Abstract
Nicotine, a major toxic alkaloid in tobacco wastes, is degraded by bacteria, mainly via pyridine and pyrrolidine pathways. Previously, we discovered a new hybrid of the pyridine and pyrrolidine pathways in Agrobacterium tumefaciens S33 and characterized its key enzyme 6-hydroxy-3-succinoylpyridine (HSP) hydroxylase. Here, we purified the nicotine dehydrogenase initializing the nicotine degradation from the strain and found that it forms a complex with a novel 6-hydroxypseudooxynicotine oxidase. The purified complex is composed of three different subunits encoded by ndhAB and pno, where ndhA and ndhB overlap by 4 bp and are ∼26 kb away from pno. As predicted from the gene sequences and from chemical analyses, NdhA (82.4 kDa) and NdhB (17.1 kDa) harbor a molybdopterin cofactor and two [2Fe-2S] clusters, respectively, whereas Pno (73.3 kDa) harbors an flavin mononucleotide and a [4Fe-4S] cluster. Mutants with disrupted ndhA or ndhB genes did not grow on nicotine but grew well on 6-hydroxynicotine and HSP, whereas the pno mutant did not grow on nicotine or 6-hydroxynicotine but grew well on HSP, indicating that NdhA and NdhB are responsible for initialization of nicotine oxidation. We successfully expressed pno in Escherichia coli and found that the recombinant Pno presented 2,6-dichlorophenolindophenol reduction activity when it was coupled with 6-hydroxynicotine oxidation. The determination of reaction products catalyzed by the purified enzymes or mutants indicated that NdhAB catalyzed nicotine oxidation to 6-hydroxynicotine, whereas Pno oxidized 6-hydroxypseudooxynicotine to 6-hydroxy-3-succinoylsemialdehyde pyridine. These results provide new insights into this novel hybrid pathway of nicotine degradation in A. tumefaciens S33.
INTRODUCTION
Agrobacterium tumefaciens is well known for its ability to induce crown gall tumors in dicotyledonous plants and mediate interkingdom genetic transfer, for that it is widely used in plant molecular biology and biotechnology (1). Interestingly, some strains of this species are also able to degrade xenobiotics such as cyanuric acid, iminodisuccinate, and methylene urea (2–4). We isolated A. tumefaciens strain S33, which has the strong ability to degrade the natural alkaloid nicotine from the rhizospheric soil of a tobacco plant (5, 6).
Nicotine is a major alkaloid in tobacco, which causes tobacco addiction and may result in diseases such as pulmonary disease and cancer (7, 8), and it is the primary toxic compound in tobacco wastes. The tobacco-manufacturing process and all activities using tobacco produce a large amount of solid or liquid waste containing high concentrations of nicotine, which are classified as “toxic and hazardous wastes” by the European Union (9). Therefore, detoxification of tobacco wastes is a major concern for public health and the environment. The discovery of nicotine degradation by microorganisms provides an alternative way to dispose of such wastes (10–12).
Microbial degradation of nicotine attracts attention because it represents a method to treat the tobacco wastes without causing significant harm to the environment (11–17). Many microorganisms, including bacteria, actinomycetes, and fungi degrade nicotine, and most of these grow using nicotine as the sole source of carbon and nitrogen. The biochemical pathways that decompose nicotine have been investigated in some species during the past 50 years. Three types of degradation are mediated through the pyridine pathway of the Gram-positive bacterium Arthrobacter sp., the pyrrolidine pathway of the Gram-negative bacterium Pseudomonas sp., and the demethylation pathway present in fungi such as Aspergillus oryzae (13, 14, 18, 19). The biochemical mechanisms involved in the pyridine and pyrrolidine pathways have been characterized in detail (13, 15, 16, 20).
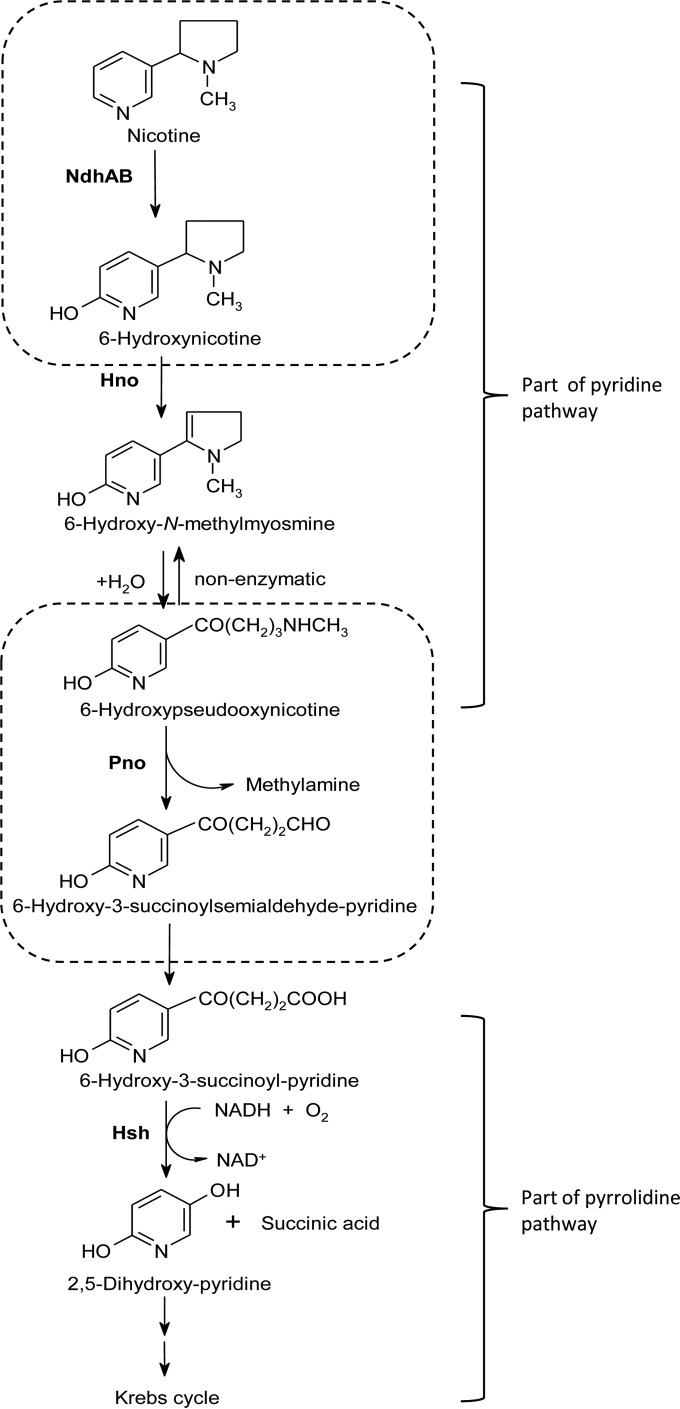
We discovered that A. tumefaciens S33 catabolizes nicotine via a hybrid between the pyridine and pyrrolidine pathways by investigating the intermediates and the key enzymes activities in cell extracts (5). In the hybrid pathway (Fig. 1), nicotine is first degraded to 6-hydroxypseudooxynicotine via the pyridine pathway through 6-hydroxynicotine and 6-hydroxy-N-methylmyosmine, and 6-hydroxy-3-succinoylpyridine (HSP) and 2,5-dihydroxypyridine are produced by the pyrrolidine pathway. The same pathway present in Shinella sp. strain HZN7 (21, 22) and Ochrobactrum sp. strain SJY1 (23, 24) was found recently using genome sequencing, intermediates analysis, and characterization of key enzymes such as nicotine hydroxylase, 6-hydroxynicotine oxidase (Hno), HSP hydroxylase (Hsh), and 2,5-dihydroxypyridine dioxygenase (Hpo), which are similar to the corresponding enzymes from Arthrobacter nicotinovorans and Pseudomonas putida S16. However, other enzymes involved in the hybrid pathway are still unknown. Previously, we partially enriched two of the key enzymes, nicotine dehydrogenase (Ndh) and Hsh from A. tumefaciens S33 (5) that serve as the enzymes in the pyridine pathway in Arthrobacter nicotinovorans (25, 26) and the pyrrolidine pathway in Pseudomonas putida S16 (27), respectively. Recently, we purified Hsh from A. tumefaciens S33 and characterized its biochemical properties and gene (28) with 62% amino acid sequence identity to the enzyme from P. putida S16 (27) and 99.7% identity to the enzyme from Ochrobactrum sp. strain SJY1 (23). This verifies at the biochemical level that partial steps of pyrrolidine pathway are present in the hybrid pathway of strain S33. In the pyridine pathway of A. nicotinovorans, the Ndh catalyzing the initial step hydroxylates the C-6 position of pyridine ring of nicotine (25, 26) and is a heterotrimeric oxidoreductase comprising a subunit (NdhM, 30.0 kDa) that binds a flavin adenine dinucleotide (FAD), as well as a subunit (NdhS, 14.9 kDa) harboring two [2Fe-2S] clusters and a subunit (NdhL, 87.7 kDa) with a molybdopterin cytosine dinucleotide cofactor (29). However, the Ndh from A. tumefaciens S33 is not identified yet (Fig. 1).
FIG 1.
Proposed hybrid pathway of nicotine degradation by A. tumefaciens S33. Ndh, nicotine dehydrogenase; Hno, 6-hydroxynicotine oxidase; Pno, 6-hydroxypseudooxynicotine oxidase; Hsh, 6-hydroxy-3-succinoylpyridine hydroxylase.
In the present study, we purified the key Ndh that initiates nicotine degradation in the hybrid pyridine and pyrrolidine pathways of A. tumefaciens S33. The purified Ndh comprises three proteins that were identified by matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS) analysis. Gene disruption and complementation experiments and biochemical analysis show that the purified Ndh is a complex of two enzymes, which catalyze the first and fourth steps of nicotine degradation, respectively. The results present new evidence for the novel hybrid pathway of nicotine degradation in A. tumefaciens S33.
MATERIALS AND METHODS
Chemicals and reagents.
(S)-Nicotine (>99%) was obtained from Fluka (Buchs, Switzerland). 6-Hydroxynicotine and Hno from A. nicotinovorans were a gift from Roderich Brandsch (University of Freiburg, Freiburg, Germany). HSP was purified from the broth of the cultures of the nicotine degrading P. putida S16 (30). All chromatography materials were purchased from GE Healthcare. All other chemicals were commercially available.
Bacterial strains, plasmids, and culture conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. A. tumefaciens S33, deposited in the China Center for Type Culture Collection (accession number M206131), was grown in nicotine medium or nicotine medium plus 1.0 g/liter glucose, 0.2 g/liter ammonium sulfate, and 1.0 g/liter yeast extract at 30°C as described previously (5, 28). Nicotine was added to a final concentration of 1.0 g/liter before inoculation. HSP and 6-hydroxynicotine media contained 0.5 g/liter HSP or 6-hydroxynicotine instead of nicotine as the sole source of carbon and nitrogen. Lysogeny broth (LB) was used for the routine propagation of Escherichia coli strains. Terrific broth (TB) was used for gene expression. Antibiotics were added depending on the strains and plasmids harbored (gentamicin [Gm], 50 mg/liter; kanamycin [Km], 50 mg/liter; ampicillin [Ap], 100 mg/liter; chloramphenicol [Cm], 25 mg/liter; and tetracycline [Tet], 10 mg/liter).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference(s) |
|---|---|---|
| Strains | ||
| A. tumefaciens | ||
| S33 | Wild type, nicotine degrader; Gram negative | 5, 6 |
| S33-ΔndhA, S33-ΔndhB, and S33-Δpno | Gmr; ndhA, ndhB, and pno mutants of strain S33, respectively | This study |
| S33ΔndhA-C, S33ΔndhB-C, and S33Δpno-C | Gmr; strains S33ΔndhA, S33ΔndhB and S33Δpno containing pBBR-ndhA, pBBR-ndhB, and pBBR-pno, respectively | This study |
| E. coli | ||
| DH5α | F− ϕ80dlacZΔM15, Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | TaKaRa |
| HB101 | Δ(mcrC-mrr) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 leuB6 thi-1; helper strain of triparental filter mating supE44 | TaKaRa |
| Mach 1-T1 | ΔrecA1398 endA1 tonA ϕ80ΔlacM15 ΔlacX74 hsdR(rK− mK+) | Invitrogen |
| C41(DE3) harboring pCodonPlus and pRKISC | F− ompT hsdSB(rB− mB−) gal dcm(DE3); harboring pCodonPlus and pRKISC | Y. Takahashi |
| Plasmids | ||
| pJQ200SK | Gmr; mob+ oriP15A lacZα+ sacB; suicide plasmid | 33 |
| pRK2013 | Kmr; helper plasmid for conjugation | Clontech |
| pBBR1-MSC5 | Gmr; broad-host-range cloning vector | 35 |
| pJQ-ΔndhA, pJQ-ΔndhB, and pJQ-Δpno | Gmr; ndhA, ndhB, and pno, respectively, were disrupted and inserted into pJQ200SK | This study |
| pBBR-ndhA, pBBR-ndhB, and pBBR-pno | Gmr; ndhA, ndhB, and pno, respectively, inserted into pBBR1-MSC5 | This study |
| pCodonPlus | Cmr; pACYC containing extra copies of the argU, ileY, and leuW tRNA genes | Stratagene |
| pRKISC | Tetr; pRK415 containing isc gene cluster | 36 |
| pEASY-Blunt | Apr Kmr; cloning vector | TransGen Biotech |
| pETDuet-1 | Apr; expression vector | Novagen |
| pEASY-Blunt-pno | Apr Kmr; pEASY-Blunt containing pno gene | This study |
| pETDuet-1-pno | Apr; pETDuet-1 containing pno gene | This study |
Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance; Tetr, tetracycline resistance; Apr, ampicillin resistance.
Purification of Ndh from A. tumefaciens S33.
Cells grown in nicotine medium plus glucose, ammonium sulfate, and yeast extract were resuspended in 50 mM sodium phosphate buffer (pH 7.0) and disrupted using a Vibra-Cell VCX 500 ultrasonic liquid processor (amplitude, 39%; 20 min; pulse on, 6 s; pulse off, 6 s) in an ice-water bath. The supernatant obtained by centrifugation at 30,000 × g at 4°C for 30 min was used for enzyme purification. All chromatography steps were performed using an ÄKTA Basic 10 chromatography system (GE Healthcare) at 16°C. The supernatant was subjected to ammonium sulfate precipitation by slowly adding saturated ammonium sulfate solution to a final concentration of 50% at 4°C. The precipitate was removed by centrifugation at 30,000 × g and 4°C for 20 min, and the supernatant was added with a saturated ammonium sulfate solution to 65% saturation. Ndh activity was detected in the fraction precipitated in 50 to 65% saturated ammonium sulfate. The precipitate obtained by centrifugation was dissolved in 50 mM sodium phosphate buffer (pH 7.0) containing 1.5 M ammonium sulfate. After removing insoluble proteins using centrifugation, the supernatant was applied to a Phenyl Sepharose 6 Fast Flow column (high-sub, 16 mm × 10 cm, 20 ml) pre-equilibrated with 50 mM sodium phosphate buffer (pH 7.0) containing 1.5 M ammonium sulfate. The column was eluted using the same buffer containing an ammonium sulfate gradient (1.0, 0.6, 0.37, 0.15, and 0 M; one column volume per concentration) at a 4 ml/min. Ndh activity was eluted at 0.15 M ammonium sulfate, and the fractions were concentrated and applied to a DEAE-Sepharose Fast Flow column (16 mm × 10 cm, 20 ml) equilibrated with 50 mM sodium phosphate buffer (pH 7.0). The column was eluted at 4 ml/min with five column volumes each of 0.1, 0.2, 0.25, 0.4, and 0.5 M NaCl in the same buffer. The Ndh activity was eluted at 0.4 M NaCl, and the fractions were concentrated and applied to a Superdex 200 column (10 mm by 30 cm, 24 ml) equilibrated with 50 mM sodium phosphate buffer (pH 7.0) containing 150 or 300 mM NaCl. The flow rate was set as 0.5 ml/min. Ndh was eluted with the same buffer as a single peak at 13.6 ml.
Assay of Ndh activity and determination of the reaction products.
Ndh activity was determined as previously described by monitoring the reduction of 2,6-dichlorophenolindophenol (DCIP) with nicotine at 600 nm (ε = 21 mM−1 cm−1) (5). The assay mixture contained 1 mM nicotine, 0.05 mM DCIP, and 50 mM sodium phosphate buffer (pH 7.0). Reduction of 1 μmol of DCIP per min was defined as one unit. The assay was performed using quartz cuvettes (1-cm light path) filled with 1-ml reaction mixture at 30°C using a UV-visible Ultrospec 2100 Pro spectrophotometer (GE Healthcare, USA) and initiated by adding enzyme. In some cases, 0.5 mM phenazine methosulfate (PMS) was added to enhance electron transfer. To identify the reaction products of Ndh, the reaction was performed in 50 mM sodium phosphate buffer (pH 7.0) containing 1 U of Ndh, 2 mM nicotine, and 0.1 mM DCIP for 60 min at 30°C and spectrophotometrically monitored as indicated above. Products were determined by using liquid chromatography-mass spectrometry (LC-MS).
Protein mass spectra determination and identification of the encoding genes.
The purified enzyme was digested with trypsin and analyzed using MALDI-TOF MS (Beijing Genomics Institute, Shenzhen, China). To identify potential genes, we searched the annotated genome draft sequence of A. tumefaciens S33 (GenBank accession number JFFS00000000) (28). Three open reading frames (ORFs) matched the protein MS data, and the genes were designated ndhA, ndhB, and pno, respectively.
Reverse transcription-PCR (RT-PCR) analysis.
The experiments were performed as described previously (28). Briefly, A. tumefaciens S33 was grown in nicotine medium containing nicotine as the sole source of carbon and nitrogen or glucose and ammonium medium and then harvested at the early exponential phase (optical density at 620 nm [OD620] = 0.4) in nicotine medium, with an OD620 of 0.7 in glucose and ammonium medium. Total RNA was extracted from the cells using an RNAprep pure cell/bacteria kit (Tiangen Biotech, China) according to the manufacturer's protocol. DNA was digested using RNase-free DNase I. Total cDNA was synthesized using TransScript first-strand cDNA synthesis supermix (Beijing TransGen Biotech, China). The ndhA, ndhB, and pno genes were amplified according to a published procedure (28). The primers used for PCR were 5′-TCTAAGTATGGGTATGTC-3′ and 5′-CTTCGTCTATCTTGTTTG-3′ for ndhA, 5′-ATGAAAGTCGATTTTACTGTTAATGGC-3′ and 5′-TCATTGAGCTGCTCCTTTCAGCATAG-3′ for ndhB, and 5′-TCAGATAAGTTGAAGACAG-3′ and 5′-CGTAGCCAAGGTAATAAG-3′ for pno. Genomic DNA and the total RNA template using the same PCR procedure served as positive and negative controls, respectively.
Construction of strains with markerless deletions of ndhA, ndhB, and pno.
The in-frame deletion of ndhA, ndhB, and pno of A. tumefaciens S33 was performed using the suicide plasmid pJQ200SK and a two-step homologous recombination method (22, 31). First, the in-frame-deleted gene fragments were obtained using crossover PCR as described previously (32). Upstream and downstream flanking sequences of the target fragment were obtained using PCR with primers A and B and primers C and D, respectively (see Table S1 in the supplemental material), where the 5′ ends of primers B and C were designed to contain 21-bp complementary sequences. The purified PCR products were mixed in equal amounts and used as the template for seven cycles of PCR with primers A and D. The final PCR step was performed using 2 μl of the products of seven cycles of PCR as the template and primers A and D. The final PCR products were digested with BamHI and XhoI and ligated to the suicide vector pJQ200SK, which was treated with the same restriction enzymes. The recombinant plasmid pJQ200SK with shortened target-gene fragment was used to transform E. coli DH5α. The vector pJQ200SK replicates using a p15A origin and contains sucB, which imparts sucrose sensitivity. Moreover, the vector encodes Gm resistance and contains the lacZα system to provide efficient screening of the strains (33). The recombinant pJQ200SK was introduced into A. tumefaciens S33 by triparental mating, with E. coli HB101(pRK2013) as the helper strain (34). Single-crossover mutants were screened using HSP medium plates containing 50 mg of Gm/liter. Double-crossover mutants were selected on HSP plates containing 20% sucrose. The deletion of the target genes in the mutants was confirmed by PCR and DNA sequencing. The mutants with disrupted ndhA, ndhB, and pno sequences were designated S33-ΔndhA, S33-ΔndhB, and S33-Δpno, respectively.
Construction of complementation strains.
To obtain complementation plasmids, the complete DNA sequences of ndhA, ndhB, and pno were synthesized using PCR with the primers listed in Table S2 in the supplemental material and ligated to pBBR1-MCS5 (35). Insert and plasmid sequences were first digested using XhoI and EcoRI (or BamHI). The recombinant plasmids verified using restriction-enzyme digestion and nucleotide sequencing were designated pBBR-ndhA, pBBR-ndhB, and pBBR-pno, respectively, and used to electroporate into the competent cells of S33-ΔndhA, S33-ΔndhB, and S33-Δpno, respectively (34). The complementation strains were designated S33-ΔndhA-C, S33-ΔndhB-C, and S33-Δpno-C, respectively.
Growth of mutants and complementation strains on nicotine, 6-hydroxynicotine, and HSP.
The mutant strains S33-ΔndhA, S33-ΔndhB, and S33-Δpno and their complementation strains were grown in LB to exponential phase and then inoculated 2% (vol/vol) into 50-ml HSP or nicotine medium, which were incubated at 30°C and 200 rpm. The cell concentrations of the cultures were determined by their OD620 values. For the utilization of 6-hydroxynicotine, tests were performed using an agar plate containing 6-hydroxynicotine medium, and the growth of complementation strains was similarly tested in nicotine medium.
Analysis of nicotine transformation by resting cells of S33-Δpno.
The strain S33-Δpno was grown in LB supplemented with 1 g of nicotine/liter and harvested at late exponential phase by centrifugation at 7,000 × g for 20 min at 4°C. The cells were washed three times with 50 mM sodium phosphate buffer (pH 7.0) and stored at 4°C for use. The transformation reaction was performed in a 250-ml flask containing 1.64 g/liter dry cell weight (DCW) of resting cells (∼4 OD620, 1 OD620 U = 0.41 g/liter DCW) and 2 g of nicotine/liter in 50 ml of sterilized 50 mM sodium phosphate buffer (pH 7.0) and then incubated overnight at 30°C and 180 rpm. The reaction was stopped by centrifuging the cells, and the supernatant was analyzed using LC-MS.
Heterologous expression and purification of Pno.
The genomic DNA of A. tumefaciens S33 was extracted from the cells and purified using a Wizard genomic DNA purification kit (Promega Corp., Madison, WI). The pno gene was amplified using PCR with High-Fidelity FastPfu DNA polymerase (ShineGene Molecular Bio-Technologies, Inc., Shanghai, China) and A. tumefaciens S33 genomic DNA as the template. The primers used were as follows: 5′-TTGGCGCGCCTGATGCGAGATCCACGTTATGACATCCT-3′ (forward, the AscI recognition site is underlined) and 5′-CCCAAGCTTCTAACGACCGGTACCGCAAAATTCAAT-3′ (reverse, the HindIII recognition site is underlined). The blunt-end PCR product was ligated to the pEASY-Blunt cloning vector (TransGen Biotech, Inc., Beijing, China), which was subsequently used to transform E. coli Mach 1-T1 (Invitrogen, Carlsbad, CA). After amplification, the construct was digested with AscI and HindIII, and the target fragment was ligated to the expression vector pETDuet-1 digested using the same restriction endonucleases. After amplification in E. coli Mach 1-T1, the new construct was verified by DNA sequencing. The construct was then used to transform E. coli C41(DE3) harboring pCodonPlus and pRKISC (36). The pRKISC plasmid contains the E. coli isc locus (37) and is used to express iron-sulfur proteins (36, 38, 39).
To induce protein expression, a 10 ml of LB overnight starter culture was inoculated into 1.0 liter of TB containing antibiotics (Ap, 100 mg/liter; Cm, 25 mg/liter; and Tet, 10 mg/liter), sources of iron and sulfur (0.12 g/liter cysteine, 0.1 g/liter ferrous sulfate, 0.1 g/liter ferric citrate, and 0.1 g/liter ferric ammonium citrate), and 50 mg of riboflavin/liter at 37°C with stirring at 200 rpm. When the OD620 reached 0.7, 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to induce gene expression. After incubation for 10 h, the cells were harvested by centrifugation at 10,000 × g for 15 min, washed twice with 50 mM sodium phosphate buffer (pH 7.4), and stored at −20°C.
To purify His-tagged Pno, recombinant E. coli cells were resuspended in 30 ml of 20 mM sodium phosphate buffer containing 0.5 M NaCl (pH 7.4) and disrupted using sonication, and cell debris was removed by centrifugation at 30,000 × g at 4°C for 30 min. The clarified supernatant was applied to a 5-ml HisTrap HP column (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) and eluted at 4 ml/min with a linear gradient imidazole (5 to 100 mM) in 20 mM sodium phosphate buffer (pH 7.4) containing 0.5 M NaCl. The fractions containing target proteins were pooled and concentrated using an Amicon filter (30-kDa cutoff). After a washing step with 50 mM sodium phosphate (pH 7.0), the protein sample was applied to a 5-ml HiTrap Q HP column (GE Healthcare) equilibrated with 50 mM sodium phosphate (pH 7.0), which was then eluted at 4 ml/min with a linear gradient of 5 to 100 mM NaCl in 50 mM sodium phosphate buffer (pH 7.0). The fractions containing target protein were pooled and concentrated using an Amicon filter (30-kDa cutoff). After dialysis against 50 mM sodium phosphate buffer (pH 7.0), the protein was stored at 4°C.
Assay of Pno activity and identification of the reaction products.
Pno activity was determined by coupling with 6-hydroxynicotine oxidation using Hno from A. nicotinovorans because the substrate 6-hydroxypseudooxynicotine was not commercially available. Hno from A. nicotinovorans converts 6-hydroxynicotine to 6-hydroxy-N-methylmyosmine, which hydrolyzes spontaneously into 6-hydroxypseudooxynicotine (40, 41). The activity of Hno was measured as described previously (5). The reaction was measured by monitoring the reduction of DCIP at 600 nm in the presence of PMS at 30°C. The reaction mixture contained 100 mM glycine-NaOH (pH 9.2), 100 mM NaCl, 0.56 mM 6-hydroxynicotine, 0.5 mM PMS, and 0.05 mM DCIP. One unit was defined as the reduction of 1 μmol of DCIP per min. In addition, 1.0 mM nicotine or 0.32 mM histamine dihydrochloride was tested as a substrate of Pno in the absence of 6-hydroxynicotine and Hno. The reaction products were analyzed and identified using LC-MS. For estimation of the apparent Km, 6-hydroxypseudooxynicotine was produced from the oxidation of 6-hydroxynicotine (3 mM) by Hno, and quantified according to its absorbance at 334 nm (ε = 20.7 mM−1 cm−1) (42) without isolation from the mixture of the transformation reaction due to its instability. The apparent Km may represent an overestimate because the residual 6-hydroxynicotine and the product 6-hydroxy-N-methylmyosmine both show absorbance at 334 nm.
Analytical methods.
Growth of the cultures was monitored by measuring their OD620 values. Protein content was determined using the Bradford assay (43) with bovine serum albumin as the standard.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a 12.5 or 6.0% gel with a Bio-Rad Mini-Protean III cell (44). The subunit stoichiometry of the enzyme was estimated using Fluorchem 8800 digital-imaging system software (Alpha Innotech, Santa Clara, CA) after the gels were stained with Coomassie brilliant blue, and the bands were measured using a densitometer. To determine Ndh activity during purification, native PAGE was also performed. Gel preparation, sample treatment, and electrophoresis conditions were the same for SDS-PAGE, except that SDS and the reducing agents were omitted. Equal amounts of the protein samples were loaded in duplicate in two halves of the gel. After electrophoresis, the gel was cut into two, and each half was stained at room temperature with Coomassie brilliant blue, or 1 mM nicotine and 0.5 mM 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide dissolved in 50 mM sodium phosphate buffer (pH 7.0).
The apparent molecular mass of the purified enzyme was measured by gel filtration on Superdex 200 (1.0 by 30 cm) calibrated with a high-molecular-weight gel filtration calibration kit (GE Healthcare). The column was equilibrated and eluted with 50 mM sodium phosphate buffer (pH 7.0) containing 150 mM NaCl at a flow rate of 0.5 ml/min.
To identify the flavin in the proteins, the purified protein was boiled for 10 min in the dark to release the flavin into solution. After the removal of the precipitate by centrifugation at 30,000 × g and 4°C for 20 min, the supernatant was used for further determination. For the Ndh sample, the type of flavin and its amount was determined using high-pressure liquid chromatography (HPLC; Agilent 1100 series; Hewlett-Packard, USA) equipped with an Eclipse XDB-C18 column (4.6 by 150 mm; particle size, 5 μm). A mixture of methanol and 25 mM ammonium carbonate mixture (20:80) was used as the mobile phase, which was delivered at 0.6 ml/min. Authentic FAD and flavin mononucleotide (FMN) were used as standards. For Pno, the type of flavin was identified using thin-layer chromatography, and the amount was calculated using a molar extinction coefficient at 446 nm of 12,200 M−1 cm−1 after determining the absorbance spectrum between 200 and 600 nm (45).
For elemental analysis, the purified Ndh sample from A. tumefaciens S33 was digested in 1 ml of nitric acid for 10 h at 55°C, and 50 mM sodium phosphate instead of Ndh was used as a control. The metal elements in the samples were determined inductively coupled plasma optical emission spectrometry (ICP-OES) (IRIS Intrepid II XSP; Thermo Scientific, USA). The iron content of Pno was determined colorimetrically using 3-(2-pyridyl)-5,6-bis(5-sulfo-2-furyl)-1,2,4-triazinedisodium trihydrate (Ferene), and Mohr's salt served as the standard (46).
The products of the enzyme reactions or resting cell reactions were determined using LC-MS. For reactions catalyzed by Ndh, the analysis was performed using a Finnigan Surveyor MSQ single-quadrupole electrospray ionization mass spectrometer coupled to a Finnigan Surveyor HPLC apparatus (Finnigan/Thermo Scientific, San Jose, CA). The HPLC system was equipped with a 20RBAX Eclipse XDB-C18 column (250 by 4.6 mm; particle size, 5 μm; Agilent) and a photodiode array detector. The mobile phase was a mixture of methanol and 1 mM formic acid (85:15, vol/vol), and the flow rate was set at 0.5 ml/min. For other reactions, we used a Bruker's Impact HD high-resolution mass spectrometer coupled to a Dionex's UltiMate 3000 ultrahigh-performance liquid chromatograph (UHPLC) system (Thermo Scientific). The same column and chromatography conditions described above were applied.
Nucleotide sequence accession number.
The GenBank accession number of the A. tumefaciens S33 draft genome sequence is JFFS00000000.
RESULTS
Purification of Ndh from A. tumefaciens S33.
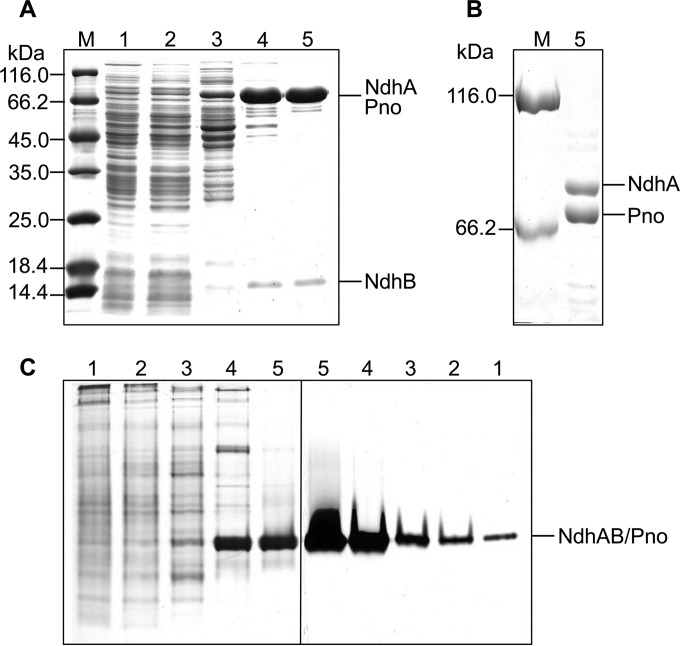
We previously reported the partial purification of Ndh from A. tumefaciens S33 grown on nicotine as the sole source of carbon and nitrogen. Using DCIP as an artificial electron acceptor to monitor nicotine oxidation, Ndh was enriched for 33.2-fold with a specific activity of 0.51 U/mg (5). Here, the bacterium was cultured in nicotine medium supplemented with glucose, ammonium sulfate, and yeast extract in order to obtain a higher yield of enzyme (28). The DCIP reduction activity with nicotine of such cell extracts was approximately 0.01 U/mg compared to approximately 0.017 U/mg for cells cultured with nicotine as the sole source of carbon and nitrogen. Through ammonium sulfate precipitation and three chromatography steps, the yellow-brown Ndh was purified from these cells and enriched 74.1-fold with a yield of 9.1% and a specific activity of 0.66 U/mg (Table 2). SDS-PAGE analysis of the purified enzyme detected 75- and 15-kDa bands using 12.5% gel (Fig. 2A), and the 75-kDa band was separated into 75- and 80-kDa bands using 6% gel (Fig. 2B). Gel filtration determined that the enzyme complex had a molecular mass of 180 kDa (see Fig. S1 in the supplemental material), which did not separate by varying the salt concentration or using a high-resolution column HiPrep Sephacryl HR S200 (16 mm by 60 cm, 120 ml; GE Healthcare). Densitometry of Coomassie blue-stained SDS-PAGE gels showed a stoichiometry of 1:1.2:1. Native-PAGE analysis comparing Coomassie brilliant blue staining and the specific activity staining (Fig. 2C) showed that the enzyme was active during purification and appeared as a single band with the same mobility.
TABLE 2.
Purification of Ndh from A. tumefaciens S33
| Step | Total protein (mg) | Sp act (U/mg)a | Total activity (U) | Yield (%) | Purification factor |
|---|---|---|---|---|---|
| Cell extract | 1,454.70 | 0.0089 | 12.95 | 100 | 1 |
| Ammonium sulfate precipitation | 379.97 | 0.024 | 9.12 | 70.42 | 2.70 |
| Phenyl Sepharose | 53.09 | 0.068 | 3.61 | 27.88 | 7.64 |
| DEAE-Sepharose | 2.92 | 0.54 | 1.58 | 12.20 | 60.67 |
| Superdex 200 | 1.79 | 0.66 | 1.18 | 9.11 | 74.16 |
The activity was measured using DCIP as the artificial electron acceptor, which would increase 25-fold when PMS was added.
FIG 2.
SDS-PAGE (A and B) and native-PAGE (C) analysis of the purification of Ndh from A. tumefaciens S33 (A, 12.5% gel; B, 6.0% gel). Lanes: M, protein marker; 1, cell extract; 2, ammonium sulfate precipitate; 3, Phenyl Sepharose; 4, DEAE-Sepharose; 5, Superdex 200. The left part of panel C was stained by Coomassie brilliant blue, and the right part shows specific activity staining with a solution of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and nicotine.
Identification of the purified enzyme and its encoding genes.
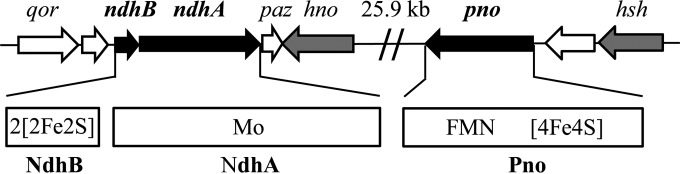
To identify the genes encoding Ndh, a tryptic digest of purified Ndh from A. tumefaciens S33 was analyzed using MALDI-TOF MS. The MS data were used to search the genome draft sequence of A. tumefaciens S33, which was automatically annotated on the RAST online server (28), and revealed three protein species as follows: 82,408 Da (NdhA, 749 amino acids), 17,068 Da (NdhB, 155 amino acids), and 73,349 Da (Pno, 671 amino acids). The sum of these values was consistent with molecular masses of 180 kDa determined using gel filtration and of 170 kDa determined using SDS-PAGE. The corresponding genes ndhA (2,259 bp) and ndhB (468 bp) overlap by 4 bp in the genome of S33 (Fig. 3). The two ORFs flanking the ndhAB genes are predicted to encode a quinone oxidoreductase (Qor) and an electron carrier protein pseudoazurin (Paz). However, whether they participate in the nicotine degradation is still unknown. These genes form a gene cluster with an ORF encoding a protein with 99% identity to 6-hydroxynicotine oxidase (Hno) from Shinella sp. strain HZN7 (22). The sequence of pno (2,016 bp) is near that of hsh encoding HSP hydroxylase (28), which is located approximately 25.9 kb from the ndhAB-hno gene cluster. Between hno and pno, 6 of 12 ORFs were predicted to encode mobile element proteins. Conserved domain analysis shows that NdhB contains two conserved binding motives for two [2Fe-2S] clusters, and NdhA harbors a conserved binding site for molybdopterin cofactor. Pno harbors conserved binding sites for one FMN and one [4Fe-4S] cluster.
FIG 3.
Region of A. tumefaciens S33 genome around the ndhAB and pno genes encoding nicotine dehydrogenase and 6-hydroxypseudooxynicotine oxidase, respectively. Open arrows without labels indicate genes encoding hypothetical proteins. Cofactor binding sites were deduced from the amino acid sequences of the proteins. Mo, molybdopterin that binds molybdenum. qor, quinone oxidoreductase; paz, pseudoazurin; hno, 6-hydroxynicotine oxidase; hsh, 6-hydroxy-3-succinoylpyridine hydroxylase. Between hno and pno, there are 12 ORFs predicted to encode six mobile element proteins and six hypothetical proteins.
The ndhA and ndhB genes are annotated to encode the isoquinoline 1-oxidoreductase alpha and beta subunits, respectively, which is a molybdenum-containing hydroxylase catalyzing the hydroxylation of isoquinoline to 1-oxo-1,2-dihydroisoquinoline from Brevundimonas diminuta 7 (formerly Pseudomonas diminuta 7) (47, 48) with the same conserved domains of NdhAB from S33. Based on BLAST analysis of protein sequence, NdhA is 99% identical to the large subunit of nicotine hydroxylase from Ochrobactrum sp. strain SJY1 (24), 27% identical to the large subunit of isoquinoline 1-oxidoreductase from B. diminuta 7 (47, 48), 14.0% identical to NdhL, and 14.4% identical to KdhL (the large subunit of 6-hydroxypseudooxynicotine dehydrogenase, also named ketone dehydrogenase) from A. nicotinovorans, which contain the conserved molybdopterin-binding domain. NdhB is 100% identical to the small subunit of nicotine hydroxylase from Ochrobactrum sp. strain SJY1 (24), 27% identical to the small subunit of isoquinoline 1-oxidoreductase from B. diminuta 7 (47, 48), 36.4% identical to the smallest subunit of NdhS, and 32.3% identical to KdhS from A. nicotinovorans that bind two [2Fe-2S] clusters. Like nicotine hydroxylase from Ochrobactrum sp. strain SJY1 and isoquinoline 1-oxidoreductase from B. diminuta 7, the intermediate size subunit containing the flavin-binding motif similar to those of NdhM and KdhM from A. nicotinovorans was not detected in the genome sequence of A. tumefaciens S33. These results indicate that the two subunits of Ndh from A. tumefaciens S33 are similar to the respective subunits of molybdopterin-containing heterotrimeric hydroxylases from other bacteria, such as Ndh and Kdh from A. nicotinovorans (25, 49, 50), except for the missing of flavin-binding subunit. The pno gene is annotated as histamine dehydrogenase or NADH:flavin oxidoreductase in RAST or GenBank, respectively, and is 48% identical to the histamine dehydrogenase of Pimelobacter simplex (formerly Nocardioides simplex) (51–53), 46% identical to histamine dehydrogenase of Rhizobium sp. strain 4-9 (54), 40% identical to trimethylamine dehydrogenase of Methylophilus methylotrophus W3A1 (55, 56), and 39% identical to dimethylamine dehydrogenase of Hyphomicrobium sp. strain X (57). The alignment of the protein sequences of the five enzymes shows that cysteine 35 in trimethylamine dehydrogenase from M. methylotrophus W3A1, which binds FMN covalently through the sulfhydryl group and is conserved in the other three enzymes, is replaced by an alanine in Pno of S33. Further, because the other amino acid residues for binding FMN are conserved in all five enzymes, this suggests that FMN of Pno from S33 may bind noncovalently to the enzyme. The [4Fe-4S]-cluster-binding sites in Pno and the three amine dehydrogenases are highly conserved. Further, three amine dehydrogenases bind an ADP with unknown function (51–53) with binding sites that are conserved in Pno. Thus, whether ADP is a cofactor for Pno remains to be determined.
The predicted cofactors of the proteins were verified by biochemical analyses of purified Ndh. ICP-OES of the purified protein (molecular mass of 180 kDa) detected 1.32 mol of molybdenum and 8.8 mol of Fe per mol of protein complex, and tungsten was undetectable. These data are consistent with an enzyme complex comprising NdhA, NdhB, and Pno with a molybdopterin-binding domain, two [2Fe-2S] clusters, and one [4Fe-4S] cluster, respectively. Further, approximately 1.4 mol of FMN was detected after heat treatment using a UV-visible spectrophotometer and an HPLC. FAD and ADP were not detected, which is consistent with the presence of an FMN-binding motif in Pno. The binding of iron-sulfur cluster and flavin in NdhAB-Pno complex was also indicated by the UV-visible spectrum of the enzyme (see Fig. S2 in the supplemental material).
Nicotine transformation catalyzed by NdhAB-Pno complex.
In order to identify the reaction catalyzed by the purified enzyme, the product of nicotine transformation was determined. LC-MS analysis (see Fig. S3 in the supplemental material) detected two peaks eluting at 3.37 and 3.95 min, respectively (see Fig. S3A in the supplemental material), and the values of m/z of their main fragments in the mass spectra were 179.14 (see Fig. S3C in the supplemental material) and 163.19 (see Fig. S3B in the supplemental material), which were consistent with the calculated molecular masses of 6-hydroxynicotine (C10H14ON2, 178.1106) and nicotine (C10H14N2, 162.1157), respectively. No other compound was detected. These results indicate that NdhAB-Pno complex catalyzes the oxidation of nicotine to produce 6-hydroxynicotine and has the same function to Ndh in the pyridine pathway of A. nicotinovorans (25, 58–60).
The purified enzyme catalyzed nicotine oxidation with low activity (0.66 U/mg). One of the problems we found is its poor thermal stability (see Fig. S4 in the supplemental material). When the enzyme was kept at 25°C for 30 min, the total activity was found to decrease ca. 50%. The enzyme presented higher stability at a temperature lower than 15°C. Under the best conditions at 16°C that we evaluated, the enzyme was purified, which showed an apparent Km for nicotine of 0.88 μM and a kcat of 2.0/s at the growth temperature of 30°C and in 50 mM phosphate buffer (pH 7.0) (see Fig. S5 in the supplemental material). Further, we tried to supplement 0.5 M PMS into the reaction mixture, and the activity greatly increased to 16.5 U/mg, suggesting that PMS efficiently mediated the electron transfer from nicotine to DCIP.
Transcriptional analysis of the genes ndhA, ndhB, and pno.
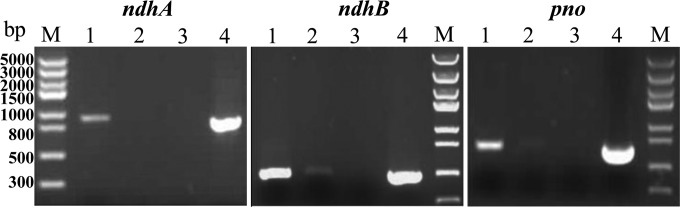
To determine whether ndhA, ndhB, and pno mediate the nicotine degradation by A. tumefaciens S33, total RNAs were isolated from cultures grown in medium with nicotine or glucose and ammonium sulfate as the sources of carbon and nitrogen, respectively. RT-PCR detected amplicons specific for the genes encoding NdhA (895 bp), NdhB (468 bp), and Pno (695 bp) when the strain was grown in medium containing nicotine (Fig. 4). In contrast, PCR products either were not detected or were detected in very small amounts when the strain was grown in glucose and ammonium medium, indicating that the transcription of the genes encoding NdhA, NdhB, and Pno of A. tumefaciens S33 is induced in the presence of nicotine.
FIG 4.
RT-PCR analysis of ndhAB and pno transcription in A. tumefaciens S33. Lanes 1 and 2, cDNAs synthesized from total RNAs isolated from cells cultured in nicotine medium and in glucose and ammonium medium, respectively; lane 3, negative control (total RNA isolated from cells cultured in nicotine medium); lane 4, positive control (genomic DNA from the culture grown in glucose and ammonium medium).
Deletion and complementation of ndhA, ndhB, and pno genes.
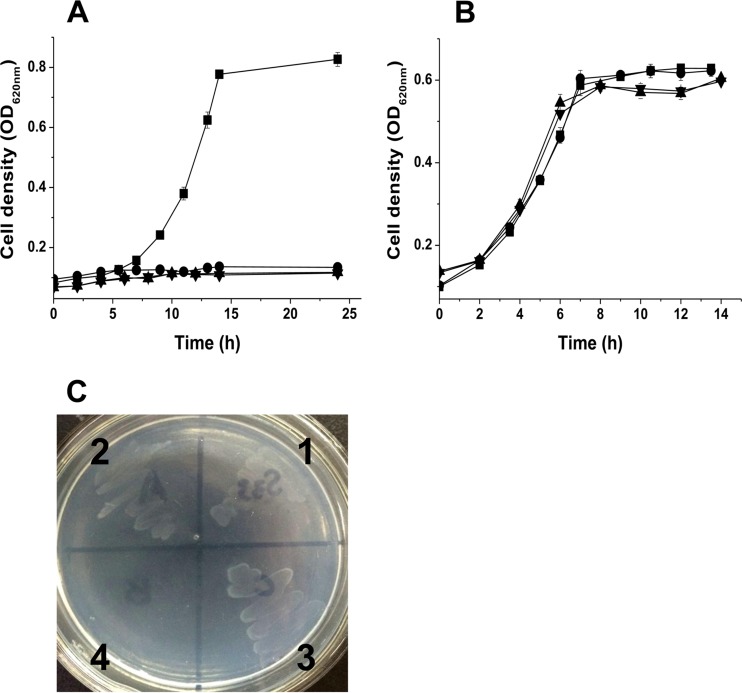
To determine the functions of ndhA, ndhB, and pno, we disrupted them individually with suicide vector plasmid pJQ200SK harboring truncated genes and then determined whether function was complemented with a broad-host-range plasmid pBBRMCS-5 harboring complete copies of the target genes. The mutant strains S33-ΔndhA, S33-ΔndhB, and S33-Δpno grew in HSP medium (Fig. 5B) but did not utilize nicotine as the sole source of carbon and nitrogen (Fig. 5A). Further, we determined whether they grew in the presence of 6-hydroxynicotine as the sole source of carbon and nitrogen (Fig. 5C). Strain S33-Δpno failed to grow, whereas both mutant strains S33-ΔndhA and S33-ΔndhB could grow well. The complemented strains S33-ΔndhA-C, S33-ΔndhB-C, and S33-Δpno-C grew in nicotine medium, and their ability to utilize nicotine was the same as for the wild type (see Fig. S6 in the supplemental material). Therefore, we concluded that the three genes are involved in the conversion of nicotine to HSP in the nicotine degradation pathway of strain S33 (Fig. 1), that ndhA and ndhB encode Ndh catalyzing the first step in the pathway, and that the product of pno mediates the steps from 6-hydroxynicotine to HSP. Because the gene encoding Hno, which catalyzes the oxidation of 6-hydroxynicotine to 6-hydroxy-N-myosmine and 6-hydroxypseudooxynicotine (Fig. 1), is adjacent to ndhAB (Fig. 2), pno must function in the reactions from 6-hydroxypseudooxynicotine to HSP. This is also confirmed by the activity assays of Ndh and Hno in the cell extracts of the three mutants (Table 3). Only the mutant strain S33-Δpno presented a very low DCIP reduction activity with nicotine of 0.0017 U/mg. Three mutants had an Hno activity of around 0.22 U/mg.
FIG 5.
Growth of strain S33 mutants in the presence of nicotine (A), 6-hydroxy-3-succinoylpyridine (B), and 6-hydroxynicotine (C) as the sole source of carbon and nitrogen. Symbols (A and B) and numbers (C): ■ and 1, wild-type strain; ● and 2, S33-ΔndhA; ▲ and 3, S33-ΔndhB; ▼ and 4, S33-Δpno.
TABLE 3.
Activities in the cell extracts of the mutants with disrupted ndhA, ndhB, or pno mutants and wild-type strain S33a
| Enzyme | Sp act (U/mg) |
|||
|---|---|---|---|---|
| S33 | S33-ΔndhA | S33-ΔndhB | S33-Δpno | |
| Ndh | 0.014 | <0.0001 | <0.0001 | 0.0017 |
| Hno | 0.52 | 0.22 | 0.21 | 0.22 |
| Pno | 0.44 | 0.31 | 0.42 | <0.01 |
The cells used for preparing the cell extracts were grown for 20 h in HSP medium supplemented with 1 g of nicotine/liter.
Moreover, we performed the nicotine transformation reaction by resting cells of the mutant strain S33-△pno. LC-MS analysis (see Fig. S7 in the supplemental material) revealed four major compounds in the reaction mixture as follows: nicotine (c, 6.3 min, m/z 163.1259; C10H14N2; calculated molecular weight [MW], 162.1157), 6-hydroxynicotine (b, 5.6 min, m/z 179.1203; C10H14N2O; calculated MW, 178.1106), 6-hydroxy-N-methylmyosime (a, 4.9 min, m/z 177.1052; C10H12N2O; calculated MW, 176.0950), and 6-hydroxypseudooxynicotine (d, 6.9 min, m/z 195.1156; C10H14N2O2; calculated MW, 194.1055). The results indicate that nicotine was converted to 6-hydroxynicotine, 6-hydroxy-N-methylmyosmine, and 6-hydroxypseudooxynicotine, which are the intermediates in the first three steps of nicotine degradation in S33 (Fig. 1). No other compound was detected. These results verified that Pno participates in the reactions leading from 6-hydroxypseudooxynicotine to HSP.
Heterologous expression and characterization of Pno.
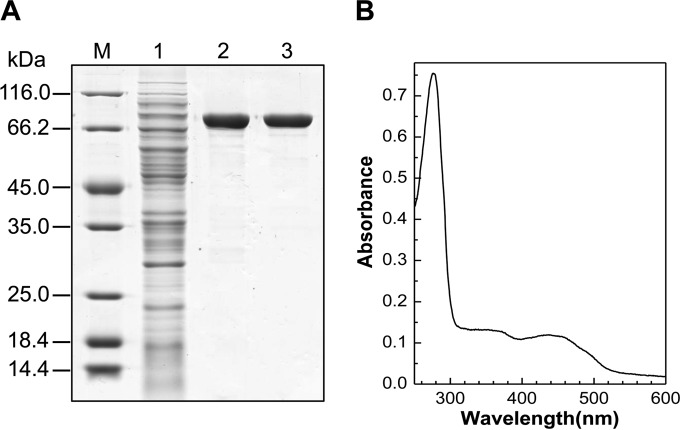
To identify the function of pno, we expressed it in E. coli C41(DE3) harboring pCodonPlus and pRKISC with a His tag at N terminus in TB medium containing extra iron and sulfur sources and riboflavin, considering the protein binding of one [4Fe-4S] cluster and one FMN. The purified His-tagged protein was yellow brown with a molecular mass of 73 kDa, determined using SDS-PAGE (Fig. 6A). The recombinant Pno contained 1.2 mol of FMN and 3.3 mol of Fe per mol of protein, which fits the prediction of one FMN and one [4Fe-4S] cluster per enzyme molecule. The UV-visible spectrum of Pno (Fig. 6B) revealed two broad peaks typical of the absorption of flavin and an iron-sulfur cluster.
FIG 6.
Purification (A) and UV-visible absorption spectrum (B) of recombinant Pno from A. tumefaciens S33. M, marker; lane 1, cell extract of recombinant E. coli; lane 2, HisTrap HP column; lane 3, HiTrap Q HP column. The sample used for UV-visible absorption spectrum analysis contained 0.38 mg of purified protein/ml in 50 mM sodium phosphate buffer (pH 7.0).
Coupling the reaction of 6-hydroxynicotine oxidation by Hno from A. nicotinovorans, the enzyme catalyzed the oxidation of 6-hydroxypseudoxynicotine (32.3 U/mg) at pH 8.5 and 30°C using DCIP-PMS as an electron acceptor. When PMS was omitted, the activity was ∼55-fold lower. These data are consistent with those from assays of NdhAB-Pno complex purified from wild-type S33 (36.2 U/mg using DCIP-PMS as an electron acceptor and 0.74 U/mg using only DCIP as an electron acceptor), where the activity was around 0.34 U/mg in cell extract using DCIP-PMS as an electron acceptor. Further, we tried to estimate the apparent Km for 6-hydroxypseudooxynicotine of both recombinant Pno and wild-type NdhAB-Pno complex. The values were approximately 0.37 and 0.18 mM for recombinant Pno and wild-type NdhAB-Pno complex, respectively (see Fig. S8 in the supplemental material). In the assays, the 6-hydroxypseudooxynicotine prepared by the conversion of 6-hydroxynicotine using Hno from A. nicotinovorans was quantified according to its absorbance at 334 nm (ε = 20.7 mM−1 cm−1) (42). The apparent Km may represent an overestimate because 6-hydroxypseudooxynicotine was not isolated from the mixture of the transformation reaction due to its instability, where the rest substrate 6-hydroxynicotine and the product 6-hydroxy-N-methylmyosmine may interfere its quantification at 334 nm. Despite all this, these results clearly showed that the wild-type enzyme complex have higher affinity to the substrate. The reaction product of the oxidation of 6-hydroxypseudooxynicotine by Pno was determined to be 6-hydroxy-3-succinoylsemialdehyde-pyridine (m/z 180.0656; C9H9NO3; calculated MW, 179.0582) using LC-MS (see Fig. S9 in the supplemental material), indicating that Pno catalyzes the fourth step of nicotine degradation in S33 (Fig. 1). Although the predicted amino acid sequence of Pno is 48% identical to that of histamine dehydrogenase, we did not detect a reduction in DCIP reduction activity using histamine as the substrate. Further, the recombinant Pno did not show any detectable activity using nicotine as the substrate.
DISCUSSION
The hybrid of the pyridine and pyrrolidine pathways for nicotine degradation was discovered by investigating nicotine catabolism in A. tumefaciens S33 (5), and the same pathway was also found in Shinella sp. strain HZN7 (21, 22) and Ochrobactrum sp. strain SJY1 (23). However, the biochemical mechanism of the novel pathway is still not clear. Here, we purified an enzyme complex of NdhAB and Pno from A. tumefaciens S33 and verified that they catalyze the first and fourth steps of the hybrid pathway of nicotine degradation, respectively (Fig. 1). BLAST analysis showed that NdhAB is almost identical to the most recently reported nicotine hydroxylase from Ochrobactrum sp. strain SJY1 (24), which mediates the first step in the same pathway, too. However, the function of the nicotine hydroxylase from Ochrobactrum sp. strain SJY1 was demonstrated with partially purified recombinant protein and recombinant P. putida KT2440 harboring its genes. Its kinetic properties are lacking. In this study, we showed that the NdhAB-Pno complex from S33 catalyzes nicotine oxidation with an activity of 0.66 U/mg using DCIP as electron acceptor, an apparent Km for nicotine of 0.88 μM, and a kcat of 2.0/s at a growth temperature of 30°C and in 50 mM phosphate buffer (pH 7.0). PMS can efficiently enhance the electron transfer from nicotine to DCIP, mediated by the enzyme with an activity of 16.5 U/mg. Notably, the Ndh activity in a cell extract of S33-Δpno was much lower than that of wild-type S33 (Table 3), which hinders the purification of the NdhAB from the pno-disrupted mutant. We tried to heterologously express it in E. coli. Just like heterologous expression of nicotine hydroxylase from Ochrobactrum sp. strain SJY1 in E. coli (24), ndhAB was not functionally expressed (data not shown), likely because the molybdopterin cofactor in NdhAB was not synthesized by E. coli (29), which normally synthesizes the molybdopterin guanosine dinucleotide (29, 61). In contrast, NdhAB from strain S33 may bind a molybdopterin cytosine dinucleotide like Ndh from A. nicotinovorans (29) and isoquinoline 1-oxidoreductase from B. diminuta 7 (48). However, this hypothesis requires experimental verification. Thus, we could not present the kinetic constants of purified NdhAB. Like isoquinoline 1-oxidoreductase from B. diminuta 7 (47, 48), NdhAB is a heterodimeric molybdenum-containing hydroxylase, and the subunit binding flavin similar to the intermediate size subunit of NdhLMS from A. nicotinovorans was missing. The heterotrimeric NdhLMS from A. nicotinovorans catalyzes the same hydroxylation reaction at the C-6 position of the pyridine ring of nicotine (25, 26) to NdhAB from S33. Why only two components of this type of enzyme can also function in the same catalysis is still a puzzle.
Pno is a novel iron-sulfur flavoprotein with conserved domains similar to those of histamine dehydrogenase, dimethylamine dehydrogenase, and trimethylamine dehydrogenase for binding an FMN and a [4Fe-4S] cluster, except that FMN may be noncovalently bound to the protein. We successfully expressed it in soluble form by using E. coli C41(DE3) harboring pCodonPlus and pRKISC, which is used specially to express iron-sulfur proteins (36). When we used E. coli BL21(DE3), most of the target protein formed inclusion bodies lacking activity. Recombinant Pno was yellow brown, consistent with the presence of an FMN and a [4Fe-4S] cluster, which may prevent proper protein folding if not correctly assembled. Coupled with the 6-hydroxynicoitne oxidation by Hno, it presented a DCIP reduction activity with 6-hydroxypseudooxynicoitne. Although it has highest identity (48%) to histamine dehydrogenase, it did not show any detectable activity for histamine. BLAST analysis showed that Ochrobactrum sp. strain SJY1 also harbors a gene encoding the same protein (GenBank accession number AIH15773) as Pno from S33, whose function has not been identified yet. In this study, we demonstrated its role in the nicotine degradation.
Because the draft genome of S33 is not complete, we could not compare the genomes of S33 and Ochrobactrum sp. strain SJY1. But the available data of S33 showed that the organization and sequence of a ndhAB-hno-pno-hsh gene cluster in S33 (Fig. 2) is similar to that from Ochrobactrum sp. strain SJY1 (23, 24). In S33, ndhA and ndhB overlap 4 bp, which is common for the genes of multiple subunits enzyme in microorganisms and is helpful for rapid cotranscription and translation and their regulation (62, 63). Interestingly, there is a 26-kb distance between ndhAB-hno and pno-hsh, where 6 of 12 ORFs encode mobile element proteins, suggesting that these genes might come from other bacteria by the way of lateral gene transfer.
Another question is whether it is necessary for functionality that NdhAB and Pno form a complex, which catalyze two different reactions and whose encoding genes are far away from each other. The results of SDS-PAGE and gel filtration clearly showed that the NdhAB-Pno complex was purified with a stoichiometry of 1:1:1.2 and a molecular mass of 180 kDa, which did not separate during purification as indicated by native PAGE. We did not separate them, even by using different elution conditions and chromatography columns (data not shown), indicating that the interaction of NdhAB and Pno was not weak. The comparison of the kinetic properties of the NdhAB-Pno complex and recombinant Pno may give the answer. The purified NdhAB-Pno complex presented a Pno activity of 36.2 U/mg and had an apparent Km of 0.18 mM for 6-hydroxypseudooxynicotine. The recombinant Pno had an activity (32.3 U/mg) similar to that of the purified wild-type NdhAB-Pno complex, but a higher apparent Km of 0.37 mM for 6-hydroxypseudooxynicotine, indicating that the NdhAB-Pno complex has greater affinity for the substrate than the single component Pno. For NdhAB, unfortunately, we failed to heterologously express it or purify it from S33-Δpno. However, the fact that the Ndh activity in cell extract of S33-Δpno (0.0017 U/mg) was much lower than that in the cell extract of wild-type S33 (0.014 U/mg, Table 3) also showed that the complex has greater activity than the two-component NdhAB. As controls, all mutants with disrupted ndhA, ndhB, or pno presented Pno activities of around 0.36 U/mg in cell extracts, values which were close to that in the cell extract of wild-type S33 (0.44 U/mg). A low activity of nicotine hydroxylation was also observed in the resting cells of recombinant P. putida KT2440 harboring the nicotine hydroxylase genes from Ochrobactrum sp. strain SJY1 (24). All of these data indicate that the NdhAB-Pno complex has better catalytic efficiency than the individual NdhAB and Pno. Thus, a complex of NdhAB and Pno is necessary for efficient nicotine degradation in vivo. Notably, a yellow fraction with Hno activity was found next to the fraction containing Ndh-Pno complex during elution of a DEAE column (H. Li, W. Yu, and S. Wang, unpublished data), which gave us a hint that Hno might loosely interact with NdhAB-Pno complex in vivo. If this is correct, this kind of organization will be very helpful for the quick detoxification and degradation of nicotine by sequential catalytic reactions like the well-known pyruvate dehydrogenase complex and fatty acid beta-oxidation multienzyme complex (64–67).
In summary, we characterized a novel enzyme complex of NdhAB and Pno, which catalyzes the first and fourth steps in the hybrid pathway of nicotine degradation in A. tumefaciens S33. The results provide new biochemical and molecular evidence for the novel hybrid pathway of nicotine degradation in A. tumefaciens S33. Our future studies will focus on the biochemical properties of the other enzymes that contribute to the hybrid pathway.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (grant 30970027), the Excellent Middle-Aged and Youth Scientist Award Foundation of Shandong Province (grant BS2009SW006), and the Fundamental Research Funds of Shandong University (grant 2014JC023).
We thank Roderich Brandsch from the University of Freiburg for the gifts of 6-hydroxynicotine and 6-hydroxynicotine oxidase and Yasuhiro Takahashi from Osaka University for the gift of E. coli C41(DE3) harboring pCodonPlus and pRKISC. We also acknowledge Ping Xu from Shanghai Jiao Tong University and Luying Xun from Washington State University for their valuable discussion and support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03909-15.
REFERENCES
- 1.Gelvin SB. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cokesa Z, Knackmuss HJ, Rieger PG. 2004. Biodegradation of all stereoisomers of the EDTA substitute iminodisuccinate by Agrobacterium tumefaciens BY6 requires an epimerase and a stereoselective C-N lyase. Appl Environ Microbiol 70:3941–3947. doi: 10.1128/AEM.70.7.3941-3947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galindez-Najera SP, Llamas-Martinez MA, Ruiz-Ordaz N, Juarez-Ramirez C, Mondragon-Parada ME, Ahuatzi-Chacon D, Galindez-Mayer J. 2009. Cyanuric acid biodegradation by a mixed bacterial culture of Agrobacterium tumefaciens and Acinetobacter sp. in a packed bed biofilm reactor. J Ind Microbiol Biotechnol 36:275–284. doi: 10.1007/s10295-008-0496-5. [DOI] [PubMed] [Google Scholar]
- 4.Koivunen ME, Morisseau C, Horwath WR, Hammock BD. 2004. Isolation of a strain of Agrobacterium tumefaciens (Rhizobium radiobacter) utilizing methylene urea (urea formaldehyde) as nitrogen source. Can J Microbiol 50:167–174. doi: 10.1139/w04-001. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, Huang H, Xie K, Xu P. 2012. Identification of nicotine biotransformation intermediates by Agrobacterium tumefaciens strain S33 suggests a novel nicotine degradation pathway. Appl Microbiol Biotechnol 95:1567–1578. doi: 10.1007/s00253-012-4007-2. [DOI] [PubMed] [Google Scholar]
- 6.Wang SN, Liu Z, Xu P. 2009. Biodegradation of nicotine by a newly isolated Agrobacterium sp. strain S33. J Appl Microbiol 107:838–847. doi: 10.1111/j.1365-2672.2009.04259.x. [DOI] [PubMed] [Google Scholar]
- 7.Benowitz NL. 2010. Nicotine addiction. N Engl J Med 362:2295–2303. doi: 10.1056/NEJMra0809890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecht SS. 1999. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst 91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 9.Novotny TE, Zhao F. 1999. Consumption and production waste: another externality of tobacco use. Tobacco Control 8:75–80. doi: 10.1136/tc.8.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Civilini M, Domenis C, Sebastianutto N, de Berfoldi M. 1997. Nicotine decontamination of tobacco agro-industrial waste and its degradation by micro-organisms. Waste Manag Res 15:349–358. doi: 10.1006/wmre.1997.0091. [DOI] [Google Scholar]
- 11.Wang JH, He HZ, Wang MZ, Wang S, Zhang J, Wei W, Xu HX, Lu ZM, Shen DS. 2013. Bioaugmentation of activated sludge with Acinetobacter sp. TW enhances nicotine degradation in a synthetic tobacco wastewater treatment system. Bioresour Technol 142:445–453. doi: 10.1016/j.biortech.2013.05.067. [DOI] [PubMed] [Google Scholar]
- 12.Zhong W, Zhu C, Shu M, Sun K, Zhao L, Wang C, Ye Z, Chen J. 2010. Degradation of nicotine in tobacco waste extract by newly isolated Pseudomonas sp. ZUTSKD. Bioresour Technol 101:6935–6941. doi: 10.1016/j.biortech.2010.03.142. [DOI] [PubMed] [Google Scholar]
- 13.Brandsch R. 2006. Microbiology and biochemistry of nicotine degradation. Appl Microbiol Biotechnol 69:493–498. doi: 10.1007/s00253-005-0226-0. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Li X, Duan Y, Zhang KQ, Yang J. 2010. Biotransformation of nicotine by microorganism: the case of Pseudomonas spp. Appl Microbiol Biotechnol 86:11–17. doi: 10.1007/s00253-009-2427-4. [DOI] [PubMed] [Google Scholar]
- 15.Qiu J, Ma Y, Zhang J, Wen Y, Liu W. 2013. Cloning of a novel nicotine oxidase gene from Pseudomonas sp. strain HZN6 whose product nonenantioselectively degrades nicotine to pseudooxynicotine. Appl Environ Microbiol 79:2164–2171. doi: 10.1128/AEM.03824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang H, Wang L, Wang W, Yu H, Zhang K, Yao Y, Xu P. 2013. Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet 9:e1003923. doi: 10.1371/journal.pgen.1003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang SN, Xu P, Tang HZ, Meng J, Liu XL, Huang J, Chen H, Du Y, Blankespoor HD. 2004. Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol Lett 26:1493–1496. doi: 10.1023/B:BILE.0000044450.16235.65. [DOI] [PubMed] [Google Scholar]
- 18.Meng XJ, Lu LL, Gu GF, Xiao M. 2010. A novel pathway for nicotine degradation by Aspergillus oryzae 112822 isolated from tobacco leaves. Res Microbiol 161:626–633. doi: 10.1016/j.resmic.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 19.Wang SN, Liu Z, Tang HZ, Meng J, Xu P. 2007. Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology 153:1556–1565. doi: 10.1099/mic.0.2006/005223-0. [DOI] [PubMed] [Google Scholar]
- 20.Qiu J, Ma Y, Wen Y, Chen L, Wu L, Liu W. 2012. Functional identification of two novel genes from Pseudomonas sp. strain HZN6 involved in the catabolism of nicotine. Appl Environ Microbiol 78:2154–2160. doi: 10.1128/AEM.07025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y, Wei Y, Qiu J, Wen R, Hong J, Liu W. 2014. Isolation, transposon mutagenesis, and characterization of the novel nicotine-degrading strain Shinella sp. HZN7. Appl Microbiol Biotechnol 98:2625–2636. doi: 10.1007/s00253-013-5207-0. [DOI] [PubMed] [Google Scholar]
- 22.Qiu J, Wei Y, Ma Y, Wen R, Wen Y, Liu W. 2014. A novel (S)-6-hydroxynicotine oxidase gene from Shinella sp. strain HZN7. Appl Environ Microbiol 80:5552–5560. doi: 10.1128/AEM.01312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu H, Tang H, Zhu X, Li Y, Xu P. 2015. Molecular mechanism of nicotine degradation by a newly isolated strain, Ochrobactrum sp. strain SJY1. Appl Environ Microbiol 81:272–281. doi: 10.1128/AEM.02265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, Tang H, Li Y, Xu P. 2015. Molybdenum-containing nicotine hydroxylase genes in a nicotine degradation pathway that is a variant of the pyridine and pyrrolidine pathways. Appl Environ Microbiol 81:8330–8338. doi: 10.1128/AEM.02253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freudenberg W, König K, Andreesen JR. 1988. Nicotine dehydrogenase from Arthrobacter oxidans: a molybdenum-containing hydroxylase. FEMS Microbiol Lett 52:13–17. doi: 10.1111/j.1574-6968.1988.tb02564.x. [DOI] [Google Scholar]
- 26.Grether-Beck S, Igloi GL, Pust S, Schilz E, Decker K, Brandsch R. 1994. Structural analysis and molybdenum-dependent expression of the pAO1-encoded nicotine dehydrogenase genes of Arthrobacter nicotinovorans. Mol Microbiol 13:929–936. doi: 10.1111/j.1365-2958.1994.tb00484.x. [DOI] [PubMed] [Google Scholar]
- 27.Tang H, Yao Y, Zhang D, Meng X, Wang L, Yu H, Ma L, Xu P. 2011. A novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida. J Biol Chem 286:39179–39187. doi: 10.1074/jbc.M111.283929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Xie K, Huang H, Wang S. 2014. 6-Hydroxy-3-succinoylpyridine hydroxylase catalyzes a central step of nicotine degradation in Agrobacterium tumefaciens S33. PLoS One 9:e103324. doi: 10.1371/journal.pone.0103324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sachelaru P, Schiltz E, Brandsch R. 2006. A functional mobA gene for molybdopterin cytosine dinucleotide cofactor biosynthesis is required for activity and holoenzyme assembly of the heterotrimeric nicotine dehydrogenases of Arthrobacter nicotinovorans. Appl Environ Microbiol 72:5126–5131. doi: 10.1128/AEM.00437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang SN, Xu P, Tang HZ, Meng J, Liu XL, Ma CQ. 2005. “Green” route to 6-hydroxy-3-succinoyl-pyridine from (S)-nicotine of tobacco waste by whole cells of a Pseudomonas sp. Environ Sci Technol 39:6877–6880. doi: 10.1021/es0500759. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q, Qin D, Zhang S, Wang L, Li J, Rensing C, McDermott TR, Wang G. 2015. Fate of arsenate following arsenite oxidation in Agrobacterium tumefaciens GW4. Environ Microbiol 17:1926–1940. doi: 10.1111/1462-2920.12465. [DOI] [PubMed] [Google Scholar]
- 32.Link AJ, Phillips D, Church GM. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 34.Wise AA, Liu Z, Binns AN. 2006. Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol Biol 343:43–53. [DOI] [PubMed] [Google Scholar]
- 35.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura M, Saeki K, Takahashi Y. 1999. Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster. J Biochem 126:10–18. doi: 10.1093/oxfordjournals.jbchem.a022409. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi Y, Nakamura M. 1999. Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J Biochem 126:917–926. doi: 10.1093/oxfordjournals.jbchem.a022535. [DOI] [PubMed] [Google Scholar]
- 38.Hamann N, Mander GJ, Shokes JE, Scott RA, Bennati M, Hedderich R. 2007. A cysteine-rich CCG domain contains a novel [4Fe-4S] cluster binding motif as deduced from studies with subunit B of heterodisulfide reductase from Methanothermobacter marburgensis. Biochemistry 46:12875–12885. doi: 10.1021/bi700679u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang H, Wang S, Moll J, Thauer RK. 2012. Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2. J Bacteriol 194:3689–3699. doi: 10.1128/JB.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai VD, Decker K, Sund H. 1968. Purification and properties of l-6-hydroxynicotine oxidase. Eur J Biochem 4:95–102. doi: 10.1111/j.1432-1033.1968.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 41.Decker K, Dai VD. 1967. Mechanism and specificity of l- and d-6-hydroxynicotine oxidase. Eur J Biochem 3:132–138. doi: 10.1111/j.1432-1033.1967.tb19507.x. [DOI] [PubMed] [Google Scholar]
- 42.Brühmüller M, Mohler H, Decker K. 1972. Covalently bound flavin in d-6-hydroxynicotine oxidase from Arthrobacter oxidans: purification and properties of d-6-hydroxynicotine oxidase. Eur J Biochem 29:143–151. [DOI] [PubMed] [Google Scholar]
- 43.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 44.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 45.Wang S, Huang H, Kahnt J, Mueller AP, Köpke M, Thauer RK. 2013. An NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J Bacteriol 195:4373–4386. doi: 10.1128/JB.00678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, Huang H, Moll J, Thauer RK. 2010. NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri. J Bacteriol 192:5115–5123. doi: 10.1128/JB.00612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehmann M, Tshisuaka B, Fetzner S, Lingens F. 1995. Molecular cloning of the isoquinoline 1-oxidoreductase genes from Pseudomonas diminuta 7, structural analysis of iorA and iorB, and sequence comparisons with other molybdenum-containing hydroxylases. J Biol Chem 270:14420–14429. doi: 10.1074/jbc.270.24.14420. [DOI] [PubMed] [Google Scholar]
- 48.Lehmann M, Tshisuaka B, Fetzner S, Roger P, Lingens F. 1994. Purification and characterization of isoquinoline 1-oxidoreductase from Pseudomonas diminuta 7, a novel molybdenum-containing hydroxylase. J Biol Chem 269:11254–11260. [PubMed] [Google Scholar]
- 49.Baitsch D, Sandu C, Brandsch R, Igloi GL. 2001. Gene cluster on pAO1 of Arthrobacter nicotinovorans involved in degradation of the plant alkaloid nicotine: cloning, purification, and characterization of 2,6-dihydroxypyridine 3-hydroxylase. J Bacteriol 183:5262–5267. doi: 10.1128/JB.183.18.5262-5267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Igloi GL, Brandsch R. 2003. Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. J Bacteriol 185:1976–1986. doi: 10.1128/JB.185.6.1976-1986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujieda N, Satoh A, Tsuse N, Kano K, Ikeda T. 2004. 6-S-Cysteinyl flavin mononucleotide-containing histamine dehydrogenase from Nocardioides simplex: molecular cloning, sequencing, overexpression, and characterization of redox centers of enzyme. Biochemistry 43:10800–10808. doi: 10.1021/bi049061q. [DOI] [PubMed] [Google Scholar]
- 52.Limburg J, Mure M, Klinman JP. 2005. Cloning and characterization of histamine dehydrogenase from Nocardioides simplex. Arch Biochem Biophys 436:8–22. doi: 10.1016/j.abb.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 53.Reed T, Lushington GH, Xia Y, Hirakawa H, Travis DM, Mure M, Scott EE, Limburg J. 2010. Crystal structure of histamine dehydrogenase from Nocardioides simplex. J Biol Chem 285:25782–25791. doi: 10.1074/jbc.M109.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bakke M, Sato T, Ichikawa K, Nishimura I. 2005. Histamine dehydrogenase from Rhizobium sp.: gene cloning, expression in Escherichia coli, characterization, and application to histamine determination. J Biotechnol 119:260–271. doi: 10.1016/j.jbiotec.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 55.Boyd G, Mathews FS, Packman LC, Scrutton NS. 1992. Trimethylamine dehydrogenase of bacterium W3A1: molecular cloning, sequence determination and overexpression of the gene. FEBS Lett 308:271–276. [DOI] [PubMed] [Google Scholar]
- 56.Lim LW, Shamala N, Mathews FS, Steenkamp DJ, Hamlin R, Xuong NH. 1986. Three-dimensional structure of the iron-sulfur flavoprotein trimethylamine dehydrogenase at 2.4-Å resolution. J Biol Chem 261:15140–15146. [PubMed] [Google Scholar]
- 57.Yang CC, Packman LC, Scrutton NS. 1995. The primary structure of Hyphomicrobium X dimethylamine dehydrogenase. Relationship to trimethylamine dehydrogenase and implications for substrate recognition. Eur J Biochem 232:264–271. [DOI] [PubMed] [Google Scholar]
- 58.Decker K, Bleeg H. 1965. Induction and purification of stereospecific nicotine oxidizing enzymes from Arthrobacter oxidans. Biochim Biophys Acta 105:313–324. doi: 10.1016/S0926-6593(65)80155-2. [DOI] [PubMed] [Google Scholar]
- 59.Hochstein LI, Rittenberg SC. 1959. The bacterial oxidation of nicotine. II. The isolation of the first oxidative product and its identification as (1)-6-hydroxynicotine. J Biol Chem 234:156–160. [PubMed] [Google Scholar]
- 60.Hochstein LI, Rittenberg SC. 1959. The bacterial oxidation of nicotine. I. Nicotine oxidation by cell-free preparations. J Biol Chem 234:151–155. [PubMed] [Google Scholar]
- 61.Neumann M, Mittelstadt G, Seduk F, Iobbi-Nivol C, Leimkuhler S. 2009. MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in Escherichia coli. J Biol Chem 284:21891–21898. doi: 10.1074/jbc.M109.008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fukuda Y, Washio T, Tomita M. 1999. Comparative study of overlapping genes in the genomes of Mycoplasma genitalium and Mycoplasma pneumoniae. Nucleic Acids Res 27:1847–1853. doi: 10.1093/nar/27.8.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson ZI, Chisholm SW. 2004. Properties of overlapping genes are conserved across microbial genomes. Genome Res 14:2268–2272. doi: 10.1101/gr.2433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishikawa M, Tsuchiya D, Oyama T, Tsunaka Y, Morikawa K. 2004. Structural basis for channeling mechanism of a fatty acid beta-oxidation multienzyme complex. EMBO J 23:2745–2754. doi: 10.1038/sj.emboj.7600298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Izard T, Aevarsson A, Allen MD, Westphal AH, Perham RN, de Kok A, Hol WG. 1999. Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes. Proc Natl Acad Sci U S A 96:1240–1245. doi: 10.1073/pnas.96.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsuchiya D, Shimizu N, Ishikawa M, Suzuki Y, Morikawa K. 2006. Ligand-induced domain rearrangement of fatty acid beta-oxidation multienzyme complex. Structure 14:237–246. doi: 10.1016/j.str.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 67.Zhou ZH, McCarthy DB, O'Connor CM, Reed LJ, Stoops JK. 2001. The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes. Proc Natl Acad Sci U S A 98:14802–14807. doi: 10.1073/pnas.011597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.