Abstract
In humans, the bulk of iron in the body (over 75%) is directed towards heme- or Fe-S cluster cofactor synthesis, and the complex, highly regulated pathways in place to accomplish biosynthesis have evolved to safely assemble and load these cofactors into apoprotein partners. In eukaryotes, heme biosynthesis is both initiated and finalized within the mitochondria, while cellular Fe-S cluster assembly is controlled by correlated pathways both within the mitochondria and within the cytosol. Iron plays a vital role in a wide array of metabolic processes and defects in iron cofactor assembly leads to human diseases. This review describes progress towards our molecular-level understanding of cellular heme and Fe-S cluster biosynthesis, focusing on the regulation and mechanistic details that are essential for understanding human disorders related to the breakdown in these essential pathways.
Keywords: Iron, Heme, Fe-S Clusters, Fe Homeostasis, Fe-Cofactor Biosynthesis
1. The Role of Iron in Biology
Iron’s abundance and unique chemical characteristics are often exploited by nature to drive the complex chemistry required by cells to maintain life. Iron is the fourth most abundant element on the earth’s crust1, so its high prevalence during early evolution is certainly a factor for its current ubiquitous presence in nature. The human body contains 3 to 4 grams of the metal and absorbs 1 to 2 mg of it each day2. The reactivity and ability of the metal to cycle between the Fe(II), (III) and (IV) oxidation states makes it extremely useful for driving intricate reactions in biology that include substrate activation, electron transfer, and oxidation-reduction reactions. Because of this chemical versatility, iron plays a role in nearly every biological pathway. While its utility within biology is apparent, the Achilles heel of this essential metal is its tendency to precipitate in aqueous solutions3. Through coordination to biomolecules, however, the solubility of the metal can be controlled and its reactivity attenuated. At the elemental level, first coordination sphere ligands stabilize the solubility of the metal while at the same time tune its chemical properties for selective participation in only desired reactions 4. Ligand variability in this first coordination sphere helps control how the metal behaves, however at the cellular level a complex network of proteins, controlled at the genetic level, helps ensure metal availability to specific protein partners in the cell5.
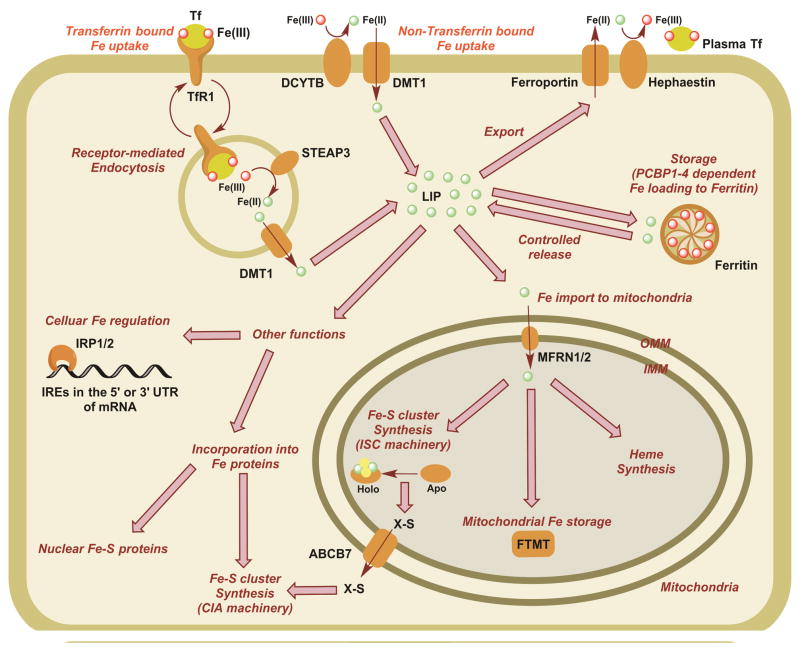
Pathways related to eukaryotic iron homeostasis are shown in Figure 16. Iron is brought into cells either via the transferrin Fe uptake pathway or through direct membrane transporter (ex. the DMT1 pathway)7. Once in the cell, imported iron can bind to iron regulatory proteins where it serves to control concentration, it can be stored within the ferritin complex, it can be exported, or it can be incorporated as cofactors into apoprotein partners. In humans, the bulk of iron in the body (over 75%) is directed towards heme-2 or Fe-S cluster cofactor8 biosynthesis. Assembly of the Fe-cofactors follows highly regulated pathways that have evolved to safely build and load these inorganic Fe-containing cofactors into the apoprotein partners while controlling metal reactivity. In eukaryotes, heme biosynthesis is initiated and completed within the mitochondria, while Fe-S cluster assembly is controlled by separate but correlated pathways within both the mitochondria and the cytosol. Iron plays a central role in a wide array of metabolic processes, so it is not surprising that defects in cofactor assembly leads to human diseases. This review details progress towards elucidating the molecular details of cellular heme and Fe-S cluster biosynthesis. A focus on both regulation and mechanism will be critical for understanding human disorders related to the breakdown in these essential pathways.
Figure 1.
Major cellular Fe utilization pathways in humans. Under physiological conditions, most Fe is internalized as the Tf-bound form, which undergoes receptor-mediated endocytosis via binding to TfR1. Decrease in endosomal pH releases Fe(III) from Tf, which is then reduced by the endosomal reductase STEAP3 to Fe(II). Fe(II) is then transported to the cytosol via DMT1 to join the LIP. Fe in LIP can be utilized for storage in ferritin, import to mitochondria for storage and heme and Fe-S cluster synthesis, export to the cytosol via ferroportin, cellular Fe regulation via IRPs and incorporation into other Fe proteins in the cytosol. Under non-physiological conditions Fe(III) can enter the cell after being reduced to Fe(II) and imported by DMT1. Tf: Transferrin, TfR1: Transferrin receptor 1, DCYTB: duodenal cytochrome b, DMT1: divalent metal transporter 1, STEAP3: six transmembrane epithelial antigen of the phosphate 3, LIP: labile iron pool, PCBP: poly (rC)-binding proteins, OMM: outer mitochondrial membrane, IMM: inner mitochondrial membrane, MFRN: Mitoferrins, FTMT: mitochondrial specific ferritin, ABCB7: transporter for unknown source of sulfur X-S from mitochondria to cytoplasm, IRP: iron regulatory proteins, IRE: iron-responsive elements of mRNA.
2. Heme Cofactors
2.1 Introduction
Among the many metalloporphyrins found in nature, heme is one of the most abundant. It is utilized in many vital biological processes including photosynthesis, oxygen transport, biological oxidation and reduction, and many more. Most of the total iron content in human body is incorporated into heme-containing proteins such as hemoglobin, myoglobin, catalases, peroxidases, nitric oxide synthases, and cytochromes9. The unique structure of heme, which consists of the iron cation coordinated to four nitrogen atoms from a tetrapyrrole ring, allows for fine tuning of the metal’s reactivity to carry out the function of the biomolecule to which it is attached. Here we discuss the different heme types, their biosynthesis and regulation, their function in biology, and finally the diseases linked to a loss of heme function.
2.2 Heme Structure and Types of Heme in Nature
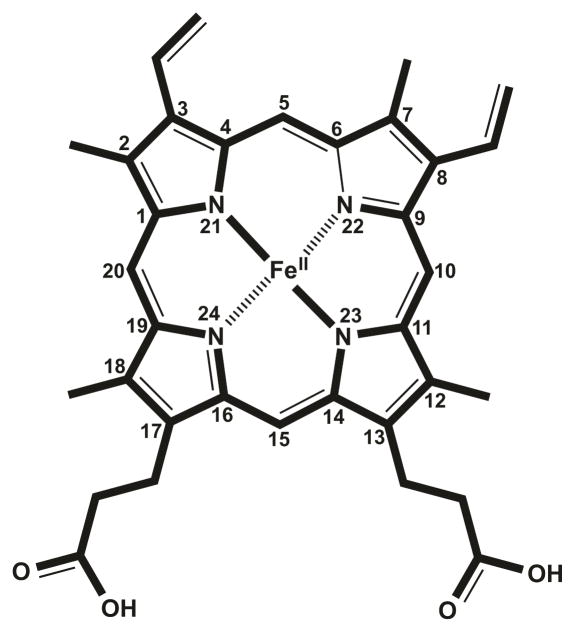
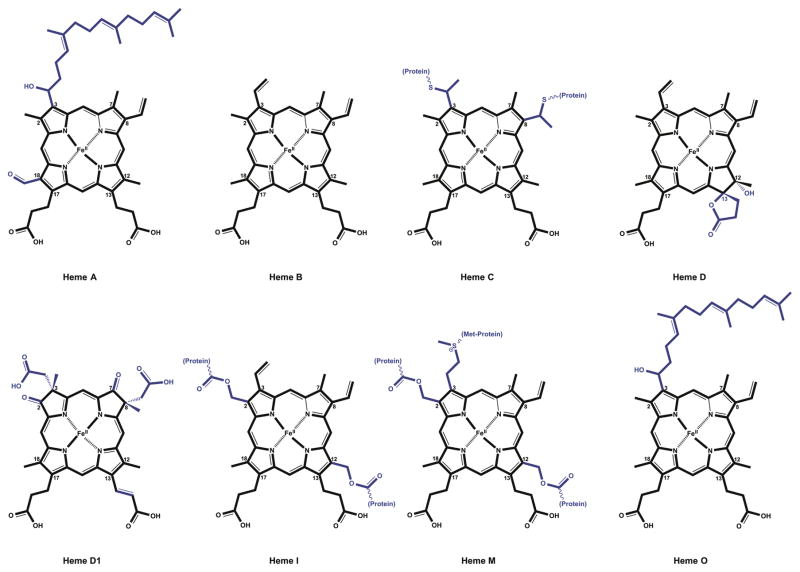
Modifications to the basic heme molecule allow for the diverse array of heme functions found in nature. The parent heme molecule, also known as heme b, protoheme IX or protoporphyrin IX, serves as the platform in each case. The tetrapyrrole unit of heme consists of four pyrrole units linked by four methine bridges. A nitrogen atom from each pyrrole coordinates the iron atom in the center of the planar tetrapyrrole ring. Distortions from planarity can be critical to heme function and reactivity (examples include hemoglobin10 and myeloperoxidase, MPO11). Fifth and sixth ligands to the Fe can be provided by amino acid side chains from the apoprotein or by small inorganic molecules coordinating above and below the plane of the ring. Eight positions in the tetrapyrrole carry side chain modifications, particularly methyl groups on carbons 2, 7, 12, and 18, vinyl groups on carbons 3 and 8, and propionyl groups on carbons 13 and 17 (Figure 2). Substitutions to the same carbons with various other side chains create several biologically important heme types found in nature (Figure 3). Heme A, B, and C are found in a wide spectrum of organisms and take part in vital biochemical processes such as respiration, photosynthesis and oxygen transport. The additional cofactor types (Heme D, D1, I, M, and O) are species-specific and carry out highly specialized functions. Table 1 summarizes the structural features, occurrences, and known functions of each heme type.
Figure 2.
Structure of heme. Standard 1–24 IUPAC numbering system is used to number the carbon atoms of the tetrapyrrole183.
Figure 3.
Important types of heme found in nature. Side chain differences with respect to the parent heme molecule (heme B) are shown in blue.
Table 1.
Comparison of different heme types in nature.
| Type | Structure Remarks | Organisms | Proteins | Function | Reference |
|---|---|---|---|---|---|
| Heme A | C3= hydroxyethylfarnesyl group C18= formyl group Conversion of heme to heme A is catalyzed by Heme A synthase. |
Bacteria, archaea, plants and animals. | Cytochrome a containing heme-Cu oxidases. Ex: Cytochrome c oxidase/Complex IV in mammalian mitochondria. |
Terminal reaction of aerobic respiration. Reduction of oxygen to water. | 193 |
| Heme B | Parent heme molecule also known as protoheme IX, Fe-protoporphyrin IX or heme. Binds to apoproteins non-covalently. |
In a variety of organisms. | Cytochromes b and hemoglobins. | Diverse array of functions associated with heme b such as electron transport and oxygen carrier. | |
| Heme C | C3 and C8= thioether bonds with cysteine residues. Binds to apoproteins covalently. |
In a variety of organisms. | Cytochromes c Ex: bacterial alcohol dehydrogenase, bacterial caa3 and cbb3 oxidases, mitochondrial and bacterial bc1 complex, and cytochrome cd1 nitrite reductase. |
Serve as electron carriers to perform single electron transfers in energy transduction processes such as photosynthesis, respiration and N and S cycles. Other functions include apoptosis, detoxification of ROS. | 194 |
| Heme D | C12= hydroxylated, in a trans conformation to the γ-spirolactone. C13= hydroxylated, propionyl group forms a γ-spirolactone with the hydroxyl group. |
In diverse bacteria including Azotobacter, Proteus, Acetobacter, Salmonella, Bacillus, Pseudomonas, Haemophilus and E. coli. | Terminal respiratory oxidases. Ex: Cytochrome d oxidase. |
Terminal reaction of aerobic respiration. Reduction of oxygen to water at low oxygen levels. | 195–197 |
| Heme D1 | C3 and C8= an acetyl group and a methyl group on each carbon. C2 and C7= keto groups on each carbon. C13 propionyl side chain has reduced to incorporate a vinyl group between second and third carbons. |
In Pseudomonas perfectomarinus, Alcaligenes faecalis, Paracoccus denitrificans, Thiobacillus denitrificans, and Thiosphaera pantotropa. | Nitrite reductase that functions in dissimilatory nitrogen metabolism. Ex: Cytochrome cd1 nitrite reductase. |
Catalyzes the one-electron reduction of nitrite to nitric oxide and fourelectron reduction of oxygen to water. | 198,199 |
| Heme I | C2 and C12= hydroxymethyl groups, they form ester bonds with carboxyl side chains of amino acids. Binds to apoproteins covalently. | In mammalian secretory fluids. | Peroxidases such as bovine milk enzyme lactoperoxidase, eosinophil peroxidase and thyroid peroxidase | Peroxide-driven oxidation of halide and pseudohalide ions as a nonspecific antimicrobial defense mechanism for the protection of mucosal surfaces. | 200 |
| Heme M | C2 and C12= hydroxymethyl groups, they form ester bonds with carboxyl side chains of amino acids. C3= vinyl group on C3 forms a sulfonium ion linkage with the sulfur of a methionyl residue. |
In mammalian neutrophils | Myeloperoxidase | Peroxide-driven oxidation of chloride and bromide ions as an antimicrobial defense mechanism. | 11,200 |
| Heme O | C3= hydroxyethylfarnesyl group. | Bacteria such as E. coli. | Cytochrome o containing quinol oxidases. Ex: Cytochrome bo3 |
Terminal reaction of aerobic respiration. Reduction of oxygen to water. | 201,202 |
2.3 Heme Biosynthesis Pathway
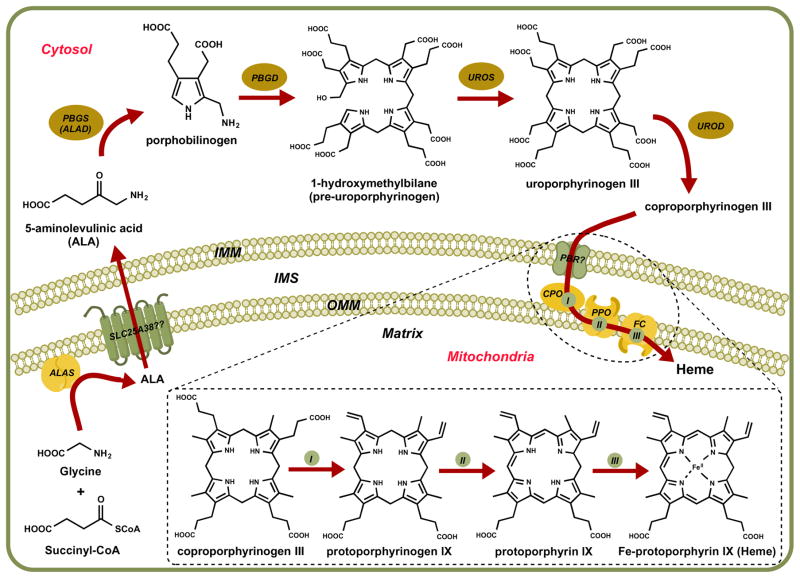
Since heme serves as an essential cofactor to several proteins involved in central metabolic processes, all organisms have established a conserved biosynthetic pathway to synthesize the cofactor. Atomic detail is available for many of the enzymes involved in bioassembly of heme, and these enzyme structures have provided key insights into reaction mechanisms. The general assembly process consists of four stages: the synthesis of a single pyrrole, the assembly of four pyrroles to make the tetrapyrrole ring, modification of the side chains, and the insertion of iron into the ring (Figure 4)12. Specific details for this conserved pathway are outlined below.
Figure 4.
Heme biosynthesis pathway. ALA: 5-aminolevulinic acid, ALAS: ALA synthase, PBGS: porphobilinogen synthase, ALAD: ALA dehydratase, PBGD: porphobilinogen deaminase, UROS: uroporphyrinogen synthase, UROD: uroporphyrinogen decarboxylase, PBR: peripheral-type benzodiazepine receptor, CPO: coproporphyrinogen III oxidase, PPO: protoporphyrinogen IX oxidase, FC: Ferrochelatase, IMM: inner mitochondrial membrane, IMS: intermembrane space, OMM: outer mitochondrial membrane.
2.3.1 Important Steps in Heme Biosynthesis
The first step in eukaryotic heme biosynthesis is the mitochondrial formation of 5-aminolevulinic acid (ALA), which is the precursor that serves as the only source of carbon and nitrogen atoms required to build the basic heme unit. Depending on species, one of two major pathways directs formation of this precursor leading to production of tetrapyrroles. In the Shemin pathway, the enzyme ALA synthase (ALAS) that resides on the matrix side of the inner mitochondrial membrane catalyzes the condensation of glycine and succinyl-CoA13–16. ALAS is found in animals, fungi, non-photosynthetic eukaryotes, and α-proteobacteria (organisms that resemble bacterial ancestors of mitochondria). Mammals have two ALAS isoforms; the first isoform (ALAS1) provides housekeeping functions and a second erythroid specific isoform (ALAS2) provides for the more robust heme requirements in erythrocytes17. Both forms of ALAS are homodimers and require pyridoxal 5′-phosphate (PLP) bound as a Schiff base covalent adduct to a catalytic lysine. During the joining of glycine and succinyl CoA on the PLP scaffold, both CO2 and Coenzyme A are released as byproducts18. Upon formation, ALA exits the mitochondria to serve as the substrate for the subsequent four-enzymatic conversions that occur in the cytosol. Little is known about the specifics of export, but it has been suggested that SLC25A38, a member of the SLC25 family of inner mitochondrial membrane transporters, facilitates import of glycine into the mitochondria in exchange for ALA transfer across mitochondrial inner membrane19.
In the second major pathway for ALA synthesis, also known as the C5 pathway, glutamylt-RNA reductase (GluTR) converts glutamyl-tRNA to glutamate-1-semialdehyde (GSA) in the first of the two-step ALA production pathway20. The second step involves the conversion of GSA to ALA by the enzyme glutamate-1-semialdehyde-2, 1-amino mutase (GSAM)21–23. This second major pathway is found in plants, archaea and most bacteria.
Once exported to the cytosol, ALA serves as the building block to synthesize uroporphyrinogen III following three consecutive steps. The first step involves condensation of two ALA molecules to form porphobilinogen (PBG) by porphobilinogen synthase (PBGS), also known as ALA dehydratase (ALAD)24,25. PBGS is a homooctamer in which each dimer contains one catalytic site26. Each active site binds an ALA molecule at two distinct sites and each subunit binds one zinc atom27. Of the eight Zn atoms on PBGS, four zinc atoms stabilize the enzyme structure while the other four engage in catalytic activity28,29. Four molecules of porphobilinogen undergo polymerization to form 1-hydroxymethylbilane in a reaction catalyzed by porphobilinogen deaminase (PBGD). PBGD uses a covalently attached dipyrromethane cofactor (made of two linked PBG molecules) to prime the polymerization of four PBG molecules. Six PBG molecules form a linear hexapyrrole covalently bound to PBGD, which is then cleaved to yield 1-hydroxymethylbilane and the protein bound cofactor30. This unstable tetrapyrrole serves as the substrate for uroporphyrinogen synthase (UROS), the enzyme that synthesizes uroporphyrinogen III. UROS functions as a monomer and completes ring inversion and the subsequent closure of the tetrapyrrole, yielding uroporphyrinogen III31,32. Spontaneous ring closure is also possible, but the resulting product uroporphyrinogen I cannot be converted to heme. Uroporphyrinogen III also serves as the branching point for the synthesis of chlorophylls and corrins. In heme biosynthesis, this cyclic intermediate is converted to coproporphyrinogen III, a product that lacks four acetic side chains. Subsequent decarboxylation steps are carried out by uroporphyrinogen decarboxylase (UROD) within the cytosol to yield four methyl groups in place of the acetic side chains33. UROD is a homodimer with each subunit carrying an active site cleft that faces the other in the dimer interface34.
Three enzymes associated with the mitochondrial inner membrane complete the terminal steps in the eukaryotic heme biosynthesis. Their arrangement suggests they act as a multiprotein complex that facilitates substrate channeling35. Coproporphyrinogen III entry into the mitochondria is likely mediated by the peripheral-type benzodiazepine receptor (PBR), located on the outer mitochondrial membrane36. In the first of the three terminal steps, coproporphyrinogen III undergoes oxidative decarboxylation of its propionyl side chains on two pyrrole rings to form protoporphyrinogen IX. This reaction is completed in higher eukaryotes and in a few bacterial species by the oxygen-dependent coproporphyrinogen III oxidase (CPO), an enzyme located in mitochondrial intermembrane space37. CPO requires molecular oxygen as the terminal electron acceptor38.
In the next step, protoporphyrinogen IX is oxidized to protoporphyrin IX, following the removal of six hydrogen atoms from the porphyrinogen ring, to provide an alternating double bond structure to the macrocycle; this reaction is catalyzed by protoporphyrinogen IX oxidase (PPO)39. In eukaryotes, the oxygen-dependent PPO is a homodimer that utilizes molecular oxygen as the terminal electron acceptor and functions as an integral membrane protein located in the inner mitochondrial membrane40. The active site of the protein faces the intermembrane space and uses a FAD cofactor for electron transfer. The complete reaction uses three O2 molecules that are reduced to three H2O2 molecules.
Ferrochelatase (FC) is responsible for the insertion of ferrous iron into protoporphyrin IX to form Fe-protoporphyrin IX or heme, in the next step of heme biosynthesis. FC is an inner mitochondrial membrane protein with its catalytic site located on the mitochondrial matrix side. Channeling of protoporphyrin IX from PPO to FC may occur via direct interaction between the two proteins that allows efficient substrate transfer40. FC functions as a homodimer and each monomer contains a 2Fe-2S cluster whose function is unknown41,42. The reaction mechanism involves distortion of the planar porphyrin molecule into a saddle conformation to facilitate the insertion of ferrous iron43. Once synthesized, heme can undergo side chain modifications (ex: heme A and heme O synthesis) or covalent attachments (ex: heme C in cytochrome c biogenesis) to produce additional heme types depending on cellular needs.
2.3.2 Regulation of Heme Biosynthesis
In humans, heme is synthesized at two locations, in erythroid progenitors within bone marrow to provide for developing red cells, and in the liver to provide for numerous heme-containing enzymes. Liver responds to various metabolic states in the body, hence heme synthesis in liver is regulated accordingly. Erythroid progenitors on the other hand maintain heme production at a steady pace to meet the demand of red blood cells. Mechanisms for regulation of heme synthesis in these two origins therefore differ.
The first step, catalyzed by two different isoforms of ALAS in liver (ALAS1) and bone marrow (ALAS2), is the rate-limiting step of heme synthesis. Both isoforms are post-transcriptionally regulated by two different mechanisms. Heme has a negative feedback effect on ALAS1 transcripts and two alternate splice forms of ALAS1 facilitate this regulatory mechanism. One splice form is subjected to heme-mediated decay44–46 but the other is resistant to this effect and requires translation in order to be regulated by heme-mediated decay47,48. Heme itself blocks translocation of a precursor form of ALAS1 from cytoplasm to mitochondria contributing to its downregulation49. Transcription of ALAS1 is upregulated by the peroxisome proliferator activated coactivator 1α (PGC-1α) in correlation to cellular glucose levels50. In contrast, erythroid-specific transcriptional factors such as GATA1 regulate ALAS2 transcription by binding to the promoter region51. In translational regulation of ALAS2, iron responsive elements (IRE) in ALAS2 transcripts can bind iron regulatory proteins (IRP). This prevents ALAS2 mRNA translation, allowing heme synthesis in differentiating red cells to be regulated in relation to cellular iron availability and mitochondrial function52,53.
In addition, three enzymes within the heme biosynthesis pathway (PBGS, PBGD, and UROS) are transcriptionally regulated, yet they utilize a dual promoter system that allows erythroid specific or non-erythroid specific regulation of a single gene. Alternative splicing of 10 exons creates two different PBGS transcripts, either with a housekeeping promoter or with an erythroid–specific promoter that binds erythroid-specific transcription factors including GATA154. Both PBGS transcripts encode identical proteins because they share the same translational start site despite their difference in lengths. On the contrary, alternative splicing of transcripts of the PBGD gene produces proteins of different lengths55,56. Erythroid-specific promoters of PBGD contain several specific cis-acting sequences (including GATA1, NF-E2 and CACCC motifs) that are not seen in the housekeeping promoter57. Similarly, alternative splicing of UROS gene creates two transcripts. Erythroid-specific promoters of UROS contain eight GATA binding sites while the non-erythroid promoter contains NF1, AP1, Oct1, Sp1 and NRF2 binding sites. Both transcripts produce identical proteins58. The remaining enzymes within the pathway have single promoters; regardless, they exhibit erythroid and non-erythroid expression differences. The UROD gene carries a promoter of non-erythroid origin but UROD levels in erythroid tissue are significantly elevated compared to ubiquitous tissue and the specific mechanism resulting in this upregulation has yet to be determined59. The human CPO gene contains a single active promoter with six Sp1 sites, four GATA binding sites and a novel regulatory element named CPRE that aids in the elevated expression in erythroid tissue60. The PPO gene contains a GATA1 binding site in its single promoter, suggesting potential erythroid-specific regulation36. Cis-elements like NF-E2, GATA1 and the Sp1 binding sites are present in the human FC gene promoter and they have been found to induce FC expression during erythroid differentiation while the GC box maintains housekeeping expression of the gene61. Expression of ferrochelatase is also regulated by iron availability of the cells, related to the Fe-S cluster of FC 62.
It is evident that the heme biogenesis enzymes in the erythroid pathway are transcriptionally induced by erythroid-specific transcription factors in coordination with iron uptake. Liver on the other hand maintains sufficient heme levels by combining synthesis and degradation in response to changes in cellular heme pools. Both systems are important however for maintaining cellular iron homeostasis and they are therefore areas of interest for understanding heme related diseases.
2.4 Incorporation of Heme into Apoproteins Recipients
Although diverse in their function, heme-containing proteins share a structurally conserved element, the heme cofactor. Primary factors contributing to the functional diversity of heme proteins are the protein ligands coordinating Fe in the proximal/distal positions and the covalent linkage of the heme to the biomolecule. Insertion and stabilization of the heme unit within a heme containing protein has been studied extensively within cytochrome c. Heme C forms two thioether bonds between C3 and C8 vinyl groups of heme and the cysteine residues of CXXCH motifs in the apocytochrome c. Three systems driving this association have been studied extensively (reviewed in63,64). System I/CCM (Cytochrome C Maturation) is most prevalent in α and γ proteobacteria, all plant mitochondria, some protozoal mitochondria and red algae, and typically involves nine assembly proteins including CcmA through CcmH65. Although system II/CCS (Cytochrome C Synthesis) was originally studied in green algae Chlamydomonas reinhardtii, it can also be found in chloroplasts, most Gram-positive bacteria, cyanobacteria and some β, δ, and ε proteobacteria. Several of the components in system II vary in different organisms but the major components include CcsA, CcsB and CcsX. System III/CCHL (Cytochrome C Heme Lyase) is mostly restricted to fungal, vertebrate and invertebrate mitochondria and some protozoal mitochondria. This system employs a cytochrome c heme lyase enzyme, which is now known as holocytochrome c synthase (HCCS)66 to convert apocytochrome c to holocytochrome c. These three systems carry out a similar function: to keep both Fe in heme and sulfur in cysteine residues of the apoproteins in the reduced state to facilitate correct covalent attachment. Mechanisms of incorporation of additional heme types 12 into different heme binding proteins can be quite diverse and species specific: hence their discussion was eliminated from this review but is discussed in additional sources67–70.
2.5 Heme Function
Heme proteins are ubiquitous in nature and perform a wide variety of functions. One abundant class of heme proteins is the photosynthetic and respiratory cytochromes. Other classes include globins, catalases, peroxidases, cytochrome P450s, oxygenases and others. Here we discuss the versatile chemistry of the heme unit in several categories of heme proteins.
Peroxidases use H2O2 to oxidize substrates without oxygen transfer. Their catalytic cycle includes three steps: 1) H2O2 oxidizes Fe3+ and porphyrin to generate a Fe(IV)oxo porphyrin π cation radical with water as a product; 2) oxidation of substrate reduces the Fe(IV)oxo porphyrin π cation radical; and 3) a second substrate reduces Fe4+ to Fe3+ 71. Examples from mammalian peroxidases include myeloperoxidase (MPO), eosinophil peroxidase (EPO), and lactoperoxidase (LPO)72, and these enzymes can oxidize a wide variety of substrates due to their high reduction potentials. The covalent vinyl sulfonium heme linkage in heme M of MPO enables heme distortion and the resulting reduction in electron density in the heme best explains the unusually high reduction potential of MPO73. These peroxidases are capable of generating oxidants such as hypohalous acids, hypothiocyanous acid, reactive nitrogen species, singlet oxygen, phenoxyl and hydroxyl radicals, all which are key components in antimicrobial properties exerted by the immune system72.
Cytochromes P450 represent another subgroup of heme proteins in high abundance in nature; humans alone carry more than fifty P450 enzymes. Despite the immense diversity of P450s in nature, as per published structures, the overall fold in these biomolecules is basically the same. The typical electron transfer catalytic cycle of a P450 requires electrons derived from redox protein partners such as flavin and Fe-S containing proteins. Substrate binding shifts low-spin hexacoordinate heme to high-spin heme while displacing the axial water ligand, which allows electron transfer from the redox partner and oxygen binding to heme to form the oxy complex. A second electron transfer step is responsible for generating the heme Fe(III)dihydroperoxy species which finally undergoes heterolytic cleavage to give heme Fe(IV)oxo species71. Fe(IV)oxo porphyrin π cation radicals have been proposed as reactive species in catalytic oxygenation reactions. Although both peroxidases and cytochromes P450 form Fe(IV)oxo intermediates during their catalytic cycles, profound differences in protein environments create different products. Cytochromes P450 are mostly known for xenobiotics detoxification in liver, where drugs and other xenobiotics are hydroxylated and made more soluble, facilitating their conversion to easily eliminated products. In addition, P450s also participate in the biosynthesis of steroids, highlighting their importance in metabolism.
Additional noteworthy heme enzymes include nitric oxide synthase (NOS) and heme oxygenase (HO). NOS catalyzes the oxidation of L-arginine to L-citrulline and nitric oxide (NO), a signaling molecule important for regulation of the cardiovascular and nervous systems as well as participating in immune responses. NOS catalysis is similar to P450 mechanism in the first step in which L-arginine is converted to Nω-L-hydroxyarginine except that an unusual cofactor, tetrahydrobiopterin or BH4 is required. Oxidation of Nω-L-hydroxyarginine to L-citrulline in the second step is proposed to be carried out via peroxy or superperoxy species as opposed to the Fe(IV)oxo intermediates discussed before. Heme oxygenase is responsible for the degradation of free heme, resulting in its efficient elimination and the recycling of iron. This conversion is carried out in three distinct steps; heme is first converted to α–meso-hydroxyheme, which in turn gets converted to verdoheme and subsequently to billiverdin. Studies show it is highly unlikely for an Fe(IV)oxo species to serve as an oxidant in the catalytic action of heme oxygenase since it uses a histidine as an axial ligand71.
The complete list of heme proteins is extensive and exceeds the limits of this review. Instead, we will focus on human disorders that result from loss of heme protein function, many of which are severe, and merit detailed discussion.
2.6 Diseases of Heme Synthesis
Several human diseases are associated with disruption of the heme biosynthetic pathway. Many of these diseases are associated with inherited mutations in heme biosynthesis genes, however some are caused by environmental factors affecting their enzyme products. Along with nine major porphyrias, we will focus on diseases associated with disruption of the heme biosynthesis pathway.
2.6.1 Porphyrias
The clinical presentation of porphyria includes skin lesions and acute neurovisceral attacks which are related to the accumulation of specific intermediates in the heme biosynthetic pathway. Nine such diseases have been identified and they are categorized as hepatic or erythropoietic, pertaining to the organ in which the intermediates accumulate. However, a more clinical classification of porphyria divides these into three groups: acute, cutaneous and rare recessive.
Even though no disease causing mutations for ALAS1 has ever been found in humans, many mutations affecting the function of the ALAS2 isoform have been identified. Although not commonly seen as loss of function mutations in the heme biosynthesis pathway, gain of function deletions in ALAS2 are found to be causative of a cutaneous porphyria called X-linked dominant erythropoietic protoporphyria (XLDPP). This disease is characterized by increased ALAS2 activity and excessive protoporphyrin production. ALA production is increased such that the final step catalyzed by ferrochelatase is rate-limiting, resulting in the accumulation of protoporphyrin. Liver damage and photosensitivity are the clinical manifestations of this disease74.
Deficiency of ALAD causes a rare recessive porphyria named ALAD porphyria, for which only less than ten cases have been reported. In this autosomal disorder, loss of ALAD activity in liver and erythroid precursors leads to excretion of ALA and coproporphyrinogen III into the urine. Patients suffer from intermittent acute neurovisceral attacks and/or chronic neuropathy, and onset of this disorder ranges from childhood to adulthood75.
An additional disease related to the first steps in heme biosynthesis is acute intermittent porphyria (AIP). As in all acute porphyrias, acute life-threatening complications occur mostly in the adulthood, however in rare cases severe attacks can occur in the childhood. Urinary excretion of ALA and porphobilinogen is increased due to decreased PBGD activity. Activity loss is caused by mutations in the PBGD gene and over 200 mutations have been identified. This disorder is seen in homozygous dominants for the trait and a majority of heterozygotes remain disease-free76.
Congenital erythropoietic porphyria (CEP) is another rare recessive type of porphyria caused by the deficiency of UROS. This autosomal recessive disease that is a result of loss of UROS function leads to the spontaneous formation of uroporphyrinogen I, the isomer of uroporphyrinogen III that cannot be converted to heme, so it is accumulated and excreted77. Clinical features of this disease include chronic hemolysis and cutaneous photosensitivity caused by diffusion of uroporphyrinogen I to plasma, and these conditions begin to manifest in early infancy. There have been over twenty UROS mutations that cause this disease identified to date78.
Porphyria cutanea tarda (PCT) is the most common porphyria and it falls under the category of cutaneous porphyrias. This disease only shows symptoms in the skin, including lesions in areas exposed to sun, skin fragility leading to secondary infections, and hypertrichosis. Ocular pain and photophobia have also been reported rarely79. The cause of these symptoms is accumulation of excessive porphyrin in the skin. Tetrapyrroles are highly photoreactive and hence they absorb energy from the visible region of electromagnetic spectrum. Excited ring structures reach ground state by transferring energy, which drives peroxidation and oxidation of biological macromolecules such as membrane lipids, nucleic acids and proteins. Familial PCT, one of the two forms of PCT, is an autosomal dominant trait similar to AIP in that only a minority of heterozygotes is affected. In familial PCT, uroporphyrinogen III is accumulated and excreted in urine due to UROD mutations. Patients with sporadic PCT, although not associated with UROD mutations, show decreased UROD activity and hepatic Fe overload, similar to that seen in familial PCT80. Hepatoerythropoietic porphyria (HEP) is a rare porphyria associated with UROD deficiency. Symptoms are similar to that seen in PCT (skin lesions, red urine, hypertrichosis and scarring) however they can be much more severe. This rare recessive porphyria often onsets in infancy or childhood and shows high porphyrin concentrations in erythrocytes and less than 10% UROD activity81.
Hereditary coproporphyria (HCP), another acute porphyria, is caused by mutations that destabilize the enzyme CPO. Clinical characteristics are very similar to other acute porphyrias (i.e., AIP) and show a high coproporphyrinogen III content in urine and feces, as well as increased photosensitivity. Also similar to other acute porphyrias, HCP is mostly seen in dominant homozygotes for the trait, with only some heterozygotes developing the disease82.
An additional porphyria caused by mutations in a heme biosynthesis enzyme includes variegate porphyria (VP), characterized by dysfunctional PPO. Symptoms are again very similar to any acute porphyria, showing acute neurovisceral attacks and cutaneous photosensitivity. Onset of the symptoms is in adulthood and over 100 PPO gene mutations (including nonsense, missense, deletion, insertion and splice mutations) have been identified83. As in AIP, urinary excretion of ALA and porphobilinogen and fecal content of protoporphyrinogen IX and coproporphyrinogen III is increased during acute attacks. In addition, PPO activity is dramatically reduced (50%) in tissues of VP patients84.
The last of the nine porphyrias associated with heme synthesis is another cutaneous porphyria named erythropoietic protoporphyria (EPP). Mutations in the ferrochelatase gene cause a deficiency in mitochondria leading to accumulation of free protoporphyrin IX, primarily in erythrocytes. Excess protoporphyrin IX is behind the predominant clinical feature of photosensitivity, which begins in childhood85,86.
2.6.2 Additional Diseases
Loss of function mutations of ALAS2 is the cause of the disorder X-linked sideroblastic anemia (XLSA). A group of point mutations alter the protein’s ability to bind the PLP cofactor, however other mutations affect protein domains outside the cofactor-binding region. The decrease in heme synthesis efficiency prevents normal erythroblast development in the bone marrow. The resulting overload of iron results in characteristic iron granules surrounding the nucleus (a sideroblast). This disease is characterized by microcytic hyperchromic anemia, the presence of mature but pale and smaller than normal erythrocytes and Fe overloaded mitochondria in erythroblasts in the bone marrow87.
ALAD requires Zn for function, and Zn deficiency makes ALAD susceptible to Pb inhibition. Pb replaces Zn in ALAD and the inhibited protein leads to high levels of ALA in blood, which is responsible for neurological manifestations due to its toxicity at high concentrations88. A similar disease, named hepatorenal tyrosinemia, shows neurotoxicity due to high ALA levels, with abnormal liver and kidney function. This disease is caused by mutations in fumarylacetoacetate hydrolase gene, which hydrolyzes fumarylacetoacetate to fumarate and acetoacetate during the tyrosine catabolism pathway. In the event of enzyme deficiency, fumarylacetoacetate is metabolized to succinylacetone, which is structurally similar to ALA, hence it acts as a potent inhibitor for ALAD89. Excess ALA and succinylacetone can be seen in patients’ urine and blood samples.
3 Fe-S Clusters
3.1 Introduction
Iron-sulfur (Fe-S) clusters are the second major form of complex iron cofactors found in biology. Due to the high abundance of iron and sulfur on the earth’s surface, and the easy association of these atoms under anaerobic conditions, Fe-S clusters likely developed early in evolution before the earth’s transition to an aerobic atmosphere. Consequently, these cofactors are ubiquitous in all organisms and play a role in almost every biological pathway. Here we provide an overview of the structure, formation, and function of Fe-S cluster cofactors.
3.2 Fe-S Cluster Structure
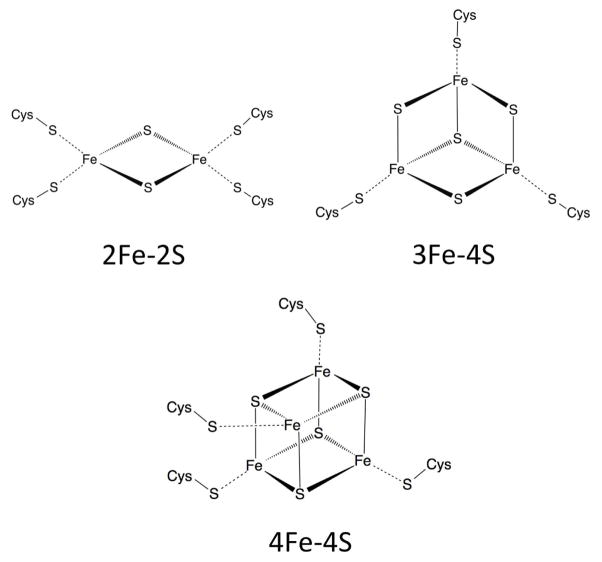
In many ways, Fe-S clusters are simpler than heme. While heme is a mixture of organic (protoporphyrin) and inorganic (iron) components, Fe-S clusters are strictly inorganic. Iron atoms in biological Fe-S clusters interact directly with protein residues and sulfur atoms bridge neighboring iron atoms. Fe-S clusters exist in a variety of configurations depending on their respective number of iron and sulfur atoms. The three most common forms (2Fe-2S, 3Fe-4S and 4Fe-4S) are illustrated in Figure 5. More complex Fe-S clusters have also been observed, including the 7Fe-8S and 8Fe-8S clusters identified in ferredoxins from Desulfovibrio africanus90.
Figure 5.
Structure for the 3 most common forms of Fe-S Cluster: 2Fe-2S, 3Fe-4S, and 4Fe-4S.
The Fe atoms within the Fe-S cluster can exist in either ferric or ferrous forms and cycling between these redox states allows the transfer of electrons for redox reactions. The tendency of the oxidized FeS cluster to gain an electron is termed the “reduction potential”. By convention, this potential is expressed in comparison to a reference standard hydrogen electrode, which is assigned a potential of 0V. Depending on Fe-S cluster type, interactions with neighboring amino acids, and solvent accessibility, a single Fe-S cluster can up to two electrons with a reduction potential spanning ~1000 mV91. This remarkable range of accessible reduction potentials can largely explain the biological utility of the FeS cluster.
Fe-S clusters do not exist freely but are intimately connected to their apoprotein partner. Free iron will form an insoluble complex when bound to sulfide, so the protein plays a critical role in solubilizing the Fe-S unit. Fe-S proteins usually bind their corresponding Fe-S cofactor via ionic interactions between cysteine thiols and iron in the Fe-S cofactor. In some cases, Fe-S clusters are alternatively ligated via histidine residues. Subsets of 2Fe2S clusters, such as those found in Rieske proteins (see section 3.5), are coordinated by two cysteine and two histidines (Cys2His2)92 and a common coordination theme of proteins involved in Fe-S cluster biogenesis is Cys3His1 coordination93.
3.3 General Fe-S Cluster Biogenesis Pathways
Production of Fe-S clusters must be highly regulated to prevent unwanted reactions of both free iron and sulfur. Similar to heme-proteins, Fe-S proteins are synthesized in their apo-state and obtain their Fe-S cluster cofactor from a dedicated Fe-S cluster formation pathway. At present, there are three known general pathways for Fe-S cluster formation: the iron sulfur cluster (ISC), cytosolic iron sulfur assembly (CIA), and sulfur assimilation (SUF) pathways. These three pathways provide Fe-S clusters for the majority of Fe-S proteins in almost all organisms. While dedicated Fe-S cluster forming pathways can exist for individual Fe-S proteins, such as the nitrogen fixation (Nif) pathway that provides an Fe-S cluster for the nitrogenase enzyme in nitrogen-fixing bacteria94, this review will focus only on the general Fe-S cluster production pathways.
3.3.1 Iron Sulfur Cluster (ISC) Pathway
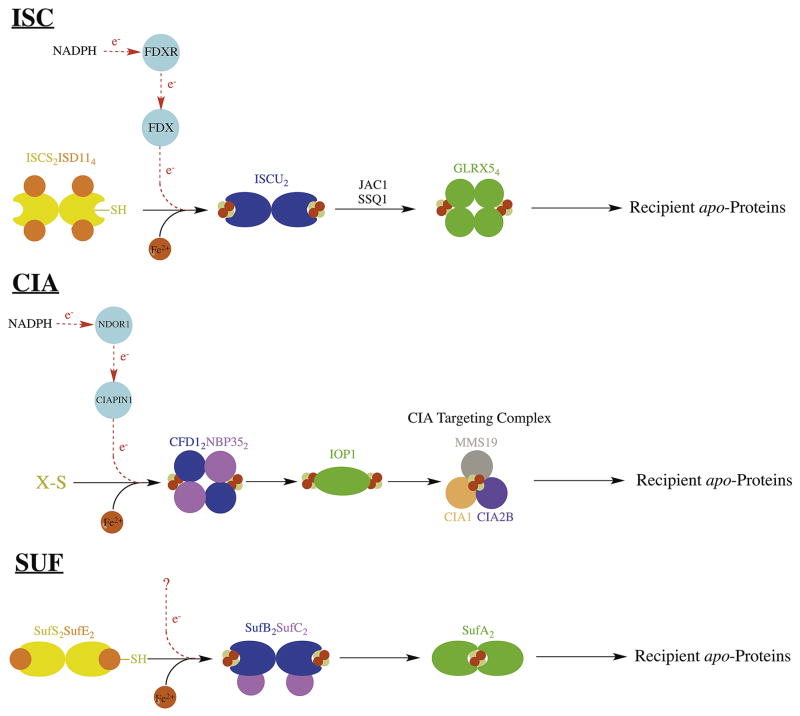
The most robust and best-characterized pathway for Fe-S cluster biosynthesis is the iron sulfur cluster (ISC) pathway. A simplified description of this pathway is provided in Figure 6. The ISC pathway, present in bacteria and in the mitochondria of eukaryotes, provides general housekeeping Fe-S clusters to a large number of Fe-S proteins. In eukaryotes, this pathway provides Fe-S clusters for several key mitochondrial Fe-S proteins. ISC was initially identified in the Azotobacter vinelandii and Escherichia coli bacterial species, where ISC genes are arranged in the isc operon. Additional early work in eukaryotes (in Saccharomyces cerevisiae and human proteins) revealed a highly homologous system localized to the mitochondria. In addition to providing for mitochondrial Fe-S proteins, the ISC system provides a component to the cytosolic and nuclear Fe-S cluster formation pathways, thus ISC is essential for the maturation of all cellular Fe-S proteins in eukaryotes95,96.
Figure 6.
Diagram depicting de novo Fe-S cluster formation and the main Fe-S cluster transfer steps in the ISC, SUF, and CIA systems. Protein names used are from the human system for ISC and CIA, and from the bacterial system for SUF.
ISC serves as a template for understanding general Fe-S cluster production. In human ISC, de novo 2Fe-2S synthesis occurs on the dedicated scaffold protein ISCU97. Sulfur for this reaction is provided by ISC’s dedicated cysteine desulfurase enzyme ISCS via a persulfide intermediate that gets transferred to ISCU98 upon formation of an ISCU-ISCS complex. In eukaryotes, ISCS has a dedicated protein co-factor ISD11 that is essential for ISCS function99. Electrons for ISCS persulfide release are provided by the 2Fe-2S cluster containing ferredoxin FDX, which in turn gets reduced by the ferredoxin reductase FDXR that uses NADPH as its final electron source100,101. FDX also interacts with ISCU, providing 2 reducing equivalents for assimilation of two 2Fe-2S clusters on an ISCU dimer to form a single 4Fe-4S cluster97. An additional Fe-binding protein Frataxin interacts with the ISCS-ISCU complex and regulates ISCS activity102. Additionally, there are alternate scaffold proteins (ISCA103 and NFU1104) that interact with ISC proteins and these are believed to be required for the maturation of a specific subset of Fe-S proteins.
Despite intense study, the physiologic source of iron for ISC remains a subject of debate. Several potential iron donors have been investigated and iron delivery to ISCU has been demonstrated from several potential sources in vitro within a variety of systems. The protein frataxin105,106 interacts with the ISCU-ISCS complex and could be the source of iron for the pathway106. In the bacterial system, two additional members of the isc operon have been investigated as potential iron donors, IscX and IscA. The small acidic protein IscX binds iron and regulates cysteine desulfurase activity in a manner very similar to Frataxin107. The alternative scaffold IscA is also an interesting candidate because of its tight (KD~1019) binding affinity for mononuclear iron108 and its capability of delivering iron to IscU109,110. An additional interesting hypothesis is that iron may come from a glutathione-glutaredoxin complex111. The lack of conclusive evidence for a specific iron source suggests that in vivo, there could be multiple iron sources or that the mode of iron delivery may be atypical.
A detailed mechanism for Fe-S cluster delivery from ISCU to downstream targets is currently under investigation. The Fe-S cluster bound to ISCU is transferred to the glutaredoxin GLRX5112,113. Efficient transfer from ISCU to GLRX5 requires involvement of the ATPase SSQ1, which binds to both ISCU and GLRX5114. Binding of SSQ1 to ISCU, along with the interaction of a DnaJ-like co-chaperone JAC1115, destabilizes the Fe-S cluster on ISCU facilitating its transfer from ISCU to GLRX5116. GLRX5 is considered the end of the ISC pathway because it is the last common Fe-S cluster carrier for all mitochondrial Fe-S proteins. GLRX5 continues the cluster transfer process, however, and it is able to interact with a variety of downstream Fe-S proteins117–119. The specific recipient depends on the ultimate destination of the Fe-S cluster. For example, 4Fe-4S cluster conversion and delivery is facilitated by GLRX5’s interaction with two other Fe-S proteins, ISCA and IBA57120.
Despite being mostly localized to mitochondria121, ISC is required for maturation of all cellular Fe-S proteins96. The mechanism by which cytosolic Fe-S proteins depend on mitochondrial ISC is actively being investigated. Because Fe-S clusters are not able to cross the inner mitochondrial membrane122, this mechanism likely involves the transport of an Fe-S cluster precursor out of the mitochondria and into the cytosol. Recent work has identified an unknown compound produced by mitochondrial ISCS (named ‘X-S’) that may provide reduced sulfur for cytosolic Fe-S cluster formation123.
3.3.2 Cytosolic Iron-Sulfur Assembly (CIA) Pathway
Recent studies have revealed the involvement of another essential and highly conserved Fe-S biosynthetic pathway that is present within the cytosol and nucleus of eukaryotes. This pathway is called cytosolic iron-sulfur assembly (CIA)124. This pathway has been identified in many eukaryotic systems and is essential in almost all cases125. CIA is unique among Fe-S maturation pathways in that it does not obtain reduced sulfur via a dedicated cysteine desulfurase. A simplified description of the CIA pathway is provided in Figure 6. Instead of an ISCS analog, CIA relies on mitochondrial export of a sulfur-containing compound, ‘X-S’, via the mitochondrial export protein ABCB7126 and the intermembrane space protein ALR127. The identity of X-S is currently unknown, but may be glutathione-complexed to an Fe-S cluster125. In human CIA, the primary scaffold for de novo Fe-S assembly is a tetrameric complex formed between CFD1 and NBP35, which binds a bridging 4Fe-4S cluster between the CFD1 and NBP35 subunits128. Reducing equivalents for this reaction are provided by an Fe-S containing protein CIAPIN1 (similar to FDX in ISC), which in turn gets reduced by the diflavin reductase NDOR1 (similar to FDXR in ISC), utilizing reducing equivalents from NADPH129,130.
The 4Fe-4S clusters from the NBP35-CFD1 complex get transferred to another Fe-S protein IOP1, which binds two 4Fe-4S clusters per monomer131,132. IOP1, in turn, delivers its Fe-S clusters to a multi-component complex called the CIA targeting complex, which consists of at least three proteins CIA1, CIA2B, and MMS19133. The CIA targeting complex is able to interact with a variety of recipient Fe-S proteins, likely an indication of its function in downstream Fe-S cluster delivery.
3.3.3 Sulfur Assimilation (SUF) Pathway
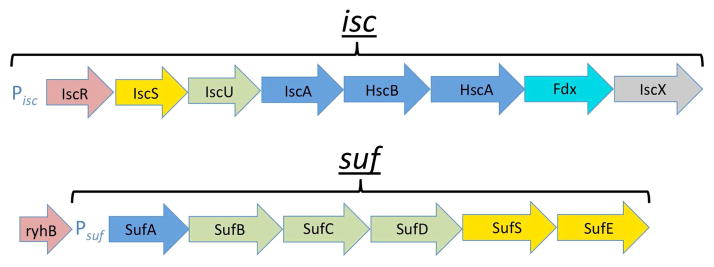
Of the three Fe-S general cluster formation pathways, the sulfur assimilation (SUF) pathway is probably the most ancient. SUF predominantly exists in prokaryotes, however it is found in specific locations in eukarya, including the chloroplasts in some plants134 and the apicoplasts in some plasmodium species135, and recently proteins homologous to bacterial SUF were discovered in the cytosol of a blastocystis species136. At present, SUF has been best characterized in the Gram-negative bacteria Escherichia coli and Erwynia chrysanthemi where its genes are organized into the suf operon (Figure 7).
Figure 7.
Illustration of suf and isc operons, with corresponding promoters (Pisc and Psuf), which play a central role in regulation of SUF and ISC at the genetic level in bacteria. Operons depicted are from the E. coli model system. Steps are colored based on the encoded protein’s function as follows: red (regulatory), yellow (sulfur delivery), green (primary scaffold), blue (downstream Fe-S cluster delivery), cyan (electron transfer), and gray (unknown).
The SUF pathway is similar to ISC in many ways. Like in ISC, SUF provides general Fe-S cluster formation to accommodate a variety of Fe-S proteins. In fact, SUF and ISC seem to be redundant in Gram-negative bacteria, as the removal of the entire isc or suf operon results in no deleterious effects. Simultaneous suf/isc operon deletion, however, is lethal137. While ISC and SUF follow the same general mechanism for Fe-S cluster formation (Figure 6), SUF seems to be favored under conditions of oxidative stress and iron limitation138 and the SUF proteins are correspondingly more stable under adverse conditions in vitro139. In E. coli, the SUF pathway centers around two heteromeric complexes called SufBC and SufSE.
The primary scaffold SufB requires a binding partner SufC for activity, forming the SufBC complex in a SufB2C2 arrangement. The SufBC complex can form a 4Fe-4S cluster on SufB that can be transferred to recipient proteins140. The exact role of SufC in this process is unknown, but it has ATPase activity that is essential for Fe-S cluster formation on SufB141. SufB, on its own is relatively unstable and prone to spontaneous oligomerization. There is also a paralogue of SufB, named SufD that is able to replace a SufB in the SufBC complex, resulting in the SufBCD complex142. However, SufBC is likely the most active form143. The SUF cysteine desulfurase SufS functions in a similar manner to ISCS, accepting sulfur from cysteine via a persulfide intermediate. SufS has an essential binding partner SufE, which is required for activity, forming the SufSE complex144. While it may seem SufE is similar to ISD11 from eukaryotic ISC, SufE functions differently from ISD11 in that it accepts the persulfide from SufS and allows the SufS enzyme to complete its turnover145.
Details of SUF’s downstream cluster delivery are not as well established as in the ISC pathway. The 4Fe-4S cluster formed by SufBC can be transferred to the A-type carrier protein SufA in vitro140,143, but SufBC also may be able to transfer directly to recipient apo-proteins. In vivo, SufA is functionally redundant with the ISCA bacterial homologue146 and possibly acts as an intermediate carrier of the 4Fe-4S cluster from SufB, passing it off to downstream apoproteins147,148. Another protein involved in this process (ErpA) has redundant function with SufA but is necessary for the development of active Fe-S proteins149.
Several important details of the SUF pathway remain to be identified. As in ISC, the in vivo source of iron is unknown. Also of interest is SufD’s incorporation in the SufBCD complex, which allows SufB to accept iron in vivo141 and facilitates binding of a FADH2 cofactor142. This cofactor may be able to reduce ferric iron, facilitating potential Fe3+ sources such as ferritins or ferric citrate150. While SufA can deliver mononuclear iron to SufBC in vitro109, it is currently believed to function downstream of de novo Fe-S formation (Figure 6). The source of electrons for SufS turnover and for Fe-S formation remains in question151.
3.4 Fe-S Cluster Biogenesis Regulation
The best-developed model for Fe-S biogenesis pathway regulation comes from work done in Gram-negative bacterial systems, where both the ISC and SUF pathways are present. This work reveals a fascinating interplay between ISC and SUF, where the necessary ISC and SUF genes are organized into their respective isc and suf operons (Figure 7). While this review has focused on eukaryotic systems, regulatory mechanisms in bacteria may provide insight into how this regulation occurs in eukaryotes. At the center of E. coli Fe-S cluster biogenesis regulation is a DNA-binding protein IscR, the first member of the isc operon, which directly regulates both the ISC and SUF systems.
Under non-stressed conditions, ISC is favored over SUF for general housekeeping of Fe-S cluster biosynthesis152. The transcriptional regulator IscR can bind a 2Fe-2S cluster (forming holo-IscR), obtaining its Fe-S cluster from the same ISC machinery utilized by other Fe-S proteins153. In the holo configuration, IscR binds to the isc promoter and prevents binding of RNA polymerase154. Thus, holo-IscR acts as a feedback regulator, inhibiting transcription of the entire isc operon when ISC activity is sufficient155. IscR is a relatively poor substrate for ISC-mediated Fe-S cluster loading156 and holo-IscR can only form after the ISC proteins have exhausted their interactions with other apo-Fe-S proteins. In addition to being a weak ISC substrate, IscR does not bind its Fe-S cluster tightly and effectively acts as a sensor of cellular iron and oxygen conditions157. Under high-oxygen or low-iron conditions, holo-IscR quickly reverts to apo-IscR. Therefore under typical aerobic conditions, high oxygen levels cause holo-IscR to revert to apo-IscR. Apo-IscR dissociates from the isc promoter and isc is uninhibited.
In the apo configuration, IscR does not bind to the isc promoter but instead favors binding to the suf promoter, activating transcription of SUF genes158. Appropriate interaction of apo-IscR with the suf promoter involves two additional transcription factors: the ferric uptake regulator (Fur) and the peroxide responsive regulator (OxyR). Suf expression is constitutively repressed by Fur, which binds Fe2+ under non-stressed conditions when iron levels are sufficient and oxidative stress is low. With its Fe2+ cofactor, Fur binds to the suf promoter at the same site as apo-IscR, inhibiting suf expression159. When the cell faces iron deficiency, Fur loses its iron cofactor, dissociates from the suf promoter, thus triggering transcription of suf. The cell’s preference for SUF over ISC in the presence of oxidative stress also reveals the involvement of another transcription factor (OxyR), as oxidized OxyR recruits RNA polymerase to the suf promoter.
There are additional regulatory mechanisms for cluster bioassembly beyond the level of gene expression. The small non-coding RNA RyhB, for example, is encoded just upstream of the SUF promoter and can bind to the iscRSUA mRNA to prevents its translation160. RyhB expression, however, is constitutively repressed by Fur-Fe2+, so RyhB effectively inhibits ISC when conditions favor SUF161.
3.5 Fe-S Cluster Function
Fe-S clusters are versatile biological cofactors found in the most fundamental biochemical pathways, including aconitase and succinate dehydrogenase of the citric acid cycle and respiratory complexes I-III of the electron transport chain121. Nuclear Fe-S proteins also have a unique role in DNA damage recognition and repair. Several forms of DNA polymerase, helicase, glycosylase, and primase all contain Fe-S clusters125. Considering their remarkable range of functions, a thorough summary of various Fe-S proteins is well beyond the limited scope of this review. New Fe-S proteins continue to be discovered but in many cases, the role of the Fe-S cluster within the Fe-S protein remains unknown, even if the cluster’s presence is essential for proper protein function.
Fe-S clusters can be found in the active site of many essential enzymes and usually are involved directly in catalysis. Being stable in a variety of redox states, Fe-S clusters are best known for their role as electron carriers. Fe-S clusters can carry usually one, but sometimes two electrons and are, subsequently, stable in various reduced states. 2Fe-2S clusters, for example, can exist in oxidized (Fe3+/Fe3+) or reduced (Fe3+/Fe2+) forms while 4Fe-4S clusters are stable in oxidized (Fe3+/Fe3+/Fe3+/Fe2+), intermediate (Fe3+/Fe3+/Fe2+/Fe2+), and reduced (Fe3+/Fe2+/Fe2+/Fe2+) forms162. The reduction potential of an Fe-S cluster is often modulated by interactions with nearest neighbor protein residues and by access to solvent, allowing for a large range of biological functions. Ferredoxins are considered the archetypical Fe-S cluster electron carriers and were the earliest Fe-S proteins to be functionally characterized163. Ferredoxins are involved in many essential biochemical pathways, transferring electrons for cellular respiration, photosynthesis, and nitrogen fixation164. A ferredoxin is even involved in the ISC iron sulfur cluster biogenesis pathway, as discussed previously (see section 3.3.1)165.
But Fe-S cluster-mediated electron transfer is not limited to ferredoxins. In fact, one of the most the fundamental electron transfer processes, the electron transport chain (ETC), utilizes numerous Fe-S clusters. Respiratory complexes I, II, and III of the ETC all contain Fe-S clusters. Respiratory complex I uses a network of 8 Fe-S clusters for step-wise electron transfer166. Similarly, complex II contains an Fe-S protein component called SDHB with a 2Fe-2S, 3Fe-4S, and 4Fe-4S cluster167. Lastly, complex III utilizes a unique Fe-S cluster called a “Rieske center”168. The Rieske center is a 2Fe-2S cluster where one of the iron atoms is coordinated by histidines instead of cysteines, resulting in a Cys2His2 coordination169.
Fe-S clusters can also be involved in non-redox reactions. The 4Fe-4S cluster in aconitase, for example, catalyzes a hydration-dehydration reaction, ligating directly to the citrate substrate170. In some cases, Fe-S clusters appear to only serve a structural function and not participate in chemistry directly, as is the case in endonuclease III171.
3.6 Fe-S Clusters in Human Disease
Unlike in Gram-negative bacteria, where ISC/SUF redundancy allows for removal of an entire pathway, in humans the absence or mutation of a single component is often incompatible with life. In select cases there are human diseases that have been linked to defective Fe-S cluster biogenesis pathways. Below we describe several diseases directly linked to dysfunctional Fe-S cluster formation.
3.6.1 Friedreich’s Ataxia
With an incidence of 1 in 50,000172,173, and a carrier prevalence of 1 in 100174, Friedreich’s ataxia (FRDA) is by far the most prevalent disease linked to defective Fe-S cluster formation. FRDA is an autosomal recessive genetic disease caused by a GAA-trinucleotide repeat expansion in an intron of the frataxin gene, a protein involved in the ISC pathway175. This trinucleotide repeat expansion leads to under-expression of the frataxin gene and subsequently, low levels of frataxin176. These insufficient frataxin levels are responsible for the pathophysiology of FRDA, but the precise role of frataxin is still unknown177. Frataxin may deliver iron to the ISC pathway178, be an allosteric activator of the ISCU-ISCS complex102, or may have a combination of roles. FRDA tissues demonstrate increased mitochondrial iron deposits179 which leads to increased oxidative stress and cell death in metabolically active tissues such as cardiomyocytes and neurons of the dorsal root ganglia. FRDA presents early in adolescence with progressive ataxia, or difficulty coordinating movement, sensory loss, weakness, and dysarthria. FRDA patients are usually wheelchair bound in their teens with a significantly reduced quality of life and life expectancy180. Median age of survival is 35 years with cardiac dysfunction usually being the cause of death181.
3.6.2 ISCU Myopathy
ISCU myopathy (IM) is an additional condition related to a defect in Fe-S cluster biogenesis. It is the 2nd most common disorder linked to defective Fe-S cluster synthesis but is much less common than FRDA with only 25 known cases. To date, all patients identified with IM have come from families of Swedish ancestry121. Similar to FRDA, the IM phenotype is inherited in an autosomal recessive pattern. IM is caused by a splicing defect during ISCU post-transcriptional processing that leads to defective ISCU protein182,183. Loss of ISCU leads to lower ISC activity and a resulting deficiency of essential Fe-S proteins, including succinate dehydrogenase and aconitase of the citric acid cycle184. Symptoms are exacerbated in cells that are metabolically active, such as the myocytes of skeletal muscle during exercise, and patients with IM experience exercise intolerance185. Prolonged activity can lead to tachycardia, tachypnea, and muscle pain186. Unlike FRDA, IM is not progressive and most cases have a normal life expectancy.
3.6.3 GLRX5 Sideroblastic Anemia
GLRX5 Sideroblastic Anemia (GSA), a disease caused by mutated GLRX5, has only been identified in a single patient to date. While GSA is exceedingly rare, this particular case study has revealed a unique mechanism linking Fe-S cluster production to general iron homeostasis52,187. GLRX5, involved in the last step of the ISC pathway, directs Fe-S cluster delivery from ISCU to downstream targets. One target is the iron-responsive protein IRP1, an Fe-S protein activated when its Fe-S cluster is absent187. Defective GLRX5, therefore, leads to constitutively active IRP1. IRP1 regulates several proteins involved in iron homeostasis188, including those involved in heme production. In particular, apo-IRP1 inhibits expression of the final enzyme in heme synthesis, aminolevulinate synthase (ALAS2). Defective GLRX5, therefore, leads to insufficient heme and impaired erythropoiesis, resulting in anemia. Iron that would be directed towards heme production accumulates in the cytosol of erythroblasts, creating the characteristic ringedsideroblasts151.
3.6.4 Additional Diseases
Succinate Dehydrogenase (SDH) subunit B, the Fe-S cluster containing protein of the succinate dehydrogenase complex, is a known tumor suppressor. Succinate, the substrate for SDH, stabilizes hypoxia-inducible factor (HIF), which regulates key processes in cell division and blood vessel growth under hypoxic conditions. Mutations in SDHB or in fact any of the other main subunits of SDH (SDHA, SDHC, and SDHD) cause susceptibility to tumor formations known as paragangliomas or phaeochromocytomas in a disorder called Hereditary Paraganglioma-Pheochromocytoma189, stemming from the accumulation of succinate and stabilization of HIF. SDHAF2, an assembly protein that flavinates SDH, is also implicated in paragangliomas. Recently, two additional Fe-S assembly proteins, SDHAF1 and SDHAF3, were found to stabilize Fe-S cluster assembly in SDH190. The later two proteins have LYR-motifs (Leu-Tyr-Arg) common to proteins involved in Fe-S cluster assembly. SDHAF1 deficiency is known to cause leukoencephalopathy191.
Lastly, mutations in either NFU1 or BOLA3, two different genes involved in Fe-S cluster biogenesis, leads to multiple mitochondrial dysfunction syndrome, a condition characterized by defects in Complexes I, II, and III and pyruvate/α-ketoglutarate dehydrogenases. NFU1 is thought to be an alternative to ISCU as a scaffold for Fe-S assembly. Both BOLA3 and NFU1 appear to be involved in lipoate synthesis, possibly related to a role in assembling Fe-S clusters in lipoic acid synthase (LIAS), thus providing an explanation for the reduced PDH and α–KGDH activities characteristic of this syndrome. The impaired energy production results in lactic acidosis, encephalopathy192 and early death.
4. Summary
The utilization of complex Fe cofactors in biology requires a tightly controlled process of cofactor assembly and of delivery to the correct apoprotein partner. Numerous ailments, some of which outlined within this review, result when there is a breakdown in the assembly process or in delivery of the cofactor. The development of treatment strategies for these disorders will require a more advanced molecular understanding of each protein malfunction outlined above. Therefore cooperation of bioinorganic chemists with the cell biologists alike will be essential to provide the broader understanding of how these complex pathways harness the power of iron in complex cofactors within the biological milieu.
Acknowledgments
DPB was supported under an NIH Cardiovascular Training grant (T32 HL120822). SPD was supported by a American Heart Association/Friedreich’s Ataxia Research Alliance pre-doctoral fellowship (14PRE18830036), and recently by the NIH (F30 DK101230). TLS is supported by the NIH (R01-DK068139 and R01-GM107542).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crichton R. Inorganic Biochemistry of Iron Metabolism. John Wiley & Sons, LTD; New York: 2001. [Google Scholar]
- 2.Wrighting DM, Andrews NC. Curr Top Dev Biol. 2008;82:141. doi: 10.1016/S0070-2153(07)00006-3. [DOI] [PubMed] [Google Scholar]
- 3.Boukhalfa H, Crumbliss AL. Biometals. 2002;15:325. doi: 10.1023/a:1020218608266. [DOI] [PubMed] [Google Scholar]
- 4.Albrecht-Gary AM, Crumbliss AL. Metal Ions in Biologival Systems Volume 35: Iron Transport and Storage in Microorganisms, Plants and Animals. Marcel Dekker; New York: 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian P, Rodrigues AV, Ghimire-Rijal S, Stemmler TL. Current opinion in chemical biology. 2011;15:312. doi: 10.1016/j.cbpa.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gitlin JD, Lill R, editors. Special Issue: Cell Biology of Metals. Vol. 1823 Elsevier; 2012. [Google Scholar]
- 7.Shawki A, Anthony SR, Nose Y, Engevik MA, Niespodzany EJ, Barrientos T, Ohrvik H, Worrell RT, Thiele DJ, Mackenzie B. Am J Physiol Gastrointest Liver Physiol. 2015 doi: 10.1152/ajpgi.00160.2015. ajpgi 00160 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson MK, SMith AD. Iron-Sulfur Proteins. Encyclopedia of Inorganic and Bioinorganic Chemistry. John Wiley & Sons, Inc; 2011. [Google Scholar]
- 9.Tsiftsoglou AS, Tsamadou AI, Papadopoulou LC. Pharmacology & therapeutics. 2006;111:327. doi: 10.1016/j.pharmthera.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Arnone A. Annual review of medicine. 1974;25:123. doi: 10.1146/annurev.me.25.020174.001011. [DOI] [PubMed] [Google Scholar]
- 11.Fenna R, Zeng J, Davey C. Archives of biochemistry and biophysics. 1995;316:653. doi: 10.1006/abbi.1995.1086. [DOI] [PubMed] [Google Scholar]
- 12.Ajioka RS, Phillips JD, Kushner JP. Biochimica et biophysica acta. 2006;1763:723. doi: 10.1016/j.bbamcr.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Shemin D, Rittenberg D. The Journal of biological chemistry. 1946;166:621. [PubMed] [Google Scholar]
- 14.Radin NS, Rittenberg D, Shemin D. The Journal of biological chemistry. 1950;184:755. [PubMed] [Google Scholar]
- 15.Shemin D, Kumin S. The Journal of biological chemistry. 1952;198:827. [PubMed] [Google Scholar]
- 16.Muir HM, Neuberger A. The Biochemical journal. 1950;47:97. doi: 10.1042/bj0470097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bishop DF, Henderson AS, Astrin KH. Genomics. 1990;7:207. doi: 10.1016/0888-7543(90)90542-3. [DOI] [PubMed] [Google Scholar]
- 18.Fanica-Gaignier M, Clement-Metral J. European journal of biochemistry / FEBS. 1973;40:13. doi: 10.1111/j.1432-1033.1973.tb03163.x. [DOI] [PubMed] [Google Scholar]
- 19.Guernsey DL, Jiang H, Campagna DR, Evans SC, Ferguson M, Kellogg MD, Lachance M, Matsuoka M, Nightingale M, Rideout A, Saint-Amant L, Schmidt PJ, Orr A, Bottomley SS, Fleming MD, Ludman M, Dyack S, Fernandez CV, Samuels ME. Nature genetics. 2009;41:651. doi: 10.1038/ng.359. [DOI] [PubMed] [Google Scholar]
- 20.Moser J, Schubert WD, Beier V, Bringemeier I, Jahn D, Heinz DW. The EMBO journal. 2001;20:6583. doi: 10.1093/emboj/20.23.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang DD, Wang WY, Gough SP, Kannangara CG. Science. 1984;225:1482. doi: 10.1126/science.6206568. [DOI] [PubMed] [Google Scholar]
- 22.Kannangara CG, Gough SP, Bruyant P, Hoober JK, Kahn A, von Wettstein D. Trends in biochemical sciences. 1988;13:139. doi: 10.1016/0968-0004(88)90071-0. [DOI] [PubMed] [Google Scholar]
- 23.Jahn D, Verkamp E, Soll D. Trends in biochemical sciences. 1992;17:215. doi: 10.1016/0968-0004(92)90380-r. [DOI] [PubMed] [Google Scholar]
- 24.Falk JE, Dresel EI, Rimington C. Nature. 1953;172:292. doi: 10.1038/172292a0. [DOI] [PubMed] [Google Scholar]
- 25.Gibson KD, Neuberger A, Scott JJ. The Biochemical journal. 1954;58:xli. [PubMed] [Google Scholar]
- 26.Bollivar DW, Clauson C, Lighthall R, Forbes S, Kokona B, Fairman R, Kundrat L, Jaffe EK. BMC biochemistry. 2004;5:17. doi: 10.1186/1471-2091-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson PM, Desnick RJ. The Journal of biological chemistry. 1979;254:6924. [PubMed] [Google Scholar]
- 28.Dent AJ, Beyersmann D, Block C, Hasnain SS. Biochemistry. 1990;29:7822. doi: 10.1021/bi00486a007. [DOI] [PubMed] [Google Scholar]
- 29.Jaffe EK, Abrams WR, Kaempfen HX, Harris KA., Jr Biochemistry. 1992;31:2113. doi: 10.1021/bi00122a032. [DOI] [PubMed] [Google Scholar]
- 30.Shoolingin-Jordan PM, Al-Dbass A, McNeill LA, Sarwar M, Butler D. Biochemical Society transactions. 2003;31:731. doi: 10.1042/bst0310731. [DOI] [PubMed] [Google Scholar]
- 31.Mathews MA, Schubert HL, Whitby FG, Alexander KJ, Schadick K, Bergonia HA, Phillips JD, Hill CP. The EMBO journal. 2001;20:5832. doi: 10.1093/emboj/20.21.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoolingin-Jordan PM. Journal of bioenergetics and biomembranes. 1995;27:181. doi: 10.1007/BF02110033. [DOI] [PubMed] [Google Scholar]
- 33.Straka JG, Kushner JP. Biochemistry. 1983;22:4664. doi: 10.1021/bi00289a009. [DOI] [PubMed] [Google Scholar]
- 34.Whitby FG, Phillips JD, Kushner JP, Hill CP. The EMBO journal. 1998;17:2463. doi: 10.1093/emboj/17.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreira GC, Andrew TL, Karr SW, Dailey HA. The Journal of biological chemistry. 1988;263:3835. [PubMed] [Google Scholar]
- 36.Taketani S, Yoshinaga T, Furukawa T, Kohno H, Tokunaga R, Nishimura K, Inokuchi H. European journal of biochemistry / FEBS. 1995;230:760. doi: 10.1111/j.1432-1033.1995.0760h.x. [DOI] [PubMed] [Google Scholar]
- 37.Phillips JD, Whitby FG, Warby CA, Labbe P, Yang C, Pflugrath JW, Ferrara JD, Robinson H, Kushner JP, Hill CP. The Journal of biological chemistry. 2004;279:38960. doi: 10.1074/jbc.M406050200. [DOI] [PubMed] [Google Scholar]
- 38.Lee DS, Flachsova E, Bodnarova M, Demeler B, Martasek P, Raman CS. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14232. doi: 10.1073/pnas.0506557102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porra RJ, Falk JE. The Biochemical journal. 1964;90:69. doi: 10.1042/bj0900069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch M, Breithaupt C, Kiefersauer R, Freigang J, Huber R, Messerschmidt A. The EMBO journal. 2004;23:1720. doi: 10.1038/sj.emboj.7600189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu CK, Dailey HA, Rose JP, Burden A, Sellers VM, Wang BC. Nature structural biology. 2001;8:156. doi: 10.1038/84152. [DOI] [PubMed] [Google Scholar]
- 42.Burden AE, Wu C, Dailey TA, Busch JL, Dhawan IK, Rose JP, Wang B, Dailey HA. Biochimica et biophysica acta. 1999;1435:191. doi: 10.1016/s0167-4838(99)00196-x. [DOI] [PubMed] [Google Scholar]
- 43.Karlberg T, Hansson MD, Yengo RK, Johansson R, Thorvaldsen HO, Ferreira GC, Hansson M, Al-Karadaghi S. Journal of molecular biology. 2008;378:1074. doi: 10.1016/j.jmb.2008.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts AG, Elder GH. Biochimica et biophysica acta. 2001;1518:95. doi: 10.1016/s0167-4781(01)00187-7. [DOI] [PubMed] [Google Scholar]
- 45.Cable EE, Miller TG, Isom HC. Archives of biochemistry and biophysics. 2000;384:280. doi: 10.1006/abbi.2000.2117. [DOI] [PubMed] [Google Scholar]
- 46.Hamilton JW, Bement WJ, Sinclair PR, Sinclair JF, Alcedo JA, Wetterhahn KE. Archives of biochemistry and biophysics. 1991;289:387. doi: 10.1016/0003-9861(91)90428-l. [DOI] [PubMed] [Google Scholar]
- 47.Roberts AG, Redding SJ, Llewellyn DH. FEBS letters. 2005;579:1061. doi: 10.1016/j.febslet.2004.12.080. [DOI] [PubMed] [Google Scholar]
- 48.Tyrrell DL, Marks GS. Biochemical pharmacology. 1972;21:2077. doi: 10.1016/0006-2952(72)90161-x. [DOI] [PubMed] [Google Scholar]
- 49.Kikuchi G, Hayashi N. Molecular and cellular biochemistry. 1981;37:27. doi: 10.1007/BF02355885. [DOI] [PubMed] [Google Scholar]
- 50.Handschin C, Lin J, Rhee J, Peyer AK, Chin S, Wu PH, Meyer UA, Spiegelman BM. Cell. 2005;122:505. doi: 10.1016/j.cell.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 51.Srivastava G, Borthwick IA, Maguire DJ, Elferink CJ, Bawden MJ, Mercer JF, May BK. The Journal of biological chemistry. 1988;263:5202. [PubMed] [Google Scholar]
- 52.Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, Paw BH, Shaw GC, Kingsley P, Palis J, Schubert H, Chen O, Kaplan J, Zon LI, Tubingen Screen C. Nature. 2005;436:1035. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 53.Rouault TA, Tong WH. Nature reviews. Molecular cell biology. 2005;6:345. doi: 10.1038/nrm1620. [DOI] [PubMed] [Google Scholar]
- 54.Kaya AH, Plewinska M, Wong DM, Desnick RJ, Wetmur JG. Genomics. 1994;19:242. doi: 10.1006/geno.1994.1054. [DOI] [PubMed] [Google Scholar]
- 55.Chretien S, Dubart A, Beaupain D, Raich N, Grandchamp B, Rosa J, Goossens M, Romeo PH. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:6. doi: 10.1073/pnas.85.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grandchamp B, De Verneuil H, Beaumont C, Chretien S, Walter O, Nordmann Y. European journal of biochemistry / FEBS. 1987;162:105. doi: 10.1111/j.1432-1033.1987.tb10548.x. [DOI] [PubMed] [Google Scholar]
- 57.Mignotte V, Wall L, de Boer E, Grosveld F, Romeo PH. Nucleic acids research. 1989;17:37. doi: 10.1093/nar/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aizencang GI, Bishop DF, Forrest D, Astrin KH, Desnick RJ. The Journal of biological chemistry. 2000;275:2295. doi: 10.1074/jbc.275.4.2295. [DOI] [PubMed] [Google Scholar]
- 59.Romeo PH, Raich N, Dubart A, Beaupain D, Pryor M, Kushner J, Cohen-Solal M, Goossens M. The Journal of biological chemistry. 1986;261:9825. [PubMed] [Google Scholar]
- 60.Takahashi S, Taketani S, Akasaka JE, Kobayashi A, Hayashi N, Yamamoto M, Nagai T. Blood. 1998;92:3436. [PubMed] [Google Scholar]
- 61.Tugores A, Magness ST, Brenner DA. The Journal of biological chemistry. 1994;269:30789. [PubMed] [Google Scholar]
- 62.Taketani S, Adachi Y, Nakahashi Y. European journal of biochemistry / FEBS. 2000;267:4685. doi: 10.1046/j.1432-1327.2000.01519.x. [DOI] [PubMed] [Google Scholar]
- 63.Hamel P, Corvest V, Giege P, Bonnard G. Biochimica et biophysica acta. 2009;1793:125. doi: 10.1016/j.bbamcr.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 64.Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. Microbiology and molecular biology reviews : MMBR. 2009;73:510. doi: 10.1128/MMBR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stevens JM, Mavridou DA, Hamer R, Kritsiligkou P, Goddard AD, Ferguson SJ. The FEBS journal. 2011;278:4170. doi: 10.1111/j.1742-4658.2011.08376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Babbitt SE, Sutherland MC, Francisco BS, Mendez DL, Kranz RG. Trends in biochemical sciences. 2015;40:446. doi: 10.1016/j.tibs.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamza I, Dailey HA. Biochimica et biophysica acta. 2012;1823:1617. doi: 10.1016/j.bbamcr.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathews AJ, Brittain T. The Biochemical journal. 2001;357:305. doi: 10.1042/0264-6021:3570305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuras R, de Vitry C, Choquet Y, Girard-Bascou J, Culler D, Buschlen S, Merchant S, Wollman FA. The Journal of biological chemistry. 1997;272:32427. doi: 10.1074/jbc.272.51.32427. [DOI] [PubMed] [Google Scholar]
- 70.Chakravarti R, Gupta K, Majors A, Ruple L, Aronica M, Stuehr DJ. Free radical biology & medicine. 2015;82:105. doi: 10.1016/j.freeradbiomed.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poulos TL. Chemical reviews. 2014;114:3919. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davies MJ, Hawkins CL, Pattison DI, Rees MD. Antioxidants & redox signaling. 2008;10:1199. doi: 10.1089/ars.2007.1927. [DOI] [PubMed] [Google Scholar]
- 73.Zederbauer M, Furtmuller PG, Brogioni S, Jakopitsch C, Smulevich G, Obinger C. Natural product reports. 2007;24:571. doi: 10.1039/b604178g. [DOI] [PubMed] [Google Scholar]
- 74.Whatley SD, Ducamp S, Gouya L, Grandchamp B, Beaumont C, Badminton MN, Elder GH, Holme SA, Anstey AV, Parker M, Corrigall AV, Meissner PN, Hift RJ, Marsden JT, Ma Y, Mieli-Vergani G, Deybach JC, Puy H. American journal of human genetics. 2008;83:408. doi: 10.1016/j.ajhg.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jaffe EK, Stith L. American journal of human genetics. 2007;80:329. doi: 10.1086/511444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nordmann Y, Puy H, Da Silva V, Simonin S, Robreau AM, Bonaiti C, Phung LN, Deybach JC. Journal of internal medicine. 1997;242:213. doi: 10.1046/j.1365-2796.1997.00189.x. [DOI] [PubMed] [Google Scholar]
- 77.Fritsch C, Bolsen K, Ruzicka T, Goerz G. Journal of the American Academy of Dermatology. 1997;36:594. doi: 10.1016/s0190-9622(97)70249-4. [DOI] [PubMed] [Google Scholar]
- 78.Desnick RJ, Glass IA, Xu W, Solis C, Astrin KH. Seminars in liver disease. 1998;18:77. doi: 10.1055/s-2007-1007143. [DOI] [PubMed] [Google Scholar]
- 79.Altiparmak UE, Oflu Y, Kocaoglu FA, Katircioglu YA, Duman S. Cornea. 2008;27:1093. doi: 10.1097/ICO.0b013e31817e905a. [DOI] [PubMed] [Google Scholar]
- 80.Elder GH. Seminars in liver disease. 1998;18:67. doi: 10.1055/s-2007-1007142. [DOI] [PubMed] [Google Scholar]
- 81.Elder GH, Roberts AG. Journal of bioenergetics and biomembranes. 1995;27:207. doi: 10.1007/BF02110035. [DOI] [PubMed] [Google Scholar]
- 82.Allen KR, Whatley SD, Degg TJ, Barth JH. Journal of inherited metabolic disease. 2005;28:779. doi: 10.1007/s10545-005-0092-z. [DOI] [PubMed] [Google Scholar]
- 83.Whatley SD, Puy H, Morgan RR, Robreau AM, Roberts AG, Nordmann Y, Elder GH, Deybach JC. American journal of human genetics. 1999;65:984. doi: 10.1086/302586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roberts AG, Puy H, Dailey TA, Morgan RR, Whatley SD, Dailey HA, Martasek P, Nordmann Y, Deybach JC, Elder GH. Human molecular genetics. 1998;7:1921. doi: 10.1093/hmg/7.12.1921. [DOI] [PubMed] [Google Scholar]
- 85.Murphy GM. Dermatologic therapy. 2003;16:57. doi: 10.1046/j.1529-8019.2003.01609.x. [DOI] [PubMed] [Google Scholar]
- 86.Went LN, Klasen EC. Annals of human genetics. 1984;48:105. doi: 10.1111/j.1469-1809.1984.tb01006.x. [DOI] [PubMed] [Google Scholar]
- 87.Fleming MD. Seminars in hematology. 2002;39:270. doi: 10.1053/shem.2002.35637. [DOI] [PubMed] [Google Scholar]
- 88.Astrin KH, Bishop DF, Wetmur JG, Kaul B, Davidow B, Desnick RJ. Annals of the New York Academy of Sciences. 1987;514:23. doi: 10.1111/j.1749-6632.1987.tb48757.x. [DOI] [PubMed] [Google Scholar]
- 89.Sassa S, Kappas A. The Journal of clinical investigation. 1983;71:625. doi: 10.1172/JCI110809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.George SJ, Armstrong FA, Hatchikian EC, Thomson AJ. The Biochemical journal. 1989;264:275. doi: 10.1042/bj2640275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bertini I, Sigel A, Sigel H. Handbook on metalloproteins. Marcel Dekker; New York: 2001. [Google Scholar]
- 92.Brown EN, Friemann R, Karlsson A, Parales JV, Couture MM, Eltis LD, Ramaswamy S. Journal of biological inorganic chemistry : JBIC : a publication of the Society of Biological Inorganic Chemistry. 2008;13:1301. doi: 10.1007/s00775-008-0413-4. [DOI] [PubMed] [Google Scholar]
- 93.Bak DW, Elliott SJ. Current opinion in chemical biology. 2014;19:50. doi: 10.1016/j.cbpa.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 94.Jacobson MR, Cash VL, Weiss MC, Laird NF, Newton WE, Dean DR. Molecular & general genetics : MGG. 1989;219:49. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- 95.Kispal G, Csere P, Prohl C, Lill R. The EMBO journal. 1999;18:3981. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gerber J, Neumann K, Prohl C, Muhlenhoff U, Lill R. Molecular and cellular biology. 2004;24:4848. doi: 10.1128/MCB.24.11.4848-4857.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Agar JN, Krebs C, Frazzon J, Huynh BH, Dean DR, Johnson MK. Biochemistry. 2000;39:7856. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- 98.Urbina HD, Silberg JJ, Hoff KG, Vickery LE. The Journal of biological chemistry. 2001;276:44521. doi: 10.1074/jbc.M106907200. [DOI] [PubMed] [Google Scholar]
- 99.Wiedemann N, Urzica E, Guiard B, Muller H, Lohaus C, Meyer HE, Ryan MT, Meisinger C, Muhlenhoff U, Lill R, Pfanner N. The EMBO journal. 2006;25:184. doi: 10.1038/sj.emboj.7600906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Webert H, Freibert SA, Gallo A, Heidenreich T, Linne U, Amlacher S, Hurt E, Muhlenhoff U, Banci L, Lill R. Nature communications. 2014;5:5013. doi: 10.1038/ncomms6013. [DOI] [PubMed] [Google Scholar]
- 101.Yan R, Adinolfi S, Pastore A. Biochimica et biophysica acta. 2015 doi: 10.1016/j.bbapap.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pandey A, Gordon DM, Pain J, Stemmler TL, Dancis A, Pain D. The Journal of biological chemistry. 2013;288:36773. doi: 10.1074/jbc.M113.525857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krebs C, Agar JN, Smith AD, Frazzon J, Dean DR, Huynh BH, Johnson MK. Biochemistry. 2001;40:14069. doi: 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- 104.Tong WH, Jameson GN, Huynh BH, Rouault TA. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9762. doi: 10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yoon T, Cowan JA. J Am Chem Soc. 2003;125:6078. doi: 10.1021/ja027967i. [DOI] [PubMed] [Google Scholar]
- 106.Bencze KZ, Kondapalli KC, Cook JD, McMahon S, Millan-Pacheco C, Pastor N, Stemmler TL. Critical reviews in biochemistry and molecular biology. 2006;41:269. doi: 10.1080/10409230600846058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim JH, Bothe JR, Frederick RO, Holder JC, Markley JL. J Am Chem Soc. 2014;136:7933. doi: 10.1021/ja501260h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ding H, Harrison K, Lu J. The Journal of biological chemistry. 2005;280:30432. doi: 10.1074/jbc.M504638200. [DOI] [PubMed] [Google Scholar]
- 109.Ding H, Clark RJ, Ding B. The Journal of biological chemistry. 2004;279:37499. doi: 10.1074/jbc.M404533200. [DOI] [PubMed] [Google Scholar]
- 110.Yang J, Bitoun JP, Ding H. The Journal of biological chemistry. 2006;281:27956. doi: 10.1074/jbc.M601356200. [DOI] [PubMed] [Google Scholar]
- 111.Qi W, Cowan JA. Chemical communications. 2011;47:4989. doi: 10.1039/c0cc05079b. [DOI] [PubMed] [Google Scholar]
- 112.Muhlenhoff U, Gerber J, Richhardt N, Lill R. The EMBO journal. 2003;22:4815. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mapolelo DT, Zhang B, Randeniya S, Albetel AN, Li H, Couturier J, Outten CE, Rouhier N, Johnson MK. Dalton transactions. 2013;42:3107. doi: 10.1039/c2dt32263c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Uzarska MA, Dutkiewicz R, Freibert SA, Lill R, Muhlenhoff U. Molecular biology of the cell. 2013;24:1830. doi: 10.1091/mbc.E12-09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hoff KG, Silberg JJ, Vickery LE. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7790. doi: 10.1073/pnas.130201997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, Rouhier N. The EMBO journal. 2008;27:1122. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E. Molecular biology of the cell. 2002;13:1109. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim KD, Chung WH, Kim HJ, Lee KC, Roe JH. Biochemical and biophysical research communications. 2010;392:467. doi: 10.1016/j.bbrc.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 119.Yeung N, Gold B, Liu NL, Prathapam R, Sterling HJ, Willams ER, Butland G. Biochemistry. 2011;50:8957. doi: 10.1021/bi2008883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brancaccio D, Gallo A, Mikolajczyk M, Zovo K, Palumaa P, Novellino E, Piccioli M, Ciofi-Baffoni S, Banci L. J Am Chem Soc. 2014;136:16240. doi: 10.1021/ja507822j. [DOI] [PubMed] [Google Scholar]
- 121.Rouault TA. Disease models & mechanisms. 2012;5:155. doi: 10.1242/dmm.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rouault TA. Nature reviews. Molecular cell biology. 2015;16:45. doi: 10.1038/nrm3909. [DOI] [PubMed] [Google Scholar]
- 123.Biederbick A, Stehling O, Rosser R, Niggemeyer B, Nakai Y, Elsasser HP, Lill R. Molecular and cellular biology. 2006;26:5675. doi: 10.1128/MCB.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sharma AK, Pallesen LJ, Spang RJ, Walden WE. The Journal of biological chemistry. 2010;285:26745. doi: 10.1074/jbc.R110.122218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Paul VD, Lill R. Biochimica et biophysica acta. 2015;1853:1528. doi: 10.1016/j.bbamcr.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 126.Cavadini P, Biasiotto G, Poli M, Levi S, Verardi R, Zanella I, Derosas M, Ingrassia R, Corrado M, Arosio P. Blood. 2007;109:3552. doi: 10.1182/blood-2006-08-041632. [DOI] [PubMed] [Google Scholar]
- 127.Lange H, Lisowsky T, Gerber J, Muhlenhoff U, Kispal G, Lill R. EMBO reports. 2001;2:715. doi: 10.1093/embo-reports/kve161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Netz DJ, Pierik AJ, Stumpfig M, Muhlenhoff U, Lill R. Nature chemical biology. 2007;3:278. doi: 10.1038/nchembio872. [DOI] [PubMed] [Google Scholar]
- 129.Netz DJ, Stumpfig M, Dore C, Muhlenhoff U, Pierik AJ, Lill R. Nature chemical biology. 2010;6:758. doi: 10.1038/nchembio.432. [DOI] [PubMed] [Google Scholar]
- 130.Banci L, Bertini I, Calderone V, Ciofi-Baffoni S, Giachetti A, Jaiswal D, Mikolajczyk M, Piccioli M, Winkelmann J. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7136. doi: 10.1073/pnas.1302378110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Song D, Lee FS. The Journal of biological chemistry. 2011;286:15797. doi: 10.1074/jbc.M110.201731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Song D, Lee FS. The Journal of biological chemistry. 2008;283:9231. doi: 10.1074/jbc.M708077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stehling O, Mascarenhas J, Vashisht AA, Sheftel AD, Niggemeyer B, Rosser R, Pierik AJ, Wohlschlegel JA, Lill R. Cell metabolism. 2013;18:187. doi: 10.1016/j.cmet.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moller SG, Kunkel T, Chua NH. Genes & development. 2001;15:90. doi: 10.1101/gad.850101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kumar B, Chaubey S, Shah P, Tanveer A, Charan M, Siddiqi MI, Habib S. International journal for parasitology. 2011;41:991. doi: 10.1016/j.ijpara.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 136.Tsaousis AD, Gentekaki E, Eme L, Gaston D, Roger AJ. Eukaryotic cell. 2014;13:143. doi: 10.1128/EC.00158-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tokumoto U, Kitamura S, Fukuyama K, Takahashi Y. Journal of biochemistry. 2004;136:199. doi: 10.1093/jb/mvh104. [DOI] [PubMed] [Google Scholar]
- 138.Takahashi Y, Tokumoto U. The Journal of biological chemistry. 2002;277:28380. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 139.Dai Y, Outten FW. FEBS letters. 2012;586:4016. doi: 10.1016/j.febslet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chahal HK, Dai Y, Saini A, Ayala-Castro C, Outten FW. Biochemistry. 2009;48:10644. doi: 10.1021/bi901518y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Saini A, Mapolelo DT, Chahal HK, Johnson MK, Outten FW. Biochemistry. 2010;49:9402. doi: 10.1021/bi1011546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wollers S, Layer G, Garcia-Serres R, Signor L, Clemancey M, Latour JM, Fontecave M, Ollagnier de Choudens S. The Journal of biological chemistry. 2010;285:23331. doi: 10.1074/jbc.M110.127449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chahal HK, Outten FW. J Inorg Biochem. 2012;116:126. doi: 10.1016/j.jinorgbio.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Loiseau L, Ollagnier-de-Choudens S, Nachin L, Fontecave M, Barras F. The Journal of biological chemistry. 2003;278:38352. doi: 10.1074/jbc.M305953200. [DOI] [PubMed] [Google Scholar]
- 145.Ollagnier-de-Choudens S, Lascoux D, Loiseau L, Barras F, Forest E, Fontecave M. FEBS letters. 2003;555:263. doi: 10.1016/s0014-5793(03)01244-4. [DOI] [PubMed] [Google Scholar]
- 146.Lu J, Yang J, Tan G, Ding H. The Biochemical journal. 2008;409:535. doi: 10.1042/BJ20071166. [DOI] [PubMed] [Google Scholar]
- 147.Gupta V, Sendra M, Naik SG, Chahal HK, Huynh BH, Outten FW, Fontecave M, Ollagnier de Choudens S. J Am Chem Soc. 2009;131:6149. doi: 10.1021/ja807551e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tan G, Lu J, Bitoun JP, Huang H, Ding H. The Biochemical journal. 2009;420:463. doi: 10.1042/BJ20090206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pinske C, Sawers RG. Journal of bacteriology. 2012;194:346. doi: 10.1128/JB.06024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. Biochimica et biophysica acta. 2013;1827:455. doi: 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 151.Lill R. Nature. 2009;460:831. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 152.Blanc B, Clemancey M, Latour JM, Fontecave M, Ollagnier de Choudens S. Biochemistry. 2014;53:7867. doi: 10.1021/bi5012496. [DOI] [PubMed] [Google Scholar]
- 153.Fleischhacker AS, Stubna A, Hsueh KL, Guo Y, Teter SJ, Rose JC, Brunold TC, Markley JL, Munck E, Kiley PJ. Biochemistry. 2012;51:4453. doi: 10.1021/bi3003204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rajagopalan S, Teter SJ, Zwart PH, Brennan RG, Phillips KJ, Kiley PJ. Nature structural & molecular biology. 2013;20:740. doi: 10.1038/nsmb.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14895. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vinella D, Loiseau L, Ollagnier de Choudens S, Fontecave M, Barras F. Molecular microbiology. 2013;87:493. doi: 10.1111/mmi.12135. [DOI] [PubMed] [Google Scholar]
- 157.Crack JC, Green J, Hutchings MI, Thomson AJ, Le Brun NE. Antioxidants & redox signaling. 2012;17:1215. doi: 10.1089/ars.2012.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yeo WS, Lee JH, Lee KC, Roe JH. Molecular microbiology. 2006;61:206. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 159.Lee KC, Yeo WS, Roe JH. Journal of bacteriology. 2008;190:8244. doi: 10.1128/JB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Desnoyers G, Morissette A, Prevost K, Masse E. The EMBO journal. 2009;28:1551. doi: 10.1038/emboj.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Masse E, Vanderpool CK, Gottesman S. Journal of bacteriology. 2005;187:6962. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Johnson DC, Dean DR, Smith AD, Johnson MK. Annual review of biochemistry. 2005;74:247. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 163.Carter CW, Jr, Kraut J, Freer ST, Alden RA, Sieker LC, Adman E, Jensen LH. Proceedings of the National Academy of Sciences of the United States of America. 1972;69:3526. doi: 10.1073/pnas.69.12.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sticht H, Rosch P. Progress in biophysics and molecular biology. 1998;70:95. doi: 10.1016/s0079-6107(98)00027-3. [DOI] [PubMed] [Google Scholar]
- 165.Lange H, Kaut A, Kispal G, Lill R. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:1050. doi: 10.1073/pnas.97.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vinothkumar KR, Zhu J, Hirst J. Nature. 2014;515:80. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Janssen S, Schafer G, Anemuller S, Moll R. Journal of bacteriology. 1997;179:5560. doi: 10.1128/jb.179.17.5560-5569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Silman HI, Rieske JS, Lipton SH, Baum H. The Journal of biological chemistry. 1967;242:4867. [PubMed] [Google Scholar]
- 169.Shergill JK, Joannou CL, Mason JR, Cammack R. Biochemistry. 1995;34:16533. doi: 10.1021/bi00051a001. [DOI] [PubMed] [Google Scholar]
- 170.Switzer RL. BioFactors. 1989;2:77. [PubMed] [Google Scholar]
- 171.Kuo CF, McRee DE, Fisher CL, O’Handley SF, Cunningham RP, Tainer JA. Science. 1992;258:434. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- 172.Leone M, Brignolio F, Rosso MG, Curtoni ES, Moroni A, Tribolo A, Schiffer D. Clinical genetics. 1990;38:161. doi: 10.1111/j.1399-0004.1990.tb03566.x. [DOI] [PubMed] [Google Scholar]
- 173.Lopez-Arlandis JM, Vilchez JJ, Palau F, Sevilla T. Neuroepidemiology. 1995;14:14. doi: 10.1159/000109774. [DOI] [PubMed] [Google Scholar]
- 174.Lodi R, Tonon C, Calabrese V, Schapira AH. Antioxidants & redox signaling. 2006;8:438. doi: 10.1089/ars.2006.8.438. [DOI] [PubMed] [Google Scholar]
- 175.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Science. 1996;271:1423. [Google Scholar]
- 176.Ohshima K, Montermini L, Wells RD, Pandolfo M. The Journal of biological chemistry. 1998;273:14588. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 177.Pastore A, Puccio H. Journal of neurochemistry. 2013;126(Suppl 1):43. doi: 10.1111/jnc.12220. [DOI] [PubMed] [Google Scholar]
- 178.Stemmler TL, Lesuisse E, Pain D, Dancis A. The Journal of biological chemistry. 2010;285:26737. doi: 10.1074/jbc.R110.118679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Whitnall M, Suryo Rahmanto Y, Huang ML, Saletta F, Lok HC, Gutierrez L, Lazaro FJ, Fleming AJ, St Pierre TG, Mikhael MR, Ponka P, Richardson DR. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20590. doi: 10.1073/pnas.1215349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Delatycki MB, Corben LA. Journal of child neurology. 2012;27:1133. doi: 10.1177/0883073812448230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tsou AY, Paulsen EK, Lagedrost SJ, Perlman SL, Mathews KD, Wilmot GR, Ravina B, Koeppen AH, Lynch DR. Journal of the neurological sciences. 2011;307:46. doi: 10.1016/j.jns.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 182.Crooks DR, Jeong SY, Tong WH, Ghosh MC, Olivierre H, Haller RG, Rouault TA. The Journal of biological chemistry. 2012;287:40119. doi: 10.1074/jbc.M112.418889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mochel F, Knight MA, Tong WH, Hernandez D, Ayyad K, Taivassalo T, Andersen PM, Singleton A, Rouault TA, Fischbeck KH, Haller RG. American journal of human genetics. 2008;82:652. doi: 10.1016/j.ajhg.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kollberg G, Melberg A, Holme E, Oldfors A. Neuromuscular disorders : NMD. 2011;21:115. doi: 10.1016/j.nmd.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 185.Mochel F, Haller RG. In: GeneReviews (R) Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, editors. Seattle (WA): 1993. [Google Scholar]
- 186.Kollberg G, Tulinius M, Melberg A, Darin N, Andersen O, Holmgren D, Oldfors A, Holme E. Brain : a journal of neurology. 2009;132:2170. doi: 10.1093/brain/awp152. [DOI] [PubMed] [Google Scholar]
- 187.Ye H, Jeong SY, Ghosh MC, Kovtunovych G, Silvestri L, Ortillo D, Uchida N, Tisdale J, Camaschella C, Rouault TA. The Journal of clinical investigation. 2010;120:1749. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wilkinson N, Pantopoulos K. Frontiers in pharmacology. 2014;5:176. doi: 10.3389/fphar.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Benn DE, Robinson BG, Clifton-Bligh RJ. Endocrine-related cancer. 2015;22:T91. doi: 10.1530/ERC-15-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Na U, Yu W, Cox J, Bricker DK, Brockmann K, Rutter J, Thummel CS, Winge DR. Cell metabolism. 2014;20:253. doi: 10.1016/j.cmet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ghezzi D, Goffrini P, Uziel G, Horvath R, Klopstock T, Lochmuller H, D’Adamo P, Gasparini P, Strom TM, Prokisch H, Invernizzi F, Ferrero I, Zeviani M. Nature genetics. 2009;41:654. doi: 10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- 192.Cameron JM, Janer A, Levandovskiy V, Mackay N, Rouault TA, Tong WH, Ogilvie I, Shoubridge EA, Robinson BH. American journal of human genetics. 2011;89:486. doi: 10.1016/j.ajhg.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hederstedt L. Biochimica et biophysica acta. 2012;1817:920. doi: 10.1016/j.bbabio.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 194.Bowman SE, Bren KL. Natural product reports. 2008;25:1118. doi: 10.1039/b717196j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Timkovich R, Cork MS, Gennis RB, Johnson PY. Journal of the American Chemical Society. 1985;107:6069. [Google Scholar]
- 196.Sotiriou C, Chang CK. Journal of the American Chemical Society. 1988;110:2264. [Google Scholar]
- 197.Murshudov GN, Grebenko AI, Barynin V, Dauter Z, Wilson KS, Vainshtein BK, MelikAdamyan W, Bravo J, Ferran JM, Ferrer JC, Switala J, Loewen PC, Fita I. Journal of Biological Chemistry. 1996;271:8863. doi: 10.1074/jbc.271.15.8863. [DOI] [PubMed] [Google Scholar]
- 198.Walsh TA, Johnson MK, Barber D, Thomson AJ, Greenwood C. Journal of inorganic biochemistry. 1981;14:15. doi: 10.1016/s0162-0134(00)80011-2. [DOI] [PubMed] [Google Scholar]
- 199.Fulop V, Moir JW, Ferguson SJ, Hajdu J. Cell. 1995;81:369. doi: 10.1016/0092-8674(95)90390-9. [DOI] [PubMed] [Google Scholar]
- 200.Rae TD, Goff HM. The Journal of biological chemistry. 1998;273:27968. doi: 10.1074/jbc.273.43.27968. [DOI] [PubMed] [Google Scholar]
- 201.Wu W, Chang CK, Varotsis C, Babcock GT, Puustinen A, Wikstrom M. Journal of the American Chemical Society. 1992;114:1182. [Google Scholar]
- 202.Puustinen A, Wikstrom M. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:6122. doi: 10.1073/pnas.88.14.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]