Abstract
Iron is an essential micronutrient for all eukaryotic organisms. However, the low solubility of ferric iron has tremendously increased the prevalence of iron deficiency anemia, especially in women and children, with dramatic consequences. Baker's yeast Saccharomyces cerevisiae is used as a model eukaryotic organism, a fermentative microorganism, and a feed supplement. In this report, we explore the genetic diversity of 123 wild and domestic strains of S. cerevisiae isolated from different geographical origins and sources to characterize how yeast cells respond to elevated iron concentrations in the environment. By using two different forms of iron, we selected and characterized both iron-sensitive and iron-resistant yeast strains. We observed that when the iron concentration in the medium increases, iron-sensitive strains accumulate iron more rapidly than iron-resistant isolates. We observed that, consistent with excess iron leading to oxidative stress, the redox state of iron-sensitive strains was more oxidized than that of iron-resistant strains. Growth assays in the presence of different oxidative reagents ruled out that this phenotype was due to alterations in the general oxidative stress protection machinery. It was noteworthy that iron-resistant strains were more sensitive to iron deficiency conditions than iron-sensitive strains, which suggests that adaptation to either high or low iron is detrimental for the opposite condition. An initial gene expression analysis suggested that alterations in iron homeostasis genes could contribute to the different responses of distant iron-sensitive and iron-resistant yeast strains to elevated environmental iron levels.
INTRODUCTION
Iron is an essential micronutrient for all eukaryotic organisms because it participates as a redox active cofactor in the biogenesis of important cellular macromolecules, such as DNA, proteins, lipids, and various vitamins and cofactors. Iron is also indispensable for generating energy through respiration. Despite iron being abundant in the Earth's crust, its availability to living organisms is restricted as a result of the low solubility of ferric iron at a physiological pH. Thus, iron deficiency has become the most widespread nutritional disorder in the world. In humans, iron deficiency anemia leads to reduced physical capacity, learning problems, and increased morbidity, especially in women and children. Multiple approaches, including iron supplementation and food fortification with iron, are currently used to prevent and treat iron deficiency. The budding yeast Saccharomyces cerevisiae, which is responsible for the generation of multiple fermentative foods and beverages, is also utilized as a feed supplement because it is rich in essential amino acids, fiber, and B-type vitamins. Interestingly, iron-enriched yeasts are currently being evaluated as promising sources of iron to prevent and palliate iron deficiency in humans and animals (1). In fact, recent reports have indicated that iron-enriched baker's yeasts are efficient for helping animals recover from iron deficiency, while maintaining their fermentative and bakery properties (2, 3).
Over the last 2 decades, the response of S. cerevisiae to fluctuating environmental levels of iron has been studied at the molecular level (4, 5). This yeast has served as a model eukaryotic organism to identify and decipher the functioning of multiple components required for the acquisition, distribution, utilization, and storage of iron. Under iron-sufficient conditions, iron enters yeast cells mainly through the low-affinity iron transporters Fet4 and Smf1, located at the plasma membrane (6–8). Yeast cells activate sophisticated regulatory mechanisms in response to imbalances in environmental iron levels (reviewed in reference 9). Upon iron scarcity, transcriptional factor Aft1 activates the expression of a group of genes known as the iron regulon (10), which includes the genes encoding cuproferroxidase Fet3, required for high-affinity iron uptake (11), and Cth2, an RNA-binding protein that remodels cellular iron metabolism (12). When present in excess, iron engages in Fenton redox reactions that accelerate the formation of reactive oxygen species (ROS), such as hydroxyl radicals, that damage cells at the membrane, protein, and nucleic acid levels. A recent report has suggested that elevated iron concentrations also cause toxicity to yeast cells by activating sphingolipid signaling and synthesis (13). When environmental iron levels rise, the yeast Yap5 transcription factor activates specific genes that protect cells from the potential danger of high iron (14). The most relevant transcriptional activation by Yap5 is exerted on CCC1, which encodes a protein that protects cells from excess iron by transporting the metal from the cytoplasm to the vacuole, where it is stored (15, 16). Recent studies have demonstrated that S. cerevisiae does not directly sense extracellular or cytosolic iron concentrations. Instead, the mitochondrial iron-sulfur synthesis rate is transduced through a currently unknown signal to transcription factors Aft1 and Yap5 (reviewed in references 17 and 18).
Iron homeostasis has been studied primarily in a few laboratory strains, whereas very little is currently known about the response of S. cerevisiae strains isolated in other sources and environments to alterations in iron bioavailability. The completion of the genome sequence of many wild and domestic S. cerevisiae strains isolated from different geographical locations has uncovered extensive differences in genomic and phenotypic variations that could allow genetic determinants to be identified (19). A recent population genomics study has been performed with yeast strains obtained from the nectar of flowers of bertam palm trees in a Malaysian rainforest, a reproductively isolated environment (20–22). This work revealed that Malaysian yeast isolates have diverged from the canonical iron homeostasis network, probably as a result of a change in selective pressure (21). Molecular analyses have revealed that the sensitivity of Malaysian strains to toxic levels of iron is a recessive trait that is attributed to loss of function of the iron resistance YAP5 gene and especially to their defective CCC1 allele. Malaysian strains also activate the iron deficiency response during growth under iron-sufficient conditions, which is consistent with an AFT1 gain-of-function allele (21). These results suggest that Malaysian strains adapt to a low-iron environment, probably by lowering the range of iron concentrations at which they respond. Their short exposure to high-iron conditions could relax the strength of the selection of the iron toxicity response, which would allow mutations to accumulate. In this report, we took advantage of the genetic diversity of more than 100 yeast strains isolated in different environments to explore how yeast cells respond to excess iron. For this purpose, we selected both iron-resistant and iron-sensitive strains, and we characterized their growth rate and viability, their endogenous iron levels, and the expression of iron homeostasis genes.
MATERIALS AND METHODS
Yeast strains and growth conditions.
All the yeast strains used in this study are listed in Table 1. Yeasts were cultivated at 30°C in rich yeast extract-peptone-dextrose (YPD) medium or in synthetic complete (SC) medium. Iron was added to SC from either a 250 mM FeCl3 (Sigma) or a 100 mM Fe(NH4)2(SO4)2 (FAS; Sigma) stock solution. Both stock solutions were prepared in 0.1 M HCl to facilitate iron solubility and were used fresh. Iron solutions were added to the autoclaved medium at the desired final concentration. A control with the equivalent HCl concentration was always prepared. Fe2+-specific chelator bathophenanthroline disulfonic acid (BPS; Sigma) was added to SC plates from a 100 mM stock solution prepared in water. For the oxidative stress experiments, methylene blue (Sigma) was added to SC or to SC with FeCl3 plates from a 1% stock solution, whereas H2O2, tert-butyl hydroperoxide (t-BOOH), and menadione were added to YPD plates from the 9.7 mM, 7.0 M, and 50 mM stocks, respectively. To grow yeast cells in liquid media, overnight SC precultures were reinoculated at an optical density at 600 nm (OD600) of 0.2 and then incubated for 24 h at 30°C and 190 rpm in 50-ml flasks that contained 5 ml of SC supplemented with the corresponding iron concentration. Depending on the experiment, these cultures were used to determine the OD600, number of cells per milliliter, cellular volume, dry weight (DW), and endogenous iron level. To perform growth assays in solid media, cells from overnight precultures were diluted to an OD600 of 0.1 and then spotted directly and after 1:10 and 1:100 dilutions in sterile water. Plates were incubated at 30°C for 3 to 6 days and were then photographed.
TABLE 1.
Saccharomyces cerevisiae strains used in this work
| Strain designationa | Geographic origin | Isolation source | Growth in high-iron mediumb |
|---|---|---|---|
| BY4741 | Laboratory strain | = | |
| Gb-FlorC | Europe (Spain) | Fermentation (flor wine) | −− |
| T73 | Europe (Spain) | Fermentation (red wine) | = |
| CBS 2992 | Asia (Pakistan) | Fermentation (palm wine) | −− |
| G1 | Europe (Belgium) | Fermentation (beer) | −− |
| CBS 8857 | Africa (Burkina Faso) | Fermentation (sorghum beer) | − |
| BAY1 | Europe (Ireland) | Fermentation (cider) | + |
| MERTY | Europe (Ireland) | Fermentation (cider) | = |
| Kyokai 6 (CECT 10711) | Asia (Japan) | Fermentation (sake) | ++ |
| MI 26 | America (Mexico) | Fermentation (agave) | − |
| Cinta Roja | Oceania (Australia) | Baking | − |
| D4 | Unknown | Dietetic (Altana Pharma tablet, beer) | − |
| D7 | Unknown | Dietetic (Levadiet Dietisa tablet, beer) | − |
| D8 | Unknown | Dietetic (Terra Verda tablet, beer) | − |
| Ultra-Levura | Europe (Spain) | Dietetic (UPSA, Biocodex) | + |
| D14 | Unknown | Dietetic (Phytodepur Intersa tablet) | = |
| D24 | Unknown | Dietetic (L'hirondelle, Lasafre, bread) | − |
| D20 | Unknown | Dietetic (Phytonorm Intersa tablet) | = |
| 60 | Europe (Spain) | Clinical (vagina) | − |
| 102 | Europe (Spain) | Clinical (bronchial aspirated) | + |
| Sc 9 | Europe (Hungary) | Wild (soil) | −− |
| YPS128 | America | Wild | −− |
| NCAIM Y.00925 | Europe (Hungary) | Wild (apricot pulp) | − |
| CBS 400 | Africa (Ivory Coast) | Fermentation (palm wine) | − |
| CBS 405 | Africa (West Africa) | Fermentation (“Bili” wine) | − |
| CBS 1544 | Europe (Netherlands) | Wild (fruit juice) | + |
| CBS 1460 | Asia (Indonesia) | Wild (fermenting fruit) | + |
| CBS 1591 | Asia (Indonesia) | Wild (fermenting cacao) | − |
| CBS 2421 | Asia (Japan) | Fermentation (kefir grains) | = |
| CECT 10120 | Europe (Spain) | Wild (fruit of Arbutus unedo) | −− |
| CECT 10109 | Europe (Spain) | Wild (prickly pear) | = |
| AQ 909 | America (Mexico) | Fermentation (sotol) | = |
| AQ 2431 | America (Peru) | Wild (skin seed) | − |
| AQ 2908 | America (Mexico) | Fermentation (raicilla) | = |
| CECT 10131 | Europe (Spain) | Wild (flower of Centaurea alba) | ++ |
| CBS 7764 | Europe (Sweden) | Wild (rainbow trout intestine) | + |
| AQ 2458 (B2) | America (Peru) | Wild (wasp) | = |
| CBS 7993 | America (Brazil) | Wild (freshwater) | = |
| CBS 8292 | Europe (Sweden) | Wild (seawater) | + |
| AQ 1559 | Europe (Spain) | Wild (olive) | + |
| AQ 1560 | Europe (Spain) | Wild (olive) | − |
| AQ 1561 | Europe (Spain) | Wild (olive) | − |
| AQ 931 (LB 1 M) | America (Peru) | Fermentation (masato) | − |
| AQ 2348 (PE 1 M) | America (Peru) | Fermentation (masato) | −− |
| AQ 885 (1LM) | America (Peru) | Fermentation (chicha de jora) | + |
| AQ 898 | America (Peru) | Fermentation (chicha de jora) | = |
| AQ 2470 (V3) | America (Peru) | Fermentation (chicha de jora) | = |
| CBS 436 | Asia (Japan) | Fermentation (SakeMoto) | = |
| Kyokai 7 | Asia (Japan) | Fermentation (sake moromi) | −− |
| A35 | Asia (Japan) | Fermentation (sake) | = |
| CBS 1198 | Asia (Japan) | Fermentation (sake) | = |
| CBS 1199 | Asia (Japan) | Fermentation (sake moromi) | + |
| CLIB215 | Oceania (New Zealand) | Baking | − |
| Plus Vital | Unknown | Baking | = |
| F27 | Europe (Spain) | Clinical (blood) | −− |
| YJM975 | Europe | Clinical | + |
| YJM978 | Europe | Clinical | ++ |
| YJM981 | Europe | Clinical | = |
| YJM320 | America | Clinical (blood) | = |
| YJM326 | America | Clinical | − |
| SSI4 | Europe (Denmark) | Clinical | = |
| 273614N | Europe | Clinical | ++ |
| F3 | Europe (Spain) | Clinical (blood) | = |
| Chr 98.2 | Europe (Spain) | Wild (oak) | − |
| DBVPG 6765 | Europe | Fermentation (wine) | = |
| CBS 2087 | Asia (China) | Wild (flower de lychee) | − |
| NCAIM Y.00678 | Europe (Hungary) | Fermentation (drink) | ++ |
| CBS 1201 | Asia (Japan) | Fermentation (sorghum brandy) | − |
| Y12 | Africa (Ivory Coast) | Fermentation | − |
| Gb49 | Europe (Spain) | Fermentation (flor wine) | − |
| EC-1118 | Europe (France) | Fermentation (Champagne wine) | = |
| ALG804 | Europe (France) | Fermentation (Champagne wine) | = |
| FT134L PE-2 | America (Brazil) | Wild (sugar cane) | = |
| FT280L CAT-1 | America (Brazil) | Wild (sugar cane) | = |
| FT013L | America (Brazil) | Wild (sugar cane) | = |
| FT119L BG-1 | America (Brazil) | Wild (sugar cane) | − |
| UWOPS03-461.4 | Asia (Malaysia) | Wild | −− |
| UWOPS03-217.3 | Asia (Malaysia) | Wild | − |
| UWOPS03-227.2 | Asia (Malaysia) | Wild | − |
| CECT 1384 | Europe (UK) | Fermentation (beer) | + |
| CECT 1387 | Europe (UK) | Fermentation (beer) | = |
| CECT 11001 | Europe (Belgium) | Fermentation (beer) | − |
| CBS 8858 | Africa (Burkina Faso) | Fermentation (sorghum beer) | − |
| CBS 8855 | Africa (Ghana) | Fermentation (sorghum beer) | − |
| CBS 4455 | Africa (South Africa) | Fermentation (kaffir beer) | − |
| CBS 4454 | Africa (South Africa | Fermentation (kaffir beer) | − |
| C10 | Europe (Ireland) | Fermentation (Clonmel cider) | − |
| Sc 42 | Europe (Ireland) | Fermentation (Clonmel cider) | = |
| Sc 97 | Europe (Ireland) | Fermentation (Clonmel cider) | ++ |
| AQ 902 | America (Mexico) | Fermentation (tequila) | = |
| AQ 903 | America (Mexico) | Fermentation (mezcal) | = |
| AQ 904 | America (Mexico) | Fermentation (tequila) | − |
| AQ 905 | America (Mexico) | Fermentation (tequila) | − |
| AQ 1887 | America (Mexico) | Fermentation (Temohaya Mezquital) | − |
| AQ 1850 | America (Mexico) | Fermentation (Temohaya Mezquital) | − |
| No. 308 (CECT 11032) | Europe (Italy) | Wild (grape must) | ++ |
| CECT 10233 | Africa (South Africa) | Wild (grape must) | − |
| ZIM 2139 | Europe (Slovenia) | Wild (grape must) | + |
| CECT 12681 | Europe (Spain) | Wild (grape must) | − |
| CBS 5287 | Europe (Russia) | Wild (grape) | −− |
| AQ 2281 | Europe (Spain) | Wild (grape) | = |
| YJM 269 | Europe (Portugal) | Wild (grape) | − |
| 322134S | Europe | Clinical | = |
| RM11 1A | America | Fermentation (wine) | = |
| YIIc17 E5 | Europe | Fermentation (wine) | + |
| BC187 | America | Fermentation (wine) | − |
| L-1374 | America | Fermentation (wine) | + |
| Tokai 9 | Europe (Hungary) | Fermentation (wine) | − |
| CECT 1477 | Europe (France) | Fermentation (wine) | + |
| CECT 1882 | Europe (Spain) | Fermentation (wine) | − |
| YJM332 | America | Fermentation (wine) | + |
| ZA9 | Africa | Fermentation (wine) | + |
| ZA26 | Africa | Fermentation (wine) | − |
| ZA4 | Africa | Fermentation (wine) | − |
| L-1334 | Europe (France) | Fermentation (wine) | + |
| L-1338 | Europe (France) | Fermentation (wine) | + |
| L-246 | America (Chile) | Fermentation (wine) | ++ |
| L-720 | America (Chile) | Fermentation (wine) | = |
| L-958 | America (Argentina) | Fermentation (wine) | = |
| L-1528 | America | Fermentation (wine) | − |
| CBS 423 | Europe (Switzerland) | Fermentation (wine) | + |
| Y-9 | Asia (Japan) | Fermentation (wine) | − |
| HA 1931 | Europe (Austria) | Fermentation (wine) | = |
CECT, Colección Española de Cultivos Tipo; CBS, CBS-KNAW Culture Collection.
Growth compared to that of laboratory strain BY4741: +, better growth; −, worse growth; =, similar growth. The strains labeled ++ and −− were selected for further studies as being representative of iron-resistant and iron-sensitive strains, respectively.
Endogenous iron measurements.
Yeast cells that corresponded to a total OD600 × ml of 5.0 were collected by centrifugation and washed twice with 1 mM EDTA and once with Milli-Q water. Cells were dissolved in 500 μl of 3% HNO3 and incubated at 98°C for 16 h with agitation. After digestion, extracts were used to determine iron levels. For the yeast cells grown in SC, iron levels were determined by inductively coupled plasma-mass spectrometry (ICP-MS) at the Universitat Jaume I (Castelló, Spain). A previously described colorimetric assay (23) was run for the other samples. The two methods provided similar results.
RNA analyses.
To determine the expression levels of specific mRNAs, total RNA was extracted and quantitative reverse transcription-PCR (qPCR) was performed as previously described (24). For each gene analyzed, a standard curve was made with serial dilutions of the cDNA sample. The ACT1 mRNA values were used to normalize data. Data and error bars represent the averages and standard deviations of the results from at least three independent biological samples. All the primers used in this study are listed in Table 2.
TABLE 2.
Oligonucleotides used in this work
Miscellaneous.
The yeast cells incubated in liquid cultures were properly diluted and sonicated before determining their concentration (numbers of cells/milliliter) and average volume in a Beckman Coulter Counter Z. To determine yeast dry weight, the cells that corresponded to a total OD600 × ml of 30 were collected by centrifugation and washed with distilled water. Then, cells were transferred to a previously weighed Eppendorf tube and were dried at 50°C for 3 days. Finally, yeast dry weight was obtained by subtracting the total weight from the Eppendorf tube weight. At least three independent biological samples were used to determine yeast cellular concentration, volume, and dry weight.
RESULTS
Selection of Saccharomyces cerevisiae strains resistant or sensitive to elevated iron concentrations.
The response of laboratory S. cerevisiae strains to excess iron has been studied at the molecular level (4, 5, 9). However, very little is currently known about how yeast strains of different geographical origins and isolation sources behave when challenged with elevated extracellular iron concentrations. To explore the genetic diversity of S. cerevisiae for this trait, the growth of 123 yeast strains isolated from different environments was analyzed. In addition to laboratory strain BY4741, we here included 67 strains obtained from different types of fermentations (23 wine, 9 beer, 5 cider, 6 sake, and 24 from other fermentative sources), 33 strains found in wild environments, 12 clinical strains, 7 dietetic strains, and 3 baking yeasts (see Table 1 for details). Two independent colonies of each strain were incubated overnight in synthetic complete (SC) liquid medium and were then assayed for growth in solid SC medium. We performed spot assays that consisted in 10-fold dilutions of yeast drops by starting at an OD600 of 0.1. We used laboratory strain BY4741 as a reference for these assays. To challenge yeast cells with iron, we added increasing FeCl3 concentrations to the solid SC medium, starting at 3 mM and reaching 7 mM. To keep FeCl3 soluble, we freshly prepared 250 mM FeCl3 stock solutions in 0.1 M HCl. We always performed an additional spot assay with the equivalent HCl used to prepare the plate with the maximum iron concentration, but HCl did not alter yeast growth at these concentrations (Fig. 1; see also Fig. S1 to S5 in the supplemental material). Reference strain BY4741 grew properly on SC plates supplemented with 3 mM FeCl3 but displayed a marked growth defect at 4 mM FeCl3 (Fig. 1; see also Fig. S1 to S5 in the supplemental material). At 5 mM FeCl3, the laboratory strain barely grew, and it did not tolerate higher FeCl3 concentrations (Fig. 1; see also Fig. S1 to S5 in the supplemental material). As for the remaining 122 strains, 56 strains (46%) grew better than the reference strain in media with high iron concentrations, 29 (24%) exhibited growth defects, and the remaining 37 strains (30%) did not substantially differ from the laboratory strain (Table 1; see also Fig. S1 to S5 in the supplemental material). Then, we compared resistance to iron-containing media with the isolation source or geographic origin of the S. cerevisiae strains. We observed a correlation only for the strains isolated from wild environments, which were more prone to exhibit increased sensitivity to high iron levels. Whereas 24% (8 of 33) of wild strains were more resistant to high iron than the laboratory strain, 55% (18 of 33) were more sensitive (Table 1; see also Fig. S1 to S5 in the supplemental material). These results suggest that the yeast cells isolated in wild environments may be more prone to display iron sensitivity than iron resistance.
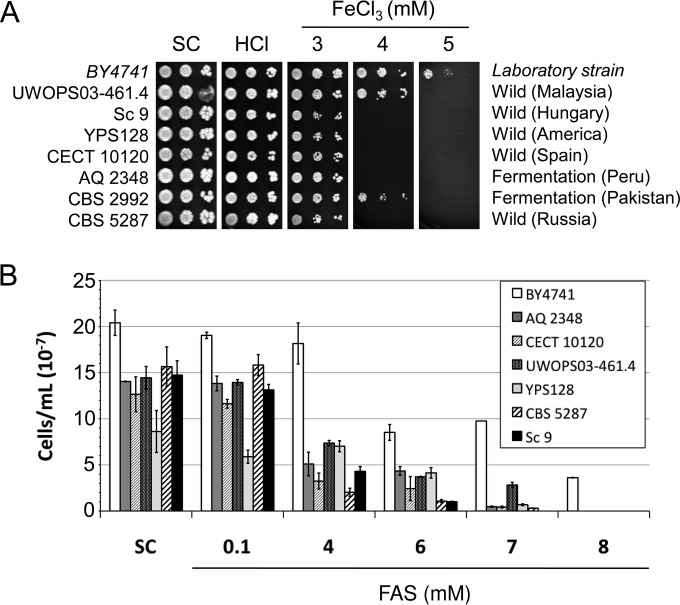
FIG 1.
Growth of a selection of yeast strains resistant to iron in media with high iron concentrations. (A) The indicated S. cerevisiae strains were grown overnight in liquid SC medium and then spotted in 1:10 dilutions, starting at an OD600 of 0.1 on solid SC plates that contained increasing FeCl3 concentrations. Given that HCl was used to prepare iron stock solutions, a control with the equivalent final HCl concentration was always used. Plates were incubated at 30°C for 3 days and then photographed. (B) After overnight growth in the liquid SC medium, yeast cells were reinoculated at an OD600 of 0.2 and incubated at 30°C with agitation in liquid SC supplemented with the indicated FAS concentrations. The number of cells per milliliter after 24 h of incubation is presented. Data and error bars represent the averages and standard deviations of the results from at least three independent biological samples.
A selection of yeast strains resistant to FeCl3, including four strains isolated from fermentation sources (Kyokai 6, NCAIM Y.00678, L-246, and Sc 97), two strains isolated from wild environments (CECT 10131 and CECT 11032), and two clinical yeast strains (YJM978 and 273614N), were studied in more detail. We observed that all the selected strains, except for NCAIM Y.00678, grew when 6 mM FeCl3 was added to the SC plates (Fig. 1A). We also noticed that sake strain Kyokai 6 was extremely resistant to excess iron since it was able to grow at 7 mM FeCl3 in the solid medium (Fig. 1A). Such adaptation to high iron concentrations was specific to strain Kyokai 6, as it was not shared by other S. cerevisiae sake strains (see Fig. S2 in the supplemental material). To further ascertain whether these phenotypes were due to the high iron concentrations and were not unique to FeCl3, we assayed growth in liquid SC medium that contained increasing concentrations of an alternative iron source: Fe(NH4)2(SO4)2 (ferrous ammonium sulfate [FAS]). Overnight precultures from at least three independent colonies were diluted to 2.5 × 106 cells/ml (OD600 of approximately 0.2), and culture growth was determined after 24 h of incubation at 30°C and 190 rpm. Despite our initial selection of iron-resistant strains also including fermentation strain BAY1 (see Fig. S1 in the supplemental material), the growth assays performed in liquid media did not corroborate its iron resistance under solid medium conditions. This strain was, therefore, ruled out (data not shown). We observed that all the selected strains displayed reduced fitness in liquid SC medium compared to BY4741 (Fig. 1B). This result is probably due to the different cellular volume of each strain (data not shown), since in most cases the differences in liquid SC growth disappeared when we presented OD600 values (see Fig. S6 in the supplemental material). As observed, when growth per strain is referred to its value under the SC conditions (see Fig. S7A in the supplemental material), the interpretation of the results presented in Fig. 1B is not biased by the differences observed among the strains under the aforementioned conditions. The addition of 100 μM FAS to liquid SC did not considerably affect the growth of any of the strains, whereas FAS concentrations of 6 mM, or higher, limited the growth of all the strains (Fig. 1B). Reference strain BY4741 grew to 8 mM FAS, whereas no growth was observed at higher iron concentrations (Fig. 1B). The additional five strains included in this experiment still grew at 10 mM FAS, which confirms their greater resistance to the high iron concentrations obtained in solid media (Fig. 1). Strain L-246 displayed even more resistance to excess iron when grown in liquid than it displayed in solid medium (Fig. 1). Sake strain Kyokai 6 was the strain most resistant to excess iron, with either FeCl3 or FAS, and also under both liquid and solid conditions. Therefore, by using two different iron sources, we were able to isolate iron-resistant strains among many other yeast strains.
For further studies, we also selected four strains isolated from wild environments (Sc 9, YPS128, CECT 10120, and CBS 5287) and two strains obtained from fermentations (AQ 2348 and CBS 2992) as being representative of iron-sensitive yeast strains. In addition to laboratory strain BY4741, we used as a control strain for these assays Malaysian strain UWOPS03-461.4, which has been previously shown to be sensitive to iron, mostly due to defects in the iron transporter to the vacuole known as Ccc1 (21). All the selected strains, except for CBS 2992, were more sensitive to FeCl3 than the laboratory and Malaysian strains, which were unable to grow at 4 mM FeCl3 (Fig. 2A). The selected iron-sensitive strains were also assayed for growth in liquid medium. These strains exhibited slower growth than the laboratory strain in liquid SC medium (Fig. 2B). Once again, the different cellular volume of each strain may account for the growth differences (data not shown), which disappeared when we represented the OD600, with the exception of the wild CECT 10120 strain (see Fig. S6 in the supplemental material). The laboratory strain was able to grow at 8 mM FAS in liquid SC, but none of the iron-sensitive strains were (Fig. 2B). This finding supports our data on solid medium. In order to directly compare the sensitivities of all the strains, we also represented the growth for each culture in relation to its SC condition (see Fig. S7B in the supplemental material). With this relative representation, we observed that wild strain YPS128 showed more sensitivity to iron than BY4741 only at 7 mM FAS or higher, whereas the other strains exhibited iron sensitivity at 4 mM FAS. These data demonstrate that the yeast strains selected here are sensitive to elevated iron concentrations.
FIG 2.
Growth of a selection of yeast strains sensitive to iron in media with high iron concentrations. (A) The indicated S. cerevisiae strains were grown overnight in liquid SC medium and then spotted in 1:10 dilutions, starting at an OD600 of 0.1 on solid SC medium plates that contained increasing FeCl3 concentrations. Given that HCl was used to prepare iron stock solutions, a control with the equivalent final HCl concentration was always used. Plates were incubated at 30°C for 3 days and then photographed. (B) After overnight growth in liquid SC medium, yeast cells were reinoculated at an OD600 of 0.2 and incubated at 30°C with agitation in the liquid SC supplemented with the indicated FAS concentrations. The number of cells per milliliter after 24 h of incubation is presented. Data and error bars represent the averages and standard deviations of the results from at least three independent biological samples.
Yeast iron accumulation inversely correlates with growth in high-iron-containing media.
With the aim of characterizing the bases of the S. cerevisiae response to elevated extracellular iron levels, we determined the endogenous iron content of both the iron-resistant and iron-sensitive strains. For this purpose, at least three independent colonies of each chosen strain were cultivated overnight in liquid SC medium and were then reinoculated at 2.5 × 106 cells/ml in fresh liquid SC medium with different FAS concentrations. After 24 h, an aliquot of cells was collected and digested with HNO3. Its iron content was determined by either a colorimetric assay or ICP-MS (see Materials and Methods). A second aliquot was used to normalize the data by determining the dry weight of every yeast strain at each iron-containing medium (see Materials and Methods). Thus, yeast iron content was displayed as the weight of iron per yeast dry weight (grams Fe/grams DW). Our study showed that the iron content of all the analyzed yeast strains gradually increased as the environmental FAS concentration rose (Fig. 3 and 4). We observed that laboratory strain BY4741 accumulated around 70 μg Fe/g DW in SC and reached 5 mg Fe/g DW when grown for 1 day in an 8 mM FAS medium (Fig. 3 and 4). By determining the cellular volume of each strain after a 1-day incubation in each iron-containing medium, we were able to calculate the yeast cellular iron concentration in molar units. We thus noted that the laboratory strain accumulated approximately 440 μM iron when incubated in SC and around 50 mM when grown in SC supplemented with 8 mM FAS for 1 day (see Fig. S8 in the supplemental material). These endogenous iron levels fell within a range similar to those previously reported (1–3, 25, 26).
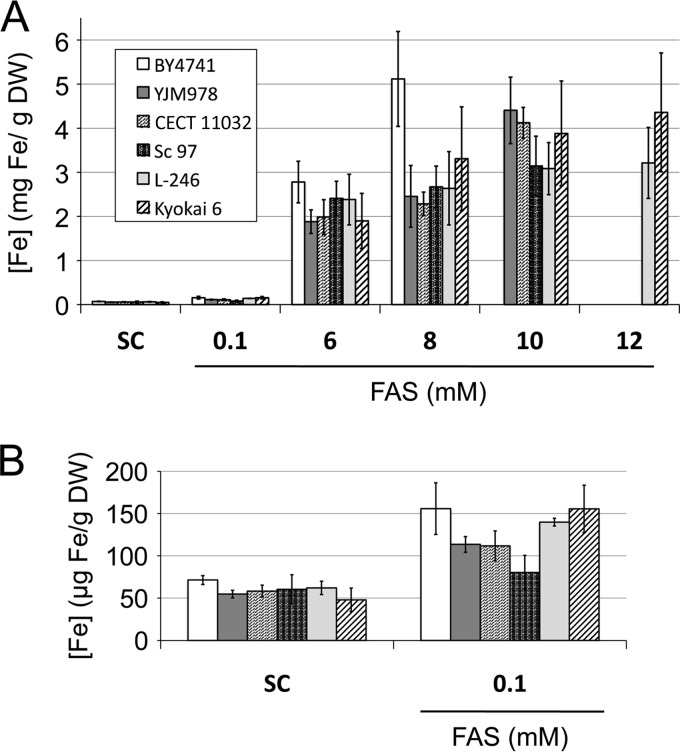
FIG 3.
Iron accumulation in a selection of iron-resistant yeast strains in media with high iron concentrations. (A) The indicated S. cerevisiae strains were grown as detailed in the legend to Fig. 1B. Then, cells were collected and digested, and endogenous iron was determined as indicated in Materials and Methods. (B) Iron accumulation is shown in more detail for the cells grown in SC or in SC with 0.1 mM FAS. Data and error bars represent the averages and standard deviations of the results from at least three independent biological samples.
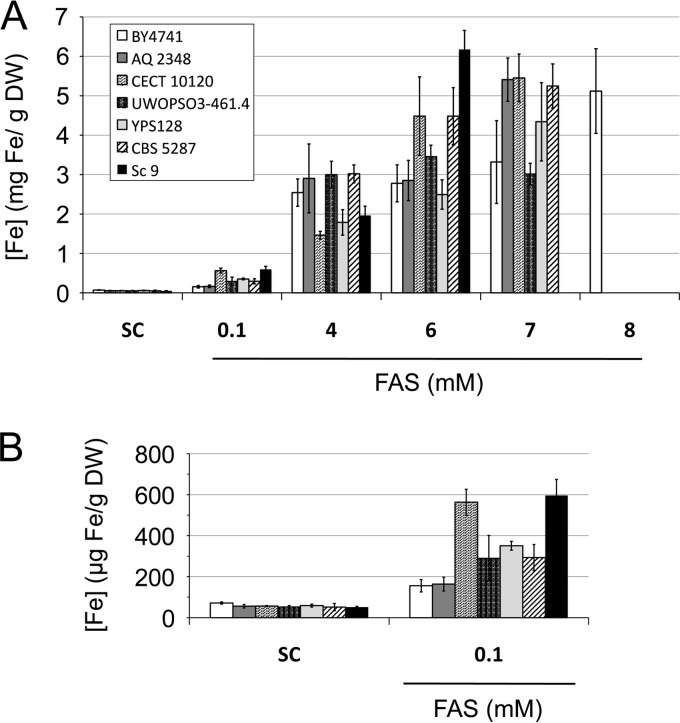
FIG 4.
Iron accumulation in a selection of iron-sensitive yeast strains in media with high iron concentrations. (A) The indicated S. cerevisiae strains were grown as detailed in the legend to Fig. 2B. Then, cells were collected and digested, and endogenous iron was determined as indicated in Materials and Methods. (B) Iron accumulation is shown in more detail for the cells grown in SC or in SC with 0.1 mM FAS. Data and error bars represent the averages and standard deviations of the results from at least three independent biological samples.
When we looked at the five iron-resistant strains selected for this experiment, the most general remarkable feature was that their endogenous iron levels were below those of the laboratory reference strain in most cases (Fig. 3). For instance, at 8 mM FAS, the endogenous iron levels of all the resistant strains were lower than those of the laboratory strain, which suggests that iron-resistant strains limit iron accumulation (Fig. 3A). Similar results were obtained when the molar iron concentrations were compared (see Fig. S8 in the supplemental material). At 10 mM FAS, the laboratory strain was unable to grow, but the iron content of the resistant strains further increased to levels similar to those reached by the laboratory strain at 8 mM FAS (Fig. 3A; see also Fig. S8A in the supplemental material). These results suggest that the iron-resistant yeast strains selected here tolerate elevated exogenous iron concentrations by limiting its accumulation and die when endogenous iron levels reach a common threshold.
In order to determine iron accumulation in iron-sensitive yeasts, we included the Malaysian strain UWOPS03-461.4 as an additional control. We observed that the Malaysian strain accumulated amounts of iron similar to or slightly larger than those of the laboratory strain (Fig. 4). However, growth of the Malaysian strain ceased at 3 mg Fe/g DW when grown in 7 mM FAS, whereas the laboratory strain accumulated up to 5 mg Fe/g DW when grown in 8 mM FAS (Fig. 4). These results indicate that the Malaysian strain tolerates lower endogenous iron concentrations than the laboratory strain, probably due to its defect in iron accumulation in the vacuole (21). When we analyzed the iron-sensitive strains selected in this study, we observed that they all accumulated more endogenous iron (around 5 mg Fe/g DW) than the laboratory and Malaysian strains (3 mg Fe/g DW) when grown in 7 mM FAS (Fig. 4). Furthermore, the iron-sensitive strains accumulated even more iron at 7 mM FAS than the iron-resistant strains at 8 mM FAS (from 3 to 4 mg Fe/g DW) (Fig. 3). Wild yeast strain Sc 9, which was even more sensitive to iron in liquid medium than the other strains (it was unable to grow at 7 mM FAS), accumulated more endogenous iron when grown at 6 mM FAS (up to 6 mg Fe/g DW) (Fig. 4). Taken together, these results suggest that the analyzed iron-sensitive yeast strains reached the maximum tolerable threshold of endogenous iron at lower exogenous iron concentrations than other more resistant strains, probably due to defects in inhibiting iron accumulation.
Iron-sensitive strains display an altered cellular redox state in the presence of excess iron that is not due to exacerbated sensitivity to oxidative stress.
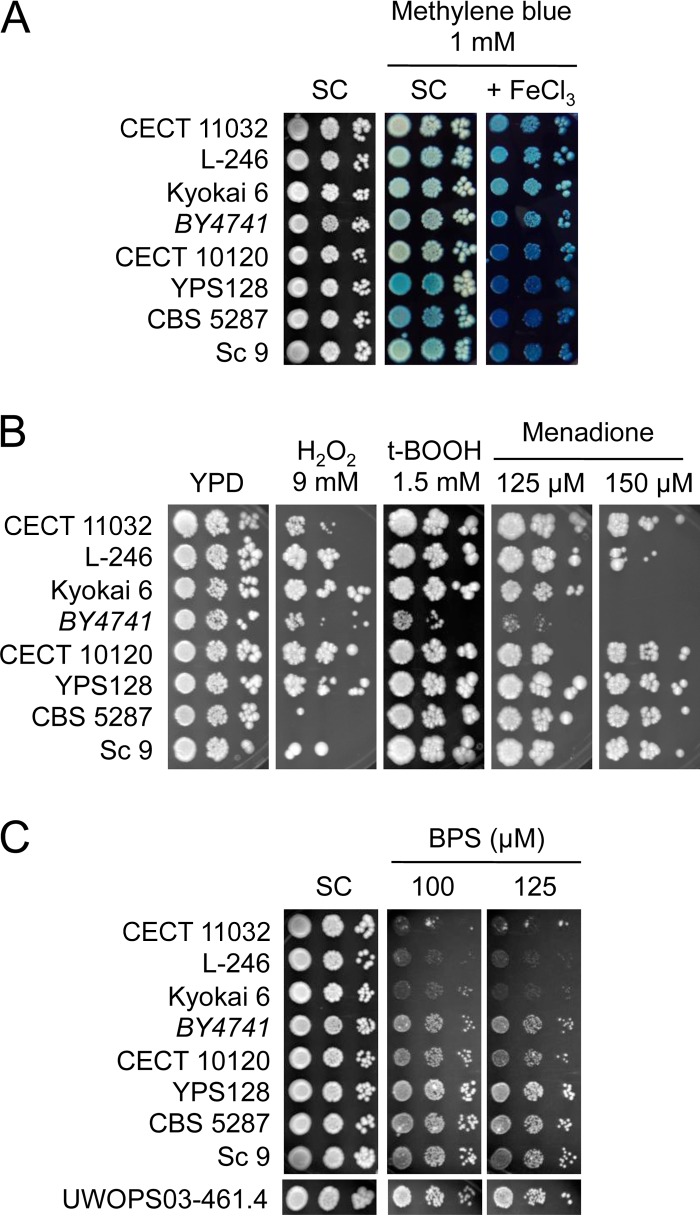
Iron is a redox active metal that generates cytotoxic ROS when present in excess. Therefore, we decided to determine how the high iron concentrations used in this study affected the cellular redox state of both iron-sensitive and iron-resistant strains. For this purpose, we assayed yeast growth in the plates that contained the biocompatible redox indicator methylene blue, which loses its blue color when reduced (27). We observed that iron-sensitive strains YPS128, CBS 5287, and Sc 9 acquired a blue color, which is characteristic of methylene blue oxidation, when grown in SC medium (Fig. 5A). No significant coloration was observed for the laboratory and iron-resistant strains. Then, we ascertained colony staining in the presence of 3 mM FeCl3, which did not alter the growth of any strain (Fig. 1). It was noteworthy that when FeCl3 was added to the methylene blue medium, iron-resistant strains CECT 11032, L-246, and Kyokai 6 displayed a faint blue tone, in contrast to the dark blue color associated with iron-sensitive strains (Fig. 5A). The laboratory strain exhibited an intermediate blue tone (Fig. 5A). These results indicate that when environmental iron levels increase, the redox state of iron-sensitive strains becomes more oxidized than that of iron-resistant strains, probably due to greater iron accumulation.
FIG 5.
Growth of iron-resistant and -sensitive strains under different conditions. The indicated yeast strains were grown overnight in the liquid SC medium and then assayed for growth under different conditions. (A) Methylene blue was added at the 1 mM concentration to SC to elucidate the redox state of yeast cells' growth at different iron concentrations. (B) Yeast cells were grown in YPD plates that contained different reagents that induce oxidative stress, including 9 mM H2O2, 1.5 mM t-BOOH, and 125 μM and 150 μM menadione. (C) Growth under the iron-deficient conditions was achieved by the addition of 100 μM or 125 μM BPS, a specific iron chelator. Plates were incubated at 30°C for 3 days and photographed.
The different behaviors of iron-sensitive and iron-resistant strains in iron-containing media could be attributed to their respective general sensitivity to the oxidative stress caused by accumulated iron levels. To discriminate between sensitivity to oxidative stress and specific iron homeostasis alterations, we decided to determine growth levels when challenging these cells with different oxidative agents, including peroxides H2O2 and tert-butyl hydroperoxide (t-BOOH) and menadione, which induces superoxide anion production. As shown in Fig. 5B, the behaviors of the assayed yeast strains were dissimilar. Laboratory strain BY4741 was the strain most sensitive to all the different sources of oxidative stress (Fig. 5B). The growth of iron-sensitive strains CBS 5287 and Sc 9 and iron-resistant CECT 11032 cells was affected by H2O2 (Fig. 5B). Iron-resistant strains L-246 and Kyokai 6 were more sensitive to menadione than the rest of the strains. These results rule out iron-resistant strains tolerating higher exogenous iron concentrations than iron-sensitive strains because they possess more-efficient mechanisms to protect them from general oxidative stress. Instead, we suggest that the observed differences are the result of alterations in iron accumulation (Fig. 3 and 4).
Yeast strains resistant to high iron are sensitive to iron-deficient conditions.
It has been previously proposed that the adaptation of Malaysian yeast strains to a constant wild iron-deficient environment and their rare exposure to high iron levels could have led to the accumulation of detrimental mutations in the yeast iron detoxification machinery (21). To explore whether the iron-sensitive strains selected in this study gained fitness under low-iron conditions, we decided to grow them under iron-deficient conditions, which we achieved by adding the Fe2+-specific chelator bathophenantholine disulfonic acid (BPS). We observed that when iron was scarce, iron-sensitive strains, except for CECT 10120, grew slightly better than reference strain BY4741 and similarly to Malaysian strain UWOPS03-461.4 (Fig. 5C). We also explored how iron-resistant strains CECT 11032, L-246, and Kyokai 6 behaved when iron availability was limited. In this case, we observed that all three iron-resistant strains were very sensitive to low-iron conditions (Fig. 5C), which indicates that adaptation to elevated iron concentrations could have detrimental effects on the iron deficiency response. Taken together, these results suggest that yeast strains that adapt to either high- or low-iron conditions frequently lose fitness under the opposite condition, that is, low or high iron, respectively.
The expression of iron-related genes is altered in iron-sensitive strains.
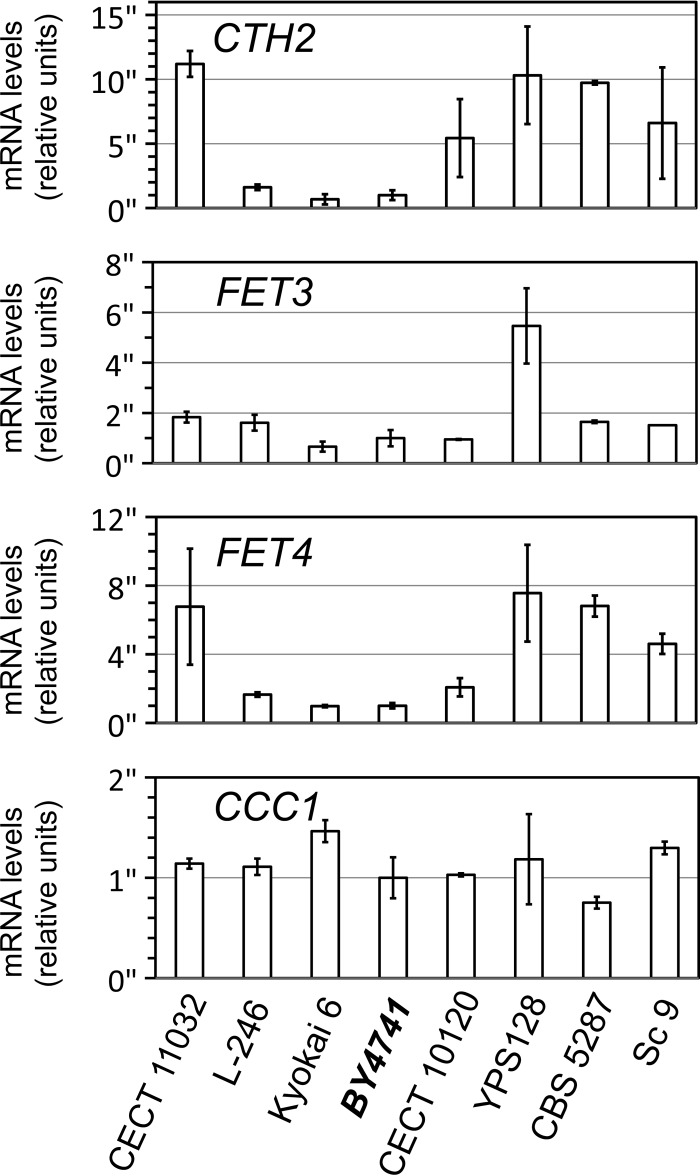
The molecular basis of the different performances displayed by the iron-sensitive and iron-resistant yeast strains selected in this study could be due to multiple factors, which could differ among strains. Despite this complexity, we decided to perform a first approach to analyze iron homeostasis in these strains at the molecular level. For this purpose, we cultivated yeast cells in liquid SC medium to reach the exponential growth phase and used quantitative reverse transcription-PCR (qPCR) to determine the mRNA levels of four genes that are crucial for iron uptake, distribution, and utilization: CTH2, which optimizes iron utilization within cells by regulating multiple iron-dependent metabolic pathways (12); FET3, a copper-dependent ferroxidase essential for high-affinity iron uptake (11); FET4, a low-affinity iron transporter located at the plasma membrane (7, 8); and CCC1, responsible for iron transport and storage in the vacuole (15, 16). It is worth highlighting that transcriptional activator Aft1 upregulates the expression of genes CTH2, FET3, and FET4 when yeast cells sense low iron (12, 28), whereas Yap5 transcription factor increases CCC1 mRNA levels upon iron excess (14). Our results showed that iron-sensitive strains YPS128, CBS 5287, and Sc 9 upregulate CTH2 and FET4 expression (Fig. 6). FET3 expression for YPS128 cells also increased (Fig. 6). These results suggest that these strains perceive less iron than the laboratory BY4741 strain and, consequently, activate the iron regulon, which makes cells more prone to accumulate iron. However, this was not the case for iron-sensitive strain CECT 10120, which showed a marked increase only in the mRNA levels of the CTH2 gene compared to the laboratory strain (Fig. 6). Iron-resistant strains L-246 and Kyokai 6 exhibited CTH2, FET3, and FET4 mRNA levels similar to those of the laboratory strain. Surprisingly, iron-resistant strain CECT 11032 activated the expression of genes CTH2 and FET4, a pattern that resembled more that of the iron-sensitive than that of the iron-resistant strains (Fig. 6). Finally, CCC1 expression was markedly upregulated only in iron-resistant strain Kyokai 6 and was downregulated in iron-sensitive strain CBS 5287 (Fig. 6). This is consistent with the greater resistance of the sake strain and suggests that changes in the capacity of iron accumulation in the vacuole could contribute to different strain sensitivities to excess iron. Altogether, these results suggest that alterations in the expression of the iron-related genes could contribute to the different responses of these strains to elevated environmental iron levels.
FIG 6.
Expression of iron homeostasis genes in iron-resistant and iron-sensitive yeast strains. The yeast strains indicated were collected during exponential growth in the liquid SC medium. Total RNA was extracted, and the CTH2, FET3, FET4, CCC1, and ACT1 mRNA levels were determined by qPCR. ACT1 was used to normalize the mRNA values. The averages and standard deviations for at least three independent biological experiments are represented.
DISCUSSION
Cultivation in elevated iron concentrations leads to iron-enriched yeasts.
The utilization of biophysical spectroscopies has recently allowed the detailed examination of yeast iron contents in terms of growth phase, chemical form, and distribution within organelles (25, 26, 29). In the exponential growth phase, the intracellular yeast iron level remains roughly constant due to a balance between iron import and cellular division (26). This equilibrium is lost when the growth rate slows down toward the end of the exponential phase because iron continues to be imported into the cell and accumulates as Fe3+ nanoparticles and vacuolar species (26). In this work, we cultivated yeast for 24 h in synthetic medium supplemented with different iron concentrations. We observed that when grown in synthetic medium with no additional iron, all the cell cultures depleted glucose and shifted to the diauxic phase, in which they utilized ethanol as a carbon source, and the growth rate lowered (data not shown). It has been previously demonstrated that yeast cells partially activate the expression of the iron acquisition machinery at the diauxic shift, probably due to iron requirements during respiration (30). The lack of synchronization between growth and iron uptake, the accumulation of iron in nanoparticles, which may not be perceived by the cellular iron-sensing system (26), and the activation of the high-affinity iron uptake system upon diauxic shift contribute to iron accumulation in this stage. Here we observe that laboratory strain BY4741 accumulated approximately 440 μM iron (∼70 μg Fe/g DW) after 1 day of incubation in synthetic medium (Fig. 3; see also Fig. S8 in the supplemental material). Other groups have obtained similar results, with around 100 μg Fe/g DW when a yeast strain was incubated for 20 h in rich medium (31). We also observed that iron accumulation gradually increased when the iron concentration in the medium rose (Fig. 3 and 4). For instance, when 100 μM FAS was added to SC, iron accumulation duplicated and reached around 950 μM iron; that is, ∼155 μg Fe/g DW (Fig. 3; see also Fig. S8 in the supplemental material). Other studies have reported similar results, with 1.5 mM iron accumulation after 24 h of cultivation in minimal medium supplemented with 40 μM ferric citrate (26). In the last case, the authors showed that endogenous iron can reach 5 mM when incubation is extended to 120 h (26). This indicates that iron accumulation depends both on the iron concentrations in the medium and on incubation time. We show here that laboratory strain BY4741 reached a maximum of 5 mg Fe/g DW (around 50 mM) when incubated in 8 mM FAS, which is a >100-fold-higher iron accumulation than when iron is not added to the medium. These intracellular iron concentrations fell within a range similar to those in recent reports in which a yeast strain grown in rich medium accumulated 3 or 8 mg Fe/g DW when incubated for either 20 h in 500 μM FeSO4 or 12 h in 1.8 mM FeSO4, respectively (2, 31). We also observed that after 24 h of incubation in the same medium, iron-sensitive strains accumulated more iron than iron-resistant isolates (discussed below). Thus, for iron-sensitive strains, the maximum endogenous iron levels were obtained after cultivation in SC with 6 or 7 mM FAS, whereas iron-resistant strains required 10 to 12 mM FAS (Fig. 3 and 4). Considering our data and other reports (2, 3), we conclude that an efficient way to obtain iron-enriched yeasts would be to cultivate yeasts for a considerable time (24 h or more) in a medium with iron concentrations that fell within the millimolar range. These iron-enriched yeasts could be used as a dietary iron supplement to help animals recover from dietary iron deficiency (3). Furthermore, iron-enriched baker's yeasts that maintain their biotechnological properties could be potentially used to enrich bread with iron (2).
Differences between iron-sensitive and iron-resistant strains of Saccharomyces cerevisiae.
The analysis of the genes of S. cerevisiae strains isolated from different sources and, more recently, their whole-genome comparison have revealed wide genomic and phenotypic variations (19). Although the forces and mechanisms that drive the evolution of yeast populations are not fully understood, we can assume that adaptation to wild environments or human domestication accounts for the majority of phenotypic differences. Therefore, this genetic variety could be exploited to characterize the major determinants of yeast response to environmental stresses, such as alterations in iron bioavailability. In fact, a recent study has shown how yeast strains isolated in a wild Malaysian niche have diverged to become markedly sensitive to elevated iron concentrations, which is mostly due to a partial loss of function of vacuolar iron transporter Ccc1 (21). In this work, we found that wild Malaysian strains are more sensitive to excess iron than laboratory strain BY4741, which we used as a reference (Fig. 2; see also Fig. S3 in the supplemental material). Interestingly, the fitness of Malaysian strains in high iron diminished despite the fact that they accumulated roughly the same endogenous iron levels as the laboratory strain (Fig. 4). As mentioned above, we believe that this is due to the specific defect in the iron detoxification system by the Malaysian CCC1 vacuolar transporter allele (21), which would mean that similar endogenous iron concentrations would lead to more deleterious damage.
After analyzing more than 100 different strains, we concluded that yeasts isolated in wild environments are more prone to display iron sensitivity than strains of other origins. The iron-sensitive strains that we selected in this study were even more sensitive to excess iron than the Malaysian strains (Fig. 2; see also Fig. S3 in the supplemental material). If we look at cellular iron content, we observe that the maximum endogenous iron levels reached by our iron-sensitive strains were similar to those of the laboratory strain (5 mg Fe/g DW) but were markedly higher than those accumulated in the Malaysian strain (3 mg Fe/g DW) (Fig. 4). These results suggest that, unlike the Malaysian strains, the iron-sensitive yeasts selected here may possess a functional iron detoxification mechanism that allows them to accumulate intracellular iron concentrations similar to those of the reference strain. However, our iron-sensitive yeasts reached their maximum endogenous iron levels at lower experimental iron concentrations than the laboratory strain. As yeast cells do not possess specialized iron export mechanisms, these results suggest that the iron-sensitive strains studied here probably have a higher iron accumulation rate than that of the laboratory strain and consequently die at lower extracellular iron concentrations.
We also selected yeast strains that are more resistant to elevated iron levels in the environment than the reference strain (Fig. 1). This was especially remarkable for sake strain Kyokai 6 (Fig. 1). However, we were unable to establish a direct correlation between the origin of the yeast isolates and their resistance to excess iron. In fact, none of the other sake yeast strains analyzed here displayed such strong resistance to excess iron (see Fig. S2 in the supplemental material). Unlike iron-deficient environments, which are very common in wild niches, conditions with excessive iron concentrations are not associated with wild or domestic locations; they are rather linked to the specific conditions encountered by particular niches. Notwithstanding, we attempted to decipher common features among these iron-resistant strains. When we looked at the maximum endogenous iron accumulated by our five iron-resistant strains, we observed that three of them (YJM978, CECT 11032, and Kyokai 6) reached intracellular iron concentrations similar to those of the reference strain, between 4 and 5 mg Fe/g DW, whereas two of the strains (Sc-97 and L-246) took up only 3 mg Fe/g DW (Fig. 3A). These data indicate that sensitivities to endogenous iron differed among these strains. It is important to note that at any given extracellular iron concentration, laboratory strain BY4741 always accumulated more iron than iron-resistant strains (Fig. 3). These results suggest that a common mechanism to tolerate high iron concentrations, which these yeast strains share, consists in limiting iron accumulation.
The redox-active properties of iron can be deleterious to cells if excess iron is present, as it participates in Fenton reactions, which generate hydroxyl radicals and other ROS that damage most cellular macromolecules. Therefore, we determined whether the yeast redox state correlated with iron accumulation within strains. Consistently with iron generating oxidative stress, the biocompatible redox indicator methylene blue indicated that iron-sensitive strains, which accumulated more iron, were more oxidized than iron-resistant strains, which had less endogenous iron (Fig. 5A). Seeing that our results could be due to alterations in the oxidative stress protection mechanisms that these strains possess, we also examined the sensitivity of these strains to different oxidative agents. However, no correlation was observed between iron and oxidative stress sensitivity (Fig. 5B). Therefore, we conclude that the diverse sensitivities of these yeast isolates to deleterious iron concentrations are not due to the efficiency of their oxidative stress protection machinery but to differences in their iron accumulation rates.
It has been proposed that Malaysian strains adapted to a wild iron-deficient niche but lost fitness in environments with toxic iron levels, which are rarely encountered (21). Therefore, we wondered how the iron-sensitive and iron-resistant yeast strains isolated here would behave under low-iron conditions. We observed that three of the four assayed wild iron-sensitive strains (YPS128, CBS 5287, Sc 9) grew slightly better than the reference strain under iron-deficient conditions (Fig. 5C). Given that these isolates displayed increased iron accumulation, we suggest that these strains may have adapted to an iron-deficient wild environment by increasing iron uptake. However, this potential increase in iron acquisition became detrimental when the environmental iron levels rose. What is even more striking is the parallel behavior of the three iron-resistant strains assayed (CECT 11032, L-246, Kyokai 6). In addition to being more resistant to toxic iron levels, these strains exhibited a major growth defect under iron-deficient conditions (Fig. 5C). We propose that these strains have decreased iron acquisition efficiency, which could be an advantage when elevated environmental iron levels occur but could be detrimental under iron-limiting conditions. Iron transport assays would be necessary to demonstrate this hypothesis. We also observed here that the yeast strains isolated from different geographical origins and isolation sources had adapted to either low-iron or high-iron conditions by altering iron accumulation, which simultaneously diminished their fitness for the opposite condition. The molecular bases that underlie these phenotypes could be complex and diverse. However, a preliminary gene expression analysis of iron homeostasis genes showed alterations in the expression pattern of the genes involved in iron acquisition (FET3, FET4), iron utilization (CTH2), or iron storage (CCC1), which suggests that the iron-sensing machinery or transcriptional regulators of these strains could have diverged (Fig. 6). Another possibility is that the same transcription factor regulates adaptation to both iron excess and iron deficiency as previously described in the fungus Aspergillus fumigatus (32). Further studies are necessary to dissect the molecular basis that underlies these changes in iron accumulation, which lead to either sensitivity or resistance to elevated iron levels.
In this report, we have uncovered that yeast strains sensitive to elevated iron concentrations in the environment accumulate more iron than iron-resistant isolates. These results suggest that the commonest difference between these two groups of strains relies on their iron accumulation pattern. Interestingly, we also observed that adaptation to low-iron conditions happens to be detrimental for growth under high-iron conditions, whereas iron-resistant strains display marked growth defects under iron-deficient conditions. Further studies are needed to elucidate the specific genetic variations that determine the molecular mechanism that has evolved in iron-sensitive and iron-resistant yeast strains.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to the University of Valencia students who have contributed to some extent to this work. We also thank the members of the Biotechnology Department at the IATA and the Department of Biochemistry and Molecular Biology of the University of Valencia for their technical and scientific assistance. We are especially grateful to Amparo Querol and Gianni Liti for yeast strains and to Agustín Aranda for oxidative reagents.
Funding Statement
This work has been funded by the AGL2011-29099 and BIO2014-56298-P grants from the Spanish Ministry of Economy and Competitiveness to S.P. and the PROMETEOII/2014/042 grant from the Generalitat Valenciana to Amparo Querol. This work has also been supported by a predoctoral fellowship from the Spanish Ministry of Economy and Competitiveness to A.M.R. and a postdoctoral JAE-Doc contract from the Spanish Research Council (CSIC) and the European Social Fund to R.D.L.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03464-15.
REFERENCES
- 1.Pas M, Piskur B, Sustaric M, Raspor P. 2007. Iron enriched yeast biomass-a promising mineral feed supplement. Bioresour Technol 98:1622–1628. doi: 10.1016/j.biortech.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Gaensly F, Brand GWD, Bonfim TM. 2011. Iron enriched Saccharomyces cerevisiae maintains its fermentative power and bakery properties. Food Sci Technol Int 31:980–983. doi: 10.1590/S0101-20612011000400025. [DOI] [Google Scholar]
- 3.Kyyaly MA, Powell C, Ramadan E. 2015. Preparation of iron-enriched baker's yeast and its efficiency in recovery of rats from dietary iron deficiency. Nutrition 31:1155–1164. doi: 10.1016/j.nut.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 4.Philpott CC, Leidgens S, Frey AG. 2012. Metabolic remodeling in iron-deficient fungi. Biochim Biophys Acta 1823:1509–1520. doi: 10.1016/j.bbamcr.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanvisens N, Puig S. 2011. Causes and consequences of nutritional iron deficiency in living organisms, p 245–276. In Merkin TC. (ed), Biology of starvation in humans and other organisms. Nova Science Publishers, Hauppage, NY. [Google Scholar]
- 6.Chen XZ, Peng JB, Cohen A, Nelson H, Nelson N, Hediger MA. 1999. Yeast SMF1 mediates H+-coupled iron uptake with concomitant uncoupled cation currents. J Biol Chem 274:35089–35094. doi: 10.1074/jbc.274.49.35089. [DOI] [PubMed] [Google Scholar]
- 7.Dix D, Bridgham J, Broderius M, Eide D. 1997. Characterization of the FET4 protein of yeast. Evidence for a direct role in the transport of iron. J Biol Chem 272:11770–11777. [DOI] [PubMed] [Google Scholar]
- 8.Dix DR, Bridgham JT, Broderius MA, Byersdorfer CA, Eide DJ. 1994. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J Biol Chem 269:26092–26099. [PubMed] [Google Scholar]
- 9.Kaplan CD, Kaplan J. 2009. Iron acquisition and transcriptional regulation. Chem Rev 109:4536–4552. doi: 10.1021/cr9001676. [DOI] [PubMed] [Google Scholar]
- 10.Shakoury-Elizeh M, Tiedeman J, Rashford J, Ferea T, Demeter J, Garcia E, Rolfes R, Brown PO, Botstein D, Philpott CC. 2004. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol Biol Cell 15:1233–1243. doi: 10.1091/mbc.E03-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552–1557. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 12.Puig S, Askeland E, Thiele DJ. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 13.Lee YJ, Huang X, Kropat J, Henras A, Merchant SS, Dickson RC, Chanfreau GF. 2012. Sphingolipid signaling mediates iron toxicity. Cell Metab 16:90–96. doi: 10.1016/j.cmet.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Bagley D, Ward DM, Kaplan J. 2008. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol Cell Biol 28:1326–1337. doi: 10.1128/MCB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Chen OS, McVey Ward D, Kaplan J. 2001. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem 276:29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- 16.Lin H, Li L, Jia X, Ward DM, Kaplan J. 2011. Genetic and biochemical analysis of high iron toxicity in yeast: iron toxicity is due to the accumulation of cytosolic iron and occurs under both aerobic and anaerobic conditions. J Biol Chem 286:3851–3862. doi: 10.1074/jbc.M110.190959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Muhlenhoff U. 2012. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta 1823:1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Outten CE, Albetel AN. 2013. Iron sensing and regulation in Saccharomyces cerevisiae: ironing out the mechanistic details. Curr Opin Microbiol 16:662–668. doi: 10.1016/j.mib.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O'Kelly MJ, van Oudenaarden A, Barton DB, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. 2009. Population genomics of domestic and wild yeasts. Nature 458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cubillos FA, Billi E, Zorgo E, Parts L, Fargier P, Omholt S, Blomberg A, Warringer J, Louis EJ, Liti G. 2011. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol Ecol 20:1401–1413. doi: 10.1111/j.1365-294X.2011.05005.x. [DOI] [PubMed] [Google Scholar]
- 21.Lee HN, Mostovoy Y, Hsu TY, Chang AH, Brem RB. 2013. Divergence of iron metabolism in wild Malaysian yeast. G3 (Bethesda) 3:2187–2194. doi: 10.1534/g3.113.008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiens F, Zitzmann A, Lachance MA, Yegles M, Pragst F, Wurst FM, von Holst D, Guan SL, Spanagel R. 2008. Chronic intake of fermented floral nectar by wild treeshrews. Proc Natl Acad Sci U S A 105:10426–10431. doi: 10.1073/pnas.0801628105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamarit J, Irazusta V, Moreno-Cermeno A, Ros J. 2006. Colorimetric assay for the quantitation of iron in yeast. Anal Biochem 351:149–151. doi: 10.1016/j.ab.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Sanvisens N, Romero AM, An X, Zhang C, de Llanos R, Martinez-Pastor MT, Bano MC, Huang M, Puig S. 2014. Yeast Dun1 kinase regulates ribonucleotide reductase inhibitor Sml1 in response to iron deficiency. Mol Cell Biol 34:3259–3271. doi: 10.1128/MCB.00472-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes-Hampton GP, Jhurry ND, McCormick SP, Lindahl PA. 2013. Iron content of Saccharomyces cerevisiae cells grown under iron-deficient and iron-overload conditions. Biochemistry 52:105–114. doi: 10.1021/bi3015339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J, McCormick SP, Chakrabarti M, Lindahl PA. 2013. The lack of synchronization between iron uptake and cell growth leads to iron overload in Saccharomyces cerevisiae during post-exponential growth modes. Biochemistry 52:9413–9425. doi: 10.1021/bi4010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishida K, Silver PA. 2012. Induction of biogenic magnetization and redox control by a component of the target of rapamycin complex 1 signaling pathway. PLoS Biol 10:e1001269. doi: 10.1371/journal.pbio.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waters BM, Eide DJ. 2002. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J Biol Chem 277:33749–53377. doi: 10.1074/jbc.M206214200. [DOI] [PubMed] [Google Scholar]
- 29.Cockrell A, McCormick SP, Moore MJ, Chakrabarti M, Lindahl PA. 2014. Mossbauer, EPR, and modeling study of iron trafficking and regulation in Δccc1 and CCC1-up Saccharomyces cerevisiae. Biochemistry 53:2926–2940. doi: 10.1021/bi500002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haurie V, Boucherie H, Sagliocco F. 2003. The Snf1 protein kinase controls the induction of genes of the iron uptake pathway at the diauxic shift in Saccharomyces cerevisiae. J Biol Chem 278:45391–45396. doi: 10.1074/jbc.M307447200. [DOI] [PubMed] [Google Scholar]
- 31.Gaensly F, Picheth G, Brand D, Bonfim TM. 2014. The uptake of different iron salts by the yeast Saccharomyces cerevisiae. Braz J Microbiol 45:491–494. doi: 10.1590/S1517-83822014000200016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gsaller F, Hortschansky P, Beattie SR, Klammer V, Tuppatsch K, Lechner BE, Rietzschel N, Werner ER, Vogan AA, Chung D, Muhlenhoff U, Kato M, Cramer RA, Brakhage AA, Haas H. 2014. The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J 33:2261–2276. doi: 10.15252/embj.201489468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.