Abstract
Although the mature dental biofilm composition is well studied, there is very little information on the earliest phase of in vivo tooth colonization. Progress in dental biofilm collection methodologies and techniques of large-scale microbial identification have made new studies in this field of oral biology feasible. The aim of this study was to characterize the temporal changes and diversity of the cultivable and noncultivable microbes in the early dental biofilm. Samples of early dental biofilm were collected from 11 healthy subjects at 0, 2, 4, and 6 h after removal of plaque and pellicle from tooth surfaces. With the semiquantitative Human Oral Microbiome Identification Microarray (HOMIM) technique, which is based on 16S rRNA sequence hybridizations, plaque samples were analyzed with the currently available 407 HOMIM microbial probes. This led to the identification of at least 92 species, with streptococci being the most abundant bacteria across all time points in all subjects. High-frequency detection was also made with Haemophilus parainfluenzae, Gemella haemolysans, Slackia exigua, and Rothia species. Abundance changes over time were noted for Streptococcus anginosus and Streptococcus intermedius (P = 0.02), Streptococcus mitis bv. 2 (P = 0.0002), Streptococcus oralis (P = 0.0002), Streptococcus cluster I (P = 0.003), G. haemolysans (P = 0.0005), and Stenotrophomonas maltophilia (P = 0.02). Among the currently uncultivable microbiota, eight phylotypes were detected in the early stages of biofilm formation, one belonging to the candidate bacterial division TM7, which has attracted attention due to its potential association with periodontal disease.
INTRODUCTION
The microbial diversity in the oral cavity is among the largest so far characterized in the human body (1). Of specific interest is the dental biofilm, which forms first by selective adsorption of bacteria from saliva onto the tooth surface, followed by bacterial growth. It is well known that biofilm microbes interact with each other and thus show characteristics significantly different from those of their planktonic counterparts (2). While the biofilm contains beneficial, as well as harmful, bacteria, their relative proportions have a tendency to change as the dental plaque matures (3). These changes depend on bacterial interactions, as well as host-derived factors, which are all responsible for the ensuing development and biological effects of the structure (4). The very first microbial settlers of tooth surfaces are critical for the maturation process of dental plaque. As such, they are likely to play an unanticipated role in pathological conditions associated with oral biofilm formation, such as caries and periodontal disease. Understanding the earliest but most critical steps in the progression of disease involves identification, timing, and quantitation of the total dental microbiome, an important goal yet to be achieved.
It has been well established that enamel tooth surfaces are immediately covered with a layer of salivary proteins upon exposure to the oral environment. This layer, which is called the acquired enamel pellicle, is several micrometers thick (5–7). It is formed by the selective adsorption of mostly phosphorylated salivary proteins (8–10). The earliest phase of bacterial biofilm formation is the attachment of oral bacteria, via specific molecular interactions, to the acquired enamel pellicle (11–13). This permits the attached bacteria to remain attached to tooth surfaces despite the mechanical forces of salivary flow, tongue movements, and rinsing with water. The first insights into the early biofilm composition, obtained with culture-based techniques, have shown that streptococci, as well as Neisseria and Rothia species, are the predominant early colonizers. Streptococci express adhesins, specifically α-amylase-binding protein A, antigen I/II, SspA/SspB, and surface lectins, that recognize receptors on proteins in the acquired enamel pellicle (14). As plaque matures, the proportions of facultative and anaerobic filamentous genera, such as Actinomyces, Corynebacterium, Fusobacterium, and Veillonella, increase gradually (15–20). Streptococci coaggregate within and between species involving, e.g., receptor polysaccharides and type 2 fimbriae expressed by Actinomyces (21–23). The multiple-affinity properties of streptococci confer advantageous characteristics on the genus and explain their dominance as the initial colonizing bacteria of the tooth surface (24, 25).
While valuable insights were obtained with the relatively few biofilm bacteria that could be cultured at that time, inevitably, the true microbial complexity of the biofilm structure could not be fully established. More recently developed molecular techniques have expanded our ability not only to uncover the complexity of colonizing microbial communities, but also to identify the noncultivable species (26, 27). We have previously used the “checkerboard assay” employing whole-genomic probes, limited to detecting cultivable bacteria (24). The successive-adhesion pattern of 40 species was characterized, and the different species contributions were quantitated. Porphyromonas gingivalis and Treponema denticola were found to be among the early colonizers but were rapidly superseded by streptococci and Actinomyces spp. The microbial complexity of oral biofilm has also been studied in plaque formed in situ on retrievable enamel chips using a 16S rRNA gene-based pyrosequencing approach, identifying at least 97 different species and some uncultivated phylotypes. Of special note is the fact that noncultivable species were found for the first time in this study, and they were tentatively assigned to Clostridia and Flavobacteria (26).
The knowledge of mature in vivo-formed oral biofilm gained, using cultivation and DNA-based approaches, is considerable (27–29). Much less information, however, is available on the earliest phases of biofilm formation and its noncultivable bacterial fraction. In the present study, we harvested in vivo biofilms formed during the very early phases (<6 h) of microbial attachment to tooth surfaces. Bacterial growth during this time interval is limited, and the focus of this study is on the very first bacteria that interact with the pellicle proteins. The Human Oral Microbiome Identification Microarray (HOMIM) was employed, using 407 microbial probes distinguishing over 300 cultivable, as well as noncultivable, species. The data obtained revealed the microbial composition of the very early stage of biofilm formation and provided in-depth information regarding its temporal development.
MATERIALS AND METHODS
Subject population.
Early dental biofilms were collected from 11 healthy subjects. All the subjects provided informed consent prior to their participation. The study was conducted according to the principles outlined in the Declaration of Helsinki on experimentation involving human subjects. The study protocol was approved by the Institutional Review Board of Boston University Medical Center. The subjects were screened using oral and systemic health histories. Exclusion criteria were (i) overt signs of gingivitis, periodontal disease, active dental caries, or any other oral condition that could affect oral fluid/biofilm composition; (ii) fewer than 14 teeth; (iii) history of antibiotic use in the past 3 months; (iv) long-term use of anti-inflammatory medication; (v) current smoking; (vi) pregnancy; (vii) presence of systemic diseases that could affect oral health; and (viii) systemic medications and treatments that could affect salivary function. Clinical examinations took place on different days of the same week at the Clinical Research Center at the School of Dental Medicine of Boston University. All clinical examinations were performed once at baseline by the same trained periodontist using the calibrated method. Measurements were taken at 6 sites per tooth (mesiobuccal, buccal, distobuccal, mesiolingual, lingual, and distolingual) for all teeth except third molars and included probing depth (PD) and clinical attachment level (CAL), measured to the nearest millimeter with a periodontal probe (UNC-15; Hu-Friedy, Chicago, IL, USA), and presence or absence of bleeding on probing (BOP), supragingival visible plaque (PL), gingival marginal bleeding (GI), and suppuration. Clinical diagnosis of periodontal health (PH) was established for all the subjects based on the following criteria: ≤10% of sites with BOP and no PD or CAL of >3 mm, although PD or CAL of 4 mm in not more than 5% of the sites without BOP was allowed.
Sample collection and processing.
The buccal tooth surfaces in both arches, excluding second and third molars, were thoroughly cleaned to remove the acquired enamel pellicle and dental plaque, using a prophylaxis hand piece with rubber cup and dental pumice containing no additives (Preppies; Whip Mix, Louisville, KY) (30). This was followed by in vivo exposure to the oral environment for either 0, 2, 4, or 6 h. At each of these four time points, biofilm was collected. The samples were acquired on two different days within the same week. The 0- and 6-h samples were collected on day 1, and the 2- and 4-h samples were collected on day 2. During the biofilm formation phase, subjects were asked to refrain from eating, drinking (except water), or oral hygiene. For harvesting biofilm, teeth were isolated from the buccal/labial mucosa with cotton rolls to avoid contact between tooth surfaces and the oral mucosa. The collection area was rinsed twice with distilled water and dried with air. Polyvinylidene difluoride (PVDF) membranes (45-μm pore size; 13-mm diameter; Durapore; Millipore, Bedford, MA, USA) soaked in 0.5 mol/liter sodium bicarbonate, pH 8.4, were used to swab the coronal two-thirds of buccal dental surfaces of incisors, canines, premolars, and first molars of both arches while applying mild pressure (Fig. 1). Sodium bicarbonate has previously been shown to be effective in releasing proteinaceous materials adsorbed onto tooth surfaces (24). The pooled membranes (four membranes per time point per subject) were placed into 300 μl of TE buffer (50 mM Tris-HCl, 0.1 mM EDTA, pH 7.6), followed by vortexing for 30 s and sonication for 5 min. DNA isolation was performed using Ready-Lyse Lysozyme Solution and the MasterPure DNA purification kit (both from Epicentre, Madison, WI) following the manufacturers' instructions. The samples were stored at −80°C until analysis.
FIG 1.
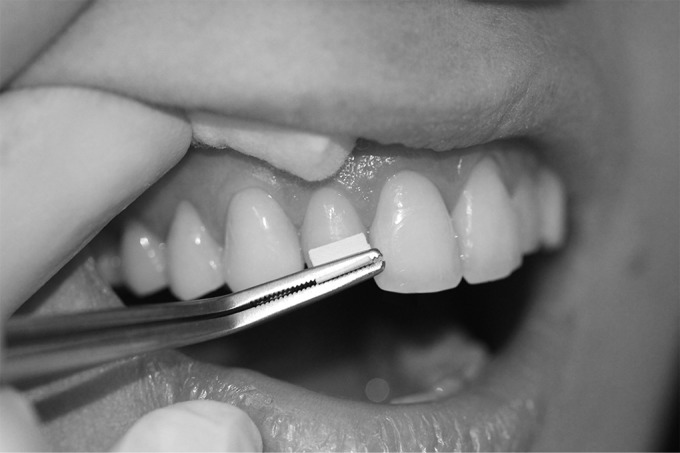
Dental biofilm collection procedure. For biofilm collection, one folded PVDF membrane presoaked in 0.5 mol/liter sodium bicarbonate, pH 8.4, was used to swab the coronal two-thirds of the buccal dental surfaces of the incisors, canines, premolars, and first molars of both arches while applying mild pressure. For each quadrant, 1 fresh membrane was used.
Microbiological assessment and quantitation.
The purified DNA samples were analyzed using HOMIM (31). For microbial identification, a library of 407 probes recognizing the most prevalent oral bacterial species was used. Briefly, 16S rRNA-based reverse-capture oligonucleotide probes (typically 18 to 20 bases) were printed on aldehyde-coated glass slides. The 16S rRNA genes were PCR amplified from DNA extracts using 16S rRNA universal forward and reverse primers and labeled via incorporation of Cy3-dCTP in a second PCR amplification. The labeled 16S rRNA amplicons were hybridized for 16 h with probes on the custom-made arrays. After washing, the microarray slides were scanned using an Axon 4000B scanner, and the raw data were extracted using GenePix Pro software (MDS Analytical Technologies, Sunnyvale, CA). The abundance of each species/phylotype interrogated by the array was then assigned an ordinal, nonlinear HOMIM score from 0 to 5 using an online analysis tool (http://bioinformatics.forsyth.org/homim), where a score of 0 indicates a fluorescent signal that is less than two times the background level and a score of 5 corresponds to the average maximum intensity of a set of universal positive-control probes.
Statistical analyses.
Statistical analyses were performed using the R software program for statistical computing (version 2.15.1). The Wilcoxon signed-rank test (as defined in the wilcox.test R function) was used to determine across all subjects whether the abundance of a given species/phylotype was significantly different between two time points. Spearman's correlation coefficient (rho) and its associated P value (as computed using the cor.test R function) were used to identify species whose abundances were significantly associated with specific time points, both within each subject and across all subjects. The rho value measures how well the amount of a given species in a given subject correlates with the amount of time that has elapsed, e.g., a rho value of +1 or −1 means that the HOMIM scores increased or decreased, respectively, at every time point during the 6-h biofilm development time. A species with a rho of 0 (no correlation) would have discrete HOMIM scores that showed high levels of variation over time, whereas a species with an undefined rho would have HOMIM scores that were unchanged over time (e.g., always 0, always 1, etc.). Correction for multiple-hypothesis testing was accomplished using the Benjamini-Hochberg false-discovery rate (FDR). The level of significance for each test was set at 5%.
RESULTS
Demographic and clinical parameters.
The demographics and clinical parameters determined for the enrolled subjects are shown in Table S1 in the supplemental material. The periodontal measures obtained, CAL, PD, and BOP, were consistent with periodontal health according to reported criteria (32).
Microbial characterization and prevalence.
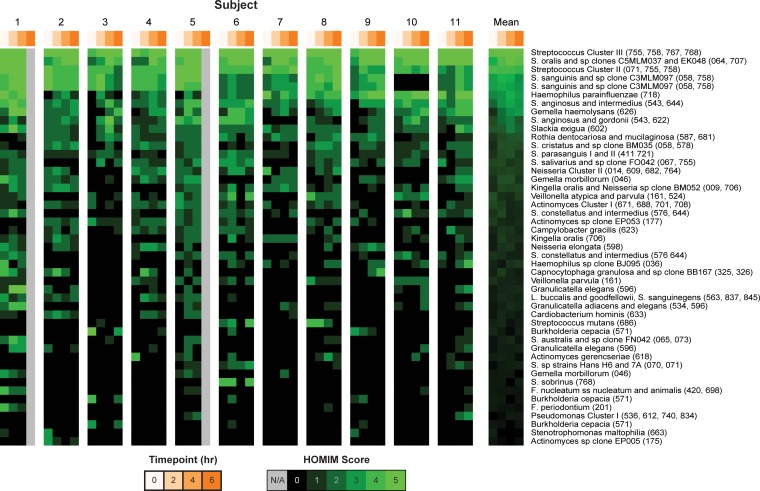
Early biofilms from the 11 subjects were obtained after 0-, 2-, 4-, and 6-h biofilm maturation times and evaluated by HOMIM. Of the possible 407 target probes, 124 hybridized with PCR products from the DNA samples obtained (HOMIM score, ≥1) (see Table S2 in the supplemental material). In view of the multitargeting by some probes, it was established that the 124 probes represented at least 92 different oral species. Among the 124 probes, 47 showed a HOMIM score of >2 in at least 1 of the 11 subjects (Fig. 2). The identified bacteria primarily belonged to the phylum Firmicutes (42.2%), followed by Protobacteria (25.6%) and Actinobacteria (16.5%). Less prevalent bacteria belonged to the phyla Bacteroidetes (8.26%), Fusobacteria (4.96%), and Synergistetes (0.83%) and the candidate bacterial division TM7 (0.83%). As expected, streptococci (Streptococcus oralis, Streptococcus anginosus/Streptococcus intermedius, and Streptococcus mitis) were the most abundant across all time points in all subjects. Together with the streptococci, the species Gemella haemolysans, Haemophilus parainfluenzae, Actinomyces cluster I (Actinomyces meyeri, Actinomyces viscosus, Actinomyces odontolyticus, and Actinomyces oricola), Rothia dentocariosa/Rothia mucilaginosa, Neisseria cluster II (Neisseria oralis, Neisseria flava, Neisseria mucosa, and Neisseria sicca), Kingella oralis, Slackia exigua, and Veillonella atypica/Veillonella parvula were the 16 predominant bacteria identified in dental biofilm formed during the first 6 h (Fig. 2; see Table S2 in the supplemental material).
FIG 2.
Intensity map of the distribution of bacterial species in each subject and the mean across all subjects after 0, 2, 4, and 6 h of biofilm formation. The image shows the intensities of the 47 probes (rows) with a maximum HOMIM score of >2 in at least one sample. The species are sorted in descending order by mean HOMIM score. The different intensities of green correspond to the signal intensities of the arrays, quantitated by HOMIM scores of 0 to 5. Gray indicates missing data for the 6-h samples in 2 subjects. The probes are labeled with species descriptors, followed by Human Oral Microbiome Database (HOMD) oral taxa in parentheses. F., Fusobacterium; L., Leptotrichia; S. sanguinegens, Sneathia sanguinegens; all other S., Streptococcus.
Species belonging to the “orange complex,” (33) comprising Fusobacterium nucleatum, Fusobacterium periodontium, and Parvimonas micra, were also found to be present in the early dental biofilm. This is consistent with previous studies (24), although we could not confirm the presence of well-established periodontal pathogens, such as Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans. Fastidious and not-yet-cultivated species more recently associated with periodontal disease, Filifactor alocis, Dialister invisus, and TM7 [G-1] oral taxon (OT) 347 (34, 35), were present in low numbers. The typical cariogenic bacterium S. mutans was detected in the early biofilm of 2 of the 11 individuals.
Changes in abundance over time.
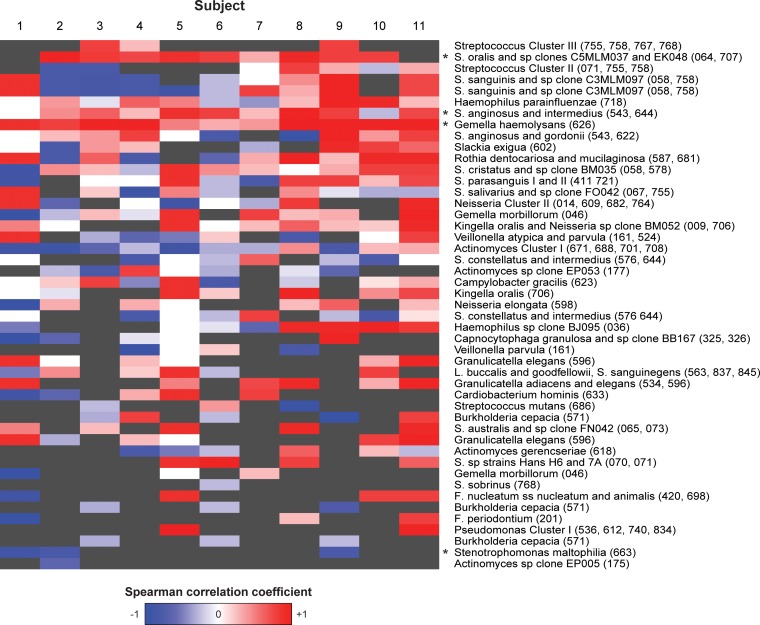
Temporal changes in bacterial prevalence and proportions during the first 6 h of biofilm formation were assessed by computing the Spearman correlation coefficient (rho), which measures how well the amount of a given species correlates with the time of enamel exposure to the oral environment (see Table S3 in the supplemental material). Among these species, significant increases in rho values over time were noted for G. haemolysans (P = 0.0005), S. anginosus or S. intermedius (P = 0.02), S. mitis bv. 2 (P = 0.0002), S. oralis (P = 0.0002), and Streptococcus cluster I (P = 0.003). Significant decreases over time were observed for Stenotrophomonas maltophilia (P = 0.02). When corrected for multiple-hypothesis testing across all probes (see Table S3 in the supplemental material), the rho values for G. haemolysans and S. oralis remained strongly significant (FDR q [corrected P value] < 0.25), indicating that these species are likely to play important roles during dental biofilm maturation and possibly microbial coaggregation. A heat map of the rho values of the 47 most abundant species is shown in Fig. 3, and the values are shown in Table S3 in the supplemental material. While trends in increases and decreases could clearly be observed for some of the bacteria listed, they were significant for only a few species.
FIG 3.
Heat map showing changes in bacterial intensities in the early biofilm over time. Shown are the results for the 47 probes with the highest level of abundance (maximum HOMIM score of >2 across all samples). Each column represents one subject, and each cell indicates Spearman's correlation coefficient (rho) of the HOMIM scores for a given species in a given subject with the four time points of biofilm formation. Red and blue represents rho values of +1 and −1, respectively, indicating that the proportion of a given species continuously increased or decreased, respectively. White represents a rho value of 0, indicating that the scores showed no pattern of increase or decrease over time. Gray indicates that all HOMIM scores for a given species in a given subject were identical and rho is undefined. All of the available time points for each subject (3 or 4) were used to compute the correlation coefficient within that subject. The probes are labeled with species descriptors, followed by HOMD oral taxa in parentheses. F., Fusobacterium; L., Leptotrichia; S. sanguinegens; Sneathia sanguinegens; all other S., Streptococcus. Spearman's correlation coefficients were also computed for each species with respect to time points across all samples in each row, and probes with across-subject rho values significantly different from zero (P < 0.05) are indicated by asterisks.
Identification of not-yet-cultivated phylotypes.
The HOMIM technology offers the opportunity to detect not only those species that can be cultured, but also those that have escaped identification so far by in vitro culturing approaches. Table 1 shows the prevalences of uncultivable species found among the 11 subjects (HOMIM score of ≥1). Eight not-yet-cultivated phylotypes were detected, including species belonging to the candidate bacterial division TM7. All noncultivable phylotypes detected exhibited low HOMIM scores (≤2), except Actinomyces sp. strain OT 177, which showed a HOMIM score of 3 at one time point (see Table S2 in the supplemental material). It was also the most prevalent, since it was detected at all time points in at least half of the subjects (Table 1). The other phylotypes were detected in less than 25% of the subjects at all time points examined. Overall, the data reveal that these noncultivable species constitute a small but integral part of the early biofilm.
TABLE 1.
Not-yet-cultivated species identified in early dental biofilm samplesa
| Phylum | Class | Not-yet-cultivated species | Identification at sampling time (h)b |
|||
|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 6 | |||
| Bacteroidetes | Flavobacteria | Bergeyella sp. strain OTc 322 | 1/11 | 1/11 | ||
| Proteobacteria | Gammaproteobacteria | Haemophilus sp. strain OT 035d | 1/11 | 1/11 | ||
| Synergistetes | Synergistetes [C-1] | Fretibacterium sp. strain OT 359d | 1/11 | 1/11 | 1/11 | |
| Fretibacterium sp. strain OT 360d | 1/11 | 1/11 | 1/11 | |||
| TM7 | TM7 [C-1] | TM7 [G-1] sp. strain OT 347 and TM7 [G-2] sp. strain OT 350 | 1/11 | 3/11 | 2/11 | |
| Actinobacteria | Actinobacteria | Actinomyces sp. OT 177 | 8/11 | 6/11 | 5/11 | 5/9 |
| Firmicutes | Clostridia | Megasphaera sp. strain OT 123d | 1/11 | |||
| Firmicutes | Clostridia | Stomatobaculum sp. strain OT 097 | 1/11 | |||
Uncultivability as reported by Dewhirst et al. (53) and at www.homd.org.
Identification of phylotypes among the subjects is depicted as follows: number of subjects/total number of subjects.
OT, oral taxon.
Recent cultivability reported by Thompson et al. (64).
DISCUSSION
The results obtained show that the early in vivo dental biofilm exhibits considerable bacterial diversity. A total of 124 probes reacted positively, providing evidence for the presence of a minimum of 92 bacterial species belonging to 40 genera and 7 phyla. Furthermore, for the first time, we report on the presence of noncultivable species in this native tooth surface biofilm.
Overall, very good consistency was observed in oral colonization among subjects, whether the species were prevalent or not, as evidenced by the abundancy gradient of the mean values among subjects (Fig. 2, right column). As expected, the streptococci were the most abundant at all time points. Within the bacteria that showed significant increases over time were the well-known early colonizers S. oralis, S. anginosus, and S. intermedius (Fig. 3). For these streptococci, the mechanism of attachment to the acquired enamel pellicle has been related to acidic proline-rich proteins, α-amylase, and various glycoproteins (36–40). Importantly, these streptococci are able to proliferate in the presence of oxygen, a characteristic of only the early biofilm environment.
At the 2-h time point, and at subsequent time points, G. haemolysans was among the 10 most prevalent bacteria. What sets this species apart from most of the other prevalent early colonizers is the fact that its rho value for increase over time was highly significant (FDR q < 0.25). G. haemolysans has previously been identified by classic cultivation methods (19) and is considered to be among the core microbial colonizers of in situ dental biofilm of healthy individuals (26). In this study, we identified this species in in vivo biofilm and in the very earliest phases of biofilm formation. The potential role that Gemella plays in biofilm processes, such as coaggregation and symbiosis, or even in disease promotion, remains to be established.
At the 4-h time point, H. parainfluenzae was among the first nonstreptococcal species to appear, with a high mean HOMIM score of 3 (Fig. 2; see Table S2 in the supplemental material). The early appearance of H. parainfluenzae can be explained by its adherence characteristics (41, 42), since it displays high affinity for salivary mucin MG1 (43) present in the acquired enamel pellicle (44). Furthermore, H. parainfluenzae has been shown to coaggregate with S. sanguis and S. oralis due to its outer membrane adhesin that recognizes specific receptor polysaccharides (45–48).
The 6-h samples revealed what could be considered the key colonizers of early in vivo-formed biofilm. In order of abundance, these colonizers are S. oralis, H. parainfluenzae, G. haemolysans, S. sanguinis, S. anginosus/S. intermedius, S. exigua, Streptococcus gordonii, and R. dentocariosa/R. mucilaginosa. Surprisingly, S. exigua, which is not well studied, was found to be among the most abundant colonizers of all subjects at all time points, with a mean HOMIM score across time points of 1.9 (see Table S2 in the supplemental material). Typically associated with endodontic (49) and periodontal lesions (50), the species is fastidious and grows poorly (51) and may therefore have been easily overlooked in previous culture-based studies. The key colonizers identified here form the substratum for the developing biofilm architecture. This structure has been shown to form in an orchestrated fashion (48) and is capable of impacting oral health adversely.
It has been estimated that at least 35% of the 700 bacterial species in the oral cavity cannot be grown in culture at this time (52, 53). The identification of both cultivable and not-yet-cultivable bacterial species in biofilm structures is equally important and critical for understanding the complete bacterial “interactome” of dental colonizers. The elegant work of Diaz and coworkers, although using not an in vivo but an in situ approach, provided evidence for uncultivable species belonging to the phyla Firmicutes and Bacteroidetes in the early biofilm, with tentative class assignments to Clostridia and Flavobacteria, respectively (26). In our in vivo investigation, we found eight uncultivable phylotypes and were able to make a more definitive genus level assignment (Table 1). They are Bergeyella sp., Haemophilus sp., Fretibacterium sp. (2 strains), TM7 sp., Actinomyces sp., Megasphaera sp., and Stomatobaculum sp.
It is well recognized that there are many qualities of oral biofilm that can lead to the development or suppression of biofilm-induced pathogenicity (54–56). The presence of a few uncultivable species, such as those of the candidate bacterial division TM7, could be highly significant for the manifestation of oral diseases, particularly periodontal disease (57). In the recently proposed polymicrobial synergy and dysbiosis model of periodontitis, the disease is caused ultimately by a dysbiotic change in the biofilm microbiota that triggers an inflammatory host response (58). This model contends that commensal oral microbes can be “accessory pathogens” (54), since such nonpathogenic species can provide biofilm conditions favoring an increase of periodontal pathogens, leading to disease manifestation. Uncultivable dental biofilm microbes, such as Actinomyces sp. OT 177, identified here and shown to be present at all time points, may play hitherto-unappreciated roles in dysbiotic mechanisms.
While the HOMIM approach has significantly expanded our knowledge of the temporal relationships of bacterial attachment, it does not allow us to decipher directly the spatial relationships among the bacterial species investigated. Some insight into the spatial relationships of mature biofilm structures have been obtained using electron microscopy techniques and, more recently, confocal laser scanning microscopy (CLSM), fluorescence in vitro hybridization (FISH), and the recent state-of-the-art combinatorial labeling and spectral imaging (CLASI)-FISH methodology (59–61). The use of such techniques will be necessary in order to gain insight into the 3-dimensional architecture of the biofilm.
In summary, this study has provided an in-depth microbial characterization of the in vivo dental biofilm formed over the first 6 h of development. It has yielded the first microbial identifications at the genus level of not-yet-cultivable microbes and has provided semiquantitative insights into their abundance in the incipient oral biofilm. It is obvious that the functional properties of these microorganisms in the oral biofilm environment are of high interest from the clinical and disease prevention perspectives. While one strategy would be to prevent growth conditions favoring detrimental bacteria, other pursuits will require a more detailed knowledge pertaining to the cultivation and cocultivation of all biofilm inhabitants (62–65). Such insights will be helpful in designing target-specific approaches for the prevention of and/or intervention in diseases exhibiting an oral-biofilm-based etiology.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants DE05672 (F.G.O.), DE07652 (F.G.O.), AI087803 (E.J.H.), AI101067 (E.J.H.), and DE021565 (B.J.P.); CTSA award U54-TR001012 (A.C.G.); Natural Sciences and Engineering Research Council of Canada grant 371813 (W.L.S.); Canadian Institutes of Health Research grants 106657 and 97577 (W.L.S.); and CIHR New Investigator Award grant 113166 (W.L.S.).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03984-15.
REFERENCES
- 1.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marsh PD, Bradshaw DJ. 1995. Dental plaque as a biofilm. J Ind Microbiol 15:169–175. doi: 10.1007/BF01569822. [DOI] [PubMed] [Google Scholar]
- 3.Haffajee AD, Teles RP, Patel MR, Song X, Veiga N, Socransky SS. 2009. Factors affecting human supragingival biofilm composition. I. Plaque mass. J Periodont Res 44:511–519. doi: 10.1111/j.1600-0765.2008.01154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosan B, Lamont RJ. 2000. Dental plaque formation. Microbes Infect 2:1599–1607. doi: 10.1016/S1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- 5.Meckel AH, Griebstein WJ, Neal RJ. 1965. Structure of mature human dental enamel as observed by electron microscopy. Arch Oral Biol 10:775–783. doi: 10.1016/0003-9969(65)90131-7. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong WG. 1968. Origin and nature of the acquired pellicle. Proc R Soc Med 61:923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meckel AH. 1965. The formation and properties of organic films on teeth. Arch Oral Biol 10:585–598. doi: 10.1016/0003-9969(65)90004-X. [DOI] [PubMed] [Google Scholar]
- 8.Hay DI. 1973. The isolation from human parotid saliva of a tyrosine-rich acidic peptide which exhibits high affinity for hydroxyapatite surfaces. Arch Oral Biol 18:1531–1541. doi: 10.1016/0003-9969(73)90128-3. [DOI] [PubMed] [Google Scholar]
- 9.Hay DI. 1973. The interaction of human parotid salivary proteins with hydroxyapatite. Arch Oral Biol 18:1517–1529. doi: 10.1016/0003-9969(73)90127-1. [DOI] [PubMed] [Google Scholar]
- 10.Oppenheim FG, Hay DI, Franzblau C. 1971. Proline-rich proteins from human parotid saliva. I. Isolation and partial characterization. Biochemistry 10:4233–4238. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons RJ, Etherden I, Moreno EC. 1983. Association of neuraminidase-sensitive receptors and putative hydrophobic interactions with high-affinity binding sites for Streptococcus sanguis C5 in salivary pellicles. Infect Immun 42:1006–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbons RJ, Moreno EC, Spinell DM. 1976. Model delineating the effects of a salivary pellicle on the adsorption of Streptococcus miteor onto hydroxyapatite. Infect Immun 14:1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scannapieco FA. 1994. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med 5:203–248. [DOI] [PubMed] [Google Scholar]
- 14.Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol Mol Biol Rev 73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritz HL. 1967. Microbial population shifts in developing human dental plaque. Arch Oral Biol 12:1561–1568. doi: 10.1016/0003-9969(67)90190-2. [DOI] [PubMed] [Google Scholar]
- 16.Socransky SS, Manganiello AD, Propas D, Oram V, van Houte J. 1977. Bacteriological studies of developing supragingival dental plaque. J Periodont Res 12:90–106. doi: 10.1111/j.1600-0765.1977.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 17.Syed SA, Loesche WJ. 1978. Bacteriology of human experimental gingivitis: effect of plaque age. Infect Immun 21:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theilade E, Theilade J, Mikkelsen L. 1982. Microbiological studies on early dento-gingival plaque on teeth and Mylar strips in humans. J Periodont Res 17:12–25. doi: 10.1111/j.1600-0765.1982.tb01127.x. [DOI] [PubMed] [Google Scholar]
- 19.Nyvad B, Kilian M. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand J Dent Res 95:369–380. [DOI] [PubMed] [Google Scholar]
- 20.Nyvad B, Kilian M. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res 24:267–272. doi: 10.1159/000261281. [DOI] [PubMed] [Google Scholar]
- 21.Kolenbrander PE, Palmer RJ Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. 2006. Bacterial interactions and successions during plaque development. Periodontol 2000 42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 22.Palmer RJ Jr, Gordon SM, Cisar JO, Kolenbrander PE. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J Bacteriol 185:3400–3409. doi: 10.1128/JB.185.11.3400-3409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruhl S, Eidt A, Melzl H, Reischl U, Cisar JO. 2014. Probing of microbial biofilm communities for coadhesion partners. Appl Environ Microbiol 80:6583–6590. doi: 10.1128/AEM.01826-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Helmerhorst EJ, Leone CW, Troxler RF, Yaskell T, Haffajee AD, Socransky SS, Oppenheim FG. 2004. Identification of early microbial colonizers in human dental biofilm. J Appl Microbiol 97:1311–1318. doi: 10.1111/j.1365-2672.2004.02420.x. [DOI] [PubMed] [Google Scholar]
- 25.Kolenbrander PE, Ganeshkumar N, Cassels FJ, Hughes CV. 1993. Coaggregation: specific adherence among human oral plaque bacteria. FASEB J 7:406–413. [DOI] [PubMed] [Google Scholar]
- 26.Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ Jr, Kolenbrander PE. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol 72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langfeldt D, Neulinger SC, Heuer W, Staufenbiel I, Kunzel S, Baines JF, Eberhard J, Schmitz RA. 2014. Composition of microbial oral biofilms during maturation in young healthy adults. PLoS One 9:e87449. doi: 10.1371/journal.pone.0087449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teles FR, Teles RP, Uzel NG, Song XQ, Torresyap G, Socransky SS, Haffajee AD. 2012. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. J Periodont Res 47:95–104. doi: 10.1111/j.1600-0765.2011.01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeshita T, Yasui M, Shibata Y, Furuta M, Saeki Y, Eshima N, Yamashita Y. 2015. Dental plaque development on a hydroxyapatite disk in young adults observed by using a barcoded pyrosequencing approach. Sci Rep 5:8136. doi: 10.1038/srep08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Y, Grogan J, Zehnder M, Lendenmann U, Nam B, Wu Z, Costello CE, Oppenheim FG. 2001. Compositional analysis of human acquired enamel pellicle by mass spectrometry. Arch Oral Biol 46:293–303. doi: 10.1016/S0003-9969(00)00134-5. [DOI] [PubMed] [Google Scholar]
- 31.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. 2009. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol 80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armitage GC. 1999. Development of a classification system for periodontal diseases and conditions. Ann Periodontol 4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 33.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 34.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. 2003. New bacterial species associated with chronic periodontitis. J Dent Res 82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Chaparro PJ, Goncalves C, Figueiredo LC, Faveri M, Lobao E, Tamashiro N, Duarte P, Feres M. 2014. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res 93:846–858. doi: 10.1177/0022034514542468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibbons RJ, Hay DI, Schlesinger DH. 1991. Delineation of a segment of adsorbed salivary acidic proline-rich proteins which promotes adhesion of Streptococcus gordonii to apatitic surfaces. Infect Immun 59:2948–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abeygunawardana C, Bush CA, Cisar JO. 1991. Complete structure of the cell surface polysaccharide of Streptococcus oralis ATCC 10557: a receptor for lectin-mediated interbacterial adherence. Biochemistry 30:6528–6540. doi: 10.1021/bi00240a025. [DOI] [PubMed] [Google Scholar]
- 38.Scannapieco FA, Torres GI, Levine MJ. 1995. Salivary amylase promotes adhesion of oral streptococci to hydroxyapatite. J Dent Res 74:1360–1366. doi: 10.1177/00220345950740070701. [DOI] [PubMed] [Google Scholar]
- 39.Murray PA, Prakobphol A, Lee T, Hoover CI, Fisher SJ. 1992. Adherence of oral streptococci to salivary glycoproteins. Infect Immun 60:31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruhl S, Sandberg AL, Cisar JO. 2004. Salivary receptors for the proline-rich protein-binding and lectin-like adhesins of oral actinomyces and streptococci. J Dent Res 83:505–510. doi: 10.1177/154405910408300614. [DOI] [PubMed] [Google Scholar]
- 41.Whittaker CJ, Klier CM, Kolenbrander PE. 1996. Mechanisms of adhesion by oral bacteria. Annu Rev Microbiol 50:513–552. doi: 10.1146/annurev.micro.50.1.513. [DOI] [PubMed] [Google Scholar]
- 42.Liljemark WF, Bloomquist CG, Fenner LJ. 1985. Characteristics of the adherence of oral Haemophilus species to an experimental salivary pellicle and to other oral bacteria, p 92–102. In Mergenhagen SE, Rosan B (ed), Molecular basis of oral microbial adhesion. American Society for Microbiology, Washington, DC. [Google Scholar]
- 43.Veerman EC, Ligtenberg AJ, Schenkels LC, Walgreen-Weterings E, Nieuw Amerongen AV. 1995. Binding of human high-molecular-weight salivary mucins (MG1) to Hemophilus parainfluenzae. J Dent Res 74:351–357. doi: 10.1177/00220345950740011101. [DOI] [PubMed] [Google Scholar]
- 44.Al-Hashimi I, Levine MJ. 1989. Characterization of in vivo salivary-derived enamel pellicle. Arch Oral Biol 34:289–295. doi: 10.1016/0003-9969(89)90070-8. [DOI] [PubMed] [Google Scholar]
- 45.Liljemark WF, Bloomquist CG, Coulter MC, Fenner LJ, Skopek RJ, Schachtele CF. 1988. Utilization of a continuous streptococcal surface to measure interbacterial adherence in vitro and in vivo. J Dent Res 67:1455–1460. doi: 10.1177/00220345880670120301. [DOI] [PubMed] [Google Scholar]
- 46.Liljemark WF, Bloomquist CG, Lai CH. 1992. Clustering of an outer membrane adhesin of Haemophilus parainfluenzae. Infect Immun 60:687–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai CH, Bloomquist C, Liljemark WF. 1990. Purification and characterization of an outer membrane protein adhesin from Haemophilus parainfluenzae HP-28. Infect Immun 58:3833–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ Jr. 2002. Communication among oral bacteria. Microbiol Mol Biol Rev 66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashimura T, Sato M, Hoshino E. 2001. Detection of Slackia exigua, Mogibacterium timidum and Eubacterium saphenum from pulpal and periradicular samples using the Polymerase Chain Reaction (PCR) method. Int Endod J 34:463–470. doi: 10.1046/j.1365-2591.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 50.Moore WE, Holdeman LV, Cato EP, Smibert RM, Burmeister JA, Ranney RR. 1983. Bacteriology of moderate (chronic) periodontitis in mature adult humans. Infect Immun 42:510–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim KS, Rowlinson MC, Bennion R, Liu C, Talan D, Summanen P, Finegold SM. 2010. Characterization of Slackia exigua isolated from human wound infections, including abscesses of intestinal origin. J Clin Microbiol 48:1070–1075. doi: 10.1128/JCM.01576-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. 2005. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitmore SE, Lamont RJ. 2011. The pathogenic persona of community-associated oral streptococci. Mol Microbiol 81:305–314. doi: 10.1111/j.1365-2958.2011.07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heller D, Silva-Boghossian CM, do Souto RM, Colombo AP. 2012. Subgingival microbial profiles of generalized aggressive and chronic periodontal diseases. Arch Oral Biol 57:973–980. doi: 10.1016/j.archoralbio.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 56.Lourenco TG, Heller D, Silva-Boghossian CM, Cotton SL, Paster BJ, Colombo AP. 2014. Microbial signature profiles of periodontally healthy and diseased patients. J Clin Periodontol 41:1027–1036. doi: 10.1111/jcpe.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camelo-Castillo AJ, Mira A, Pico A, Nibali L, Henderson B, Donos N, Tomas I. 2015. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front Microbiol 6:119. doi: 10.3389/fmicb.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG. 2011. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci U S A 108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmur R, Harmsen HJ. 2010. Oral biofilm architecture on natural teeth. PLoS One 5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dige I, Nilsson H, Kilian M, Nyvad B. 2007. In situ identification of streptococci and other bacteria in initial dental biofilm by confocal laser scanning microscopy and fluorescence in situ hybridization. Eur J Oral Sci 115:459–467. doi: 10.1111/j.1600-0722.2007.00494.x. [DOI] [PubMed] [Google Scholar]
- 62.Soro V, Dutton LC, Sprague SV, Nobbs AH, Ireland AJ, Sandy JR, Jepson MA, Micaroni M, Splatt PR, Dymock D, Jenkinson HF. 2014. Axenic culture of a candidate division TM7 bacterium from the human oral cavity and biofilm interactions with other oral bacteria. Appl Environ Microbiol 80:6480–6489. doi: 10.1128/AEM.01827-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jenkinson HF, Lamont RJ. 2005. Oral microbial communities in sickness and in health. Trends Microbiol 13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 64.Thompson H, Rybalka A, Moazzez R, Dewhirst FE, Wade WG. 2015. In vitro culture of previously uncultured oral bacterial phylotypes. Appl Environ Microbiol 81:8307–8314. doi: 10.1128/AEM.02156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oberhardt MA, Zarecki R, Gronow S, Lang E, Klenk HP, Gophna U, Ruppin E. 2015. Harnessing the landscape of microbial culture media to predict new organism-media pairings. Nat Commun 6:8493. doi: 10.1038/ncomms9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.