Abstract
Background
Vibrio parahaemolyticus is a major foodborne pathogen, particularly in Asian countries. Increased occurrence of outbreaks of V. parahaemolyticus gastroenteritis in China indicates the need to evaluation of the prevalence of this pathogenic species. V. parahaemolyticus distribution in shellfish from the eastern coast of China has been reported previously. However, to date, the prevalence of V. parahaemolyticus in retail aquatic products in North China has not been determined. To investigate the prevalence of V. parahaemolyticus in aquatic products in North China, 260 aquatic product samples were obtained from retail markets in 6 provinces of North China from November to December in 2012 and July to August in 2013.
Results
V. parahaemolyticus was detected in 94 (36.2 %) of the samples by the most probable number method. The density of V. parahaemolyticus ranged from 1.50 to 1100 MPN/g. V. parahaemolyticus was detected at a rate of 50.0 % and 22.7 % in summer and in winter, respectively. The density of V. parahaemolyticus was significantly higher in summer than in winter, with mean levels of 16.5 MPN/g and 5.0 MPN/g, respectively. Among 145 V. parahaemolyticus isolates examined, none of the isolates possessed tdh and trh. In multiplex PCR-based O-antigen serotyping of these 145 isolates, all serotypes, other than O6, O7, and O9, were detected, and serotype O2 was found to be the most prevalent (detected in 54 isolates). The 145 isolates were grouped into 7 clusters by enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) at a similarity coefficient of 0.66. The antimicrobial resistance patterns of these 145 isolates to 12 antimicrobial agents revealed that most of the isolates were resistant to streptomycin (86.2 %), while fewer were resistant to ampicillin (49.6 %), cefazolin (43.5 %), cephalothin (35.9 %), and kanamycin (22.1 %). All of the examined isolates were susceptible to azithromycin and chloramphenicol.
Conclusions
The findings of this study will help in defining appropriate monitoring programs, understanding of the dissemination of antibiotic resistant strains, and providing information for the assessment of exposure to this microorganism at the consumption level.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-016-0650-6) contains supplementary material, which is available to authorized users.
Keywords: Antimicrobial sensitivity, Aquatic products, ERIC-PCR, Prevalence, Quantitative analysis, Serotype, Vibrio parahaemolyticus, Virulence gene
Background
Vibrio parahaemolyticus is a human pathogen that has been associated with gastroenteritis worldwide [1–4], and outbreaks have been reported in many countries such as the USA, France, and New Zealand [5–7]. Moreover, in recent years, V. parahaemolyticus has been reported as a significant cause of foodborne bacterial poisoning in China [8, 9].
V. parahaemolyticus has been isolated from samples of a variety of aquatic products, including fish, shrimp, oyster, and clam [10, 11], and it is among the most common causative agents of aquatic product-associated gastroenteritis in the world [3, 12–16]. With the vigorous development of the Chinese economy, there has been a rapid increase in aquatic product consumption, not only along the coast of China, but also in mainland China. V. parahaemolyticus distribution in shellfish from the eastern coast of China has been reported previously [17]. Our previous studies have also shown that shrimp in Chinese retail markets are contaminated with V. parahaemolyticus [18]. However, to date, the presence of V. parahaemolyticus in retail aquatic products in North China has received less attention, and little information is available on the prevalence and contamination levels of V. parahaemolyticus in such aquatic products. Although V. parahaemolyticus is frequently present in aquatic products, most strains of this species are nonpathogenic to humans [19]; however, virulent V. parahaemolyticus strains are clearly a concern for aquatic product safety.
Detection of pathogenic V. parahaemolyticus isolates is typically based on molecular biological analysis that amplify tdh and trh sequences [20, 21]. These 2 genes, encoding the thermostable direct hemolysin (TDH) and the homologous thermostable direct hemolysin-related hemolysin (TRH), respectively, have been implicated in V. parahaemolyticus virulence [22–26]. However, a recent study showed that pathogenesis of V. parahaemolyticus does not appear to rely solely on a given virulence function; rather, virulence is a complex trait and different strains may employ somewhat different strategies [1].
To date, on the basis of somatic (O) and capsular (K) antigens, V. parahaemolyticus is classified into 13 O-serogroups and 71 K-serogroups [27, 28]. Serotyping has been widely used for identifying isolates in epidemiological studies. Furthermore, certain V. parahaemolyticus serotypes have been considered to be more virulent than others [29, 30]. A multiplex PCR-based O-antigen serotyping method for V. parahaemolyticus has been successfully developed [31]. Therefore, PCR-based serotyping is considered a convenient method for the rapid and accurate identification of a wide array of V. parahaemolyticus isolates. However, serotyping offers limited information about the genetic relatedness of strains.
In addition to serotyping, a variety of molecular typing methods have been applied to characterization of V. parahaemolyticus. Molecular typing of V. parahaemolyticus was shown to be a useful tool for providing information about the genetic relatedness of strains and for detection of virulent strains [32]. In recent years, a number of typing methods such as pulsed-field gel electrophoresis (PFGE) [33], ribotyping [34], random amplified polymorphic DNA (RAPD) analysis [35, 36], multi-locus sequence typing (MLST) [37], and enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) [38, 39] have been applied in the typing of V. parahaemolyticus. ERIC-PCR has previously proven useful for subtyping V. parahaemolyticus [33, 38, 39], and has been successfully used for genotyping different bacterial pathogens in previous studies [40–42].
Antimicrobials are commonly used in the treatment of infectious diseases in the aquaculture industry; however, the extensive use of antimicrobials has led to the development of antimicrobial resistance among pathogens in aquatic products and has rendered many known antimicrobials ineffective. V. parahaemolyticus has been reported to have resistance to ampicillin, streptomycin, kanamycin, tetracycline, and ciprofloxacin [43–46]. Antimicrobial resistance, particularly multi-drug resistance, is among the most important public health concerns because it is directly related to disease management and control [47, 48]. Therefore, it is necessary to establish a monitoring system for the objective evaluation of the antimicrobial-resistance profile.
Therefore, the objective of this study was to investigate the seasonal prevalence and levels of V. parahaemolyticus in retail aquatic products in North China. The virulence, serological types, and ERIC types were focused on, and the antibiotic resistance patterns of the isolated strains were determined.
Results
V. parahaemolyticus in aquatic products
The prevalence of V. parahaemolyticus in the 260 aquatic product samples examined in this study is shown in Table 1. V. parahaemolyticus was detected in 94 (36.2 %) of the 260 samples. Among the positive samples, the prevalence of V. parahaemolyticus were 23.4 % (22/94) in fish samples and 43.4 % (72/166) in shrimp samples. The density of V. parahaemolyticus varied from 1.50 to 1100 MPN/g. The mean levels of the pathogen in fish and shrimp samples were 14.0 MPN/g and 8.7 MPN/g, respectively. Independent-samples t-test analysis of V. parahaemolyticus levels versus 2 kinds of aquatic product samples indicated no statistically significant differences (P = 0.190).
Table 1.
Prevalence and levels of Vibrio parahaemolyticus in retail aquatic products from North China
| Aquatic products samples | No. of samples analyzed | No. of samples positive (%) | No. of samples containing the pathogen (MPN/g) | |||
|---|---|---|---|---|---|---|
| 3 to 10 | >10 to 102 | >102 to 103 | >103 | |||
| Fish | 94 | 22 (23.4) | 18 | 3 | 0 | 1 |
| Shrimp | 166 | 72 (43.4) | 50 | 19 | 3 | 0 |
| Total | 260 | 94 (36.2) | 68 | 22 | 3 | 1 |
In seasonal distribution, the maximum isolation rate of V. parahaemolyticus in aquatic products was in summer, and reached 50.0 %, while it was 22.7 % in winter (Table 2). The mean levels of V. parahaemolyticus in samples collected during summer and winter were 16.5 MPN/g and 5.0 MPN/g, respectively, which was significantly different (P = 0.040).
Table 2.
Prevalence and levels of Vibrio parahaemolyticus in retail aquatic products from North China during different seasons
| Season | No. of samples analyzed | No. of samples positive (%) | No. of samples containing the pathogen (MPN/g) | |||
|---|---|---|---|---|---|---|
| 3 to 10 | >10 to 102 | >102 to 103 | >103 | |||
| Winter | 132 | 30 (22.7) | 25 | 4 | 1 | 0 |
| Spring | 128 | 64 (50.0) | 43 | 18 | 2 | 1 |
| Total | 260 | 94 (36.2) | 68 | 22 | 3 | 1 |
Detection of tdh and trh genes in V. parahaemolyticus isolates
In total, 145 V. parahaemolyticus isolates were confirmed and tested for the presence of trh and tdh. None of the isolates possessed these genes.
O-serogroup typing by multiplex PCR
With the exception of serotypes O6, O7, and O9, all other serotypes were detected among the isolates. Serotype O2 was the most prevalent (54 isolates), followed by serotype O1 (25 isolates). The results of the O-antigen serotyping for all 145 isolates are shown in Table 3 and Additional file 4: Table S1.
Table 3.
Results of the PCR-based O-antigen serotyping of 145 Vibrio parahaemolyticus isolates
| Serogroups | Product sizes (bp) | No. of isolates analyzed | |
|---|---|---|---|
| Group 1 | O1 | 474 | 25 |
| O2 | 238 | 54 | |
| O4 | 671 | 5 | |
| O5 | 852 | 6 | |
| O10 | 343 | 2 | |
| Group 2 | O3a | 868 | 8 |
| O8 | 680 | 11 | |
| O11 | 524 | 17 | |
| O12 | 256 | 6 | |
| Uncertain | 11 | ||
| Total | 145 | ||
aO3 or O13
ERIC-PCR
The results of ERIC-PCR analysis of the 145 isolates are shown in Fig. 1. ERIC-PCR resulted in 4 – 10 amplification bands, with a size ranging from 130 bp to about 6000 bp. Bands with molecular sizes of 500, 1500, and 2500 bp were common to most isolates (Additional file 1: Figure S1, Additional file 2: Figure S2, and Additional file 3: Figure S3). Only 1 strain was represented in the figure if more than 2 strains of the same isolate type were analyzed. At a relative similarity coefficient of 0.66, the 145 isolates were classified into 7 clusters (designated as A, B, C, D, E, F, and G). Most isolates were distributed between the B and E clusters. One isolate (NO. 109) and a reference strain (ATCC 33847) were grouped into the same cluster; and some isolates (NOs. 53, 88, and 192) and clinical strains (NOs. SZ43, SZ53, and SZ51) were grouped into the same cluster, respectively.
Fig 1.
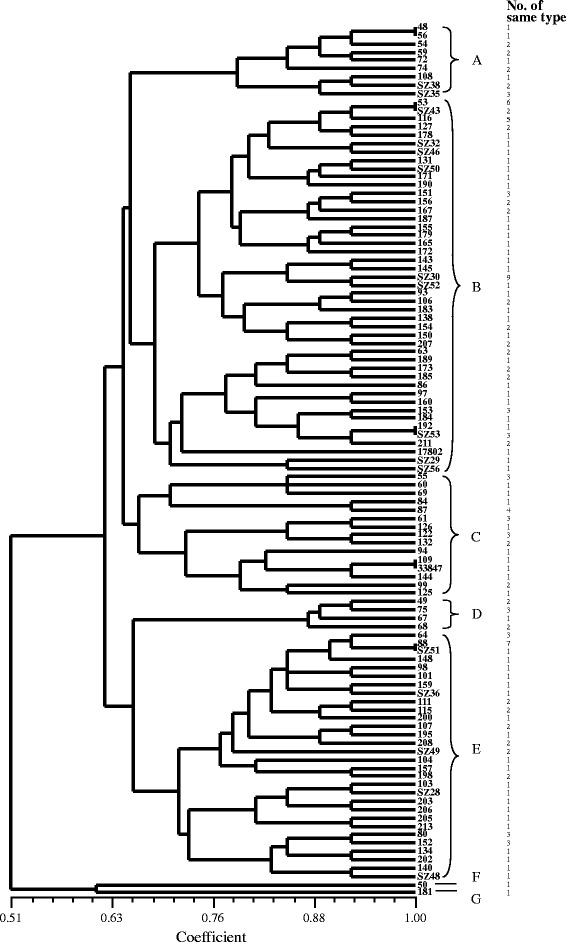
ERIC-PCR DNA fingerprint analysis of Vibrio parahaemolyticus isolates in retail aquatic products from North China
Antimicrobial susceptibility
Isolates of V. parahaemolyticus were tested for different levels of antibiotic resistance. The isolates were most resistant to streptomycin, with resistance and intermediate rates of 86.2 % and 11.7 %, respectively. In addition, the isolates exhibited relatively high resistance rates, of 49.6 %, 43.5 %, 35.9 %, and 22.1 %, for ampicillin, cefazolin, cephalothin, and kanamycin, respectively. However, it was surprising to note that strain 58, isolated from a fish sample, was a multi-drug-resistant strain, which showed resistance to 7 antibiotics: streptomycin, cephalothin, ampicillin, tetracycline, kanamycin, trimethoprim-sulfamethoxazole, and cefazolin. All of the examined isolates were susceptible to azithromycin and chloramphenicol. Among the remaining tested antibiotics, the next-highest susceptibility rates were observed for nalidixic acid (97.2 %), ciprofloxacin (91.7 %), tetracycline (83.4 %), trimethoprim-sulfamethoxazole (75.2 %), and gentamicin (62.8 %). The susceptibility, intermediate resistance, and resistance rates of the 145 examined V. parahaemolyticus isolates with respect to 12 antibiotics are shown in Table 4 and Additional file 4: Table S1.
Table 4.
Antimicrobial resistance profiles of 145 Vibrio parahaemolyticus isolates from North China
| Antimicrobial agent | Vibrio parahaemolyticus (n = 145) | ||
|---|---|---|---|
| NO. (%) of Ra | NO. (%) of Ia | NO. (%) of Sa | |
| Ampicillin (AMP) | 72 (49.6) | 40 (27.6) | 33 (22.8) |
| Azitromycin (AZM) | 0 (0.0) | 27 (18.6) | 118 (81.4) |
| Cefazolin (KZ) | 63 (43.5) | 75 (51.7) | 7 (4.8) |
| Cephalothin (KF) | 52 (35.9) | 81 (55.9) | 12 (8.2) |
| Chloramphenicol (C) | 0 (0.0) | 6 (4.1) | 139 (95.9) |
| Ciprofloxacin (CIP) | 3 (2.1) | 9 (6.2) | 133 (91.7) |
| Gentamicin (CN) | 1 (0.7) | 53 (36.5) | 91 (62.8) |
| Kanamycin (K) | 32 (22.1) | 108 (74.5) | 5 (3.4) |
| Nalidixic acid (NA) | 3 (2.1) | 1 (0.7) | 141 (97.2) |
| Streptomycin (S) | 125 (86.2) | 17 (11.7) | 3 (2.1) |
| Trimethoprim-sulfamethoxazole (SXT) | 17 (11.7) | 19 (13.1) | 109 (75.2) |
| Tetracycline (TE) | 2 (1.4) | 22 (15.2) | 121 (83.4) |
a R; resistant, I; intermediate resistance, S; susceptibility
Discussion
In this study, we analyzed 260 aquatic product samples and detected V. parahaemolyticus contamination in 94 samples. Thus, the overall prevalence of V. parahaemolyticus in aquatic product samples was determined to be 36.2 %, which was in accordance with the results reported in a study from France [49] and in our previous study [18]. Notably, the prevalence of V. parahaemolyticus in summer (50.0 %) was higher than that in winter (22.7 %), and the levels of V. parahaemolyticus in the summer were significantly different from those in the winter. These results may be related to the differences in the average temperature of the two seasons. These observations were in agreement with the results of previous studies that showed a seasonal variation in the occurrence of this pathogen [50–52]. These results, which also confirm the conclusions of the WHO risk assessment for V. parahaemolyticus [53], can be useful for defining efficient monitoring programs in harvesting areas, based on temperature values for control of V. parahaemolyticus.
As the presence of tdh- and/or trh-positive V. parahaemolyticus strains in aquatic products represents a public health risk, their detection would be of paramount importance. In the present study, tdh- and/or trh-positive V. parahaemolyticus strains were not detected in any of the aquatic product samples. This finding is consistent with the findings of a previous study reported in India [54]. However, it is in contrast to the findings of other previous studies [55, 56]. The occurrence rate of these genes in pathogenic V. Parahaemolyticus isolates is high, as has been proven for clinical isolates. Isolates obtained from the environment and food contain much less tdh or trh than clinical isolates [55, 57]. However, it has also been shown recently that some clinical V. parahaemolyticus isolates do not possess tdh and trh. Even in the absence of these two hemolysins, V. parahaemolyticus remains pathogenic, indicating the existence of other virulence factors [29, 58].
As mentioned above, 13 O-serogroups and 71 K-serogroups have been identified in V. parahaemolyticus. The pathogenicity of V. parahaemolyticus strains varies and is associated with the serotype. Recently, a multiplex PCR-based O-antigen serotyping method was developed for detection and identification of V. parahaemolyticus [31]. This assay can effectively distinguish all V. parahaemolyticus O-serogroups, except O3 and O13. In the present study, nine O-serogroups were detected among the isolates. Our data indicated that serovar O2 was the predominant serotype among the strains isolated from the aquatic product samples, a finding that was in agreement with that of a study previously conducted by our group [18]. However, our findings were in disagreement with those of a previous study that identified the O3 serotype as the predominant serotype from shellfish from the eastern coast of China [17]. Previous study demonstrated that most V. parahaemolyticus outbreaks were caused by multiserovars of strains, mainly including O3:K6, O1:KUT, and O4:K68 [29, 30]. O3:K6, O1:Kut, O4:K8, and O2:K3 were also the dominant serovars of V. parahaemolyticus, that caused outbreaks in China [16, 59]. The relationship of serotype between the food poisoning isolates and the aquatic food isolates are of concern.
Recently, highly discriminatory molecular typing methods such as PFGE and ERIC-PCR have been developed for differentiation of pathogenic bacteria. ERIC-PCR is a relatively simple, cost-effective method. It is easier to perform than PFGE and is very useful for the analysis of large numbers of strains [38]. Using this approach in this study, the isolates were classified into 7 clusters, at 66 % similarity. This result is similar to those of other studies, confirming the genetic diversity within V. parahaemolyticus strains [60–62]. Some V. parahaemolyticus isolates were of the same types as the clinical strains and a reference strain, which may indicate that these strains are genetically related.
Susceptibility tests revealed that the isolates were resistant to some antibiotics. The highest resistance rate (86.2 %) was observed for streptomycin, followed by ampicillin (49.6 %), cefazolin (43.5 %), cephalothin (35.9 %), and kanamycin (22.1 %). Similarly, previous studies have shown that the occurrence of streptomycin- and ampicillin-resistance in V. parahaemolyticus isolates is common [63]. In the present study, a small number of isolates showed resistance to ciprofloxacin, gentamicin, nalidixic acid, and tetracycline, while none of the isolates demonstrated resistance to azithromycin, indicating that these antimicrobials were still highly effective against V. parahaemolyticus isolates. Based on our findings, these antibiotics could be prescribed by doctors for the treatments of V. parahaemolyticus. In our study, half of the isolates were resistant to more than three antibiotics. Increasingly, resistant strains are being reported [63, 64], which may be explained as follow: On one hand, along with the steady expansion of the Asian aquaculture industry, aquaculture farmers use many different antibiotics to prevent (prophylactic use) and treat (therapeutic use) pathogenic bacterial infections in aquatic produce [64, 65]. On the other hand, a wide range of antibiotics used in humans contaminate water, leading to resistance in pathogenic bacteria. In general, infection emergence of microbial resistance to multiple drugs is a serious clinical problem and can lead to an increase in fatality rates [65].
Conclusions
This study showed that the levels of V. parahaemolyticus in retail aquatic products were relatively low and that none of the isolates possessed tdh and trh. Furthermore, serotype O2 was found to be the most prevalent; the isolates showed genetic diversity, as determined by ERIC-PCR typing, and the antimicrobial-resistance patterns showed that most of the isolates were resistant to streptomycin (86.2 %). The findings provided in this study may be useful in defining appropriate monitoring programs, understanding of the dissemination of antibiotic-resistant strains, and providing information for the assessment of exposure to this microorganism at the consumption level.
Methods
Bacterial strains
Two V. parahaemolyticus reference strains (ATCC 33847 and ATCC 17802) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). ATCC 33847 is tdh+, ATCC 17802 is trh+. Thirty-one clinical isolates were gifted by the Nanshan Shenzhen Center for Disease Control and Prevention (Shenzhen, China). All strains mentioned above were grown on Tryptone Soy Agar (TSA, Huankai Co. Ltd, Guangzhou, China) supplemented with 3 % (w/v) NaCl and incubated at 37 °C for 18 h.
Sample collection of aquatic products
In total, 260 aquatic product samples, including 94 fish samples and 166 shrimp samples were collected in retail markets from 6 different cities in North China, belonging to 6 provinces, i.e., Harbin (n = 22 [winter], n = 22 [summer]), Lanzhou (n = 23 [winter], n = 22 [summer]), Xi’an (n = 22 [winter], n = 22 [summer]),Taiyuan (n = 22 [winter], n = 22 [summer]), Jinan (n = 21 [winter], n = 19 [summer]), and Beijing (n = 22 [winter], n = 21 [summer]). Samples were collected from November to December in 2012 and from July to August in 2013. In this region, the climate is cold from November to December (winter), and it is hot from July to August (summer). The samples were placed in sterile sealed plastic bags and transported to the laboratory in a cold box below 4 °C and were analyzed immediately.
Most probable number (MPN) method for quantitative analysis
In this study, the MPN method was conducted accordance with the Bacteriological Analytical Manual standard and our previously study [18, 66]. Briefly, samples weighing 25 g were homogenized and combined with 225 mL of alkaline peptone water (APW) containing 3 % NaCl (Huankai, Guangzhou, China). Serial 10-fold dilutions were prepared up to a 1:103 dilution, and 3 x 1 mL portions of each dilution were inoculated into 9 mL of APW with 3 % NaCl. Dilutions were incubating at 37 °C for 16–18 h. After incubation, the collected samples were streaked onto thiosulfate-citrate-bile salts-sucrose (TCBS) agar plates (Huankai, Guangzhou, China) with an inoculation loop and incubated at 37 °C for 18–24 h. Three to five (if have) presumptive V. parahaemolyticus colonies (green or blue green colonies, 2–3 mm in diameter) were selected from each plate, streaked onto Chromogenic Vibrio Medium (Huankai, Guangzhou, China) and incubated at 37 °C for 24 h. One (if have) mauve colony from each Chromogenic Vibrio Medium plate was selected for identification tests including halophilism tests, oxidase activity assessment, gram staining, the 3.5 % NaCl triple-sugar-iron (TSI) test, and API 20E diagnostic strips testings (BioMerieux Company, Marcy-l’Étoile, France) test. The total numbers of V. parahaemolyticus in samples were determined by converting the numbers of culture tubes positive for V. parahaemolyticus to MPN/g using an MPN table. The V. parahaemolyticus isolates were confirmed by amplifying toxR, as described previously [67].
Detection of tdh and trh genes
Detection of the V. parahaemolyticus tdh and trh genes was performed by PCR, as described previously [68].
Multiplex serotyping PCR
The serotypes of V. parahaemolyticus isolates were identified using the PCR-based O-antigen serotyping technique. The primer concentrations and amplification conditions used were as previously described [31].
ERIC-PCR analysis
Genomic DNA was extracted from V. parahaemolyticus by using a commercial Universal DNA Extraction Kit (Sangon, Shanghai, China), according to the manufacturer’s instructions. Genomic DNA concentration was determined at 260 nm using a Nano Drop®ND-1000UVeVis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). A pair of primers, ERIC 1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC 2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) were used as previously reported [69]. ERIC-PCR typing was performed on the V. parahaemolyticus strains, using the protocol described previously with some modification [38]. More specifically, the reaction mixture (25 μL per reaction) consisted of 12.5 μL 2 × Long Taq Mix (Dongsheng Biotech, Guangzhou, China), 0.6 μmol/L of each primer, and 100 ng of template DNA. PCR was performed in a DNA thermocycler (Applied Biosystems, Foster City, CA, USA) by using the following cycling conditions: 1 cycle of denaturation at 95 °C for 5 min; followed by 35 cycles each consisting of 94 °C for 45 s, 52 °C for 1 min, and 72 °C for 3 min; and a final extension at 72 °C for 10 min. The PCR products were separated by electrophoresis in 2.0 % agarose gels, following which, they were subjected to GoldView staining (0.005 %, v/v) (SBS Genetech, Beijing, China) and photographed with a UV Imaging System (GE Healthcare, Waukesha, WI, USA). The images were captured in TIFF file format for further analysis.
Antimicrobial susceptibility
The susceptibility of the V. parahaemolyticus isolates to antibiotics was examined by the disk-diffusion method, according to the guidelines of the Clinical and Laboratory Standards Institute [70]. Muller − Hinton agar and a panel of 12 antibiotics disks were selected for the resistance tests. These 12 antibiotic disks (Oxoid, Hampshire, UK) contained ampicillin (10 μg), azithromycin (15 μg), cefazolin (30 μg), cephalothin (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), kanamycin (30 μg), nalidixic acid (30 μg), streptomycin (10 μg), trimethoprim − sulfamethoxazole (25 μg), or tetracycline (30 μg). The results were expressed as sensitive (S), intermediate (I), and resistant (R), following the methods of the CLSI. Escherichia coli ATCC 25922 and V. parahaemolyticus ATCC 17802 were used as quality control organisms.
Statistical analysis
The size of each band in the ERIC patterns was determined and the data were coded as 0 (absence) or 1 (presence). Cluster analysis was performed with NTSYS-pc (Version 2.10), a numerical taxonomy and multivariate analysis software package [71], based on Dice’s similarity coefficient (SD), with a 1 % position tolerance and the unweighted-pair group method using arithmetic averages (UPGMA).
To facilitate statistical analyzes of quantitative data, half the detection limit (1.5 MPN/g) for the total V. parahaemolyticus levels in aquatic product samples was substituted when levels were below the limit of detection [52]. Significance of differences was determined by using SPSS 11.0 (IBM, USA) to perform an independent-samples t-test.
Acknowledgements
This work was supported by research grants from the Natural Science Foundation of Guangdong Province (No. S2012030006235) and the Science and Technology Planning Project of Guangzhou (No. 201300000074). We would like to thank Editage for providing editorial assistance.
Additional files
ERIC-PCR1. M: DL2000; CK1: ATCC33847; CK2: ATCC17802. (PDF 153 kb)
ERIC-PCR2. M: DL5000; CK1: ATCC33847; CK2: ATCC17802. (PDF 142 kb)
ERIC-PCR3. M: DL5000; 1–31: SZ28-SZ58. (PDF 110 kb)
Results of serotyping, antimicrobial resistance, and ERIC-typing of Vibrio parahaemolyticus isolates in this study. (DOC 221 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XX and JC contributed equally to this work, and carried out the experimental work together. XX drafted the manuscript. QW conceived of the study, participated in its design, and provided technical support. JZ coordinated the study. TX helped to carry out ERIC-typing. All authors read and approved the final version of the manuscript.
References
- 1.Klein SL, Gutierrez West CK, Mejia DM, Lovell CR. Genes similar to the Vibrio parahaemolyticus virulence-related genes tdh, tlh, and vscC2 occur in other vibrionaceae species isolated from a pristine estuary. Appl Environ Microbiol. 2014;80(2):595–602. doi: 10.1128/AEM.02895-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paydar M, Teh CSJ, Thong KL. Prevalence and characterisation of potentially virulent Vibrio parahaemolyticus in seafood in Malaysia using conventional methods. PCR and REP-PCR Food Control. 2013;32(1):13–8. doi: 10.1016/j.foodcont.2012.11.034. [DOI] [Google Scholar]
- 3.FDA . Vibrio parahaemolyticus risk assessment: quantitative risk assessment on the public health impact of pathogenic Vibrio parahaemolyticus in raw oysters. Washington, DC: Food and Drug Administration; 2005. [Google Scholar]
- 4.Letchumanan V, Chan KG, Lee LH. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front Microbiol. 2014;5:705. doi: 10.3389/fmicb.2014.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteves K, Hervio-Heath D, Mosser T, Rodier C, Tournoud MG, Jumas-Bilak E, Colwell RR, Monfort P. Rapid proliferation of Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio cholerae during freshwater flash floods in French Mediterranean Coastal Lagoons. Appl Environ Microbiol. 2015;81(21):7600–9. doi: 10.1128/AEM.01848-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw KS, Sapkota AR, Jacobs JM, He X, Crump BC. Recreational swimmers’ exposure to Vibrio vulnificus and Vibrio parahaemolyticus in the Chesapeake Bay, Maryland, USA. Environ Int. 2015;74:99–105. doi: 10.1016/j.envint.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Cruz CD, Hedderley D, Fletcher GC. Long-term study of Vibrio parahaemolyticus prevalence and distribution in New Zealand shellfish. Appl Environ Microbiol. 2015;81(7):2320–7. doi: 10.1128/AEM.04020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu XM, Chen Y, Guo YC, Wang ZT. Foodborne diseases outbreaks in 2005----report of National Foodborne Diseases Surveillance Network in China. Chin J Food Hyg. 2008;20(6):506–9. [Google Scholar]
- 9.Chen Y, Guo YC, Wang ZT, Liu XM, Liu H, Dai Y, Tang ZZ, Wen J. Foodborne disease outbreaks in 2006---report of the National Foodborne Disease Surveillance Network. China J Hyg Res. 2010;39(3):331–4. [PubMed] [Google Scholar]
- 10.Liston J. Microbial hazards of seafood consumption. Food Technol-Chicago. 1990;44(12):58–62. [Google Scholar]
- 11.Letchumanan V, Yin WF, Lee LH, Chan KG. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front Microbiol. 2015;6:33. doi: 10.3389/fmicb.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, Luo J, Tan H, Ke B, He D, Ke C, Klena JD, Zhang Y. Phenotypic and phylogenetic analysis of Vibrio parahaemolyticus isolates recovered from diarrhea cases in Guangdong Province, China. Int J Food Microbiol. 2015;200:13–7. doi: 10.1016/j.ijfoodmicro.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan TM, Wang TK, Lee CL, Chien SW, Horng CB. Food-borne disease outbreaks due to bacteria in Taiwan, 1986 to 1995. J Clin Microbiol. 1997;35(5):1260–2. doi: 10.1128/jcm.35.5.1260-1262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePaola A, Ulaszek J, Kaysner CA, Tenge BJ, Nordstrom JL, Wells J, Puhr N, Gendel SM. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in north America and Asia. Appl Environ Microb. 2003;69(7):3999–4005. doi: 10.1128/AEM.69.7.3999-4005.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suffredini E, Mioni R, Mazzette R, Bordin P, Serratore P, Fois F, Piano A, Cozzi L, Croci L. Detection and quantification of Vibrio parahaemolyticus in shellfish from Italian production areas. Int J Food Microbiol. 2014;184:14–20. doi: 10.1016/j.ijfoodmicro.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Ma C, Deng X, Ke C, He D, Liang Z, Li W, Ke B, Li B, Zhang Y, Ng L, et al. Epidemiology and etiology characteristics of foodborne outbreaks caused by Vibrio parahaemolyticus during 2008–2010 in Guangdong province, China. Foodborne Pathog Dis. 2014;11(1):21–9. doi: 10.1089/fpd.2013.1522. [DOI] [PubMed] [Google Scholar]
- 17.Zhao F, Zhou DQ, Cao HH, Ma LP, Jiang YH. Distribution, serological and molecular characterization of Vibrio parahaemolyticus from shellfish in the eastern coast of China. Food Control. 2011;22(7):1095–100. doi: 10.1016/j.foodcont.2010.12.017. [DOI] [Google Scholar]
- 18.Xu XK, Wu QP, Zhang JM, Cheng JH, Zhang SH, Wu K. Prevalence, pathogenicity, and serotypes of Vibrio parahaemolyticus in shrimp from Chinese retail markets. Food Control. 2014;46:81–5. doi: 10.1016/j.foodcont.2014.04.042. [DOI] [Google Scholar]
- 19.Nishibuchi M, Kaper JB. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect Immun. 1995;63(6):2093–9. doi: 10.1128/iai.63.6.2093-2099.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirai H, Ito H, Hirayama T, Nakamoto Y, Nakabayashi N, Kumagai K, Takeda Y, Nishibuchi M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun. 1990;58(11):3568–73. doi: 10.1128/iai.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoh M, Kawakami N, Funakoshi Y, Okada K, Honda T. Evaluation of two assay kits for thermostable direct hemolysin (TDH) as an indicator of TDH-related hemolysin (TRH) produced by Vibrio parahaemolyticus. Microbiol Immunol. 1995;39(2):157–9. doi: 10.1111/j.1348-0421.1995.tb02183.x. [DOI] [PubMed] [Google Scholar]
- 22.Kaysner CA, DePaola AJ. Vibrio. In: Downes FP, Ito K, editors. Compendium of methods for the microbiological examination of food. Washington, D.C.: American Public Health Association; 2001. [Google Scholar]
- 23.Okuda J, Ishibashi M, Abbott SL, Janda JM, Nishibuchi M. Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J Clin Microbiol. 1997;35(8):1965–71. doi: 10.1128/jcm.35.8.1965-1971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okuda J, Nishibuchi M. Manifestation of the Kanagawa phenomenon, the virulence-associated phenotype, of Vibrio parahaemolyticus depends on a particular single base change in the promoter of the thermostable direct haemolysin gene. Mol Microbiol. 1998;30(3):499–511. doi: 10.1046/j.1365-2958.1998.01072.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang XH, Austin B. Haemolysins in Vibrio species. J Appl Microbiol. 2005;98(5):1011–9. doi: 10.1111/j.1365-2672.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 26.Raghunath P. Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front Microbiol. 2014;5:805. doi: 10.3389/fmicb.2014.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara-Kudo Y, DePaola A. Detection of Pathogenic Vibrios. In: Su YC, editor. Pathogenic Vibrios and Food Safety. NewYork: Nova Science Publishers, Inc.; 2012. pp. 179–210. [Google Scholar]
- 28.Ishibashi M, Ohta K, Shimada T, Honda T, Sugiyama J, Miwatani T, Yokoo H. Current status of OK serotype combinations of Vibrio parahaemolyticus. Nippon Saikingaku Zasshi. 2000;55:539–41. doi: 10.3412/jsb.55.539. [DOI] [Google Scholar]
- 29.Jones JL, Ludeke CH, Bowers JC, Garrett N, Fischer M, Parsons MB, Bopp CA, DePaola A. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J Clin Microbiol. 2012;50(7):2343–52. doi: 10.1128/JCM.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. Global dissemination of Vibrio parahaemolyticus serotype O3: K6 and its serovariants. Clin Microbiol Rev. 2007;20(1):39–48. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M, Guo D, Wong HC, Zhang X, Liu F, Chen H, Liu B, Wang L, Wu F, Feng L. Development of O-serogroup specific PCR assay for detection and identification of Vibrio parahaemolyticus. Int J Food Microbiol. 2012;159(2):122–9. doi: 10.1016/j.ijfoodmicro.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Olive DM, Bean P. Principles and applications of methods for DNA-based typingof microbial organisms. J Clin Microbiol. 1999;37(6):1661–9. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall S, Clark CG, Wang G, Mulvey M, Kelly MT, Johnson WM. Comparison of molecular methods for typing Vibrio parahaemolyticus. J Clin Microbiol. 1999;37(8):2473–8. doi: 10.1128/jcm.37.8.2473-2478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bag PK, Nandi S, Bhadra RK, Ramamurthy T, Bhattacharya SK, Nishibuchi M, Hamabata T, Yamasaki S, Takeda Y, Nair GB. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3: K6 associated with pandemic spread. J Clin Microbiol. 1999;37(7):2354–7. doi: 10.1128/jcm.37.7.2354-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong HC, Liu CC, Pan TM, Wang TK, Lee CL, Shih YC. Molecular typing of Vibrio parahaemolyticus isolates, obtained from patients involved in food poisoning outbreaks in Taiwan, by random amplified polymorphic DNA analysis. J Clin Microbiol. 1999;37(6):1809–12. doi: 10.1128/jcm.37.6.1809-1812.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang ZQ, Jiao XA, Zhou XH, Cao GX, Fang WM, Gu RX. Isolation and molecular characterization of Vibrio parahaemolyticus from fresh, low-temperature preserved, dried, and salted seafood products in two coastal areas of eastern China. Int J Food Microbiol. 2008;125(3):279–85. doi: 10.1016/j.ijfoodmicro.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus LA, DePaola A. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J Bacteriol. 2008;190(8):2831–40. doi: 10.1128/JB.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W, Xie Y, Xu J, Wang Q, Gu M, Yang J, Zhou M, Wang D, Shi C, Shi X. Molecular typing of Vibrio parahaemolyticus isolates from the middle-east coastline of China. Int J Food Microbiol. 2012;153(3):402–12. doi: 10.1016/j.ijfoodmicro.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Maluping RP, Ravelo C, Lavilla-Pitogo CR, Krovacek K, Romalde JL. Molecular typing of Vibrio parahaemolyticus strains isolated from the Philippines by PCR-based methods. J Appl Microbiol. 2005;99(2):383–91. doi: 10.1111/j.1365-2672.2005.02571.x. [DOI] [PubMed] [Google Scholar]
- 40.Chen M, Wu Q, Zhang J, Yan Z, Wang J. Prevalence and characterization of Listeria monocytogenes isolated from retail-level ready-to-eat foods in South China. Food Control. 2014;38:1–7. doi: 10.1016/j.foodcont.2013.09.061. [DOI] [Google Scholar]
- 41.Chen L, Zhang J, Yang X, Wu Q, Xu M. Prevalence and characterization of Salmonella spp. from foods in South China. Acta Microbiol Sin. 2013;53(12):1326–33. [PubMed] [Google Scholar]
- 42.Ye Y, Wu Q, Yao L, Dong X, Wu K, Zhang J. Analysis of a consensus fragment in ERIC-PCR fingerprinting of Enterobacter sakazakii. Int J Food Microbiol. 2009;132(2–3):172–5. doi: 10.1016/j.ijfoodmicro.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 43.Devi R, Surendran PK, Chakraborty K. Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from shrimp farms along the southwest coast of India. World J Microb Biot. 2009;25(11):2005–12. doi: 10.1007/s11274-009-0101-8. [DOI] [Google Scholar]
- 44.Raissy M, Moumeni M, Ansari M, Rahimi E. Antibiotic resistance pattern of some Vibrio strains isolated from seafood. Iran J Fish Sci. 2012;11(3):618–26. [Google Scholar]
- 45.Wong MHY, Liu M, Wan HY, Chen S. Characterization of Extended-Spectrum-beta-Lactamase-Producing Vibrio parahaemolyticus. Antimicrob Agents Ch. 2012;56(7):4026–8. doi: 10.1128/AAC.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitiyodom S, Khemtong S, Wongtavatchai J, Chuanchuen R. Characterization of antibiotic resistance in Vibrio spp. isolated from farmed marine shrimps (Penaeus monodon) Fems Microbiol Ecol. 2010;72:219–27. doi: 10.1111/j.1574-6941.2010.00846.x. [DOI] [PubMed] [Google Scholar]
- 47.Ansari M, Raissy M. In vitro susceptibility of commonly used antibiotics against Vibrio spp. isolated from Lobster (Panulirus homarus) Afr J Microbiol Res. 2010;4(23):2629–31. [Google Scholar]
- 48.Okoh AI, Igbinosa EO. Antibiotic susceptibility profiles of some Vibrio strains isolated from wastewater final effluents in a rural community of the Eastern Cape Province of South Africa. BMC Microbiol. 2010;10:143. doi: 10.1186/1471-2180-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert-Pillot A, Copin S, Himber C, Gay M, Quilici ML. Occurrence of the three major Vibrio species pathogenic for human in seafood products consumed in France using real-time PCR. Int J Food Microbiol. 2014;189:75–81. doi: 10.1016/j.ijfoodmicro.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 50.Zarei M, Borujeni MP, Jamnejad A, Khezrzadeh M. Seasonal prevalence of Vibrio species in retail shrimps with an emphasis on Vibrio parahaemolyticus. Food Control. 2012;25(1):107–9. doi: 10.1016/j.foodcont.2011.10.024. [DOI] [Google Scholar]
- 51.DePaola A, Nordstrom JL, Bowers JC, Wells JG, Cook DW. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl Environ Microb. 2003;69(3):1521–6. doi: 10.1128/AEM.69.3.1521-1526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parveen S, Hettiarachchi KA, Bowers JC, Jones JL, Tamplin ML, McKay R, Beatty W, Brohawn K, Dasilva LV, Depaola A. Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int J Food Microbiol. 2008;128(2):354–61. doi: 10.1016/j.ijfoodmicro.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 53.WHO. Risk assessment of Vibrio parahaemolyticus inseafood. In: WHO (Ed.), MRA Series, Joint FAO/WHO expert meetings on microbiological risk assessment. 2011.
- 54.Raghunath P, Acharya S, Bhanumathi A, Karunasagar I. Detection and molecular characterization of Vibrio parahaemolyticus isolated from seafood harvested along the southwest coast of India. Food Microbiol. 2008;25(6):824–30. doi: 10.1016/j.fm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 55.Rahimi E, Ameri M, Doosti A, Gholampour AR. Occurrence of toxigenic Vibrio parahaemolyticus strains in shrimp in Iran. Foodborne Pathog Dis. 2010;7(9):1107–11. doi: 10.1089/fpd.2010.0554. [DOI] [PubMed] [Google Scholar]
- 56.Sujeewa A, Norrakiah A, Laina M. Prevalence of toxic genes of Vibrio parahaemolyticus in shrimps (Penaeus monodon) and culture environment. Int Food Res J. 2009;16:89–95. [Google Scholar]
- 57.Di Pinto A, Clecarese G, De Corato R, Novello L, Terio V. Detection of pathogenic Vibrio parahaemolyticus in southern Italian shellfish. Food Control. 2008;19(11):1037–41. doi: 10.1016/j.foodcont.2007.10.013. [DOI] [Google Scholar]
- 58.Mahoney JC, Gerding MJ, Jones SH, Whistler CA. Comparison of the pathogenic potentials of environmental and clinical Vibrio parahaemolyticus strains indicates a role for temperature regulation in virulence. Appl Environ Microb. 2010;76(22):7459–65. doi: 10.1128/AEM.01450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H, Sun S, Shi W, Cui L, Gu Q. Serotype, virulence, and genetic traits of foodborne and clinical Vibrio parahaemolyticus isolates in Shanghai. China Foodborne Pathog Dis. 2013;10(9):796–804. doi: 10.1089/fpd.2012.1378. [DOI] [PubMed] [Google Scholar]
- 60.Wong HC, Chen MC, Liu SH, Liu DP. Incidence of highly genetically diversified Vibrio parahaemolyticus in seafood imported from Asian countries. Int J Food Microbiol. 1999;52(3):181–8. doi: 10.1016/S0168-1605(99)00143-9. [DOI] [PubMed] [Google Scholar]
- 61.Wong HC, Liu SH, Ku LW, Lee IY, Wang TK, Lee YS, Lee CL, Kuo LP, Shih DY. Characterization of Vibrio parahaemolyticus isolates obtained from foodborne illness outbreaks during 1992 through 1995 in Taiwan. J Food Protect. 2000;63(7):900–6. doi: 10.4315/0362-028x-63.7.900. [DOI] [PubMed] [Google Scholar]
- 62.Wong HC, Lin CH. Evaluation of typing of Vibrio parahaemolyticus by three PCR methods using specific primers. J Clin Microbiol. 2001;39(12):4233–40. doi: 10.1128/JCM.39.12.4233-4240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pazhani GP, Bhowmik SK, Ghosh S, Guin S, Dutta S, Rajendran K, Saha DR, Nandy RK, Bhattacharya MK, Mukhopadhyay AK, et al. Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. PLoS Negl Trop Dis. 2014;8(5) doi: 10.1371/journal.pntd.0002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Letchumanan V, Pusparajah P, Hern T, Loh T, Yin WF, Lee L-H, Chan KG. Occurrence and antibiotic resistance of Vibrio parahaemolyticus from shellfish in Selangor, Malaysia. Front Microbiol. 2015;6:1417. doi: 10.3389/fmicb.2015.01417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cabello FC, Godfrey HP, Tomova A, Ivanova L, Dölz H, Millanao A, Buschmann AH. Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ Microbiol. 2013;15(7):1917–42. doi: 10.1111/1462-2920.12134. [DOI] [PubMed] [Google Scholar]
- 66.Kaysner CA, DePaola AJ. U. S. Food and Drug Administration; bacteriological analytical manual; methods for specific pathogens; Chapter 9 Vibrio. Available at http://www.fda.gov/food/foodscienceresearch/laboratorymethods/ucm070830.htm. 2004.
- 67.Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37(4):1173–7. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gutierrez West CK, Klein SL, Lovell CR. High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl Environ Microb. 2013;79(7):2247–52. doi: 10.1128/AEM.03792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19(24):6823–31. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.CLSI . Clinical and Laboratory Standards Institute. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or Fastidious Bacteria. Approved Standard M45-A. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- 71.Rohlf FJ. NTSYS-pc. Numerical Taxonomy and Multi-variate Analysis System, Version 2.1. Exeter Software, Setauket, NewYork. 2000.


