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chia JH, Chu C, Su LH, Chiu CH, Kuo AJ, Sun CF, Wu TL. 2005. Development of a multiplex PCR and SHV melting-curve mutation detection system for detection of some SHV and CTX-M beta-lactamases of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae in Taiwan. J Clin Microbiol 43:4486–4491. doi: 10.1128/JCM.43.9.4486-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 28.Rashid M, Rakib MM, Hasan B. 2015. Antimicrobial-resistant and ESBL-producing Escherichia coli in different ecological niches in Bangladesh. Infect Ecol Epidemiol 5:26712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Zhou Y, Guo S, Chang W. 2015. Multidrug resistance found in extended-spectrum beta-lactamase-producing Enterobacteriaceae from rural water reservoirs in Guantao, China. Front Microbiol 6:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez J, Johansson A, Stedt J, Bengtsson S, Porczak A, Granholm S, Gonzalez-Acuna D, Olsen B, Bonnedahl J, Drobni M. 2013. Characterization and comparison of extended-spectrum beta-lactamase (ESBL) resistance genotypes and population structure of Escherichia coli isolated from Franklin's gulls (Leucophaeus pipixcan) and humans in Chile. PLoS One 8:e76150. doi: 10.1371/journal.pone.0076150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao L, Hu J, Zhang X, Ma R, Gao J, Li S, Zhao M, Miao Z, Chai T. 2014. Dissemination of ESBL-producing Escherichia coli of chicken origin to the nearby river water. J Mol Microbiol Biotechnol 24:279–285. doi: 10.1159/000365786. [DOI] [PubMed] [Google Scholar]
- 32.Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, Johnson AP, Pike R, Warner M, Cheasty T, Pearson A, Harry S, Leach JB, Loughrey A, Lowes JA, Warren RE, Livermore DM. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J Antimicrob Chemother 54:735–743. doi: 10.1093/jac/dkh424. [DOI] [PubMed] [Google Scholar]
- 33.Geser N, Stephan R, Hachler H. 2012. Occurrence and characteristics of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet Res 8:21. doi: 10.1186/1746-6148-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geser N, Stephan R, Korczak BM, Beutin L, Hachler H. 2012. Molecular identification of extended-spectrum-beta-lactamase genes from Enterobacteriaceae isolated from healthy human carriers in Switzerland. Antimicrob Agents Chemother 56:1609–1612. doi: 10.1128/AAC.05539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, Huijsdens X, Kluytmans J. 2011. Extended-spectrum beta-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg Infect Dis 17:1216–1222. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J, Liu JH, Lv L, Zong Z, Sun Y, Zheng H, Chen Z, Zeng ZL. 2012. Characterization of extended-spectrum beta-lactamase genes found among Escherichia coli isolates from duck and environmental samples obtained on a duck farm. Appl Environ Microbiol 78:3668–3673. doi: 10.1128/AEM.07507-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dierikx C, van der Goot J, Fabri T, van Essen-Zandbergen A, Smith H, Mevius D. 2013. Extended-spectrum-beta-lactamase- and AmpC-beta-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother 68:60–67. doi: 10.1093/jac/dks349. [DOI] [PubMed] [Google Scholar]
- 38.Bergeron CR, Prussing C, Boerlin P, Daignault D, Dutil L, Reid-Smith RJ, Zhanel GG, Manges AR. 2012. Chicken as reservoir for extraintestinal pathogenic Escherichia coli in humans, Canada. Emerg Infect Dis 18:415–421. doi: 10.3201/eid1803.111099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blaak H, Hamidjaja RA, van Hoek AH, de Heer L, de Roda Husman AM, Schets FM. 2014. Detection of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli on flies at poultry farms. Appl Environ Microbiol 80:239–246. doi: 10.1128/AEM.02616-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cortes P, Blanc V, Mora A, Dahbi G, Blanco JE, Blanco M, Lopez C, Andreu A, Navarro F, Alonso MP, Bou G, Blanco J, Llagostera M. 2010. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl Environ Microbiol 76:2799–2805. doi: 10.1128/AEM.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peirano G, van der Bij AK, Gregson DB, Pitout JD. 2012. Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum beta-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J Clin Microbiol 50:294–299. doi: 10.1128/JCM.06025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, Bonten MJ, Mevius DJ, National ESBL Surveillance Group . 2011. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect 17:873–880. doi: 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- 43.Hu YY, Cai JC, Zhou HW, Chi D, Zhang XF, Chen WL, Zhang R, Chen GX. 2013. Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl Environ Microbiol 79:5988–5996. doi: 10.1128/AEM.01740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding Y, Tang X, Lu P, Wu B, Xu Z, Liu W, Zhang R, Bei W, Chen H, Tan C. 2012. Clonal analysis and virulent traits of pathogenic extraintestinal Escherichia coli isolates from swine in China. BMC Vet Res 8:140. doi: 10.1186/1746-6148-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borjesson S, Bengtsson B, Jernberg C, Englund S. 2013. Spread of extended-spectrum beta-lactamase producing Escherichia coli isolates in Swedish broilers mediated by an incl plasmid carrying bla(CTX-M-1). Acta Vet Scand 55:3. doi: 10.1186/1751-0147-55-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Food and Drug Administration, Taiwan. 2013. Annual report of drug residuals in animal product of 2014. Food and Drug Administration, Taipei, Taiwan: (In Chinese.) [Google Scholar]
- 47.Al-Badaii F, Shuhaimi-Othman M. 2015. Water pollution and its impact on the prevalence of antibiotic-resistant E. coli and total coliform bacteria: a study of the Semenyih River, peninsular Malaysia. Water Qual Exp Health 7:319–330. doi: 10.1007/s12403-014-0151-5. [DOI] [Google Scholar]
- 48.Huang JJ, Hu HY, Tang F, Li Y, Lu SQ, Lu Y. 2011. Inactivation and reactivation of antibiotic-resistant bacteria by chlorination in secondary effluents of a municipal wastewater treatment plant. Water Res 45:2775–2781. doi: 10.1016/j.watres.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 49.Murray GE, Tobin RS, Junkins B, Kushner DJ. 1984. Effect of chlorination on antibiotic resistance profiles of sewage-related bacteria. Appl Environ Microbiol 48:73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng WH, Badal RE, Hsueh PR, SMART Program. 2013. Distribution of extended-spectrum beta-lactamases, AmpC beta-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: results of the Study for Monitoring Antimicrobial Resistance Trends (SMART). Antimicrob Agents Chemother 57:2981–2988. doi: 10.1128/AAC.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]