Abstract
The Adventfjorden time series station (IsA) in Isfjorden, West Spitsbergen, Norway, was sampled frequently from December 2011 to December 2012. The community composition of microbial eukaryotes (size, 0.45 to 10 μm) from a depth of 25 m was determined using 454 sequencing of the 18S V4 region amplified from both DNA and RNA. The compositional changes throughout the year were assessed in relation to in situ fjord environmental conditions. Size fractionation analyses of chlorophyll a showed that the photosynthetic biomass was dominated by small cells (<10 μm) most of the year but that larger cells dominated during the spring and summer. The winter and early-spring communities were more diverse than the spring and summer/autumn communities. Dinophyceae were predominant throughout the year. The Arctic Micromonas ecotype was abundant mostly in the early-bloom and fall periods, whereas heterotrophs, such as marine stramenopiles (MASTs), Picozoa, and the parasitoid marine alveolates (MALVs), displayed higher relative abundance in the winter than in other seasons. Our results emphasize the extreme seasonality of Arctic microbial eukaryotic communities driven by the light regime and nutrient availability but point to the necessity of a thorough knowledge of hydrography for full understanding of their succession and variability.
INTRODUCTION
Microbial eukaryotes are critically important for the functioning of marine ecosystems as primary producers (1, 2) and consumers (3, 4) of carbon, as well as maintainers of biogeochemical cycles (5–7). In Arctic waters, where marine planktonic cyanobacteria are infrequent, marine microbial eukaryotes are the predominant primary producers (8–10). In spite of their importance, our knowledge of the diversity and role of picosized (0.2 to 2 μm) and nanosized (2 to 20 μm) (11) eukaryotic plankton is still limited in extreme areas.
High-Arctic regions are characterized by extreme seasonality in light conditions, with 24 h of sunlight in summer giving way to several months of complete darkness in winter. The cold, dark polar night period at high latitudes strongly limits the activity of autotrophic organisms, and Arctic species in general have to adjust to the timing of seasonal events (12). The few studies performed during the Arctic winter-spring transition suggest a strong seasonal response by the microbial community to irradiance (13–15). Most studies of Arctic microbial eukaryotes have so far utilized traditional identification techniques, such as microscopy, and focused on bloom-forming pelagic protists (4, 16–18). The development of molecular techniques, especially high-throughput sequencing (HTS), has made it possible to study the diversity and assemblages of pico- and nanosized eukaryotic plankton as well (19–22). This has resulted in several diversity surveys of microbial eukaryotes from the Arctic Ocean and the shelf seas (23–27). Pico- and nanosized planktons are now known to govern major processes in the oceans to a larger degree than previously assumed (6, 7, 28, 29). Furthermore, 18S rRNA gene surveys have started to unravel new phylogenetic relationships within cryptic protist groups (30).
The environmental changes now in progress, reflected by increasing ocean and air temperatures and decreasing sea ice cover, are expected to impact Arctic marine ecosystems strongly (31, 32). The ongoing climate-related changes in the Arctic Ocean have already led to a shift in microbial communities (10, 33, 34), potentially altering the whole Arctic food web and benefiting smaller cells (10, 33).
The Svalbard archipelago is a unique area in which to study the Arctic marine ecosystem, since it is influenced both by the West Spitsbergen Current (WSC), an extension of the North Atlantic Current system that transports warm, saline Atlantic water (AW) along the western coast, and by colder, less saline water from the Arctic Ocean on the northern and eastern coasts (35). Adventfjorden, on the western coast of Spitsbergen, is a small fjord branch (approximately 8.3 km long and 3.4 km wide) of the large Isfjorden system. While it consists mostly of locally produced Arctic water (ArW), the fjord is periodically influenced by influxes of relatively warm AW, and such flooding events are predicted to increase both in frequency and in magnitude in a climate change scenario (36). Due to mixing with ArW on the shelf, AW often enters the fjords of West Spitsbergen as transformed Atlantic water (TAW) with reduced temperature and salinity (36, 37). Adventfjorden can also experience strong freshwater input from two glacial rivers between June and October every year (38, 39). The periodic changes in hydrography, in combination with extreme Arctic light conditions, makes Adventfjorden well suited for study of the year-round effects of climate shifts on Arctic pelagic microbial eukaryotic communities.
We used high-throughput metabarcoding of the hypervariable V4 region of the 18S rRNA gene (rDNA) and gene product (rRNA) to identify microbial eukaryotes (size, 0.45 to 10 μm) from a high-Arctic fjord (Isfjorden-Adventfjorden [IsA], West Spitsbergen, Norway) over the course of 1 year. Our aims were (i) to characterize the temporal variation in diversity and community composition in relation to environmental parameters and (ii) to identify the most abundant taxa (operational taxonomic units [OTUs]) in this Arctic marine ecosystem and determine their contribution to the microbial community throughout the year.
MATERIALS AND METHODS
Study area and sample processing.
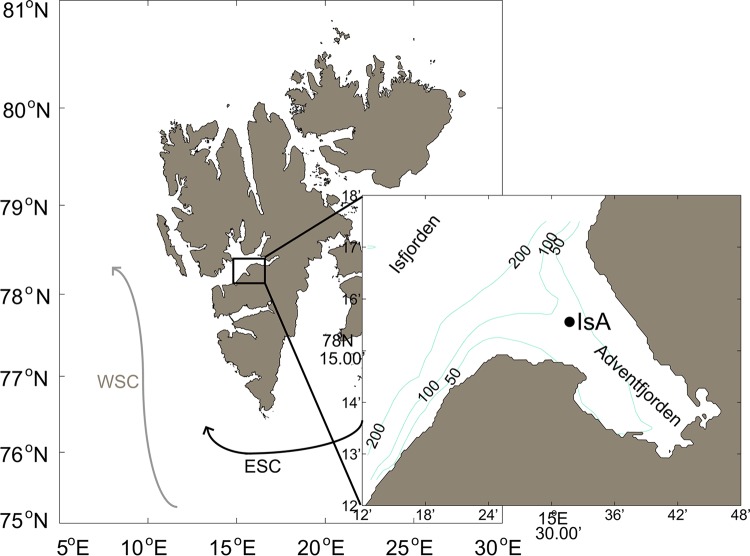
Sampling was conducted in the vicinity of a subsurface ocean observatory in Isfjorden, at the mouth of Adventfjorden (IsA time series station), close to Longyearbyen on the western coast of Spitsbergen (78°15.6′N, 15°31.8′E) (Fig. 1).
FIG 1.
Map of the IsA (Isfjorden-Adventfjorden) sampling station. (Left) The Svalbard archipelago with the large Isfjorden system at the west coast of Spitsbergen. The West Spitsbergen Current (WSC) and the East Spitsbergen Current (ESC) are indicated. (Right) Detailed map of Adventfjorden showing the position of the IsA sampling station. Depth contours (in meters) are from IBCAO (version 3.0) (101).
The IsA station was sampled weekly (February to May) or (bi)monthly (December to January and June to December) from December 2011 to December 2012 (Table 1, Sampling date [n = 26]) either with small boats (Polarcirkel boats, R/V Viking Explorer, M/S FARM) or with large vessels (R/V Helmer Hanssen, NoCGV Svalbard) when available.
TABLE 1.
Overview of sampling dates and environmental data for the DNA and RNA samples collected from the IsA stationa
| Sampling date (day.mo.yr) | Julian day | Season | Water massb | No. of PCR cycles (DNA, RNA)c |
Salinity | Temp (°C) | Fluorescenced | Density (kg m−3) | PAR (μmol m−2 s−1)e | Nutrient concn (μM)f |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NO3 and NO2 | PO4 | Si(OH)4 | ||||||||||
| 14.12.2011 | −18 | Winter | LW | 30, 23 | 34.32 | 0.87 | BD | 27.63 | 2.81 | 0.15 | 1.51 | |
| 17.01.2012 | 17 | Winter | LW | 30, 23 | 34.21 | −1.24 | 0.04 | 27.52 | ||||
| 28.01.2012 | 28 | Winter | LW | 30, 23 | 34.23 | −0.83 | BD | 27.64 | 7.22 | 0.3 | 4.48 | |
| 16.02.2012 | 47 | Winter | LW | 30, 00 | 34.31 | −0.61 | BD | 27.7 | 5.52 | 0.25 | 2.65 | |
| 01.03.2012 | 61 | Early spring | TAW | 30, 00 | 34.69 | 1.63 | BD | 27.87 | 6.38 | 0.5 | 3.2 | |
| 08.03.2012 | 68 | Early spring | 30, 00 | 3.98 | 0.33 | 1.95 | ||||||
| 19.03.2012 | 79 | Early spring | TAW | 30, 00 | 34.86 | 2.42 | BD | 27.94 | 5.56 | 9.39 | 0.66 | 5.5 |
| 22.03.2012 | 82 | Early spring | TAW | 30, 00 | 34.78 | 1.99 | BD | 27.92 | 14.43 | 4.26 | 0.39 | 1.9 |
| 11.04.2012 | 102 | Early spring | TAW | 30, 00 | 34.85 | 1.51 | 0.01 | 28.01 | 18.63 | 9.02 | 0.7 | 4.1 |
| 16.04.2012 | 107 | Early spring | TAW | 30, 00 | 34.85 | 1.51 | 0.04 | 28.01 | 25.45 | 5.06 | 0.59 | 2.2 |
| 19.04.2012 | 110 | Spring | 30, 00 | 7.66 | 0.64 | 3.3 | ||||||
| 23.04.2002 | 114 | Spring | LW | 30, 00 | 34.78 | 0.84 | 0.49 | 28 | 6.14 | 3.24 | 0.42 | 0.82 |
| 26.04.2012 | 117 | Spring | LW | 30, 00 | 34.56 | −0.23 | 0.85 | 27.88 | 6.14 | 4.49 | 0.38 | 1.65 |
| 30.04.2012 | 121 | Spring | LW | 30, 00 | 34.54 | −0.21 | 1.21 | 27.87 | 5.05 | 3.41 | 0.35 | 1.27 |
| 03.05.2012 | 124 | Spring | LW | 30, 00 | 34.53 | −0.23 | 0.92 | 27.86 | 5.7 | 1.55 | 0.26 | 0.64 |
| 07.05.2012 | 128 | Spring | LW | 30, 00 | 34.53 | 0.02 | 0.32 | 27.84 | 5.42 | 1.86 | 0.18 | 1.21 |
| 09.05.2012 | 130 | Spring | LW | 30, 00 | 34.57 | 0.28 | 1.18 | 27.86 | 0.83 | 0.21 | 1.03 | |
| 10.05.2012 | 131 | Spring | LW | 30, 18 | 34.58 | 0.28 | 0.79 | 27.87 | 6.9 | 1.54 | 0.24 | 0.28 |
| 16.05.2012 | 137 | Spring | LW | 30, 00 | 34.63 | 0.64 | 1.22 | 27.89 | 1.34 | 0.15 | 1.33 | |
| 30.05.2012 | 151 | Spring | LW | 30, 18 | 34.43 | 0.33 | 0.45 | 27.75 | 0.51 | 0 | 0.23 | 0.85 |
| 14.06.2012 | 166 | Summer/autumn | IW | 30, 00 | 34.52 | 1.93 | 0.62 | 27.71 | 10 | 0 | 0.08 | 1.37 |
| 06.07.2012 | 188 | Summer/autumn | IW | 30, 18 | 34.24 | 2.06 | 0.05 | 27.48 | 0.3 | 0.21 | 0.08 | 0.43 |
| 06.08.2012 | 219 | Summer/autumn | IW | 30, 18 | 34.1 | 2.98 | 0.09 | 27.29 | 3.4 | 0.25 | 0.12 | 1.42 |
| 18.09.2012 | 262 | Summer/autumn | IW | 30, 18 | 34.35 | 3.81 | 0.07 | 27.41 | 1.7 | 2.6 | 0.3 | 2.45 |
| 31.10.2012 | 305 | Winter | SW | 30, 00 | 33.69 | 1.4 | 0.05 | 27.08 | 0.07 | 2.03 | 0.14 | 1.24 |
| 29.11.2012 | 334 | Winter | IW | 30, 23 | 34.39 | 1.8 | BD | 27.62 | 0 | 5.42 | 0.45 | 3.03 |
Environmental data are given for samples collected at a depth of 25 m. Salinity, temperature, fluorescence, and density were measured using a SAIV CTD probe.
Identified according to references 36 and 54. LW, local water; TAW, transformed Atlantic water; IW, intermediate water; SW, surface water.
Number of PCR cycles utilized to prepare the DNA and RNA libraries.
BD, below detection.
Measured using a flat sensor (Li-Cor).
Originally reported by Kubiszyn et al. (submitted).
At each sampling date, physical environmental data from a vertical conductivity-temperature-depth (CTD) profiler were obtained using an SD204 CTD probe (SAIV A/S, Bergen, Norway) equipped with a Seapoint fluorescence sensor or the R/V Helmer Hanssen ship CTD (Sealogger CTD system [Sea-Bird Electronics] equipped with a fluorometer from Seapoint Sensors, Inc.). Light measurements were collected with a flat PAR (photosynthetically active radiation) sensor and an LI-1000 DataLogger device (Li-Cor, Inc., Lincoln, NE) from Julian day (JD) 47 onward in 2012. The different seasons at IsA were characterized predominantly by chlorophyll a biomass estimates and day length; in addition, meroplankton (grazer) abundance was considered in accordance with the work of Stübner et al. (86). The seasons identified were winter (December, January, February, October, November), early spring (March, start of April), spring (mid-April to the end of May), and summer/autumn (June to September) (Table 1). The summer and autumn samples were combined, because sampling was less frequent in these periods.
Seawater was sampled using Niskin bottles (KC Denmark) at four standard depths (5 m, 15 m, 25 m, and 60 m). Only the 25-m depth was sampled for DNA or RNA analyses. Sampling was performed as close to noon (local time) as practically possible. Immediately after collection, water for DNA and RNA extraction (usually 4 liters for each depth) was prefiltered by gravity through a 10-μm nylon mesh (KC Denmark). This process was completed in 5 to 20 min (the latter during peak bloom). Organisms were collected on 0.45-μm Durapore filters (Millipore, USA) using vacuum filtration, either in the field (RNA) or upon return to the laboratory. All filters were snap-frozen and were stored at −80°C until further analysis (see reference 40). Seawater from the same depths was used for measurement of nutrients (silicate, phosphate, nitrate, and nitrite [Table 1] [values reported in the work of A. M. Kubiszyn, J. M. Wiktor, J. M. Wiktor, Jr., C. Griffiths, S. Kristiansen, and T. M. Gabrielsen {submitted for publication}]), particulate organic carbon (POC) and nitrogen (PON) (values from seven cruises reported in reference 41), and fractionated chlorophyll a (Chl a) biomass (biomass of cells >10 μm and total Chl a [the latter reported by Stübner et al. {86}]). The Chl a biomass was quantified from triplicate subsamples of 200 to 500 ml of seawater from every depth filtered onto GF/F glass microfiber filters (Whatman, England) or 10-μm Isopore membrane polycarbonate filters (Millipore, USA). Filters were either stored at −80°C until further analysis (within the next 9 months) or immediately extracted in 10 ml of methanol. Chlorophyll was extracted in darkness at 4°C for 20 to 24 h according to the method of Holm-Hansen and Riemann (42), and Chl a biomass was measured with a 10-AU-005-CE fluorometer (Turner, USA) calibrated with Chl a (S6144; Sigma).
DNA/RNA isolation and 454 amplicon preparation.
DNA was extracted from 27 samples, while RNA was extracted from 10 of the same samples (Table 1). DNA and RNA were extracted from environmental samples, and PCRs were carried out, according to reference 40. Briefly, DNA was extracted using the DNeasy Plant minikit (Qiagen, USA) according to the manufacturer's protocol, except that a bead-beating step using a Retsch MM400 bead beater (2 times for 1 min each time at a frequency of 1/22 s) (40) was included. RNA was extracted with the RNAqueous kit (Ambion), also with a bead-beating step included. All RNA samples were DNase treated using the Turbo DNA-free kit (Ambion) and were transcribed into cDNA using Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT) and random decamer primers (both from Ambion). Negative controls (water) were included during all extractions and PCR amplifications. For all RNA preparations, PCRs were also run using DNase-treated RNA as the template (no-RT controls) to make sure that the amplification products were not due to residual DNA.
The V4 variable region of the nuclear 18S rDNA was amplified using the universal eukaryotic primers designed by Comeau et al. (24). PCRs were carried out in a total volume of 25 μl containing 1× high-fidelity (HF) buffer, 0.2 μM each deoxynucleoside triphosphate (dNTP), 0.2 μM each primer, 0.5 U Phusion Hot Start II HF DNA polymerase, and 1 μl undiluted environmental DNA or RNA. Reactions were run on an Eppendorf Mastercyler (ep gradient S) instrument. Cycling conditions were as follows: 98°C for 30 s; 18, 23, or 30 cycles of 98°C for 10 s, 55°C for 30 s, and 72°C for 30 s; and 72°C for 5 min (Table 1). The reaction product was then stored at 4°C. Products of independent triplicate PCRs were mixed, purified using AMPure XP beads at a bead/PCR product ratio of 4:5, and quantified spectrophotometrically (NanoDrop ND-2000 spectrophotometer; Thermo Scientific) prior to pooling at equimolar ratios. Six to 10 samples were pooled for each 1/8 lane of the 3 sequencing plates prepared (see Table S1 in the supplemental material). Sequencing was performed on a Roche 454 GS-FLX Titanium platform at IBIS, Plate-forme d'Analyses Génomiques de l'Université Laval.
Data analyses and statistics.
The amplicon data were analyzed with MacQIIME (version 1.7.0) (43). Quality filtering and demultiplexing were done according to the following parameters: sequence length, between 350 and 550 bp; number of ambiguous bases, 0; maximum homopolymer length, 7; bar code type, 10; minimum average quality score, 30.
Chimera checking was performed in Mothur (version 1.30.0) (44) on individual samples using the UCHIME algorithm (45) in a de novo setup. Clustering into operational taxonomic units (OTUs) was done in MacQIIME using the UCLUST algorithm (46) at a similarity level of 97%. The most abundant sequence was chosen as the representative sequence of the cluster. Global singletons were removed from the data on the assumption that they were sequencing errors (47, 48). Information on pyrosequencing raw data, quality filtering, and postfiltering is given in Table S1 in the supplemental material. A BLASTN search (E value, 0.00001) (49) for the representative sequences was run on a curated reference database of Arctic marine eukaryotes (24) and on the PR2 protist database (50–52). Taxonomy was assigned based on agreement between the two databases and a sequence identity above 98%. The DNA sample from JD 129 was dominated by one mold OTU (Eurotium sp.; 98.8% sequence identity; GenBank accession no. AB002076.1), which made up 89% of all fungus reads on that day, in contrast to the JD 128 and 130 results, and was therefore removed from the analyses on the assumption that this sample was contaminated.
OTUs with hits to Metazoa (ca. 6% of total reads [see Table S1 in the supplemental material]) were removed prior to normalization to an even sequencing depth (5,550 reads). Accumulation curves and diversity estimates according to the Chao index were done in MacQIIME. Other indices (species richness, Shannon-Wiener diversity, and Pielou's evenness), as well as statistical tests and multivariate analyses, were calculated in R (R.app GUI 1.63; S. Urbanek and H.-J. Bibiko, 2012). Tables and plots were made in Excel (version 14.4.3, 2010; Microsoft Corporation) and R. The taxon histograms presented were grouped according to taxonomy, with groups corresponding to phyla as well as families. The same plot was also generated using the number of OTUs within a group (presence/absence) instead of the read abundance, and both plots showed the same overall pattern (data not shown). The sampling map and ocean data plots were created in MATLAB (release 2013b; MathWorks, Inc.).
A global one-way-ANOSIM (an alysis o f sim ilarities) (53) was computed in R (with 999 permutations and the Bray-Curtis distance matrix) to test whether there were significant differences between two or more groups of sampling units using log10-transformed (DNA and RNA) read abundance data. The test was conducted to look for differences between seasons (factors were winter, early spring, spring, and summer/autumn), water mass (factors were local water [LW], surface water [SW], TAW, and intermediate water [IW], according to references 36 and 54), and type (factors were DNA and RNA). To detect differences between types (DNA and RNA community composition), only the 10 days when both RNA and DNA samples were available (Table 1) were used.
Detrended correspondence analyses (DCA) (55) with default settings and the Bray-Curtis distance matrix were performed to identify similarities between sampling dates with respect to changes in the community composition. The data set (number of reads for each OTU) was log10(n + 1)-transformed to downweight the influence of highly abundant OTUs. The same analysis performed with raw data or unweighted data (presence/absence) showed the same overall trends (data not shown). The DCA were created with the vegan package in R (56), and different environmental parameters were related to the observed structure of the community composition using the envfit function. Environmental parameters (abiotic and biotic) were tested for statistical dependence using the Spearman rank correlation coefficient (see Table S3 in the supplemental material), and a selection of highly covarying parameters was excluded from the DCA. Kendall's tau coefficient, calculated in R, was used to determine linear relationships within DNA and RNA diversity.
Nucleotide sequence accession numbers.
The sequences of the representative OTUs determined in this study have been submitted to GenBank (accession no. KT810188 to KT818498).
RESULTS
The Isfjorden-Adventfjorden (IsA) system. (i) Hydrography.
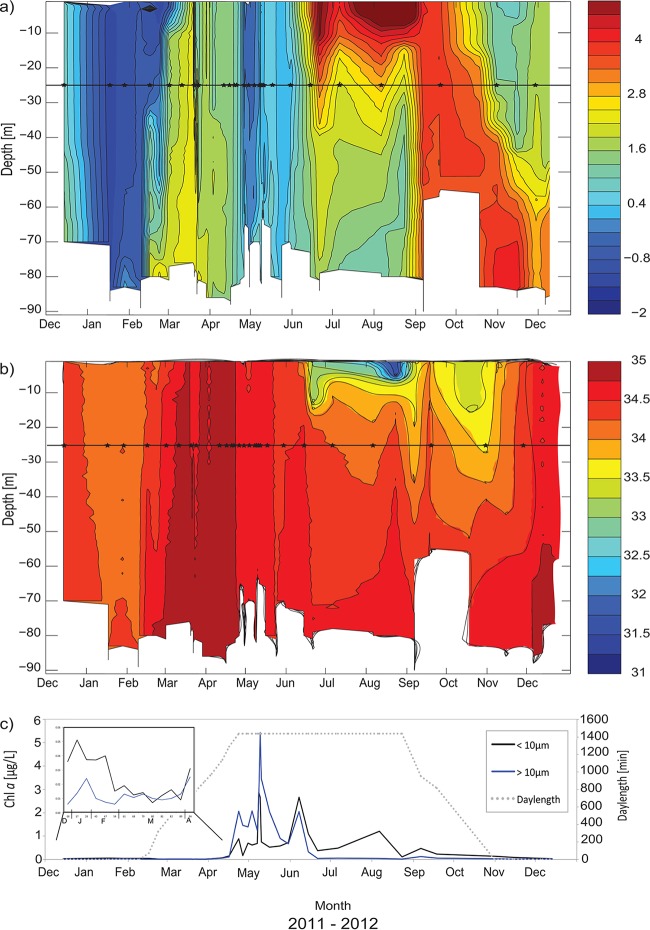
Hovmøller diagrams were prepared based on seawater temperature (T) and salinity (S) data extracted from the CTD profiles collected at the IsA station from December 2011 to December 2012 (Fig. 2). The water masses were characterized according to references 36 and 54 (Table 1). The water column was well mixed from December 2011 until mid-February 2012, consisting of local water (LW) (T < 1°C) (Fig. 2). An incursion of warmer and more saline water was identified at the bottom in late January, spread throughout the water column in February, and reached the 25-m sampling depth in mid-March (on Julian Day [JD] 79; temperature, 2.4°C; salinity, 34.9) (see Table 1 for an overview of Julian days), changing the water mass to TAW (T > 1°C; S > 34.7). The water column cooled to below 1°C in mid-April and was again characterized as LW at the 25-m sampling depth until the end of May, when surface heating started (data from the Svalbard Airport weather station). The surface salinity decreased due to freshwater input from melting snow and ice on land, and the water column was characterized as intermediate water (IW) in June (34 < S > 34.4). The upper mixed layer identified in the stratified summer water column never reached the sampling depth of 25 m (Fig. 2). In early September, and again in October, the whole water column was warm (above 4°C), and freshwater input impacted the water column down to 30 m. In November and December, warm and saline AW entered the fjord below 60 m, while the upper water column had a temperature of 0.5 to 2.5°C, with salinity from 34.5 to 35. The whole water column in December 2012 was warmer than in December 2011.
FIG 2.
Development of the water column at the IsA station from December 2011 to December 2012. (a and b) Hovmøller diagrams of seawater temperature (in degrees Celsius) (a) and salinity (b) based on CTD measurements. The horizontal line in each diagram indicates a sampling depth of 25 m, and the stars along the line mark the sampling dates. (c) Day length (provided by www.solartopo.com/daylength.so) and measured chlorophyll a (Chl a) concentrations in two size fractions (smaller than 10 μm and larger than 10 μm; measurements are from the 25-m sampling depth). (Inset) Enlargement of the period from the end of 2011 until mid-April 2012. Fractionated (>10 μm and 10 to 0.7 μm) and total Chl a concentrations at IsA at the 25-m depth are given in Table S4 in the supplemental material.
(ii) Light and chlorophyll a (Chl a).
The extreme light climate of Svalbard was represented by the day length (Fig. 2c) in all analyses. During the sampling period, the sun was below the horizon from the end of October 2011 until mid-February 2012 (from JD 300 in 2011 to JD 47 in 2012); there was midnight sun from mid-April to the end of August (JD 110 to 237 in 2012) and again no sun from the end of October (JD 301 in 2012). The level of photosynthetically active radiation (PAR) was very low at the 25-m depth for most of the year 2012 (Table 1).
Chl a biomass was detectable throughout the year, even during the dark winter months of December and January, when low values were found (e.g., the concentration of total Chl a was 0.04 μg liter−1 at JD −18) (Fig. 2c; see also Table S4 in the supplemental material). From December to the beginning of April, the Chl a biomass was dominated by the small-size fraction (<10 μm), which contributed, e.g., 84% of the total Chl a biomass on JD 47 (see Table S4). The concentrations of both size fractions of Chl a started to increase from the end of March (JD 89), when the day length was 895 min. At the end of April (JD 114), the spring bloom started, and the larger cells dominated the phototrophic biomass (70% of total Chl a). The spring bloom peak was observed in the first half of May (JD 130) (Fig. 2c), with total-Chl a concentrations of 8.1 μg liter−1 (see Table S4). At this time, the small-size fraction made up 33% of the total Chl a biomass. From the end of May, the contribution of the large cells decreased again, and the pico- and nanoflagellates(<10 μm) dominated, contributing 56 to 97% of the total Chl a biomass during several small Chl a peaks in June (JD 159), August (JD 219), and September (JD 240).
The microbial eukaryotic community at IsA. (i) Diversity.
Sequencing depths differed on different sampling dates (see Fig. S1 in the supplemental material). In general, rarefaction curves indicated that samples had not been sequenced to saturation, but some of the samples, especially those from April and May, approached a plateau phase (see Fig. S1 and Table S1 in the supplemental material). Taxonomic identities could be assigned to 4,312 of 5,006 total OTUs (operational taxonomic units) (see Table S1), with 3,664 OTUs detected in the 26 DNA libraries and 2,039 in the 10 RNA libraries.
Shannon-Wiener diversity (H′) and Pielou's evenness (J′), calculated for DNA libraries, showed that these two indices were linearly related (see Fig. S2a and Table S2 in the supplemental material) (Kendall's tau [τ]= 0.9; P < 0.05). A similar trend was observed between species richness (number of OTUs) and Pielou's evenness (see Fig. S2b in the supplemental material) (Kendall's tau = 0.7; P < 0.05). Diversity and evenness were high and fairly stable during the winter and early spring but started to decrease in mid-April (JD 107). From the end of May, both indices increased again, so that the values at the end of the year were comparable to those observed at its beginning (e.g., on JD 334, H′DNA was 4.9 and J′DNA was 0.74). The highest DNA diversity and species richness were measured on JD 61, with an H′ of 5.21 and a J′ of 0.79 (742 OTUs), and the lowest diversity and species richness on JD 128, with an H′ of 2.15 and a J′ of 0.4 (229 OTUs) (see Table S2). The RNA diversity and evenness indices displayed patterns similar (τ = 0.8; P < 0.05) to those for the DNA indices (see Table S2). The highest RNA diversity and species richness were found in January (on JD 17, H′RNA was 4.82 and J′RNA was 0.75, for 610 OTUs) and the lowest in September (on JD 262, H′RNA was 3.4 and J′RNA was 0.61, for 261 OTUs) (see Table S2).
(ii) Community composition.
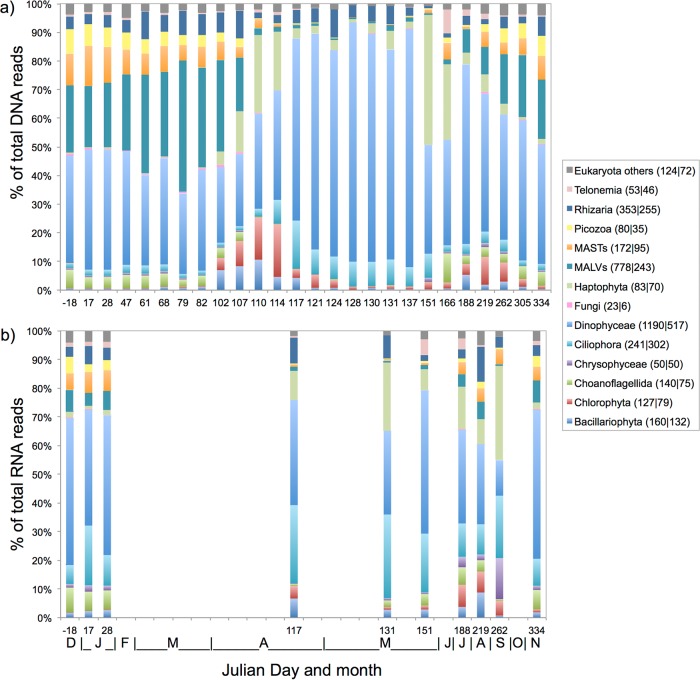
Reads with matches to Dinophyceae dominated DNA and RNA libraries in terms of both read numbers (30 to 80% of total reads) and OTU numbers (1,190 of 3,664 total OTUs in the DNA libraries) throughout the year and were particularly abundant in the spring (Fig. 3). OTUs assigned to Gyrodinium helveticum (16% of total reads; 35% of total Dinophyceae reads), Gyrodinium fusiforme (10% of total reads; 19% of total Dinophyceae reads), and an unknown Dinophyceae strain (strain 28) (4% of total reads; 8% of total Dinophyceae reads) were highly abundant over time within both the DNA and RNA libraries (Table 2). The marine alveolates (MALVs) also accounted for many reads (10 to 40% of total reads on different sampling dates) and OTUs (778 OTUs within the combined DNA libraries) in both the DNA and RNA libraries, particularly during the winter and early spring. The marine stramenopiles (MASTs), as well as the newly established phylum Picozoa (57), showed a temporal trend similar to that of the MALVs, predominating in the winter and early spring (Fig. 3). The relative abundances of MAST and Picozoa reads were similar in DNA and RNA samples from the same date; MASTs had a higher OTU richness (172 OTUs in DNA libraries) than Picozoa (80 OTUs in DNA libraries). Within the MASTs, one OTU belonging to MAST subclade 1a (Table 2) was very abundant, recruiting 47% of all MAST reads over time. The Picozoa community was dominated by three different OTUs that accounted for 77% of all Picozoa reads over time.
FIG 3.
Relative OTU read abundances of dominant taxonomic groups within the DNA (a) and RNA (b) libraries from December 2011 (Julian day −18) to November 2012 (Julian day 334). Months are indicated by the first letters of their names at the bottom of panel b. OTUs that did not belong to any of the groups listed or that had <100 reads in every sample were grouped in the “Eukaryota others” category. The numbers of OTUs assigned to each taxonomic group are given in parentheses in the key (DNA|RNA).
TABLE 2.
Abundant OTUs (>1% of read abundance in total DNA or RNA data set)
| Assigned OTU name | Sequence identity (%) | Group | % of total reads (2011–2012) | GenBank accession no. | Size (bp) |
|---|---|---|---|---|---|
| DNA OTUs | |||||
| Gyrodinium_helveticum | 99.29a | Dinophyceae | 16 | AB120004.1 | |
| Gyrodinium_fusiforme | 98.81a | Dinophyceae | 10 | AB120002.1 | |
| Phaeocystis+sp | 99.52b | Haptophyta | 6 | KC488454.1 | 1,667 |
| Dinophyceae_XXX+sp._strain28 | 99.52b | Dinophyceae | 4 | FJ000254.1 | 1,382 |
| MAST_1a;DH22_2A47 | 99.53a | MAST | 2 | FJ032662.1 | |
| Dino-Group-II-Clade-7_X+spc | 99.52b | MALV | 2 | JQ956301.1 | 1,704 |
| Micromonas_CCMP2099_Arctic | 98.8a | Chlorophyta | 2 | DQ025753.1 | |
| Dino-Group-I-Clade-1_X+spd | 99.76b | MALV | 2 | KF031743.1 | 1,755 |
| Strombidiidae_X+sp._strain37 | 100b | Ciliophora | 1 | HQ867429.1 | 866 |
| Dinophyceae_XXX+sp | 99.29b | Dinophyceae | 1 | JF698775.1 | 1,643 |
| Dino_clone_North_Pole_SW0_72 | 100a | Dinophyceae | 1 | JF826353.1 | |
| Glenodinium | 98.34a | Dinophyceae | 1 | EF058237.1 | |
| Picobiliphyta_XXXX+sp._strain5 | 99.52b | Picozoa | 1 | HQ869692.1 | 905 |
| Choreotrichia-1_X+sp | 100b | Ciliophora | 1 | HM561031.1 | 949 |
| RNA OTUs | |||||
| Gyrodinium_helveticum | 99.29a | Dinophyceae | 12 | AB120004.1 | |
| Phaeocystis+sp | 99.52b | Haptophyta | 7 | KC488454.1 | 1,667 |
| Dinophyceae_XXX+sp._strain28 | 99.52b | Dinophyceae | 7 | FJ000254.1 | 1,382 |
| Gyrodinium_fusiforme | 98.81a | Dinophyceae | 4 | AB120002.1 | |
| Choreotrichia-1_X+sp | 100b | Ciliophora | 4 | HM561031.1 | 949 |
| Strombidiidae_X+sp._strain37 | 100b | Ciliophora | 2 | HQ867429.1 | 866 |
| Syracosphaera | 99.52b | Haptophyta | 2 | AM490987.2 | |
| Dinophyceae_XXX+sp | 99.29b | Dinophyceae | 2 | JF698775.1 | 1,643 |
| Dino_clone_North_Pole_SW0_72 | 100a | Dinophyceae | 2 | JF826353.1 | |
| Dino-Group-III_XX+spe | 100b | MALV | 1 | FJ431851.1 | 820 |
| Micromonas_CCMP2099_Arctic | 98.8a | Chlorophyta | 1 | DQ025753.1 |
Bacillariophyta reads in DNA samples were persistent (<10% [Fig. 3a]) throughout the year, including the dark winter months. Read numbers increased from mid-April (JD 102) and were stable until the end of April. A slight increase was again observed during summer/autumn (JD 188 to 305). The RNA samples showed a similar pattern (Fig. 3b), except that the relative abundance of diatom reads was higher than that for DNA, especially during the winter (JD −18 to 28 and JD 334). Reads assigned to Haptophyta were found throughout the year in both the DNA and RNA libraries, and a Phaeocystis sp. was the most abundant OTU (∼85% of all Haptophyta reads). Haptophyta read numbers started to increase in April (JD 102) and peaked in late May in the DNA libraries (44% of total reads on JD 151) and in autumn in the RNA libraries (35% of total reads on JD 262). During the peak spring bloom period, Haptophyta contributed 5 to 10% of the total DNA reads.
Chlorophyta reads were abundant (Fig. 3a) in mid-April (JD 110 and 114), during the early phase of the spring bloom. In general, chlorophytes displayed high read numbers from early spring (mid-April) to the end of the year. From December 2011 to mid-March 2012, read numbers were very low (<1% of total DNA reads). More than 60% of the Chlorophyta reads, from both the DNA and RNA libraries, were assigned to the Arctic strain of Micromonas pusilla (referred to below as the Arctic Micromonas ecotype) (58) with 99% sequence identity (Table 2).
The phylum Cercozoa accounted for many of the Rhizaria OTUs, and one OTU assigned to the genus Cryothecomonas was especially abundant in the DNA samples. Rhizaria displayed high OTU richness in both the DNA (353 OTUs) and RNA (255 OTUs) libraries. Members of the phylum Telonemia also occurred at the IsA station (1 to 10% per sample [Fig. 3]), with maximum relative read numbers (∼10%) in late May (JD 151) in both the DNA and RNA libraries. Within the phylum Ciliophora, two OTUs were especially common (Table 2), accounting for 51% of all Ciliophora reads over time. Ciliophora reads were present throughout the year but were in general more abundant in the spring, when they made up 10 to 20% of the reads within the DNA libraries (Fig. 3a). Ciliophora read abundances were always higher in the RNA libraries than in the DNA libraries (Fig. 3b).
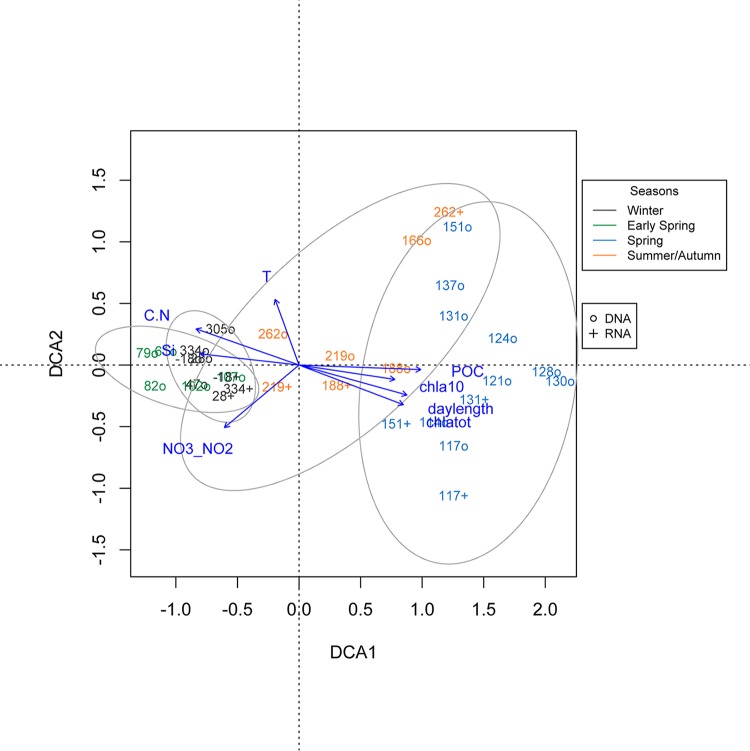
Ordination analysis clustered the IsA samples according to seasons (Fig. 4). The one-way ANOSIM used to test for similarities between types of samples (DNA and RNA) indicated no overall significant differences in the changes in community composition identified throughout the year between the DNA and RNA libraries. A major shift in community composition occurred in mid-April (between JD 107 and JD 110), corresponding to the transition from early spring to spring and the onset of the spring bloom. A tight cluster was formed by the winter and early-spring samples. This cluster also included the winter samples from 2012 (JD 305 and 334). The spring samples formed another cluster, albeit with much larger internal compositional differences, a pattern also observed with the relative read abundances (Fig. 3 and 4). Although the summer/autumn samples formed an intermediate, loose cluster, distances inside this cluster were large, in terms of both sampling dates and sample type (DNA/RNA). A shift in community composition occurred within the summer/autumn period (between JD 166 and JD 188).
FIG 4.
Detrended correspondence analysis for samples on the basis of read abundances. The read numbers were log10(n + 1) transformed. Only a selection of the significant environmental parameters (vectors) (see Table S5 in the supplemental material) is shown. Overlapping clusters show the 0.95 confidence limits of the season grouping. Julian days 17, 68, and 110 were removed from the ordination analyses due to missing environmental data. The different seasons are color coded as shown in the key. o, samples from DNA libraries; +, samples from RNA libraries. C.N, C/N ratio.
Chlorophyll a, day length, and POC were very pronounced factors influencing the axis along which spring and summer sampling dates occurred, while nutrients (NO3/NO2, Si), the C/N ratio, and, to a lesser degree, temperature (see Table S5 in the supplemental material) correlated with the opposite axis, suggesting an influence on the winter and early-spring samples. In agreement with the DCA results (see Table S5), a one-way ANOSIM test indicated significant differences in community composition with respect to water mass (R = 0.17; P = 0.012) as well as seasons (R = 0.6831; P = 0.001).
(iii) DNA versus RNA.
Of the 3,664 OTUs detected in the 26 DNA libraries, only 14 OTUs were identified as abundant (i.e., present at >1% throughout the whole year in the total data set) (cf. reference 59) (Table 2). For the 10 RNA libraries (2,039 OTUs in total), only 11 OTUs were abundant throughout the year (Table 2). Nine of the most abundant OTUs were shared by the DNA and RNA libraries (Table 2), while two OTUs, assigned to a Syracosphaera sp. and MALV III, were abundant only within the RNA libraries.
In total, 3,159 OTUs were detected on the 10 sampling dates when both DNA and RNA libraries were available. Of these, 1,191 were shared between the RNA and DNA libraries, while 1,120 and 848 were unique to the DNA and RNA libraries, respectively. For certain taxa, the number of OTUs differed greatly between the RNA and DNA libraries (see Fig. S3 in the supplemental material). Dinophyceae and MALVs had far more OTUs in the DNA libraries, while Bacillariophyceae, Cercozoa, and Ciliophora had more OTUs within the RNA libraries. In general, OTUs that were not shared between the DNA and RNA libraries were low in read abundance (on average, 2 reads per OTU).
DISCUSSION
Stable and diverse winter/early-spring communities of microbial eukaryotes.
The predominantly heterotrophic winter communities of microbial eukaryotes (0.45 to 10 μm) identified at the IsA station in 2011 and 2012 were highly diverse and even (see Table S2 and Fig. S2 in the supplemental material) and represented the main heterotrophic groups commonly found in Arctic waters (cf. reference 30). In the absence of photosynthetic activity, a baseline level of heterotrophy is sustained in Arctic waters even during the polar night period (reviewed in reference 12). Chloroplast-bearing cells of the phyla Haptophyta, Bacillariophyta, and Chlorophyta were also detected in the winter RNA libraries (Fig. 3), suggesting that these cells are present in the water column and maintain some level of activity, possibly in a vegetative stage or as resting spores (cf. references 13, 15, and 40). Diatom cells present in Kongsfjorden during the polar night period showed a rapid response to light levels as low as 0.1 to 1 μmol photons s−1 m2 (12). Thus, phototrophs seem to exist in the water column as active cells to a larger degree than previously recognized. Furthermore, mixotrophy may facilitate survival during the dark winter period, as suggested for phototrophic flagellates in high-latitude lakes (60, 61). The Arctic Micromonas ecotype, which is dominant and widespread in Arctic waters (58, 62–64), was recently shown to be capable of phagotrophy (65). Living cells of the Arctic Micromonas ecotype have been detected around Spitsbergen even during the polar night (15, 40).
Winter-spring transition and bloom dynamics: low diversity and evenness.
The transition from early spring to spring (Fig. 3), with a longer day and increasing Chl a biomass, coincided with a shift in water mass from TAW to LW at the IsA station (Fig. 2). At this time, the microbial eukaryotic community changed into a more typical Arctic spring bloom community dominated by a Phaeocystis sp. (Fig. 3). This is the only period when considerable numbers of Bacillariophyta reads were found in the 0.45- to 10-μm fraction. The abundance and richness of diatoms may be underestimated in our study due to the difficulty of breaking the diatom silicate frustules prior to DNA extraction (cf. reference 66), although the diatom spring bloom taxa are often in the nano- and microsize ranges (16). The read numbers of common winter microbial eukaryotes, in particular the parasitic MALVs (67), were strongly reduced at the initiation of the spring bloom. Possibly, their host cells became less abundant during the spring bloom, or the change in water mass characteristics influenced the viability of the MALV cells or their hosts. Whereas the Dinophyceae reads were a prominent part of the DNA libraries during the spring, the opposite trend was seen in the RNA libraries (Fig. 3), suggesting a less active role for dinoflagellates during the spring bloom. A similar trend was seen for the Phaeocystis sp. in the late-bloom phase (JD 151), when the RNA library showed a much lower relative read abundance than the DNA library (Fig. 3), indicating that Phaeocystis sp. cells were fading at this time, when the nitrate concentration was below detection (Table 1).
Alternatively, these trends could be related to the ability of heterotrophs to fill more ecological niches than autotrophs, as hypothesized by Vaulot et al. (68), or to higher grazing pressure during winter, which would lead to a top-down regulation that may increase diversity. Higher microbial diversity in the winter has also been reported from other Arctic and more-southern marine environments (69–71) and may be of biological significance. The reduced diversity and evenness of the microbial eukaryotic communities at the end of spring and in summer/autumn relative to winter/early spring may also be a sampling effect due to increased cell numbers (e.g., 12 × 106 cells liter−1 found in Kongsfjorden in April 2006 [72]) and the complete dominance of a few taxa during the spring bloom (see, e.g., references 73 and 74), in contrast to the dilute winter communities (12). In addition, read abundances may not reflect organismal abundances, due to inherent technical issues with amplicon sequences as well as to the numbers of rDNA copies present in different organisms (75, 76). Interpreting our data with these limitations in mind, and with the data available from microscopic analyses of the same samples (Kubiszyn et al., submitted), we find that the patterns described above are likely to reflect true biological changes at the transition to the spring bloom.
Postspring phase (summer and autumn).
Arctic summer pelagic communities are often dominated by pico-and nanosized flagellates as well as ciliates (63, 77, 78). Prominent members of the summer communities in the 0.45- to 10-μm fraction at IsA were Dinophyceae, Haptophyta, and Choanoflagellida (Fig. 3). In addition, heterotrophic unicellular flagellates of the phylum Telonemia (79–81) increased in relative abundance during this period. Members of the phylum Telonemia have been reported in freshwater (82, 83) and in Svalbard (e.g., in the Bayelva River [84]) previously. Although the higher contribution of Telonemia members in the summer/autumn at IsA may have originated from the two large rivers adjoining Adventfjorden, as hypothesized for Telonemia members in Kongsfjorden (85), their presence during winter suggests that marine Telonemia taxa were also present. As found by earlier studies from Arctic seas (the Canadian Arctic [reference 58 and references therein], the Fram Strait [4], and Svalbard [63]), the Arctic Micromonas strain showed higher relative abundance during the pre- and postbloom phases at IsA (Fig. 3).
A second major shift in the community composition of microbial eukaryotes was observed between July and August with a relative increase in the abundance of MALV II, MASTs, Chlorophyceae, Cryptophyceae, and Picozoa. No obvious drivers in the physical environment were identified, although the concentrations of Si, POC, PON, and chlorophyll a (total and small fraction) (Table 1; see also Table S4 in the supplemental material) increased slightly. Possibly, intrinsic factors lead to successions in the microbial eukaryote community. A concurrent shift was identified in the IsA meroplankton community, where a summer dominance of bivalve larvae changed into a more-even zooplankton composition with fewer mero- and holoplankton individuals (86). Thus, it is possible that diminished grazing pressure led to the shift in the microbial eukaryotic communities. Other potentially relevant explanatory parameters, i.e., bacterial production and growth, were not measured in this study. Finally, the reduced sampling frequency in the summer and fall (monthly from June to November) and the sampling at only one depth (25 m) in a period when the water column was stratified may have contributed to our inability to accurately identify the drivers for the major community shift in July/August. After August, a microbial eukaryotic winter community similar to that of 2011–2012 seemed to reestablish itself, even though the hydrographic conditions differed from those of the previous year. Such an annual cycling of the microbial community was identified during a 6-year study of the bacterial community of the Western English Channel (87). Perennial time series data are needed in order to further evaluate the stability of seasonal succession as well as its main biotic and abiotic drivers at the IsA station (see reference 88). Multiple sampling depths covering the whole water column will facilitate a more in-depth interpretation of succession patterns and variability.
DNA versus RNA.
The relatively high abundance of a MAST 1a OTU identified in the DNA libraries (Table 2) and not in the RNA libraries may reflect procedural biases, as discussed in reference 89, rather than a high abundance of the MAST 1a OTU. The higher abundances of Dinophyceae and MALVs in the DNA libraries than in the RNA libraries are probably due to the higher rDNA copy numbers found in these organisms than in other flagellates (66, 75, 76). Interestingly, although ciliates have extremely high rDNA copy numbers (76), they are more abundant and species rich in our RNA libraries than in our DNA libraries (Fig. 3; see also Fig. S3 in the supplemental material), suggesting that ciliates represent an active fraction of the microbial eukaryotic community at IsA, as was also found by Stecher et al. (90) for Western Arctic sea ice communities. Ciliates of the oligotrich family Strombidiidae and the spirotrich subclass Choreotrichia were the main representatives in our libraries. Kleptoplasty (the process of ingesting and using organelles of another organism) (91) is known for different ciliates of the family Strombidiidae (92, 93); they are able to switch from heterotrophy to a phototrophic mode by using ingested chloroplasts, and they may thus remain active under both light and dark conditions. The filter size used in this study should remove most of the Ciliophora described, since these are generally larger than 10 μm (94). Ciliates are, however, commonly detected on small-pore-sized filters (14, 63), probably because they have flexible cells that are able to pass through (89). The high OTU richness identified among the Alveolata could result from a lack of homogenization of their rDNA gene copies (95), although we believe that we have minimized the effects of intracellular variability, PCR errors, and sequencing errors by using a 97% cutoff during clustering.
Dinophyceae dominated at the IsA station with their high diversity, activity, and abundance; OTUs assigned to Gyrodinium fusiforme and Gyrodinium helveticum (96) were the most common throughout the year. These two taxa are commonly found in investigations in other Arctic regions, e.g., the Barents Sea, Canadian Arctic, Greenland Sea, and Central Arctic (23, 27, 97–99), as well as in regions outside the Arctic (62). Our results based on both DNA and RNA libraries show the usefulness of this combined approach, which potentially reduces the biases inherent in using only the rDNA gene (see also reference 90).
Perspectives.
In this 1-year study, changes in seasonal parameters and hydrography coincided during the spring bloom, and thus, the differential effects of the history of the water masses and the strong seasonality of the Arctic environment were difficult to separate. The extreme temporal variability in light, nutrients, and, to some degree, temperature strongly influenced the succession of the marine microbial eukaryotic communities (0.45 to 10 μm) at the IsA station. Seasonally influenced parameters, such as day length and nutrient availability in the euphotic zone, previously identified as main drivers in Arctic protist surveys (13–15, 27), showed strong correlations to community composition in the present study as well. Whereas generalist taxa (e.g., Gyrodinium fusiforme and G. helvecticum) persisted in high relative abundance throughout the year, seemingly regardless of changes in the habitat, potential specialist taxa (e.g., MALVs in our study) may have been more strongly influenced by different hydrographic conditions (cf. references 89 and 100) and thus fluctuated in Adventfjorden during our annual study. Further studies, including interannual comparisons, should enable a more thorough understanding of this dynamic system.
Supplementary Material
ACKNOWLEDGMENTS
We thank the UNIS logistics department, especially Lars F. Stangeland, and the crews of R/V Helmer Hanssen (UiT) and M/S FARM for valuable help during field sampling. Thanks to Stuart Thomson, Prasad Rao, and the UNIS AB202 students of 2012 for excellent support in the field. We appreciate the help of Stuart Thomson, Courtney D. Nadeau, and Lilith Kuckero in the laboratory. Special thanks to Marie L. Davey for instruction in next-generation sequencing analyses. Thanks to Anna Kubiszyn for making IsA data on microplankton communities available prior to publication. Special thanks to Ragnheid Skogseth for providing oceanography plots and the sampling map, and for discussions about the Adventfjorden system, and to Connie Lovejoy for providing the protist database and for discussions at early stages of this project. We thank NOTUR and Lifeportal (UiO) for making computing resources available, and we thank the three anonymous reviewers for valuable input that significantly improved the manuscript.
Funding Statement
This project is part of the MicroFun project, funded by ConocoPhillips and Lundin Petroleum via their Northern Area Program. Fieldwork was supported by the Svalbard Science Forum (grant 219843/E10). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03208-15.
REFERENCES
- 1.Falkowski PG, Barber RT, Smetacek V. 1998. Biogeochemical controls and feedbacks on ocean primary production. Science 281:200–206. doi: 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- 2.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 3.Sherr EB, Sherr BF. 2007. Heterotrophic dinoflagellates: a significant component of microzooplankton biomass and major grazers of diatoms in the sea. Mar Ecol Prog Ser 352:187–197. doi: 10.3354/meps07161. [DOI] [Google Scholar]
- 4.Seuthe L, Iversen KR, Narcy F. 2011. Microbial processes in a high-latitude fjord (Kongsfjorden, Svalbard). II. Ciliates and dinoflagellates. Polar Biol 34:751–766. doi: 10.1007/s00300-010-0930-9. [DOI] [Google Scholar]
- 5.Fogg GE. 1995. Some comments on picoplankton and its importance in the pelagic ecosystem. Aquat Microb Ecol 9:33–39. doi: 10.3354/ame009033. [DOI] [Google Scholar]
- 6.Richardson TL, Jackson G. 2007. Small phytoplankton and carbon export from the surface ocean. Science 315:838–840. doi: 10.1126/science.1133471. [DOI] [PubMed] [Google Scholar]
- 7.Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmermann AE, Keeling PJ. 13 February 2015. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science doi: 10.1126/science.1257594. [DOI] [PubMed] [Google Scholar]
- 8.Li WKW. 1998. Annual average abundance of heterotrophic bacteria and Synechococcus in surface ocean waters. Limnol Oceanogr 43:1746–1753. doi: 10.4319/lo.1998.43.7.1746. [DOI] [Google Scholar]
- 9.Vincent WF. 2000. Cyanobacterial dominance in the polar regions, p 321–340. In Whitton B, Potts M (ed), The ecology of cyanobacteria: their diversity in time and space. Kluwer Academic Publishers, Dordrecht, the Netherlands. [Google Scholar]
- 10.Li WKW, McLaughlin F, Lovejoy C, Carmack EC. 2009. Smallest algae thrive as the Arctic Ocean freshens. Science 326:539. doi: 10.1126/science.1179798. [DOI] [PubMed] [Google Scholar]
- 11.Sieburth JM, Smetacek V, Lenz J. 1978. Pelagic ecosystem structure: heterotrophic compartments of the plankton and their relationship to plankton size fractions. Limnol Oceanogr 23:1256–1263. doi: 10.4319/lo.1978.23.6.1256. [DOI] [Google Scholar]
- 12.Berge J, Renaud PE, Darnis G, Cottier C, Last K, Gabrielsen TM, Johnsen G, Seuthe L, Weslawski JM, Leu E, Moline M, Nahrgang J, Søreide JE, Varpe Ø, Lønne OJ, Daase M, Falk-Petersen S. 2015. In the dark: a review of ecosystem processes during the Arctic polar night. Prog Oceanogr 139:258–271. doi: 10.1016/j.pocean.2015.08.005. [DOI] [Google Scholar]
- 13.Sherr EB, Sherr BF, Wheeler PA, Thompson K. 2003. Temporal and spatial variation in stocks of autotrophic and heterotrophic microbes in the upper water column of the central Arctic Ocean. Deep Sea Res Part I 50:557–571. doi: 10.1016/S0967-0637(03)00031-1. [DOI] [Google Scholar]
- 14.Mundy CJ, Barber DG, Michel C. 2005. Variability of snow and ice thermal, physical and optical properties pertinent to sea ice algae biomass during spring. J Mar Syst 58:107–120. doi: 10.1016/j.jmarsys.2005.07.003. [DOI] [Google Scholar]
- 15.Terrado R, Lovejoy C, Massana R, Vincent WF. 2008. Microbial food web responses to light and nutrients beneath the coastal Arctic Ocean sea ice during the winter-spring transition. J Mar Syst 74:964–977. doi: 10.1016/j.jmarsys.2007.11.001. [DOI] [Google Scholar]
- 16.von Quillfeldt CH. 2000. Common diatom species in arctic spring blooms: their distribution and abundance. Bot Mar 43:499–516. doi: 10.1515/BOT.2000.050. [DOI] [Google Scholar]
- 17.Sukhanova IN, Flint MV, Pautova LA, Stockwell DA, Grebmeier JM, Sergeeva VM. 2009. Phytoplankton of the western Arctic in the spring and summer of 2002: structure and seasonal changes. Deep Sea Res Part II 56:1223–1236. doi: 10.1016/j.dsr2.2008.12.030. [DOI] [Google Scholar]
- 18.Dünweber M, Swalethorp R, Kjellerup S, Nielsen TG, Arendt KE, Hjorth M, Tönnesson K, Møller EF. 2010. Succession and fate of the spring diatom bloom in Disko Bay, western Greenland. Mar Ecol Prog Ser 419:11–29. doi: 10.3354/meps08813. [DOI] [Google Scholar]
- 19.Díez B, Pedrós-Alió C, Massana R. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol 67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- 21.Moon-van der Staay SY, De Wachter R, Vaulot D. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- 22.Massana R, Pedrós-Alió C. 2008. Unveiling new microbial eukaryotes in the surface ocean. Curr Opin Microbiol 11:213–218. doi: 10.1016/j.mib.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Bachy C, López-García P, Vereshchaka A, Moreira D. 2011. Diversity and vertical distribution of microbial eukaryotes in the snow, sea ice and seawater near the North Pole at the end of the polar night. Front Microbiol 2:106. doi: 10.3389/fmicb.2011.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comeau AM, Li WKW, Tremblay JE, Carmack EC, Lovejoy C. 2011. Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS One 6:e27492. doi: 10.1371/journal.pone.0027492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piquet AM-T, van de Poll WH, Visser RJW, Wiencke C, Bolhuis H, Buma AGJ. 2014. Springtime phytoplankton dynamics in Arctic Krossfjorden and Kongsfjorden (Spitsbergen) as a function of glacier proximity. Biogeosciences 11:2263–2279. doi: 10.5194/bg-11-2263-2014. [DOI] [Google Scholar]
- 26.Monier A, Terrado R, Thaler M, Comeau A, Medrinal E, Lovejoy C. 2013. Upper Arctic Ocean water masses harbor distinct communities of heterotrophic flagellates. Biogeosciences 10:4273–4286. doi: 10.5194/bg-10-4273-2013. [DOI] [Google Scholar]
- 27.Kilias E, Kattner D, Wolf C, Frickenhaus S, Metfies K. 2014. A molecular survey of protist diversity through the central Arctic Ocean. Polar Biol 37:1271–1287. doi: 10.1007/s00300-014-1519-5. [DOI] [Google Scholar]
- 28.Barber RT. 2007. Picoplankton do some heavy lifting. Science 315:777. doi: 10.1126/science.1137438. [DOI] [PubMed] [Google Scholar]
- 29.Balzano S, Marie D, Gourvil P, Vaulot D. 2012. Composition of the summer photosynthetic pico and nanoplankton communities in the Beaufort Sea assessed by T-RFLP and sequences of the 18S rRNA gene from flow cytometry sorted samples. ISME J 6:1480–1498. doi: 10.1038/ismej.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovejoy C. 2014. Changing views of Arctic protists (marine microbial eukaryotes) in a changing Arctic. Acta Protozool 53:91–100. doi: 10.4467/16890027AP.14.009.1446. [DOI] [Google Scholar]
- 31.Tremblay J-É, Bélanger S, Barber DG, Asplin M, Martin J, Darnis G, Fortier L, Gratton Y, Link H, Archambault P, Williams WJ, Sallon A, Michel C, Philippe B, Gosselin M. 2011. Climate forcing multiplies biological productivity in the coastal Arctic Ocean. Geophys Res Lett 38:L18604. doi: 10.1029/2011GL048825. [DOI] [Google Scholar]
- 32.Hoegh-Guldberg O, Bruno JF. 2010. The impact of climate change on the world's marine ecosystems. Science 328:1523–1528. doi: 10.1126/science.1189930. [DOI] [PubMed] [Google Scholar]
- 33.Daufresne M, Lengfellner K, Sommer U. 2009. Global warming benefits the small in aquatic ecosystems. Proc Natl Acad Sci U S A 106:12788–12793. doi: 10.1073/pnas.0902080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morán XAG, López-Urrutia A, Calvo-Díaz A, Li WKW. 2010. Increasing importance of small phytoplankton in a warmer ocean. Glob Change Biol 16:1137–1144. doi: 10.1111/j.1365-2486.2009.01960.x. [DOI] [Google Scholar]
- 35.Ingvaldsen R, Loeng H. 2009. Physical oceanography, p 33–65. In Sakshaug E, Johnsen GH, Kovacs KM (ed), Ecosystem Barents Sea. Tapir Academic Press, Trondheim, Norway. [Google Scholar]
- 36.Nilsen F, Cottier F, Skoseth R, Mattson S. 2008. Fjord-shelf exchanges controlled by ice and brine production: the interannual variation of Atlantic water in Isfjorden, Svalbard. Cont Shelf Res 28:1838–1853. doi: 10.1016/j.csr.2008.04.015. [DOI] [Google Scholar]
- 37.Cottier F, Nilsen F, Inall M, Gerland S, Tverberg V, Svendsen H. 2007. Wintertime warming of an Arctic shelf in response to large-scale atmospheric circulation. Geophys Res Lett 34:L10607. doi: 10.1029/2007GL029948. [DOI] [Google Scholar]
- 38.Dobrzyn P, Keck A, Tatur A. 2005. Sedimentation of chlorophylls in an Arctic fjord under freshwater discharge. Hydrobiologia 532:1–8. doi: 10.1007/s10750-004-6420-8. [DOI] [Google Scholar]
- 39.Zajaczkowski M, Wlodarska-Kowalczuk M. 2007. Dynamic sedimentary environments of an Arctic glacier-fed river estuary (Adventfjorden, Svalbard). I. Flux, deposition, and sediment dynamics. Estuar Coast Shelf Sci 74:285–296. doi: 10.1016/j.ecss.2007.04.015. [DOI] [Google Scholar]
- 40.Vader A, Marquardt M, Meshram AR, Gabrielsen TM. 2014. Key Arctic phototrophs are widespread in the polar night. Polar Biol 38:13–21. doi: 10.1007/s00300-014-1570-2. [DOI] [Google Scholar]
- 41.Wiedmann I, Reigstad M, Marquardt M, Vader A, Gabrielsen TM. 2016. Seasonality of vertical flux and sinking particle characteristics in an ice-free high Arctic fjord—different from subarctic fjords? J Mar Syst Part B 154:192–205. doi: 10.1016/j.jmarsys.2015.10.003. [DOI] [Google Scholar]
- 42.Holm-Hansen O, Riemann B. 1978. Chlorophyll a determination: improvements in methodology. Oikos 30:438–447. doi: 10.2307/3543338. [DOI] [Google Scholar]
- 43.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 47.Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, Saar I, Bahram M, Bechem E, Chuyong G, Koljalg U. 2010. 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol 188:291–301. doi: 10.1111/j.1469-8137.2010.03373.x. [DOI] [PubMed] [Google Scholar]
- 48.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 49.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 50.Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chevenet F, Croce O, Hebrard M, Christen R, Berry V. 2010. ScripTree: scripting phylogenetic graphics. Bioinformatics 26:1125–1126. doi: 10.1093/bioinformatics/btq086. [DOI] [PubMed] [Google Scholar]
- 52.Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, Boutte C, Burgaud G, de Vargas C, Decelle J, del Campo J, Dolan JR, Dunthorn M, Edvardsen B, Holzmann M, Kooistra WHCF, Lara E, Le Bescot N, Logares R, Mahé F, Massana R, Montresor M, Morard R, Not F, Pawlowski J, Probert I, Sauvadet AL, Siano R, Stoeck T, Vaulot D, Zimmermann P, Christen R. 2013. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res 41:D597–D604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clarke KR. 1993. Non-parametric multivariate analysis of changes in community structure. Austral Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 54.Svendsen H, Beszczynska-Møller A, Hagen JO, Lefauconnier B, Tverberg V, Gerland S, Ørbæk JB, Bischof K, Papucci C, Zajaczkowski M, Azzolini R, Bruland O, Wiencke C. 2002. The physical environment of Kongsfjorden-Krossfjorden, an Arctic fjord system in Svalbard. Polar Res 21:133–166. doi: 10.1111/j.1751-8369.2002.tb00072.x. [DOI] [Google Scholar]
- 55.Hill MO, Gauch HG Jr. 1980. Detrended correspondence analysis: an improved ordination technique. Vegetatio 42:47–58. doi: 10.1007/BF00048870. [DOI] [Google Scholar]
- 56.Oksanen J, Guillaume Blanchet F, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. vegan: Community Ecology Package. R package, version 2.3-0. http://cran.r-project.org/package=vegan.
- 57.Seenivasan R, Sausen N, Medlin LK, Melkonian M. 2013. Picomonas judraskeda gen. et sp. nov.: the first identified member of the Picozoa phylum nov., a widespread group of picoeukaryotes, formerly known as ‘picobiliphytes.’ PLoS One 8(3):e59565. doi: 10.1371/journal.pone.0059565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lovejoy C, Vincent WF, Bonilla S, Roy S, Martineau MJ, Terrado R, Potvin M, Massana R, Pedrós-Alió C. 2007. Distribution, phylogeny, and growth of cold-adapted picoprasinophytes in Arctic seas. J Phycol 43:78–89. doi: 10.1111/j.1529-8817.2006.00310.x. [DOI] [Google Scholar]
- 59.Logares R, Audic S, Bass D, Bittner L, Boutte C, Christen R, Claverie JM, Decelle J, Dolan JR, Dunthorn M, Edvardsen B, Gobet A, Koolstra WHCF, Mahé F, Not F, Ogata H, Pawlowski J, Pernice MC, Romac S, Shalchian-Tabrizi K, Simon N, Stoeck T, Santini S, Siano R, Wincker P, Zingone A, Richards TA, de Vargas C, Massana R. 2014. Patterns of rare and abundant marine microbial eukaryotes. Curr Biol 24:813–821. doi: 10.1016/j.cub.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 60.Bell EM, Laybourn-Parry J. 2003. Mixotrophy in the Antarctic phytoflagellate, Pyramimonas gelidicola (Chlorophyta: Prasinophyceae). J Phycol 39:644–649. doi: 10.1046/j.1529-8817.2003.02152.x. [DOI] [Google Scholar]
- 61.Laybourn-Parry J, Marshall WA, Marchant HJ. 2005. Flagellate nutritional versatility as a key to survival in two contrasting Antarctic saline lakes. Freshwater Biol 50:830–838. doi: 10.1111/j.1365-2427.2005.01369.x. [DOI] [Google Scholar]
- 62.Romari K, Vaulot D. 2004. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol Oceanogr 49:784–798. doi: 10.4319/lo.2004.49.3.0784. [DOI] [Google Scholar]
- 63.Sørensen N, Daugbjerg N, Gabrielsen TM. 2012. Molecular diversity and temporal variation of picoeukaryotes in two Arctic fjords, Svalbard. Polar Biol 35:519–533. doi: 10.1007/s00300-011-1097-8. [DOI] [Google Scholar]
- 64.Kilias E, Wolf C, Nöthig E-M, Peeken I, Metfies K. 2013. Protist distribution in the Western Fram Strait in summer 2010 based on 454-pyrosequencing of the 18S rDNA. J Phycol 49:996–1010. doi: 10.1111/jpy.12109. [DOI] [PubMed] [Google Scholar]
- 65.McKie-Krisberg ZM, Sanders RW. 2014. Phagotrophy by the picoeukaryotic green alga Micromonas: implications for Arctic oceans. ISME J 8:1953–1961. doi: 10.1038/ismej.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Medinger R, Nolte V, Pandey RV, Jost S, Ottenwälder B, Schlötter C, Boenigk J. 2010. Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol 19(Suppl 1):32–40. doi: 10.1111/j.1365-294X.2009.04478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R, Scanlan DJ, Worden AZ. 2008. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ Microbiol 10:3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- 68.Vaulot D, Eikrem W, Viprey M, Moreau H. 2008. The diversity of small eukaryotic phytoplankton (≤3 μm) in marine ecosystems. FEMS Microbiol Rev 32:795–820. doi: 10.1111/j.1574-6976.2008.00121.x. [DOI] [PubMed] [Google Scholar]
- 69.Niemi A, Michel C, Hille K, Poulin M. 2011. Protist assemblages in winter sea ice: setting the stage for the spring ice algal bloom. Polar Biol 34:1803–1817. doi: 10.1007/s00300-011-1059-1. [DOI] [Google Scholar]
- 70.Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T, Somerfield PJ, Huse S, Joint I. 2009. The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- 71.Dasilva CR, Li WKW, Lovejoy C. 2014. Phylogenetic diversity of eukaryotic marine microbial plankton on the Scotian Shelf Northwestern Atlantic Ocean. J Plankton Res 36:344–363. doi: 10.1093/plankt/fbt123. [DOI] [Google Scholar]
- 72.Hegseth EN, Tverberg V. 2013. Effect of Atlantic water inflow on timing of the phytoplankton spring bloom in a high Arctic fjord (Kongsfjorden, Svalbard). J Mar Syst 113–114:94–105. doi: 10.1016/j.jmarsys.2013.01.003. [DOI] [Google Scholar]
- 73.Wassmann P, Ratkova TN, Andreassen IJ, Vernet M, Pedersen G, Rey F. 1999. Spring bloom development in the marginal ice zone and the central Barents Sea. Mar Ecol 20:321–346. doi: 10.1046/j.1439-0485.1999.2034081.x. [DOI] [Google Scholar]
- 74.Degerlund M, Eilertsen HC. 2010. Main species characteristics of phytoplankton spring blooms in NE Atlantic and Arctic waters (68–80°N). Estuar Coast 33:242–269. doi: 10.1007/s12237-009-9167-7. [DOI] [Google Scholar]
- 75.Zhu F, Massana R, Not F, Marie D, Vaulot D. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 76.Gong J, Dong J, Liu C, Massana R. 2013. Extremely high copy number polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist 164:369–379. doi: 10.1016/j.protis.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 77.Piwosz K, Spich K, Calkiewicz J, Weydmann A, Kubiszyn AM, Wiktor JM. 2015. Distribution of small phytoflagellates along an Arctic fjord transect. Environ Microbiol 17:2393–2406. doi: 10.1111/1462-2920.12705. [DOI] [PubMed] [Google Scholar]
- 78.Seuthe L, Töpper B, Reigstad M, Thyrhaug R, Vaquer-Sunyer R. 2011. Microbial communities and processes in ice-covered Arctic waters of the northwestern Fram Strait (75 to 80°N) during the vernal pre-bloom phase. Aquat Microb Ecol 64:253–266. doi: 10.3354/ame01525. [DOI] [Google Scholar]
- 79.Klaveness D, Shalchian-Tabrizi K, Thomsen HA, Eikrem W, Jakobsen KS. 2005. Telonema antarcticum sp. nov., a common marine phagotrophic flagellate. Int J Syst Evol Microbiol 55:2595–2604. doi: 10.1099/ijs.0.63652-0. [DOI] [PubMed] [Google Scholar]
- 80.Shalchian-Tabrizi K, Eikrem W, Klaveness D, Vaulot D, Minge M, Le Gall F, Romari K, Throndsen J, Botnen A, Massana R, Thomsen HA, Jakobsen K. 2006. Telonemia, a new protist phylum with affinity to chromist lineages. Proc R Soc Lond B Biol Sci 273:1833–1842. doi: 10.1098/rspb.2006.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shalchian-Tabrizi K, Kauserud H, Massana R, Klaveness D, Jakobsen KS. 2007. Analysis of environmental 18S ribosomal RNA sequences reveals unknown diversity of the cosmopolitan phylum Telonemia. Protist 158:173–180. doi: 10.1016/j.protis.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 82.Tong S, Vørs N, Patterson DJ. 1997. Heterotrophic flagellates, centrohelid heliozoa and filose amoebae from marine and freshwater sites in the Antarctic. Polar Biol 18:91–106. doi: 10.1007/s003000050163. [DOI] [Google Scholar]
- 83.Lefèvre E, Roussel B, Amblard C, Sime-Ngando T. 2008. The molecular diversity of freshwater picoeukaryotes reveals high occurrence of putative parasitoids in the plankton. PLoS One 3:e2324. doi: 10.1371/journal.pone.0002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krawczyk WE, Lefauconnier B, Pettersson LE. 2003. Chemical denudation rates in the Bayelva Catchment, Svalbard, in the fall of 2000. Phys Chem Earth Parts A/B/C 28:1257–1271. doi: 10.1016/j.pce.2003.08.054. [DOI] [Google Scholar]
- 85.Bråte J, Køaveness D, Rygh T, Jakobsen KS, Shalchian-Tabrizi K. 2010. Telonemia-specific environmental 18S rDNA PCR reveals unknown diversity and multiple marine-freshwater colonization. BMC Microbiol 10:168. doi: 10.1186/1471-2180-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stübner EI, Søreide J, Reigstad M, Marquardt M, Blachowiak-Samolyk K. Year-round meroplankton dynamics in high-Arctic Svalbard. J Plankton Res, in press. [Google Scholar]
- 87.Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, Somerfield P, Fuhrman JA, Field D. 2012. Defining seasonal marine microbial community dynamics. ISME J 6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim DY, Countway PD, Jones AC, Schnezter A, Yamashita W, Tung C, Caron DA. 2014. Monthly to interannual variability of microbial eukaryote assemblages at four depths in the eastern North Pacific. ISME J 8:515–530. doi: 10.1038/ismej.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thaler M, Lovejoy C. 2015. Biogeography of heterotrophic flagellate populations indicates the presence of generalist and specialist taxa in the Arctic Ocean. Appl Environ Microbiol 81:2137–2148. doi: 10.1128/AEM.02737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stecher A, Neuhaus S, Lange B, Frickenhaus S, Beszteri B, Kroth PG, Valentin K. 22 October 2015. rRNA and rDNA based assessment of sea ice protist biodiversity from the central Arctic Ocean. Eur J Phycol doi: 10.1080/09670262.2015.1077395. [DOI] [Google Scholar]
- 91.Rumpho ME, Dastoor FP, Manhart JR, Lee J. 2006. The kleptoplast, p 451–473. In Wise RR, Hoober JK (ed), The structure and function of plastids. Springer, New York, NY. [Google Scholar]
- 92.Stoecker DK, Silver MW. 1990. Replacement and aging of chloroplasts in Strombidium capitatum (Ciliophora: Oligotrichida). Mar Biol 107:491–502. doi: 10.1007/BF01313434. [DOI] [Google Scholar]
- 93.Schoener DM, McManus GB. 2012. Plastid retention, use, and replacement in a kleptoplastidic ciliate. Aquat Microb Ecol 67:177–187. doi: 10.3354/ame01601. [DOI] [Google Scholar]
- 94.Lynn DH. 2008. The ciliated protozoa: characterization, classification, and guide to the literature, 3rd ed, p 129 Springer, New York, NY. [Google Scholar]
- 95.Massana R, Terrado R, Forn I, Lovejoy C, Pedrós-Alió C. 2006. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ Microbiol 8:1515–1522. doi: 10.1111/j.1462-2920.2006.01042.x. [DOI] [PubMed] [Google Scholar]
- 96.Takano Y, Horiguchi T. 2014. Surface ultrastructure and molecular phylogenetics of four unarmored heterotrophic dinoflagellates, including the type species of the genus Gyrodinium (Dinophyceae). Phycol Res 52:107–116. [Google Scholar]
- 97.Rat'kova TN, Wassmann P. 2002. Seasonal variation and spatial distribution of phyto- and protozooplankton in the central Barents Sea. J Mar Syst 38:47–75. doi: 10.1016/S0924-7963(02)00169-0. [DOI] [Google Scholar]
- 98.Richardson K, Markager S, Buch E, Lassen MF, Kristensen AS. 2005. Seasonal distribution of primary production, phytoplankton biomass and size distribution in the Greenland Sea. Deep Sea Res Part I Oceanogr Res Pap 52:979–999. doi: 10.1016/j.dsr.2004.12.005. [DOI] [Google Scholar]
- 99.Rózanska M, Gosselin M, Poulin M, Wiktor JW, Michel C. 2009. Influence of environmental factors on the development of bottom ice protist communities during the winter-spring transition. Mar Ecol Prog Ser 386:43–59. doi: 10.3354/meps08092. [DOI] [Google Scholar]
- 100.Kubiszyn AM, Piwosz K, Wiktor JM. 2014. The effect of inter-annual Atlantic water inflow variability on the planktonic protist community structure in the West Spitsbergen waters during the summer. J Plankton Res 36:1190–1203. doi: 10.1093/plankt/fbu044. [DOI] [Google Scholar]
- 101.Jakobsson M, Mayer LA, Coakley B, Dowdeswell JA, Forbes S, Fridman B, Hodnesdal H, Noormets R, Pedersen R, Rebesco M, Schenke H-W, Zarayskaya AY, Accettella D, Armstrong A, Anderson RM, Bienhoff P, Camerlenghi A, Church I, Edwards M, Gardner JV, Hall JK, Hell B, Hestvik OB, Kristoffersen Y, Marcussen C, Mohammad R, Mosher D, Nghiem SV, Pedrosa MT, Travaglini PG, Weatherall P. 2012. The International Bathymetric Chart of the Arctic Ocean (IBCAO) version 3.0. Geophys Res Lett 39:L12609. doi: 10.1029/2012GL052219. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.