Abstract
Bone morphogenetic proteins 2 and 4 (BMP2/4) are essential for osteoblast differentiation and osteogenesis. Generation of a BMP2/4 dual knock‐out (ko/ko) osteoblastic cell line is a valuable asset for studying effects of BMP2/4 on skeletal development. In this study, our goal was to create immortalized mouse deleted BMP2/4 osteoblasts by infecting adenoviruses with Cre recombinase and green fluorescent protein genes into immortalized murine floxed BMP2/4 osteoblasts. Transduced BMP2/4ko/ko cells were verified by green immunofluorescence and PCR. BMP2/4ko/ko osteoblasts exhibited small size, slow cell proliferation rate and cell growth was arrested in G1 and G2 phases. Expression of bone‐relate genes was reduced in the BMP2/4ko/ko cells, resulting in delay of cell differentiation and mineralization. Importantly, extracellular matrix remodeling was impaired in the BMP2/4ko/ko osteoblasts as reflected by decreased Mmp‐2 and Mmp‐9 expressions. Cell differentiation and mineralization were rescued by exogenous BMP2 and/or BMP4. Therefore, we for the first time described establishment of an immortalized deleted BMP2/4 osteoblast line useful for study of mechanisms in regulating osteoblast lineages. J. Cell. Physiol. 231: 1189–1198, 2016. © 2015 The Authors. Journal of Cellular Physiology Published by Wiley Periodicals, Inc.
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor‐β (TGF‐β) superfamily. BMPs are initially identified by their capability to induce bone formation when implanted subcutaneously or intramuscularly in rodents (Urist, 1965; Wozney et al., 1988). To date, about 20 unique BMP ligands have been identified and compose at least four subgroups based on their amino acid sequence similarity (Sakou, 1998; Shi and Massague, 2003; Kishigami and Mishina, 2005). BMP2 and BMP4 are most similar to decapentaplgic (Dpp) in Drosophila melonogaster and belong to the BMP2/4 subclass as both of the two ligands exhibit a high affinity for the extracellular ligand binding domains of the type I BMP receptor (Hayward et al., 2002; Shi and Massague, 2003). The capacity of BMP2 to induce osteoblast differentiation has been rigorously demonstrated (Takuwa et al., 1991; Yamaguchi et al., 1991; Kubler et al., 1998; Welch et al., 1998; Bax et al., 1999; Chung et al., 1999; Wu et al., 2011). Moreover, BMP4 also plays an important role in osteogenesis (Martinovic et al., 2006; Wang et al., 2006; Luppen et al., 2008; Miyazaki et al., 2008). However, it is difficult to decipher unique roles of BMP2 and/or BMP4 during osteogenesis because of their functional redundancy each other (Selever et al., 2004). BMP2/4 are involved in organ development (Reversade et al., 2005; Cejalvo et al., 2007; Goldman et al., 2009; Uchimura et al., 2009). Mice with BMP2/4 conditional knock‐out (cKO) exhibited severe impairments of osteogenesis and displayed different genotypic and phenotypic characteristics compared to that of BMP2 or BMP4 null mice (Bandyopadhyay et al., 2006). Furthermore, clinical investigations showed that variants in BMP2/4 genes are susceptible to otosclerosis and other diseases (Schrauwen et al., 2008; Tomlinson et al., 2011; Mu et al., 2012). Otosclerosis is a common form of adult‐onset conductive hearing loss resulting from abnormal bone remodeling of the bony labyrinth that surrounds the inner ears. Genotyping pups bred between BMP2 and BMP4 heterozygous mice revealed that the ratio of adult compound heterozygous mice for BMP2/4 is much low (Uchimura et al., 2009). Therefore, generation of a dual BMP2/4ko/ko osteoblastic cell line would be a valuable asset for studying the modulatory effects of BMP2/4 on osteoblast differentiation and relevant molecular events involved in bone‐relate gene expression and extracellular matrix remodeling.
In the present study, we established an immortalized mouse deleted BMP2/4 osteoblast cell line using Cre‐recombinase to simultaneously knock‐out BMP2 and BMP4 genes in immortalized mouse floxed BMP2/4 osteoblastic cells and observed these cell behaviors. We further examined cell growth as well as their genotypic and phenotypic characteristics. Finally, we tested whether biological functions of these BMP2/4ko/ko cells were rescued by exogenous BMP2 and/or BMP4.
Materials and Methods
Generation of immortalized deleted BMP2/4 osteoblastic cells
The immortalized mouse floxed BMP2/4 osteoblasts (iBMP2/4fx/fx ob) were maintained in alpha minimum essential medium (a‐MEM, Invitrogen, San Diego, CA) containing 10% fetal calf serum (FCS) plus penicillin (100 U/ml) and streptomycin (100 mg/ml) and cultivated in 5% CO2 atmosphere at 37°C. Detail generation of iBMP2/4fx/fx ob cells were described by our previous study ((Wu et al., 2009), Fig. 1A). For BMP2/4 double knock‐out, adenoviruses with Cre recombinase and green fluorescent protein (Ad‐Cre‐GFP, Vector Biolabs, Malvern, PA) were added to the cells at 37°C. The cells were transduced overnight and then recovered in the cultured medium. GFP positive cells were observed using a Nikon inverted fluorescent microscope. The several GFP positive cells were selectively picked up and re‐plated at low densities to obtain further cell growth. Genomic DNAs were isolated from the iBMP2/4fx/fx ob and immortalized mouse BMP2/4 knock‐out osteoblasts (iBmp2ko.ko ob) using DNA purification kit, Wizard® Genomic (Promega, Madison, WI). PCR genotyping was performed by amplification of the BMP2/4fx/fx and BMP2/4ko/ko alleles using specific primers for BMP2 and BMP4 (Table I). PCR conditions: 4 min at 94°, 35 cycles of 1 min at 94°C, 1 min at 58–64°C and 2 min at 72°C, followed by 10 min at 72°C. The amplified products were run on 1% agarose gels.
Figure 1.
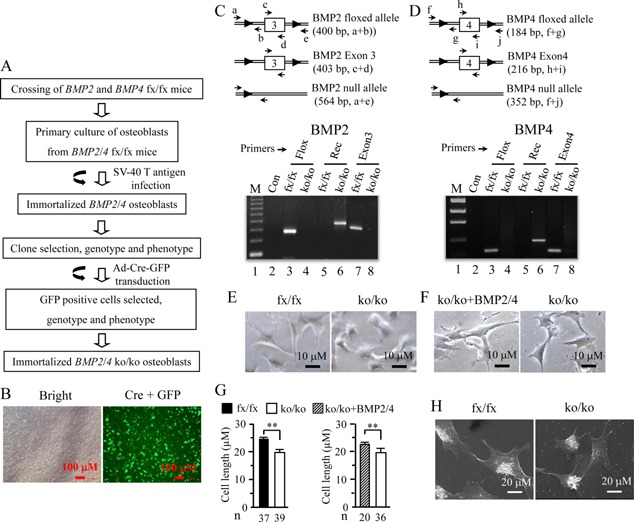
Generation of an immortalized mouse deleted BMP2/4 osteoblast cell line. (A) Strategy for generation of immortalized BMP2/4ko/ko osteoblasts. (B) The iBMP2/4fx/fx ob cells were infected with adenovirus carrying Cre recombinase and GFP genes for 14 h. The GFP positive cells were observed under a Nikon inverted fluorescent microscope. (C and D) Genotyping and PCR strategy. Genomic DNAs from the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were isolated and amplified by the BMP2/4 specific primers shown in Table I. (C) primers, a and b, c and d, amplify fragments of 400, 403 bp from iBMP2/4fx/fx osteoblasts; primers, a and e, amplify fragment of 564 bp from iBMP2/4ko/ko osteoblasts. (D) primers, f and g, h and i, amplify fragments of 184, 216 bp from iBMP2/4fx/fx osteoblasts; primers, f and j, amplify fragment of 418 bp from iBMP2/4ko/ko osteoblasts. The amplified PCR products were run on 1% agarose gels and stained with ethidium bromide. Lane 1, lower molecular DNA marker; lane 2, negative control. Lanes 3 and 4. Genomic DNAs isolated from the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were amplified using the floxed BMP2 and BMP4 primers, respectively. Lanes 5 and 6. Genomic DNAs isolated from the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were amplified using the recombinant BMP2 and BMP4 primers, respectively. Lanes 7 and 8. Genomic DNAs isolated from the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were amplified using the BMP2 exon 3 and BMP4 exon 4 primers, respectively. (E) The iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were photographed under a Nikon inverted microscope. (F) The iBmp2/4ko/ko ob cells were treated with or without BMP2 (100 ng/ml) plus BMP4 (20 ng/ml) for 48 h and cell morphology was observed under the microscope. (G) Cell length of iBMP2/4fx/fx, iBMP2/4ko/ko ob cells, and iBMP2/4ko/ko ob cells treated with BMP2/4 was quantitated. (H) Morphology of the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells was observed using scanning electron microscope. Fx. floxed; Con, control; Rec, recombinant; Exon, BMP2 exon 3; BMP4 exon 4; ko, knock‐out; n, number. **P < 0.01.
Table I.
Primer sequences used for polymerase chain reactions
| Gene | Primers | Temperature (°C) |
|---|---|---|
| Bsp | Forward: 5′‐AAAGTGAAGGAAAGCGACGA‐3′ | 58 |
| Reversed: 5′‐GTTCCTTCTGCACCTGCTTC‐3′ | ||
| Col1a1 | Forward: 5′‐CCTGACGCATGGCCAAGAAGA‐3′ | 60 |
| Reversed: 5′‐GCATTGCACGTCATCGCACA‐3′ | ||
| CREB‐2 | Forward: 5′‐GAAACCTCATGGGTTCTCCA‐3′ | 58 |
| Reversed: 5′‐AGAGCTCATCTGGCATGGTT‐3′ | ||
| Cyclo A | Forward: 5′‐GAGCTCTGAGCACTGGAGAGA‐3′ | 64 |
| Reversed: 5′‐GATGCCAGGACCTGTATGCT‐3′ | ||
| Dlx3 | Forward: 5′‐GCGACACTCAGGAATCATTG‐3′ | 55 |
| Reversed: 5′‐CGGTCCATGCATTTGTTATC‐3′ | ||
| Dmp1 | Forward: 5′‐CAGTGAGGATGAGGCAGACA‐3′ | 62 |
| Reversed: 5′‐TCGATCGCTCCTGGTACTCT‐3′ | ||
| Mmp‐2 | Forward: 5′‐CATCGCCCATCATCAAGTTCC‐3′ | 62 |
| Reversed: 5′CCGAGCAAAAGCATCATCCAC‐3′ | ||
| Mmp‐9 | Forward: 5′‐TGGTGTGCCCTGGAACTCA‐3′ | 64 |
| Reversed: 5′‐TGGAAACTCACACGCCAGAAG‐3′ | ||
| Oc | Forward: 5′‐CTTGGTGCACACCTAGCAGA‐3′ | 58 |
| Reversed: 5′‐TTCTGTTTCCTCCCTGCTGT‐3′ | ||
| Opn | Forward: 5′‐TCTGATGAGACCGTCACTGC‐3′ | 54 |
| Reversed: 5′‐AGGTCCTCATCTGTGGCATC‐3′ | ||
| Osn | Forward: 5′‐AAACATGGCAAGGTGTGTGA‐3′ | 54 |
| Reversed: 5′‐TTGCATGGTCCGATGTAGTC‐3′ | ||
| Osx | Forward: 5′‐ACTCATCCCTATGGCTCGTG‐3′ | 55 |
| Reversed: 5′‐GGTAGGGAGCTGGGTTAAGG‐3′ | ||
| Runx2 | Forward: 5’‐TACAAACCATACCCAGTCCCTGTTT ‐3’ | 66 |
| Reversed: 5′‐AGTGCTCTAACCACAGTCCATGCA ‐3′ | ||
| Floxed BMP2 | Forward: 5′‐GATGATGAGGTTCTTGGCGG‐3′ | 64 |
| Reversed: 5′‐AGGGTTTCAGGTCAGTTTCCG‐3′ | ||
| Exon3 BMP2 | Forward: 5′‐CGGGAACAGATACAGGAAGC‐3′ | 58 |
| Reversed: 5′‐GCTGTTTGTGTTTGGCTTGA‐3′ | ||
| Recom BMP2 | Forward: 5′‐GATGATGAGGTTCTTGGCGG‐3′ | 64 |
| Reversed: 5′‐AGCATGAACCCTCATGTGTTGG‐3’ | ||
| Floxed BMP4 | Forward: 5′‐AGACTCTTTAGTGAGCATTTTCAAC‐3′ | 55 |
| Reversed: 5′‐AGCCCAATTTCCACAACTTC‐3′ | ||
| Exon4 BMP4 | Forward: 5′‐TTCCTGGACACCTCATCACA‐3′ | 58 |
| Reversed: 5′‐CCACTCCCTTGAGGTAACGA‐3′ | ||
| Recom BMP4 | Forward: 5′‐AGACTCTTTAGTGAGCATTTTCAAC‐3′ | 64 |
| Reversed: 5′‐AGGTGAGCAGAGCTAAGATG‐3′ |
Bsp, bone sialoprotein; Col1α1, alpha 1 collagen type; CREB‐2, cAMP responsive element binding protein 2; Cyclo A, Cyclophilin A; Dlx3, Distal‐less 3; Dmp1, dentin matrix protein 1; Mmp‐2, matrix metalloproteinase‐2; Mmp‐9, matrix metalloproteinase‐9; Oc, osteocalcin; Opn, osteopontin; Osn, osteonectin; Osx, Osterix; Recom, recombinant.
Cell proliferation and morphology assays
Cell proliferation assay was identified by 5‐bromo‐2′‐deoxyuridine (BrdU) incorporation and MTT method. Briefly, cells were plated into 6‐well glass slides and incubated with 30 μM BrdU (Sigma–Aldrich, St. Louis, MO) in culture medium for 4 h. The cells were then treated with a mouse monoclonal anti‐BrdU antibody (1:100, Santa Cruz Biotechnology Inc., Santa Cruz, CA), followed by a 1:1,000 dilution of the secondary antibody with Alexa Fluo® 488 green (Molecular Probes, Eugene, OR). For nucleus staining, the cells were incubated with a 1:5,000 dilution of Hoechst (Sigma–Aldrich). Images were obtained in a Nikon inverted fluorescent microscope and proliferative cells were expressed as a percentage of the number of BrdU positive cells relative to total number of Hoechst positive nuclei. For MTT assay, cells were seeded into 96‐well plates with 1.0 × 103 cells/per well and detected at days 1, 3, 5, 7, respectively, using MTT cell proliferation assay kit (ATCC, Manassas, VA). Morphology of the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells was observed by a light microscope and cell length measured using Image J software (ImageJ, NIH/gov/iJ). For scanning electron microscopy, the cells were rinsed with PBS, fixed in Karnowsky's fixative (4% paraformaldehyde and 1% glutaldehyde) and washed with PBS. The cells were then post‐fixed in 1% Zetterquist's osmium and dehydrated in a grade series of ethanol and hexamethydisilizane for 5 min before air drying. Specimen were hit with gold/palladium and examined on a JEOL JSM 6610 LV microscope at 20 kV (Jeol, Inc., Peabody, MA).
Cell cycle analysis
The iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were grown with a‐MEM containing with or without 10% FCS plus penicillin (100 U/ml) and streptomycin (100 mg/ml), and cultivated in 5% CO2 atmosphere at 37°C. The cells were harvested, washed with PBS, fixed and permeabilized with 70% ethanol. For detection of DNA, the cells were incubated for 5 min at room temperature in citrate/phosphate buffer (pH 7.8) and then with 50 mg/ml of propidium iodide (PI) (Sigma–Aldrich), containing RNase A without DNases in Vindelov's solution for 60 min at 37°C. The cell cycle was analyzed by dual laser BD FACSCalibur equipped with BD FACS‐Flow Supply System (BD Biosciences, San Jose, CA).
Alkaline phosphatase (ALP) and mineralization assays
For detection of ALP activity, cultures of the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were fixed with 70% ethanol for 5 min and washed in the buffer (100 mM Tris–HCl, pH 9.5; 100 mM NaCl; 50 mM MgCl2). In situ ALP staining was performed according to the supplier's instructions (Bio‐Rad Laboratories, Hercules, CA). For mineralization assay, these cells were seeded into 6‐well plates and cultured in calcifying medium [α‐MEM supplemented with 5% FCS, 100 U/ml penicillin and 100 µg/ml streptomycin, 50 µg/ml ascorbic acid, 10 nM dexamethasone and 10 mM sodium β‐glycerophosphate] at 37°C on given time points. The cells were fixed in 10% formaldehyde neutral buffer and then stained with alizarin red S dye (Sigma–Aldrich). The amount of calcium deposition was quantified by de‐staining with 10% cetylpyridinium chloride (Sigma–Aldrich) in 10 mM sodium phosphate at room temperature for 20 min. The absorbance was measured at 550 nm wavelength.
Induction of osteoblast differentiation and mineralization by recombinant BMP2 and/or BMP4
The iBMP2/4ko/ko ob cells were maintained in α‐MEM medium with 10% FCS plus 100 unit/ml penicillin and 100 µg/ml streptomycin. The cells were then grown in α‐MEM medium with 1% FCS plus 100 unit/ml penicillin, 100 µg/ml streptomycin, 50 µg/ml ascorbic acid, 10 nM dexamethasone and 10 mM sodium β‐glycerophosphate and treated either with or without recombinant BMP2 (rBMP2, 100 ng/ml) and/or rBMP4 (20 ng/ml) (R&D Systems, Minneapolis, MN). After rBMPs induction, the cell differentiation and mineralization were detected using ALP and alizarin red S analyses.
Quantitative real time polymerase chain reaction
Total RNA was extracted from the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells using RNA STAT‐60 kit (Tel‐Test, Inc., Friendswood, TX), treated with DNase I (Promega), and purified with the RNeasy Mini Kit (Qiagen Inc., Valencia, CA). RNA concentration was determined at an optical density of OD260. The RNA was transcribed into cDNA by SuperScript II reverse transcriptase (Life technologies, Grand Island, NY). Specific primers for the quantitative real time polymerase chain reaction (qRT‐PCR) were shown in Table I. qRT‐PCR amplification reaction was analyzed in real time on an ABI 7500 (Applied Biosystems, Foster City, CA) using SYBR Green chemistry, and threshold values were calculated using SDS2 software (Applied Biosystems). The ΔΔ Ct method was used to calculate gene expression levels normalized to cyclophilin A value. The results were performed in triplicate of three separate experiments and expressed as a relative fold change in gene expression compared to the control.
Western blot analysis
Cells were maintained in α‐MEM medium with 5% FCS, 100 units/ml penicillin, 100 µg/ml streptomycin, 50 μg/ml ascorbic acid, 10 nM dexamethasone and 10 mM sodium β‐glycerophosphate and were then washed with PBS and lysed with RIPA buffer (1% Nonidet P‐40, 0.5% sodium deoxycholate, 0.1% SDS, 10 mg/ml phenylmethylsulfonyl fluoride (PMSF), 50 KIU/ml aprotinin, 100 mM sodium orthovanadate; Santa Cruz Biotechnology, Inc.). Whole cell lysates were resolved by 7% SDS‐PAGE gels and transferred to Trans‐Blot membranes (Bio‐Rad, Laboratories). Antibodies directed against mouse Bsp and Dmp1 (gifts from Dr. Larry Fisher, NIDCR), CREB‐2 (ATF‐4), Col1α1, Mmp‐2, Mmp‐9, Oc, Opn, Osn (SPARC), Osx, PCNA, Rsk‐2, Runx2 (Santa Cruz Biotechnology Inc.) and Dlx3 (Abcam, Cambridge, MA) were used as primary antibodies. The membranes were blocked with 5% non‐fat milk in TBST buffer (10 mM Tris‐HCl, pH 7.5, 100 mM NaCl, 0.1% Tween‐20) for 60 min at room temperature. After washing, the membranes were incubated with primary antibodies against those proteins with appropriate dilution (1:500‐1,000) overnight at 4°C, respectively. The secondary antibody (horseradish peroxidase‐conjugated anti‐rabbit or anti‐goat IgG) was diluted to 1:5,000–10,000 at room temperature for 60 min. Immunoreactivity was determined using ECL chemiluminescence reagent (Thermo Scientific, Pittsburgh, PA). As a control, goat polyclonal anti‐mouse β‐actin antibody was used (Santa Cruz Biotechnology, Inc.). The band intensity was measured using ImageJ software (ImageJ). Protein expression level of each sample was normalized to β‐actin value. The iBMP2/4fx/fx proteins were used as control and act as onefold increase. The fold change in the iBMP2/4ko/ko proteins were calculated by dividing the control group.
Analysis of Mmps by zymography
Supernatant was harvested from the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells and briefly centrifuged at low speed to remove cell debris. Protein concentrations were measured using Bio‐Rad protein assay and equal amount of total proteins from the two cell lines were run onto 10% SDS‐PAGE gels and stained with Coomassie brilliant blue dye. Gelatinolytic activities of Mmps were analyzed using 10% SDS‐PAGE gels co‐polymerized with 150 μg/ml gelatin (Bio‐Rad Laboratories). In brief, after separation of samples, electrophoresis gels were washed in 5% Triton X‐100, equilibrated with collagenase assay buffer (50 mM Tris‐HCl, pH 7.5, 5 mM CaCl2, 100 mM NaCl, 0.01% Triton X‐100, 0.1 mM ZnCl2, 0.2% Brij 35 non‐ionic detergent and 2 mM Na3N) and incubated at 37°C overnight. Gelatin degradation was detected by Coomassie brilliant blue staining.
In situ DQ‐FITC‐collagen types I, IV, and ‐gelatin degradation assays
Glass slides were pre‐coated with DQ‐FITC‐collagen types I, IV, and DQ‐FITC‐gelatin (Life technologies) at a final concentration of 40 ng/μl for 2 h at 37°C, respectively. After washing with PBS, the slides were air dried and fixed with 2% formaldehyde. After washing with PBS, the slides coated with the DQ‐FITC‐collagen types I, IV, and DQ‐FITC‐gelatin were equilibrated with α‐MEM without serum. The iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were added to the plates containing the DQ‐FITC‐collagen type I or IV or DQ‐FITC‐gelatin coated slides and cultured for 12 h, respectively. The cells were fixed with 4% formaldehyde for 15 min and washed with PBS. Then, the cells were mounted using Vectashield mounting medium (Vector Laboratories Inc., Burlingame, CA). Images were taken using a Nikon inverted fluorescent microscope coupled to cool CCD camera and NIS‐GIEMENTS software. Spots of processing collagen type I, IV, and gelatin in the two cells were quantitated.
Statistical analysis
Quantitative data were presented as means ± S.D. with triplicate from three independent experiments and compared with the results of one‐way ANOVA using GraphPad Prism 5 (GraphPad Software, Inc. La Jolla, CA). The differences between groups were statistically significant at *P < 0.05 and **P < 0.01.
Results
Generation of immortalized mouse BMP2/4 knock‐out osteoblastic cells
To establish BMP2/4 knock‐out osteoblasts (iBMP2/4ko/ko ob), the immortalized mouse floxed BMP2/4 osteoblastic cells (iBMP2/4fx/fx) were transduced with Ad‐Cre‐GFP and then selected. The transduced cells showed a high efficiency of infection observed under a Nikon fluorescent microscope (Fig. 1B). Several GFP positive clones were selected and re‐grown. Deletion of BMP2/4 genes by Cre recombinase in the iBMP2/4fx/fx ob cells was confirmed by using PCR (Fig. 1C and D; Table I). This result showed that Cre recombinase knocks out both BMP2/4 genes in the iBMP2/4fx/fx ob cells. Cell morphology between the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells was observed using light inverted and scanning electron microscopes (Fig. 1E, G, and H). The iBMP2/4fx/fx ob cells show a spindle shape and long branches whereas the iBMP2/4ko/ko ob cells exhibit short branches. Also, size of the iBMP2/4ko/ko osteoblasts is smaller than their floxed counterparts. Morphology of the iBMP2/4ko/ko ob cells was changed by the recombinant BMP2/4 protein induction, displaying long branches similar to the iBMP2/4fx/fx ob cells (Fig. 1F and G).
Deletion of BMP2/4 leads to change of cell proliferation rate
To study the effect of BMP2/4 on mouse osteoblast proliferation, the iBMP2/4fx/fx and iBMP2/4ko/ko ob cell proliferation was analyzed using BrdU and MTT assays. This result showed that the iBMP2/4ko/ko ob cells display a slow growth rate compared to the iBMP2/4fx/fx ob cells (Fig. 2A–C). Exogenous BMP2/4 proteins were able to promote the knock‐out osteoblast cell proliferation (Fig. 2D–F). To assess which mechanisms of BMP2/4 control cell proliferation, we analyzed the cell cycle distribution of the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells. The data indicated that the G2 phase is delayed in the iBMP2/4ko/ko ob cells when these cells were cultured with 10% serum (Fig. 2Ga and b). However, when both of the cells were grown without serum condition, cell cycles in the G1 and G2 phases were interfered in the iBMP2/4ko/ko ob cells (Fig. 2Gc and d). These results suggested that the slow growth of the iBMP2/4ko/ko ob cells is involved in the G1 and G2 phase arrest. Furthermore, we tested several genes related to cell proliferation and found that expression levels of the proliferating cell nuclear antigen (PCNA) and ribosomal S6 kinase‐2 (Rsk‐2) were decreased in the iBMP2/4ko/ko ob cells, whereas overexpression of BMP2/4 in the iBMP2/4 knock‐out osteoblasts increased PCNA and Rsk‐2 expression (Fig. 2H).
Figure 2.
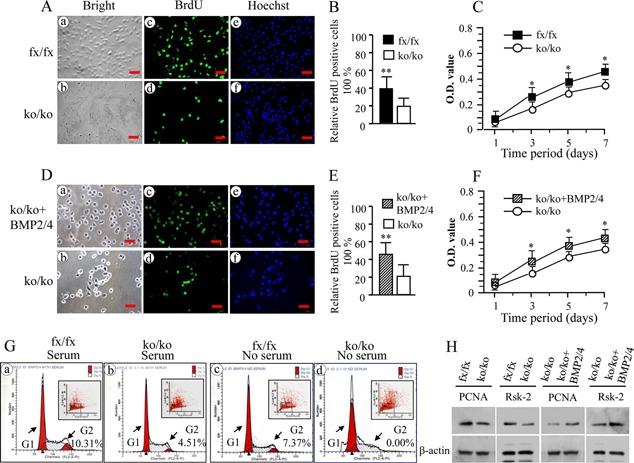
Deletion of BMP2/4 disrupts cell proliferation. (A and D) Proliferation of the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells as well as iBMP2/4ko/ko ob cells treated with BMP2 (100 ng/ml) and BMP4 (20 ng/ml) for 12 h was immunostained using the BrdU antibody after 4 h BrdU incorporation (30 µM). (c and d) The iBMP2/4fx/fx and BMP2/4 treated iBMP2/4ko/ko cells showed a higher proliferation rate (c) than that of the iBMP2/4ko/ko cells (d). (a and b) The cells were photographed under a light microscope using a Nikon camera. (e and f) The cells were stained with Hoechst for the nuclei. Scale bar, 20 µM. (B, E) A percentage of the number of BrdU positive cells relative to the total number of Hoechst positive nuclei between the iBMP2/4fx/fx, BMP2/4 treated iBMP2/4ko/ko cells, and iBMP2/4ko/ko ob cells was calculated (**P < 0.01). (C, F) Proliferation data of the iBMP2/4fx/fx, iBMP2/4ko/ko ob, and BMP2/4 treated iBMP2/4ko/ko ob cells were measured using MTT assay. The iBMP2/4fx/fx ob and BMP2/4 treated iBMP2/4ko/ko ob cells showed higher proliferation rate than the iBMP2/4ko/ko ob cells from 1‐, 3‐, 5‐, 7‐day culture. Asterisk shows significant differences between the iBMP2/4fx/fx, BMP2/4 treated iBMP2ko/ko cells, and iBMP2/4ko/ko ob cells (*P < 0.05). (G) Cell cycle distributions were measured using BD FACSCalibur cytofluorometer detection of DNA content in the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells with or without 10% fetal calf serum. Significant decrease of number of cell cycle in the G2 phase could be seen in the iBMP2/4ko/ko ob cells with 10% serum culture (a and b). Without serum, the cell cycles of iBMP2/4ko/ko cells were arrested in both the G1 and G2 phases (c and d). fx, floxed; ko, knock‐out. (H) The iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were maintained in α‐MEM containing 1% fetal calf serum plus penicillin and streptomycin, and iBMP2/4ko/ko ob cells were treated with 100 ng/ml of recombinant BMP2 and 20 ng/ml of BMP4 for 72 h. These cells were lysed and protein expressional levels were detected by Western blot assay using antibodies specific to PCNA and Rsk‐2, respectively. β‐actin was used as an internal control. The experiments were performed in triplicate of three separate samples.
Deletion of BMP2/4 causes delay of osteoblast differentiation and mineralization
To assess the effect of BMP2/4 on cell differentiation and mineralization activities, we measured ALP activity by in situ ALP histochemistry as ALP is a marker of osteoblast differentiation. Both cells were cultured in calcifying medium in given time periods. This result showed that delayed cell differentiation is seen in the iBMP2/4ko/ko ob cells (Fig. 3A). Also, deletion of BMP2/4 genes led to low activity of cell mineralization by using alizarin red S staining (Fig. 3B and C). Furthermore, when exogenous BMP2 and/or BMP4 proteins were added to the iBMP2/4ko/ko ob cells, these cell differentiation and mineralization were rescued by either recombinant BMP2 or BMP4. Exogenous BMP2/4 had synergic effects on these osteoblast differentiation and mineralization (Fig. 3D and E).
Figure 3.
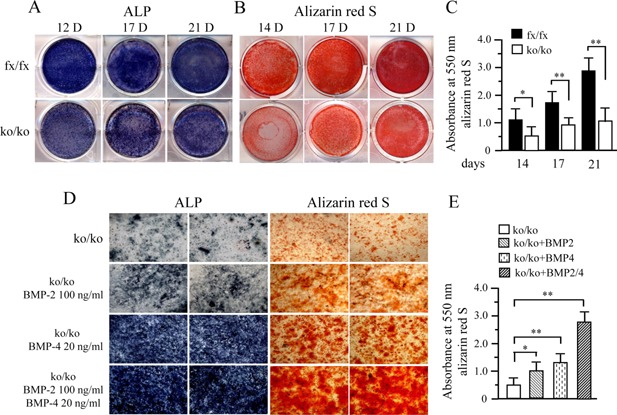
Deletion of BMP2/4 delays cell differentiation and mineralization. (A) The iBMP2/4fx/fx and iBMP2/4ko/ko cells were cultured in the calcifying medium for 12, 17, and 21 days. ALP activity was analyzed using in situ ALP staining. (B) For cell mineralization assay, both of iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were treated with calcifying medium for 14, 17, and 21 days. Mineralized nodules were visualized with alizarin red S staining. (C) Alizarin red S staining was quantified by destaining with 10% cetylpyridinium chloride in 10 mM sodium phosphate and the absorbance measured at 550 nm. Asterisks indicate significant differences (*P < 0.05; **P < 0.01). (D) The BMP2/4ko/ko ob cells were treated either recombinant BMP2 and/or BMP4 in calcifying medium. The 10‐day‐induced cells were used for ALP assay while 21‐day induced cells were assayed by alizarin red S staining. Exogenous BMP2 or BMP4 enhanced the iBMP2/4ko/ko ob cell differentiation and mineralization compared to the iBMP2/4ko/ko cells without BMPs induction. Recombinant BMP2/4 had synergic effects on the cell differentiation and mineralization. (E) Amount of the mineralization was evaluated using alizarin red S staining. The asterisks indicate the statistical significances of the iBMP2/4ko/ko ob cells induced by BMP2 or/and BMP4 compared to the control. fx, floxed; ko, knock‐out.
Knock‐out of BMP2/4 down‐regulates their downstream gene expression
To further determine which gene expression is regulated by BMP2/4 in cell differentiation and mineralization, we measured bone‐relate gene expression including transcription factors, extracellular matrix proteins and proteinases. Using qRT‐PCR, we observed decreases of bone‐relate gene expressions in the BMP2/4ko/ko ob cells (Fig. 4A). It includes Bsp, Collα1, CREB‐2, Dlx3, Dmp1, Oc, Osn, Osx, and Runx2. Also, expression of matrix metalloproteinases, Mmp‐2 and Mmp‐9, was reduced. These results were also verified by Western blot assay (Fig. 4B and C). Furthermore, these downstream gene expressions were induced in the iBMP2/4ko/ko ob cells when BMP2/4 proteins were added into the deleted iBMP2/4 osteoblastic cells (Fig. 4D).
Figure 4.
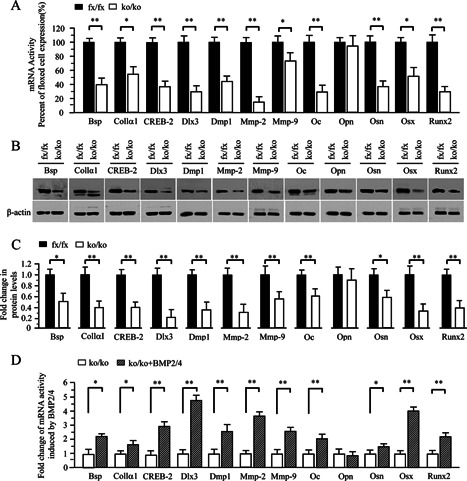
Altered expression of bone‐related and processing protein genes in the iBMP2/4 knock‐out osteoblastic cells. (A) Total RNA was isolated from the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells for measuring transcripts of Bsp, Collα1, CREB‐2, Dlx3, Dmp1, Mmp‐2, Mmp‐9, Oc, Opn, Osn, Osx, and Runx2 genes by qRT‐PCR. Cyclophilin A was used as an internal control. Expression of these mRNAs in the BMP2/4fx/fx cells acts as a 1.0‐fold increase. The bar graphs show mean ± S.D. from three independent experiments with triplicate for each transcript measurement. *P < 0.05, **P < 0.01. (B) The iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were lysed and protein expressional levels were detected by Western blot assay using antibodies specific to Bsp, Collα1, CREB‐2, Dlx3, Dmp1, Mmp‐2, Mmp‐9, Oc, Opn, Osn, Osx, and Runx2, respectively. β‐actin was used as an internal control. (C) The protein band intensity was quantitated by ImageJ software. The proteins from the iBMP2/4fx/fx ob cells were normalized to β‐actin protein as control. The fold change in the BMP2/4ko/ko cell protein expression levels was calculated by dividing the iBMP2/4fx/fx ob protein expression levels. The bar graphs show mean ± S.D. (n = 3). This result demonstrates that several protein expression levels were decreased in the iBMP2/4ko/ko ob cells. (D) The iBMP2/4ko/ko ob cells were treated with or without the recombinant BMP2/4 for 48 h and total RNA was isolated for measuring transcripts of Bsp, Collα1, CREB‐2, Dlx3, Dmp1, Mmp‐2, Mmp‐9, Oc, Opn, Osn, Osx, and Runx2 genes by qRT‐PCR. Cyclophilin A was used as an internal control. Expression of these mRNAs in the BMP2/4ko/ko cells acts as a 1.0‐fold increase. The bar graphs show mean ± S.D. (n = 3). Asterisks indicate *P < 0.05, ** P < 0.01. fx, floxed; ko, BMP2/4 knock‐out.
BMP2/4 activate extracellular matrix remodeling through Mmp‐2 and Mmp‐9
Expression of Mmp‐2 and Mmp‐9 was decreased in the iBMP2/4ko/ko ob cells. As Mmp‐2 and Mmp‐9 are involved in physiological and pathological roles including extracellular matrix remodeling (Lund et al., 2011), we then investigated these proteinase secretion. Using zymography assay, we found that secretion of Mmp‐2 and Mmp‐9 in the iBMP2/4ko/ko ob cells is lower than that of the iBmp2fx/fx ob cells (Fig. 5A). To further investigate roles of Mmp‐2 and Mmp‐9 in extracellular matrix remodeling, in situ degradation of collagen types I, IV, and gelatin was measured as collagen types I, IV, and gelatin are substrates of Mmp‐2 and Mmp‐9 (Chaussain‐Miller et al., 2006). The few and faint fluorescent spots of collagen types I, IV, and gelatin degradations were observed in the iBMP2/4ko/ko ob cells compared to that of the iBMP2/4fx/fx ob cells (Fig. 5Cc and d; Dc and d; Ec and d; F–H). This result indicates that BMP2/4 regulate extracellular matrix remodeling by regulating Mmp‐2 and Mmp‐9 activities.
Figure 5.
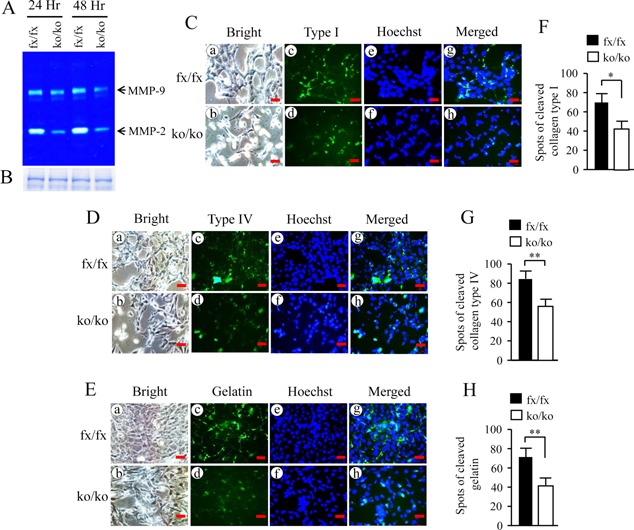
Deletion of BMP2/4 genes leads to impairment of extracellular matrix remodeling. (A) The equal amount of supernatant collected from the iBMP2/4fx/fx and iBMP2/4ko/ko cells was analyzed using gelatinolytic activities. Expression of Mmp‐2 and Mmp‐9 in the iBMP2/4ko/ko cells was decreased. (B) The equal amount of supernatant harvested from the iBMP2/4fx/fx and iBMP2/4ko/ko cells was loaded onto a 10% SDS‐PAGE gel and stained by Coomassie brilliant blue dye. (C–E) The iBMP2/4fx/fx and iBMP2/4ko/ko ob cells were grown on the DQ‐FITC‐collagen types I‐, IV‐, and DQ‐FITC‐gelatin‐coated slides for 12 h. The cells were fixed and degraded spots of the DQ‐FITC‐collagen types I, IV, and DQ‐FITC‐gelatin were observed using Nikon inverted fluorescent microscope (c and d). (a and b) The cells were photographed under a light inverted microscope. (e and f) The cells were treated with Hoechst dye for nuclei staining. (g and h) The images were merged. (F–H) Spots of the cleaved collagen type I, IV, and gelatin were quantitated from the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells. There are significant differences between the iBMP2/4fx/fx and iBMP2/4ko/ko ob cells. *P < 0.05; **P < 0.01. Scale bars, 20 µm. fx, floxed; ko, BMP2/4 knock‐out.
Discussion
The results of this study have demonstrated, after infection of the immortalized murine iBMP‐2/4fx/fx osteoblasts with Ad‐Cre‐GFP, the creation of an immortalized iBMP2/4ko/ko osteoblastic cell line. Successful infection was confirmed through GFP immunofluorescence of the cells, and BMP2/4 gene knock‐out in the iBMP2/4fx/fx ob cells was further verified using PCR assay. As the iBMP2/4fx/fx osteoblasts show similar genotypic and phenotypic characteristics to the primary mouse normal osteoblasts (Wu et al., 2009), the advantage of generation of these BMP2/4 cells is to obtain lot of BMP2/4fx/fx and BMP2/4ko/ko ob cells and used to study molecular mechanisms of BMP2/4 during osteoblast lineages and osteogenesis in vitro. In this study, we noted that the morphology of the iBMP2/4ko/ko osteoblasts in size is smaller than the iBMP2/4fx/fx ob cells and the iBMP2/4 ko/ko osteoblast growth is slow compared to the floxed iBMP2/4 osteoblasts. Further study demonstrated that the cell cycle of the iBMP2/4ko/ko ob cells is arrested in the G1 and G2 phases, resulting from decreases of PCNA and Rsk‐2 gene expression (Prosperi et al., 1994; Kawabe et al., 2002; Cude et al., 2007; Wu et al., 2014). Other studies have also shown that BMP2 and BMP4 are able to induce growth and differentiation of osteogenic cells in rats (Sakou, 1998). This finding demonstrates that BMPs are not only necessary for osteogenesis and ECM regulation, but also play a vital role in the life of the cell cycle and their ability to survive.
The iBMP2/4ko/ko osteoblasts demonstrated a diminished osteogenic phenotype as observed by lighter ALP activity and a lowered capacity to mineralize as detected by alizarin red S staining. These results coincide with previous studies in which BMP2/4 cKO genes in mouse limbs caused severe skeletal defects (Bandyopadhyay et al., 2006). Rescue of the iBMP2/4 knock‐out osteoblast differentiation and capacity for mineralization was noted with the addition of exogenous BMP2 and/or BMP4. The study further demonstrated that BMP2/4 had synergic effects on the iBMP2/4ko/ko osteoblast differentiation and mineralization.
The iBMP2/4ko/ko osteoblasts, in conjunction with a reduced capacity for osteoblast differentiation and mineralization, also have a significant reduction in the expression of osteogenic genes. These include bone‐relate extracellular matrix proteins, Bsp, Collα1, Dmp1, Oc, and Osn; transcription factors, CREB‐2, Dlx3, Osx, and Runx2; and proteinases, Mmp‐2 and Mmp‐9. Overexpression of BMP2 or/and BMP4 in the deleted iBMP2/4 osteoblast cells induced the above gene expression. Previous studies were reported that BMPs regulate expression of osteogenic transcription factors such as CREB‐2, Osx and Runx2 (Komori, 2006; Javed et al., 2008; Matsubara et al., 2008), which in turn up‐regulate the activity of osteogenic genes like Dmp1, Oc and others (Xiao et al., 2002; Jensen et al., 2010). BMP2/4 have also shown roles in regulating homoebox gene Dlx3 (Park and Morasso, 2002; Singh et al., 2012), which controls osteogenesis and limb formation. Dlx3 gene mutations are associated with autosomal dominant genetic disorder called tricho‐dento‐osseous syndrome (Price et al., 1999). This reduction in gene expression explains the loss of osteogenic cellular activity in the iBMP2/4ko/ko osteoblasts as compared to the iBMP2/4fx/fx osteoblasts.
Along with a reduction in expression of genes involve in osteogenesis, there was also a decrease in genes associated with ECM formation and remodeling, Collα1, Mmp‐2, and Mmp‐9. Expression of Mmp‐2 and Mmp‐9 was dramatically reduced in the iBMP2/4ko/ko osteoblasts as detected by qRT‐PCR, Western blot and zymography assays. In situ zymography demonstrated in the iBMP2/4ko/ko ob cells weak and less fluorescent spots of collagen types I, IV, and gelatin degradations, which are Mmp‐2 and Mmp‐9 substrates (Chaussain‐Miller et al., 2006). This control of Mmp activity by BMPs is thought through the MAP kinase and Runx2 signal pathways (Pratap et al., 2005; Kang et al., 2011), however, the mechanisms how BMP2/4 regulate ECM remodeling via Mmp signal needs to be further investigated in the future.
Conclusively, we created an immortalized mouse deleted BMP2/4 osteoblast line for utilization in in vitro studies of BMP2/4 activity in osteogenesis. When characterized, the iBMP2/4ko/ko osteoblasts demonstrated delay of cell growth capability and cell cycle is arrested in the G1 and G2 phases as well as decreased expression of cell proliferation genes, PCNA and Rsk‐2. Deletion of BMP2/4 caused decreased expression of osteogenic genes, resulting in retardation of cell differentiation and mineralization. Knock‐out of BMP2/4 genes also reduced the osteoblast ability to degrade ECM via Mmp signal, confirming BMP2/4 roles in regulating ECM formation. Therefore, the generated cell line would provide a useful tool for studies of the molecular mechanisms involved in regulating osteoblast cell proliferation, differentiation and extracellular matrix remodeling during osteogenesis and bone regeneration.
Acknowledgments
We are grateful to core facility center at The University of Texas Health Center at San Antonio, Texas performed cell cycle experiments. This research was supported by the National Institutes of Health (NIH), National Institute of Dental and Craniofacial Research (NIDCR, DE19892) and partially by the Natural Science Foundation of China (81170929).
Li‐An Wu and Feng Wang contribute equally to this work.
Literature Cited
- Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. 2006. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet 2:e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax BE, Wozney JM, Ashhurst DE. 1999. Bone morphogenetic protein‐2 increases the rate of callus formation after fracture of the rabbit tibia. Calcif Tissue Int 65:83–89. [DOI] [PubMed] [Google Scholar]
- Cejalvo T, Sacedon R, Hernandez‐Lopez C, Diez B, Gutierrez‐Frias C, Valencia J, Zapata AG, Varas A, Vicente A. 2007. Bone morphogenetic protein‐2/4 signalling pathway components are expressed in the human thymus and inhibit early T‐cell development. Immunology 121:94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussain‐Miller C, Fioretti F, Goldberg M, Menashi S. 2006. The role of matrix metalloproteinases (MMPs) in human caries. J Dent Res 85:22–32. [DOI] [PubMed] [Google Scholar]
- Chung CY, Iida‐Klein A, Wyatt LE, Rudkin GH, Ishida K, Yamaguchi DT, Miller TA. 1999. Serial passage of MC3T3‐E1 cells alters osteoblastic function and responsiveness to transforming growth factor‐beta1 and bone morphogenetic protein‐2. Biochem Biophys Res Commun 265:246–251. [DOI] [PubMed] [Google Scholar]
- Cude K, Wang Y, Choi HJ, Hsuan SL, Zhang H, Wang CY, Xia Z. 2007. Regulation of the G2‐M cell cycle progression by the ERK5‐NFkappaB signaling pathway. J Cell Biol 177:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman DC, Donley N, Christian JL. 2009. Genetic interaction between Bmp2 and Bmp4 reveals shared functions during multiple aspects of mouse organogenesis. Mech Dev 126:117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward DC, Samuel G, Pontynen PC, Catmull J, Saint R, Miller DJ, Ball EE. 2002. Localized expression of a dpp/BMP2/4 ortholog in a coral embryo. Proc Natl Acad Sci USA 99:8106–8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Bae JS, Afzal F, Gutierrez S, Pratap J, Zaidi SK, Lou Y, van Wijnen AJ, Stein JL, Stein GS, Lian JB. 2008. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem 283:8412–8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ED, Gopalakrishnan R, Westendorf JJ. 2010. Regulation of gene expression in osteoblasts. Biofactors 36:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MH, Oh SC, Lee HJ, Kang HN, Kim JL, Kim JS, Yoo YA, et al. 2011. Metastatic function of BMP‐2 in gastric cancer cells: The role of PI3K/AKT, MAPK, the NF‐kappaB pathway, and MMP‐9 expression. Exp Cell Res 317:1746–1762. [DOI] [PubMed] [Google Scholar]
- Kawabe T, Suganuma M, Ando T, Kimura M, Hori H, Okamoto T. 2002. Cdc25C interacts with PCNA at G2/M transition. Oncogene 21:1717–1726. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Mishina Y. 2005. BMP signaling and early embryonic patterning. Cytokine Growth Factor Rev 16:265–278. [DOI] [PubMed] [Google Scholar]
- Komori T. 2006. [Mechanism of transcriptional regulation by Runx2 in osteoblasts]. Clin Calcium 16:801–807. [PubMed] [Google Scholar]
- Kubler NR, Reuther JF, Faller G, Kirchner T, Ruppert R, Sebald W. 1998. Inductive properties of recombinant human BMP‐2 produced in a bacterial expression system. Int J Oral Maxillofac Surg 27:305–309. [DOI] [PubMed] [Google Scholar]
- Lund IK, Nielsen BS, Almholt K, Rono B, Hald A, Illemann M, Green KA, Christensen IJ, Romer J, Lund LR. 2011. Concomitant lack of MMP9 and uPA disturbs physiological tissue remodeling. Dev Biol 358:56–67. [DOI] [PubMed] [Google Scholar]
- Luppen CA, Chandler RL, Noh T, Mortlock DP, Frenkel B. 2008. BMP‐2 vs. BMP‐4 expression and activity in glucocorticoid‐arrested MC3T3‐E1 osteoblasts: Smad signaling, not alkaline phosphatase activity, predicts rescue of mineralization. Growth Factors 26:226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinovic S, Borovecki F, Miljavac V, Kisic V, Maticic D, Francetic I, Vukicevic S. 2006. Requirement of a bone morphogenetic protein for the maintenance and stimulation of osteoblast differentiation. Arch Histol Cytol 69:23–36. [DOI] [PubMed] [Google Scholar]
- Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, Aburatani H, Nishimura R, Yoneda T. 2008. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem 283:29119–29125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Miyauchi S, Tawada A, Anada T, Matsuzaka S, Suzuki O. 2008. Oversulfated chondroitin sulfate‐E binds to BMP‐4 and enhances osteoblast differentiation. J Cell Physiol 217:769–777. [DOI] [PubMed] [Google Scholar]
- Mu Y, Xu Z, Contreras CI, McDaniel JS, Donly KJ, Chen S. 2012. Phenotype characterization and sequence analysis of BMP2 and BMP4 variants in two Mexican families with oligodontia. Genet Mol Res 11:4110–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GT, Morasso MI. 2002. Bone morphogenetic protein‐2 (BMP‐2) transactivates Dlx3 through Smad1 and Smad4: Alternative mode for Dlx3 induction in mouse keratinocytes. Nucleic Acids Res 30:515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, Lian JB. 2005. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol 25:8581–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JA, Wright JT, Walker SJ, Crawford PJ, Aldred MJ, Hart TC. 1999. Tricho‐dento‐osseous syndrome and amelogenesis imperfecta with taurodontism are genetically distinct conditions. Clin Genet 56:35–40. [DOI] [PubMed] [Google Scholar]
- Prosperi E, Scovassi AI, Stivala LA, Bianchi L. 1994. Proliferating cell nuclear antigen bound to DNA synthesis sites: Phosphorylation and association with cyclin D1 and cyclin A. Exp Cell Res 215:257–262. [DOI] [PubMed] [Google Scholar]
- Reversade B, Kuroda H, Lee H, Mays A, De Robertis EM. 2005. Depletion of Bmp2, Bmp4, Bmp7 and Spemann organizer signals induces massive brain formation in Xenopus embryos. Development 132:3381–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakou T. 1998. Bone morphogenetic proteins: From basic studies to clinical approaches. Bone 22:591–603. [DOI] [PubMed] [Google Scholar]
- Schrauwen I, Thys M, Vanderstraeten K, Fransen E, Dieltjens N, Huyghe JR, Ealy M, Claustres M, Cremers CR, Dhooge I, Declau F, Van de Heyning P, Vincent R, Somers T, Offeciers E, Smith RJ, Van Camp G. 2008. Association of bone morphogenetic proteins with otosclerosis. J Bone Miner Res 23:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selever J, Liu W, Lu MF, Behringer RR, Martin JF. 2004. Bmp4 in limb bud mesoderm regulates digit pattern by controlling AER development. Dev Biol 276:268–279. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. 2003. Mechanisms of TGF‐beta signaling from cell membrane to the nucleus. Cell 113:685–700. [DOI] [PubMed] [Google Scholar]
- Singh M, Del Carpio‐Cano FE, Monroy MA, Popoff SN, Safadi FF. 2012. Homeodomain transcription factors regulate BMP‐2‐induced osteoactivin transcription in osteoblasts. J Cell Physiol 227:390–399. [DOI] [PubMed] [Google Scholar]
- Takuwa Y, Ohse C, Wang EA, Wozney JM, Yamashita K. 1991. Bone morphogenetic protein‐2 stimulates alkaline phosphatase activity and collagen synthesis in cultured osteoblastic cells, MC3T3‐E1. Biochem Biophys Res Commun 174:96–101. [DOI] [PubMed] [Google Scholar]
- Tomlinson IP, Carvajal‐Carmona LG, Dobbins SE, Tenesa A, Jones AM, Howarth K, Palles C, Broderick P, Jaeger EE, Farrington S, Lewis A, Prendergast JG, Pittman AM, Theodoratou E, Olver B, Walker M, Penegar S, Barclay E, Whiffin N, Martin L, Ballereau S, Lloyd A, Gorman M, Lubbe S, Howie B, Marchini J, Ruiz‐Ponte C, Fernandez‐Rozadilla C, Castells A, Carracedo A, Castellvi‐Bel S, Duggan D, Conti D, Cazier JB, Campbell H, Sieber O, Lipton L, Gibbs P, Martin NG, Montgomery GW, Young J, Baird PN, Gallinger S, Newcomb P, Hopper J, Jenkins MA, Aaltonen LA, Kerr DJ, Cheadle J, Pharoah P, Casey G, Houlston RS, Dunlop MG. 2011. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet 7:e1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura T, Komatsu Y, Tanaka M, McCann KL, Mishina Y. 2009. Bmp2 and Bmp4 genetically interact to support multiple aspects of mouse development including functional heart development. Genesis 47:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urist MR. 1965. Bone: Formation by autoinduction. Science 150:893–899. [DOI] [PubMed] [Google Scholar]
- Wang Z, Clark CC, Brighton CT. 2006. Up‐regulation of bone morphogenetic proteins in cultured murine bone cells with use of specific electric fields. J Bone Joint Surg Am 88:1053–1065. [DOI] [PubMed] [Google Scholar]
- Welch RD, Jones AL, Bucholz RW, Reinert CM, Tjia JS, Pierce WA, Wozney JM, Li XJ. 1998. Effect of recombinant human bone morphogenetic protein‐2 on fracture healing in a goat tibial fracture model. J Bone Miner Res 13:1483–1490. [DOI] [PubMed] [Google Scholar]
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. 1988. Novel regulators of bone formation: Molecular clones and activities. Science 242:1528–1534. [DOI] [PubMed] [Google Scholar]
- Wu CF, Liu S, Lee YC, Wang R, Sun S, Yin F, Bornmann WG, Yu‐Lee LY, Gallick GE, Zhang W, Lin SH, Kuang J. 2014. RSK promotes G2/M transition through activating phosphorylation of Cdc25A and Cdc25B. Oncogene 33:2385–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LA, Feng J, Wang L, Mu YD, Baker A, Donly KJ, Harris SE, MacDougall M, Chen S. 2011. Development and characterization of a mouse floxed Bmp2 osteoblast cell line that retains osteoblast genotype and phenotype. Cell Tissue Res 343:545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LA, Yuan G, Yang G, Ortiz‐Gonzalez I, Yang W, Cui Y, MacDougall M, Donly KJ, Harris S, Chen S. 2009. Immortalization and characterization of mouse floxed Bmp2/4 osteoblasts. Biochem Biophys Res Commun 386:89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Gopalakrishnan R, Jiang D, Reith E, Benson MD, Franceschi RT. 2002. Bone morphogenetic proteins, extracellular matrix, and mitogen‐activated protein kinase signaling pathways are required for osteoblast‐specific gene expression and differentiation in MC3T3‐E1 cells. J Bone Miner Res 17:101–110. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Katagiri T, Ikeda T, Wozney JM, Rosen V, Wang EA, Kahn AJ, Suda T, Yoshiki S. 1991. Recombinant human bone morphogenetic protein‐2 stimulates osteoblastic maturation and inhibits myogenic differentiation in vitro. J Cell Biol 113:681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]


