Abstract
The polymorphic products of major histocompatibility complex class I–related chain A (MICA) genes are important in solid organ transplantation rejection. MICA expression is limited to gut epithelium and may play a role in triggering acute graft-versus-host disease (aGVHD). A total of 236 recipients of unrelated donor transplantation were studied. Donor-recipient human leukocyte antigen (HLA) match was 10/10 human leukocyte antigen (HLA-A, -B, -C, -DRB1, -DQB1) in 73% and MICA mismatch in 8.4%. Because of physical vicinity of the loci, MICA mismatch was significantly associated with mismatch at HLA-B and HLA-C. A higher rate of grade II-IV aGVHD was seen in MICA-mismatched patients (80% vs 40%, P = .003) irrespective of degree of HLA matching (HLA 10/10 match: 75% vs 39%, P = .02) and HLA any mismatch (83% vs 46%, P = .003). The rate of grade II-IV gastrointestinal aGVHD was also higher in MICA-mismatched patients (35% vs 17%, P = .05). We conclude that MICA may represent novel a transplantation antigen recognized by human allogeneic T cells. This study was registered at ClinicalTrials.gov (Identifier NCT00506922).
Introduction
Graft-versus-host disease (GVHD) is a major cause of mortality after allogeneic hematopoietic stem cell transplantations (HSCT). Even with high-resolution donor-recipient human leukocyte antigen (HLA) typing and matching for major histocompatibility complex (MHC) class I (HLA-A, -B, and -C) and class II (HLA-DRB1 and -DQB1), incidence of grade III-IV acute GVHD (aGVHD) can be 30%.1 Therefore, improvement in HLA matching and evaluation of novel compatibility factors is desirable. MHC class I–related chain A (MICA) genes have evolved in parallel with human class I genes2 and are located approximately 46 kb centromeric to HLA-B locus.3 The MICA gene products have been shown to play a role in some aspects of antigen presentation and T-cell recognition and appear to be important in innate immunity as ligands to NKG2D receptor4 expressed on most γδ T cells, CD8+αβ T cells, and NK cells.5 MICA antigens are polymorphic and elicit antibody production after solid organ transplantation, which may be associated with organ allograft rejection.6,7 MICA products are expressed constitutively in gut epithelium, endothelial cells, and fibroblasts and induced by stress in other cell types.2,8,9 Because MICA gene products are augmented by stress in epithelia10 and are recognized by a subpopulation of intestinal γδ T cells,11 they may play a role in triggering aGVHD.8,12 We hypothesized that donor-recipient MICA mismatch, which may trigger a response from αβT cells, would increase the incidence of aGVHD after HSCT, and we investigated this hypothesis in a cohort of patients with myeloid leukemias undergoing HSCT.
Methods
We analyzed 236 patients with acute myeloid leukemia/myelodysplastic syndrome (82%) and chronic myeloid leukemia/myeloproliferative disorder (18%) prospectively treated in Institutional Review Board–approved unrelated-donor HSCT protocols at M. D. Anderson Cancer Center from 2002 to 2007 (Table 1). Patient informed consent was obtained in accordance with the Declaration of Helsinki. Patients who failed to engraft (n = 7) or died early (n = 11) were excluded. Median follow-up was 30 months (range, 4-117 months). All donor-recipient pairs were fully typed at high resolution1 for the alleles of HLA-A, -B, -C, -DRB1, -DRB3/4/5, -DQB1, and -DPB1. For assignment of MICA alleles, polymorphisms in exons 2, 3, 4, and 5 were evaluated by sequence-based typing.15 Matching grade is described in graft versus host direction. Patients matched at HLA-A, -B, -C, -DRB1, and DQB1 were termed “HLA 10/10,” whereas mismatches at any of these loci were indicated as “HLA < 10/10.” Analysis of chimerism, response to treatment, and aGVHD grading are detailed elsewhere.16 Median age was 50 years (range, 13-75 years). A 10/10 donor-recipient HLA match was present in 73% (n = 172), any DPB1 mismatch in 73.3% (n = 173), and MICA mismatch in 8.5% (n = 20). GVHD-relevant covariates were equally distributed in MICA-matched and -mismatched patients (Table 1), with the exception of HLA mismatch, which possibly represents the association of MICA mismatches with HLA-B (P < .001) as well as HLA-C (P = .001) mismatches, resulting from linkage disequilibrium patterns as described previously.3
Table 1.
Patient characteristics and transplantation conditions
| Characteristic | All patients (n = 236), n (%) | MICA no mismatch (n = 216), n (%) | MICA mismatch (n = 20), n (%) | P |
|---|---|---|---|---|
| Age > 60 y | 54 (23) | 49 (23) | 5 (25) | .8 |
| Male sex | 138 (58.5) | 130 (60) | 8 (40) | .1 |
| CR at transplantation | 102 (43) | 92 (43) | 10 (50) | .6 |
| Pentostatin GVHD prophylaxis | 61 (25.8) | 54 (25) | 4 (20) | .8 |
| PB stem cell source | 65 (27.5) | 59 (27) | 6 (30) | .5 |
| Fludarabine regimen | 213 (90) | 196 (91) | 17 (85) | .4 |
| Ablative regimen | 139 (59) | 126 (58) | 13 (65) | .6 |
| ATG as part of conditioning regimen | 227 (96) | 209 (97) | 18 (90) | .17 |
| DPB1 2-mismatch | 60 (25) | 52 (24) | 8 (40) | .3 |
| HLA 10/10 match | 172 (73) | 164 (76) | 8 (40) | .001 |
Preparative regimens were fludarabine-containing (fludarabine/busulfan [n = 138], fludarabine/melphalan [n = 74], fludarabine/busulfan/clofarabine [n = 1]) and non–fludarabine-containing (busulfan/cyclophosphamide [n = 20], cytarabine/total body irradiation [n = 2], and total lymphocyte irradiation/antithymocyte globulin [ATG] [n = 1]). ATG was administered as part of conditioning regimen in 96% of the cases (n = 227). Tacrolimus-based standard GVHD prophylaxis was used,13 with additional pentostatin in 25% (n = 58) of patients.14 Day 11 methotrexate was omitted if pentostatin was administered. Supportive care treatments and blood products were administered as per institutional standards.13
Statistical analysis
Kaplan-Meier estimates were used for analysis of treatment-related mortality and relapse-free survival.17 Log-rank statistics were used to compare time-to-event curves, measured from the date of transplantation to the date of death or last contact. Cox18 proportional hazards regression model was used to evaluate significance of prognostic factors on rate of aGVHD (Table 2). All factors with a P value less than .10 from univariate analysis were included in a saturated model, and backward elimination was used to remove factors based on likelihood ratio test in the multiple regression analysis. Gray test was used to compare cumulative incidence of aGVHD and relapse with death as competing event.19
Table 2.
Factors associated with grade II to IV aGVHD incidence: univariate analysis of potential prognostic factors on the incidence rate of grade II to IV aGVHD
| Factor | All patients, % (no.) |
HLA 10 of 10 match patients, % (no.) |
HLA < 10 of 10 match patients, % (no.) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| aGVHD | No aGVHD | P | aGVHD | No aGVHD | P | aGVHD | No aGVHD | P | |
| Age > 60 y | 42 (23/54) | 44 (81/182) | .5 | 42 (19/45) | 40 (51/127) | .94 | 44 (4/9) | 54 (30/55) | .4 |
| Pentostatin GVHD prophylaxis | 44 (27/61) | 44 (77/175) | .79 | 39 (17/44) | 41 (53/128) | .95 | 59 (10/17) | 51 (24/47) | .65 |
| Fludarabine-based regimen | 42 (89/213) | 65 (15/23) | .002* | 39 (62/159) | 61 (8/13) | .53 | 50 (27/54) | 70 (7/10) | .04* |
| Reduced intensity conditioning | 35 (34/97) | 50 (70/139) | .02 | 32 (24/74) | 47 (46/98) | .05* | 43 (10/23) | 58 (24/41) | .25 |
| HLA 10/10 | 40 (70/172) | 53 (34/64) | .05 | ||||||
| Two-DPB1 mismatch | 55 (33/60) | 39 (25/63) | .04* | 37 (8/52) | 51 (12/27) | .02 | 65 (11/17) | 46 (7/15) | .32 |
| MICA GvH mismatch | 80 (16/20) | 40 (88/216) | < .001* | 75 (6/8) | 39 (64/164) | .03* | 83 (10/12) | 46 (24/52) | .03* |
| Remission at transplantation | 41 (42/102) | 46 (62/134) | .55 | 32 (25/77) | 47 (45/95) | .03 | 68 (17/25) | 43 (17/39) | .07 |
P values remain significant in multivariate analysis.
Results and discussion
Treatment-related mortality and relapse
A total of 128 (54%) patients have died with median overall survival of 15.3 months. Causes of death were infection (28%), disease relapse (44%), GVHD (24%), and other (4%). Day 100 treatment-related mortality was 13.6%.
A total of 78 (33%) recipients have relapsed with median relapse-free survival of 70 months. Remission at transplantation (19% vs 44%, P < .001) and 2 DPB1 mismatches compared with zero DPB1 mismatch (19% vs 38%, P = .01) significantly correlated with improved relapse-free survival. The 3-year cumulative incidence of relapse in MICA mismatch versus match was 20% versus 35% (P = .23; Figure 1A).
Figure 1.
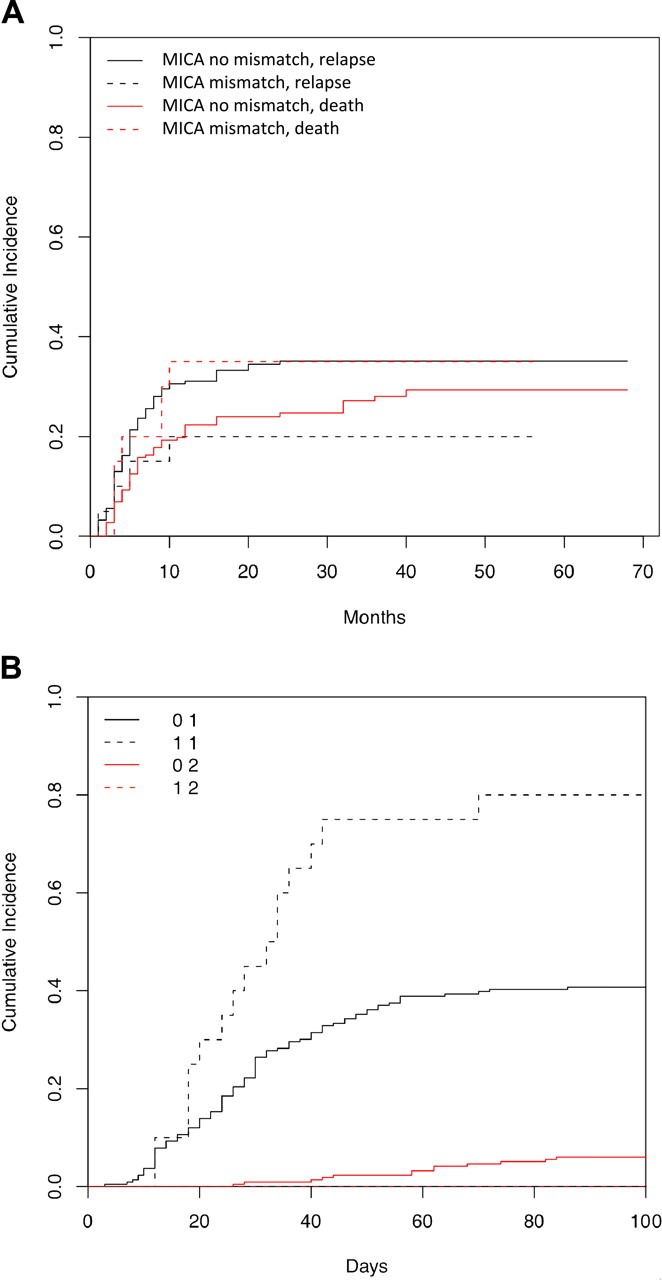
Cumulative incidence of relapse and aGVHD by MICA group. (A) Cumulative incidence of relapse with death as competing risk, by MICA group. Cumulative incidence curves for MICA match and mismatch groups are not statistically different for relapse (35% vs 20%, P = .21) or for the competing risk of death (29% vs 35%, P = .4). (B) Cumulative incidence of acute GVHD with death as competing risk, by MICA group. Cumulative incidence curves for MICA match and mismatch groups are statistically different for acute GVHD (40% vs 80%, P < .001), but they are not significantly different for the competing risk of death (6% vs 0%, P = .15).
aGVHD
Overall grade II-IV aGVHD incidence was 44% and grade III-IV was 14%. In multivariate analysis, fludarabine-based regimen (hazard ratio [HR] = 0.42), 2 DPB1 mismatches (HR = 1.69), and MICA mismatch (HR = 2.33; Figure 1B) were significantly associated with grade II-IV aGVHD. Incidence of grade III-IV aGVHD in the MICA mismatch group was increased nonsignificantly (25% vs 13.4%). Furthermore, the presence of MICA mismatch correlated with development of grade II-IV gastrointestinal aGVHD (35% vs 17%, P = .05, χ2). The effect of MICA mismatching on incidence of grade II-IV aGVHD was independent of HLA-B and -C mismatches (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Of the 63 known MICA alleles, MICA*008 is the commonest in North Americans.20,21 In our study, MICA*008 was present in 65%, reflecting a predominantly white cohort. There was no significant correlation between the presence or gene dose of this allele with any of the outcomes. A study of a more racially diverse population is needed to evaluate the full impact of MICA mismatch.
Reduced intensity regimen (HR = 0.60), remission at transplantation (HR = 0.57), and MICA mismatch (HR = 2.87) were significantly associated with grade II-IV aGVHD in HLA10/10 patients. Fludarabine-based conditioning regimen (HR = 0.35) and MICA mismatch (HR = 2.63) remained significant in HLA less than 10/10 match patients (Table 2).
Although our analysis is limited by the relatively small number of patients, the effect of MICA on increased aGVHD rate remained consistently significant among all patients as well as HLA10/10 and HLA less than 10/10 patients (Table 2).
The immune response against MICA allo-antigens may resemble the response to mismatched HLA antigens. Antibodies in transplantation recipients recognize specific epitopes present in MICA antigens of the donors, and these patterns of reactivity can be identified by conventional methods of testing by absorption and elution.22 MICA expression can be induced on dendritic cells that play an essential role in antigen presentation processes that initiate GVHD.4 Binding of Escherichia coli leads to MICA induction on epithelial cells, which in turn trigger interferon-γ release.5 Because Peyer patches are essential for GVHD,12 it is possible that MICA on the surface of intestinal cells interacts with NK cells through the NKG2D receptor as well as with γδ and αβ T cells. Thus, our finding that the presence of MICA mismatches correlated with the occurrence of gastrointestinal aGVHD may represent a clinical expression of these various interactions. In addition, reversible induction of MICA/B expression has been reported to occur in skin and liver aGVHD,8 suggesting that NKG2D ligand induction participates in the amplification loop, leading to tissue damage during aGVHD.8 MICA could be directly responsible for this effect or may in turn be a marker of differences in other yet to be identified histocompatibility loci. The observation of MICA mismatching leading to higher incidence of aGVHD correlates with Malkki et al,23 where mismatch for microsatellite markers located in the HLA region was associated with increased risk of death. In another study, improved survival was seen in MICA-matched patients.24 Petersdorf et al25 found that, among HLA-matched donor-recipient pairs, those with a haplotype mismatch were at increased risk for aGVHD. It was suggested that lack of phasing may lead to haplotype mismatches in neighboring genes, including MICA, MICB, or HLA-E, which encode for products involved in immune function. The work of other investigators23–25 and the data presented here provide evidence of the existence of other histocompatibility loci in the HLA region, including or in addition to MICA, which may induce immune responses, leading to an additive or synergistic effect in worsening of aGVHD.
In conclusion, our exploratory analysis indicates that mismatches at MICA locus in the GvH direction increase the risk of aGVHD. Therefore, MICA locus polymorphisms may represent novel transplantation antigens, and our results deserve confirmation in a larger cohort of patients.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S. Parmar wrote the manuscript and collected and analyzed data; M.d.L. collected and analyzed data, approved the final manuscript, and developed the project; Y.Z. developed the sequence-based typing procedure for high resolution of the MICA alleles and performed the MICA typing for these experiments; P.A.P., S. Pesoa, and L.d.P.S. collected and analyzed data and approved the final manuscript; P.L. performed statistical analysis; P.C. performed HLA typing and approved the final manuscript; G.R. collected data and approved and reviewed the final manuscript; M.H.Q., C.H., U.P., and P.K. contributed patients and approved and reviewed the final manuscript; E.J.S. and S.G. approved and reviewed the final manuscript and referred patients to most studies analyzed here; R.E.C. approved and reviewed the final manuscript and developed the project; P.S. developed the project, supervised the MICA typing, and approved and reviewed the final manuscript; and M.F.-V. developed the project, performed HLA and MICA typing, collected and analyzed data, and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marcos de Lima, Department of Stem Cell Transplantation and Cellular Therapy, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: mdelima@mdanderson.org.
References
- 1.Flomenberg N, Baxter-Lowe LA, Confer D, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104(7):1923–1930. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 2.Bahram S, Bresnahan M, Geraghty DE, Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci U S A. 1994;91(14):6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas D, Campillo JA, Lopez-Hernandez R, et al. Allelic diversity of MICA gene and MICA/HLA-B haplotypic variation in a population of the Murcia region in southeastern Spain. Hum Immunol. 2008;69(10):655–660. doi: 10.1016/j.humimm.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Jinushi M, Takehara T, Kanto T, et al. Critical role of MHC class I-related chain A and B expression on IFN-alpha-stimulated dendritic cells in NK cell activation: impairment in chronic hepatitis C virus infection. J Immunol. 2003;170(3):1249–1256. doi: 10.4049/jimmunol.170.3.1249. [DOI] [PubMed] [Google Scholar]
- 5.Tieng V, Le Bouguenec C, du Merle L, et al. Binding of Escherichia coli adhesin AfaE to CD55 triggers cell-surface expression of the MHC class I-related molecule MICA. Proc Natl Acad Sci U S A. 2002;99(5):2977–2982. doi: 10.1073/pnas.032668099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zwirner NW, Marcos CY, Mirbaha F, Zou Y, Stastny P. Identification of MICA as a new polymorphic alloantigen recognized by antibodies in sera of organ transplant recipients. Hum Immunol. 2000;61(9):917–924. doi: 10.1016/s0198-8859(00)00162-2. [DOI] [PubMed] [Google Scholar]
- 7.Mizutani K, Terasaki PI, Shih RN, Pei R, Ozawa M, Lee J. Frequency of MIC antibody in rejected renal transplant patients without HLA antibody. Hum Immunol. 2006;67(3):223–229. doi: 10.1016/j.humimm.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Gannage M, Buzyn A, Bogiatzi SI, et al. Induction of NKG2D ligands by gamma radiation and tumor necrosis factor-alpha may participate in the tissue damage during acute graft-versus-host disease. Transplantation. 2008;85(6):911–915. doi: 10.1097/TP.0b013e31816691ef. [DOI] [PubMed] [Google Scholar]
- 9.Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol. 2001;2(3):255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- 10.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci U S A. 1996;93(22):12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou Y, Stastny P, Susal C, Dohler B, Opelz G. Antibodies against MICA antigens and kidney-transplant rejection. N Engl J Med. 2007;357(13):1293–1300. doi: 10.1056/NEJMoa067160. [DOI] [PubMed] [Google Scholar]
- 12.Murai M, Yoneyama H, Ezaki T, et al. Peyer's patch is the essential site in initiating murine acute and lethal graft-versus-host reaction. Nat Immunol. 2003;4(2):154–160. doi: 10.1038/ni879. [DOI] [PubMed] [Google Scholar]
- 13.de Lima M, Couriel D, Thall PF, et al. Once-daily intravenous busulfan and fludarabine: clinical and pharmacokinetic results of a myeloablative, reduced-toxicity conditioning regimen for allogeneic stem cell transplantation in AML and MDS. Blood. 2004;104(3):857–864. doi: 10.1182/blood-2004-02-0414. [DOI] [PubMed] [Google Scholar]
- 14.de Padua Silva L, Anderson BS, Popat U, et al. Treatment of AML in first remission (CR1) with allogeneic hematopoietic stem cell transplantation (HSCT) using unrelated donors (UD). Blood (ASH Annual Meeting Abstracts) 2008;112 Abstract 976. [Google Scholar]
- 15.Zou Y, Han M, Wang Z, Stastny P. MICA allele-level typing by sequence-based typing with computerized assignment of polymorphic sites and short tandem repeats within the transmembrane region. Hum Immunol. 2006;67(3):145–151. doi: 10.1016/j.humimm.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Przepiorka D, Weisdorf D, Martin P, et al. 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant. 1995;15(6):825–828. [PubMed] [Google Scholar]
- 17.Kaplan MMP. Nonparametric estimation from incomplete observations. J Am Statist Assoc. 1958;53:457–481. [Google Scholar]
- 18.Cox D. Regression models and life tables. J R Statist Soc B. 1972;34:187–220. [Google Scholar]
- 19.Gray R. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16:1141–1154. [Google Scholar]
- 20.Petersdorf EW, Shuler KB, Longton GM, Spies T, Hansen JA. Population study of allelic diversity in the human MHC class I-related MIC-A gene. Immunogenetics. 1999;49(7):605–612. doi: 10.1007/s002510050655. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Lazaro AM, Lavingia B, Stastny P. Typing for all known MICA alleles by group-specific PCR and SSOP. Hum Immunol. 2001;62(6):620–631. doi: 10.1016/s0198-8859(01)00241-5. [DOI] [PubMed] [Google Scholar]
- 22.Zou Y, Qin Z, Silveus A, Fan Y, Stastny P. Polymorphisms of MICA recognized by human alloantibodies. Immunogenetics. 2009;61(2):91–100. doi: 10.1007/s00251-008-0344-9. [DOI] [PubMed] [Google Scholar]
- 23.Malkki M, Gooley TA, Horowitz MM, et al. Mapping MHC-resident transplantation determinants. Biol Blood Marrow Transplant. 2007;13(8):986–995. doi: 10.1016/j.bbmt.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitcharoen K, Witt CS, Romphruk AV, Christiansen FT, Leelayuwat C. MICA, MICB, and MHC beta block matching in bone marrow transplantation: relevance to transplantation outcome. Hum Immunol. 2006;67(3):238–246. doi: 10.1016/j.humimm.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Petersdorf EW, Malkki M, Gooley TA, Martin PJ, Guo Z. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med. 2007;4(1):e8. doi: 10.1371/journal.pmed.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]


