Summary
Eukaryotic protein kinases phosphorylate substrates at serine, threonine and tyrosine residues that fall within the context of short sequence motifs. Knowing the phosphorylation site motif for a protein kinase facilitates designing substrates for kinase assays and mapping phosphorylation sites in protein substrates. Here, we describe an arrayed peptide library protocol for rapidly determining kinase phosphorylation consensus sequences. This method uses a set of peptide mixtures in which each of the 20 amino acid residues is systematically substituted at nine positions surrounding a central site of phosphorylation. Peptide mixtures are arrayed in multiwell plates and analyzed by radiolabel assay with the kinase of interest. The preferred sequence is determined from the relative rate of phosphorylation of each peptide in the array. Consensus peptides based on these sequences typically serve as efficient and specific kinase substrates for high throughput screening or incorporation into biosensors.
Keywords: Protein kinases, Peptide libraries, Enzyme specificity, Substrate profiling, Kinase assay, Signal transduction
1. Introduction
Protein kinases target specific downstream substrates in cells through a combination of direct and indirect physical interactions (1). Kinases interact directly with their substrates through both “proximal” interactions between the kinase active site and the site of phosphorylation, as well as more “distal” non-catalytic interactions to substrate docking sites. Most kinases phosphorylate substrates in the context of specific sequence motifs that interact favorably with the kinase catalytic cleft (2). Early studies with peptides derived from the phosphorylation sites of known protein substrates established the idea of the consensus sequence, in which key residues proximal to the phosphoacceptor residue were particularly critical for efficient phosphorylation (3). These consensus sequences were shown to vary among different kinases (Table 1). More recently, the application of peptide library methods to analyzing kinase specificity, as well as mapping of large numbers of phosphorylation sites on protein substrates, has suggested that the simple notion of the consensus sequence is inadequate to define the substrate specificity of a kinase (4–7). For example, a kinase may prefer, though not absolutely require, certain residues or sets of residues at a particular position relative to the phosphorylation site. In addition, at some positions a kinase may be permissive for most residues, yet strongly disfavor others (8, 9). From this standpoint the sequence specificity of a kinase may be better described quantitatively by a weight matrix (also called a position-specific scoring matrix) that provides the relative rates of phosphorylation of each residue at multiple positions near the site of phosphorylation (4). Such matrices are often depicted graphically as heat maps or sequence logos (10, 11).
Table 1.
Consensus sequences recognized by protein kinases.
| Kinase | Consensus sequence(s) | Reference |
|---|---|---|
| cAMP-dependent protein kinase (PKA) | R-R/K-x-S-ϕ | (25) |
| AKT | R-x-R-x-x-S/T-ϕ | (26) |
| AMP-activated protein kinase (AMPK) | ϕ-x-R-x-x-S-x-x-x-I/L | (27, 28) |
| Mitogen-activated protein kinases (MAPKs) | P/ϕ-x-S/T-P | (29) |
| Cyclin-dependent kinases (CDKs) | S/T-P-x-K/R | (5) |
| Casein kinase 1 (CK1) | D/E-D/E-D/E-x-x-S/T-ϕ pS/pT-x-x-S/T-ϕ |
(30) |
| Casein kinase 2 (CK2) | S/T-D/E-x-D/E | (6) |
| NimA-related kinase (NEK) | ϕ-x-x-S/T | (31) |
x: any amino acid
underline: phosphorylated residue
ϕ: hydrophobic residue
pS/pT: priming phosphoserine/phosphothreonine
Understanding the phosphorylation site specificity of a kinase has several applications in biochemistry, biology, and drug discovery. Such information facilitates the design of peptides that are typically highly efficient substrates for the kinase (12, 13). Optimized peptides can be used as substrates in high-throughput screening to identify kinase inhibitors, and can substantially reduce the quantity of kinase required for screening in comparison to generic peptide or protein substrates, which tend to be phosphorylated with low efficiency. In addition, optimized substrate sequences can be incorporated into “biosensors” that can be used to readout the activity of kinases in living cells in spatial and temporal dimensions (14). Lastly, knowing sequences that are preferred by a kinase facilitates identification of new phosphorylation sites by scanning sequences of known protein substrates or by searching protein sequence or mass spectrometry databases to identify novel substrates. Several bioinformatics tools have been developed to identify kinase substrates by searching such databases using peptide library-derived weight matrices of substrate specificity (4, 15–17).
Multiple methods have been developed to determine sequence motifs phosphorylated by kinases. Early peptide library approaches used either mixture-based “oriented” peptide libraries that were analyzed by Edman sequencing, or peptides immobilized in arrays on cellulose membranes (5, 18, 19). Recent developments have provided advantages in more reliably identifying negatively selected residues and decreased background associated with immobilized substrates. Current methods include analysis of proteome-derived peptide libraries by mass spectrometry, and tagging peptides bound to beads by thiophosphorylation with ATP-γ-S (20–22). Here, we describe a positional scanning peptide library (PSPL) method developed in our laboratory (23, 24). This method employs one of two “universal” sets of 198 biotinylated peptide mixtures that can be used to generally profile either serine-threonine or tyrosine kinases. Peptides are phosphorylated in solution with radiolabeled ATP, and then captured on streptavidin-coated membranes. Following washing and drying of the membrane, radiolabel incorporation into each peptide mixture is quantified by phosphor imaging. This method provides the relative phosphorylation rate of peptides incorporating every amino acid at several positions surrounding the phosphoacceptor site. In this way, the method indicates which residues are “essential”, which are preferred, and which are disfavored at each position. Quantified data are readily converted into a weight matrix that can be used for protein and database scanning.
2. Materials
2.1 Positional scanning peptide library (PSPL)
The PSPL for analysis of Ser-Thr kinases is made up of 198 peptide mixtures having the general sequence Y-A-X-X-X-X-X-S/T-X-X-X-X-G-A-K-K(biotin). For each peptide mixture, 8 of the X positions represent a degenerate mixture of all amino acids (except Cys, Ser, and Thr), S/T indicates an even mixture of Ser and Thr, and K(biotin) is ε-(N-biotinyl-6-aminohexanoyl)-lysine. The remaining X is fixed as one of the twenty standard amino acids, phosphothreonine (pT), or phosphotyrosine (pY). These 22 fixed residues at 9 positions yield the 198 peptide mixtures (22 × 9 = 198) that constitute the PSPL (see Note 1). This library is commercially available from Anaspec, Inc. (sold as Kinase Substrates Library, Groups I and II).
Dimethyl sulfoxide (DMSO).
Argon gas.
20 mM HEPES, pH 7.4.
Distilled, deionized water (ddH2O).
UV-transparent 96-well plates.
Transparent, flat-bottom polystyrene 1536 well plates.
Aluminum plate seals.
2.2 PSPL screening
Purified kinase(s) to be assayed (see Note 2).
Kinase reaction buffer. If the optimal buffer is not known for the kinase, we generally use 50 mM HEPES, pH 7.4, 10 mM MgCl2, 1 mM DTT, 0.1% Tween 20 (see Note 3).
10 μCi/μL γ-[33P]-ATP, 3000 Ci/mmol (see Note 4).
Streptavidin coated membrane (SAM2 Biotin Capture Membrane, Promega), cut into 5.5″ × 1″ strips and stored at −20°C.
Transparent, flat-bottom, polystyrene 1536-well plates.
Clear adhesive plate seals.
Whatman blotting paper, cut slightly larger than a multi-well plate, approximately 6″ × 4″.
10 mM ATP, adjusted to pH 7.0 with 0.1 M NaOH (store at −20°C).
Isopropanol.
1% SDS.
0.1% Tween 20.
Pin Cleaning Solution (V&P Scientific VP110).
SDS/TBS wash solution: 0.1% SDS, 10 mM Tris-HCl, pH 7.5, 140 mM NaCl.
2M NaCl.
2M NaCl, 1% H3PO4.
2.3 Equipment
UV-visible absorbance plate reader.
1536 floating pin replicator from V&P Scientific, loaded with 240 (48 × 5 rows) 0.787 mm diameter 200 nL slot pins (V&P Scientific FP3S200). Place the 240 pins in 5 of the 7 rows closest to the guide pins, leaving the two rows closest to the guide pins empty. This allows for the pin tool to access rows 2 through 6 of each quadrant of a 1536-well plate for the transfer of a complete peptide library (see Fig. 1 for quadrant layout of the 1536-well plate) (see Note 5).
48-pin strip (also loaded with 0.787 mm diameter, 200 nL slot pins, V&P Scientific FP3S200).
Pin tool alignment frame with guide holes to allow the replicator loaded with 240 pins to access horizontal quadrants of a 1536-well plate (V&P Scientific, see Fig. 2A)
Pin tool alignment frame with guide holes to allow the 48 pin strip to access every row of a 1536-well plate (V&P Scientific, see Fig. 2B).
Several extra 0.787 mm diameter, 200 nL slot pins (type FP3S200).
Rubber mat (such as a 4″ × 6″ Speedball Speedy Carve Block).
Incubator set to 30°C.
Low speed refrigerated centrifuge with rotor capable of spinning multiwell plates at 4°C.
Storage phosphor screen and cassette (Amersham Biosciences).
Phosphorimager (for example Molecular Imager FX Pro Plus, Bio-Rad) for scanning storage phosphor screen.
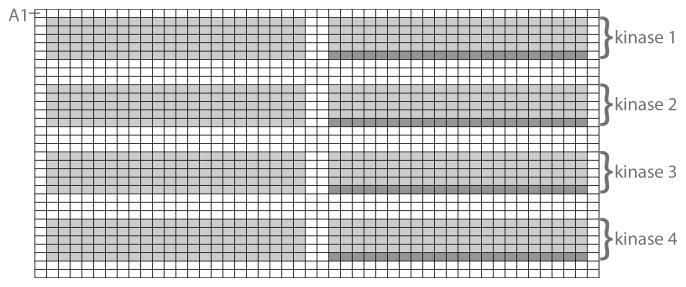
Fig. 1.
Up to four kinases can be profiled per 1536-well plate. The light gray boxes represent wells in which peptide (laid out as in Fig. 3) and kinase are present, the dark gray represent wells in which only kinase/ATP are present and are used as background controls.
Fig. 2.
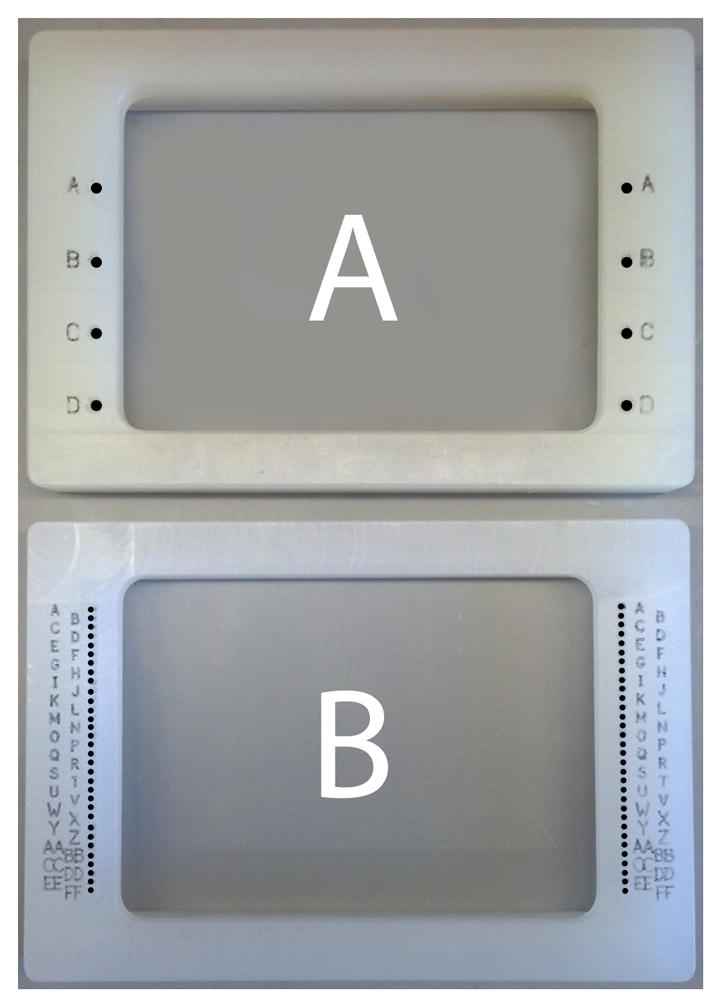
Two pin tool alignment frames used in the procedure. The A frame has four holes for guide pins that allow a pin tool loaded with 6 rows of 48 pins to transfer the peptide library into one of four quadrants of a 1536-well plate (see Fig. 1). The B frame has 32 pairs of holes to allow a 48 pin strip to transfer kinase/ATP to each row of a 1536-well plate.
3. Methods
3.1 Preparation of PSPL stock plates
Purge a sufficient quantity of DMSO with oxygen by bubbling with a steady stream of argon for at least 5 min.
Dissolve powdered peptides in DMSO (15 μL/mg).
Dilute 1 μL of each DMSO stock to 500 μL with 20 mM HEPES, pH 7.4 in duplicate, and read the absorbance at 280 nm in a plate reader (a standard cuvette reading spectrometer may also be used).
Back calculate the concentration of peptide in the DMSO solutions based on the molar absorptivity of the peptides (see Note 6).
Based on the absorbance readings, add additional DMSO to stock solutions to bring peptides to a concentration of 10 mM. Store DMSO stocks at −20 °C.
Dilute 1.5 μL of each 10 mM DMSO stock with 23.5 μL ddH2O to generate working aqueous 0.6 mM stock solutions. Aqueous stocks may be stored at −20 °C.
Array 0.6 mM aqueous solutions in a 1536-well plates by adding 5 μL per well using the template shown in Fig. 3. Seal stock plates with aluminum seals and store at −20 °C. This plate can be used to assay approximately 16 kinases.
Fig. 3.
Peptide library layout in stock plate and for kinase in the upper quadrant of reaction plate. The peptide sets with fixed residues N-terminal to the phosphosite (labeled −5 through −1) are on the left side of the plate (columns 2–23), and the peptide sets with fixed residues C-terminal to the phosphosite (labeled +1 through +4) are on the right side of the plate (columns 26–47). Letters indicate the fixed amino acid, where pT is phosphothreonine, pY is phosphotyrosine.
3.2 Peptide library screening
Both peptide stock and reaction plates should be kept cool on ice at all possible times to minimize evaporation (see Note 7). Up to 4 kinases can be profiled per 1536-well plate (Fig. 1). For the purposes of this protocol, we define four horizontal quadrants as: Quadrant I = rows A--H, Quadrant II = rows I--P, Quadrant III = rows Q--X, and Quadrant IV = rows Y--FF.
Thaw the peptide library stock plate on the benchtop. Shake or vortex the plate to mix, chill it on ice, spin plate in the refrigerated centrifuge (1400 × g for 1 min), and return to ice.
Chill a 1536 well plate to be used for the kinase reactions. Aliquot 2 μL of ice-cold kinase reaction buffer into each well of the first 7 rows of each quadrant, leaving the 8th row of the quadrant empty (see Note 8).
Centrifuge the reaction plate (1400 × g for 1 min) to remove air bubbles and bring the solution to the bottom of the wells.
Unseal the peptide stock plate, and place on the benchtop alongside the reaction plate. Place alignment frames with quadrant guide holes over both plates (Fig. 2A).
Dip the pins of the 240 pin replicator in 0.1% Tween, and then blot out the liquid on Whatman paper.
Align the replicator with the upper quadrant of the peptide stock plate by placing the guide pins into the appropriate guide holes. Insert the pins into the wells to transfer aliquots of each peptide solution to the pins. Dip the pins in the stock plate five times moving the device up and down, breaking the surface of the liquid each time (see Note 9).
Transfer the peptides from the replicator to the first quadrant of the reaction plate by placing the guide pins into the appropriate guide holes as above, and dipping the pins into the wells. Move the device up and down five times to mix thoroughly.
Blot out excess liquid from the pin tool on Whatman paper.
If more than one kinase is to be analyzed, repeat steps 5–8 to use additional quadrants of the reaction plate.
Wash the pins by dipping in the following solutions five times, blotting out the liquid in between each solution: one wash with 0.1% Tween, two washes with ddH2O, and a final wash with isopropanol. Blot out isopropanol and allow the pin tool to air dry.
Cover the stock plate with a fresh aluminum seal and return to storage at −20 °C.
Prepare 120 μL 10X kinase/ATP solutions containing the desired amount of each kinase (see Note 10), 550 μM cold ATP, and 0.33 μCi/μL γ-[33P]-ATP in kinase reaction buffer.
Place the 32 row alignment frame (Fig. 2B) on the reaction plate.
Add 10X kinase/ATP solution to a disposable reservoir.
Transfer the 10X kinase/ATP solution to the reaction wells of the each row of the plate that contains peptide solutions as follows. First prime the 48 pin strip by dipping in 0.1% Tween and blotting out the liquid. Next, dip the pins into the 10X kinase/ATP solution, ensuring that the pins are coated with solution. Using the alignment holes on the sides of the frame, place the pins into the second row of the appropriate quadrant (row B for the first quadrant). Dip the pins in the buffer five times by moving the strip up and down to ensure complete transfer of the solution to the wells. In between rows, blot out excess liquid on Whatman paper (caution: blotting paper will be radioactive), and wash pin strip twice with 0.1% Tween and four times with ddH2O by dipping the pin strip in each solution 5 times and blotting in between each wash. Repeat this step for the four other rows in the quadrant.
Wash the 48-pin strip by soaking the pins in 1% SDS for 2 min. Blot out excess liquid, and then wash twice with ddH2O and once with isopropanol. Blot out isopropanol and allow pins to air dry.
Repeat steps 14–16 to add any additional kinases to the remaining quadrants (see Fig. 1).
Seal the reaction plate with plastic adhesive tape and incubate for 2 hr at 30 °C (see Note 11). During the incubation, allow streptavidin membrane strips to warm to room temperature.
When the incubation is complete, chill reaction plate on ice, remove the seal, and place the four quadrant alignment frame (Fig. 2A) over the plate.
Tape the streptavidin membrane (labeled with a pencil if profiling more than one kinase) onto the rubber mat.
Prime the 240-pin replicator by dipping the pins into 0.1% Tween and blotting out the solution on Whatman paper.
Transfer aliquots from one reaction quadrant to the replicator pins by dipping the pins into the reaction plate, using the guide holes on the alignment frame. Raise and lower the pins 5 times to ensure efficient transfer.
In one even motion, lower the pins onto the streptavidin membrane. Hold the pins on the membrane for a few seconds, rocking the pin tool back and forth slightly.
Lift the pin tool from the membrane and inspect to confirm that all aliquots were transferred successfully. If any aliquots failed to transfer, use an individual pin to manually transfer aliquots from the reaction plate.
After 20 sec, remove the streptavidin membrane from the rubber mat and submerge it in the first wash solution (0.1% SDS/TBS) to quench the reactions.
Wash the pin device by dipping five times into and blotting between the following solutions: once in 0.1% Tween, twice in ddH2O, once in isopropanol. Blot out isopropanol and allow the device to air dry.
Repeat steps 20–26 for each of the remaining quadrants.
Swirl the membranes in the first wash solution a few times, then dispose of the solution as radioactive liquid waste according to the protocols of your institution.
Wash membranes twice with 100 mL 0.1% SDS/TBS, twice with 100 mL 2M NaCl, and twice with 2M NaCl/1%H3PO4, three minutes per wash. Rinse the membranes twice briefly with dH2O to remove excess buffer. Dispose of the wash solutions as appropriate for mildly (<1 μCi) radioactive liquid waste.
Allow membranes to air dry on a piece of aluminum foil for approximately 30 min, wrap in saran wrap, and expose to a storage phosphor screen overnight.
The following day, scan the phosphor screen with the phosphor imager. If the signal is weak, a longer exposure (generally not more than 1 week) may be required.
3.3 Data Analysis
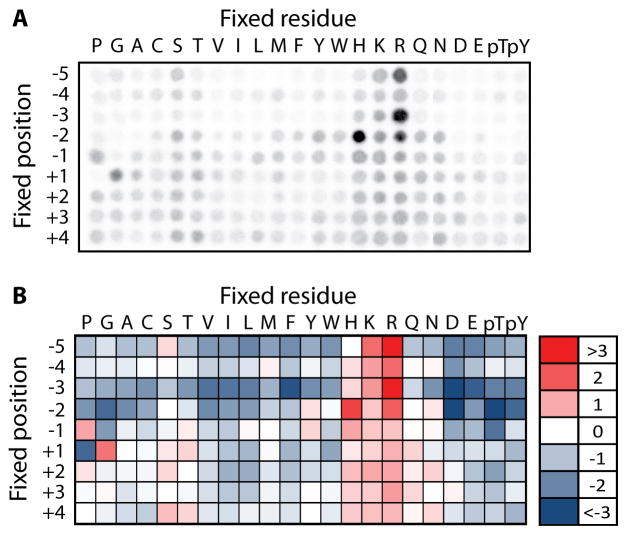
The image will appear as an array of spots (see Fig. 4A). Quantify the spot intensities using the image analysis software accompanying the phosphor imager. Export raw spot intensity data into a spreadsheet.
The wells in the 6th row, columns 26–47 of each quadrant (dark gray in Fig. 1) contain kinase and radiolabeled ATP but no peptide, and provide the background signal. Average the signal from these positions, and subtract this average background value from all the spot intensities to be quantified.
Normalize the data for each spot by dividing its background subtracted signal from the average value for all the spots in the same position. Once normalized in this way, the average value at each position is 1. Positively selected residues have values greater than 1, and negatively selected residues have values less than 1 (see Note 12).
Fig. 4.
Representative results from PSPL assay. (A) Raw autoradiogram showing array of radioactive spots for the kinase Pim3. Spots arising from wells on the left and right sides of the plate, respectively, were arranged using Adobe Photoshop so that the peptide positions are presented in ascending order. (B) Normalized spot intensities from panel A were converted into a heat map showing positively (red) and negatively (blue) selected residues at each position.
Acknowledgments
The authors are supported by NIH grants R01 GM102262 and R01 GM104047.
Footnotes
For analysis of tyrosine kinases, we have used a similar peptide library with the general sequence G-A-X-X-X-X-X-Y-X-X-X-X-A-G-K-K(biotin), in which the degenerate X positions are comprised of all the amino acids except Tyr and Cys. The 240 pin tool used to transfer the libraries should be washed if the library (e.g., tyrosine vs. serine/threonine library) is changed between runs. To clean the pin tool, allow it soak in pin cleaner for two minutes, then wash twice with ddH2O and once with isopropanol. Allow the pin tool to air dry before continuing with the protocol.
A suitable expression system for producing active kinase is required. If possible, bacterial expression systems are preferred because there is little concern that contaminating endogenous kinases will produce a signal on the peptide arrays. However, most kinases are inactive or insoluble when produced in bacteria, necessitating the use of eukaryotic cell (insect, mammalian or yeast) expression systems. These expression systems always carry the risk of having contaminating kinases produce a signal that could be spuriously attributed to the kinase of interest. We have encountered cases where kinases that appear to be highly pure on a Coomassie-stained polyacrylamide gel have contaminating kinase activities present. We recommend for kinases produced in eukaryotic expression systems that a kinase inactive mutant also be produced and subjected to PSPL screening. We tend to avoid using commercial sources of kinase because this control is generally unavailable. We have encountered cAMP-dependent protein kinase (PKA) as a common contaminant from mammalian cells. PKA contamination can be managed by including the specific peptide inhibitor PKI (100 nM) in the reactions.
In our hands, this buffer works for most kinases. If the kinase is not sufficiently active, substituting MnCl2 for MgCl2 will activate some kinases. The assay is compatible with a wide range of buffer conditions. If a different buffer is being used, it is important to add a low concentration of a non-ionic detergent (i.e. 0.1% Tween 20) so that the pin tools function properly.
γ-[32P]-ATP can also be used if appropriate safety precautions are taken. We recommend γ-[33P]-ATP because it does not require a safety shield.
Though the procedure can be performed with a single replicator, having two replicators available is useful. One is used for transferring peptides from the stock plates to the reaction plates. While the other, dedicated for radioactive use, is used for transferring aliquots of the reactions to the streptavidin membrane. Having two replicators allows many steps of the procedure to be performed in laboratory spaces not approved for radioactive use.
Because of variable solvent content, peptide weight is not a reliable parameter for determining the concentration of peptide solutions. It is preferable to use UV absorbance based on the molar extinction coefficients of Trp (5690 M−1 cm−1) and Tyr (1200 M−1 cm−1) in peptides. We assume that the mixture positions are 5.9% Trp and 5.9% Tyr, which provides a molar absorptivity value of 4560 M−1 cm−1 for most peptides, 10250 M−1 cm−1 for peptides with fixed Trp residues, 5840 M−1 cm−1 for peptides with fixed Tyr residues, and 5010 M−1 cm−1 for peptides with fixed phosphoTyr residues.
We typically place custom machined aluminum blocks (11.5 × 7.2 × 0.4 cm) underneath the multiwell plates to facilitate uniform cooling on ice. By placing the blocks on top of a larger aluminum plate (21 × 15 × 0.4 cm), all liquid transfer steps can be performed within a tray of ice rather than on the benchtop. This is useful for minimizing evaporation, particularly if multiple kinases will be screened at one time. If these are not available, re-chilling the plates by placing on ice in between transfers is important to reduce evaporation.
By adding buffer to all wells in rows 1 through 7, reaction wells containing peptide and kinase are surrounded by “blank” wells containing only buffer (see Figs. 3 and 1), which helps to prevent evaporation in the reaction wells.
The slot pins in the replicator withdraw aliquots of liquid by capillary action. Occasionally a pin will fail to pick up liquid, resulting in no liquid transfer. Dipping the pins into the liquid multiple times during transfers helps to ensure that all pins are filled. When dispensing liquid into the destination plate, dipping the pins repeatedly serves to mix the contents of the pins with buffer in the well, ensuring complete transfer of the contents.
How much kinase is required for the PSPL procedure depends on multiple factors, including the level of activity of the kinase preparation and the intrinsic ability of the kinase to phosphorylate peptides, which can vary considerably from kinase to kinase. In our experience the amount of kinase used in this protocol has ranged from 240 ng to 100 μg. We typically start with 10 μg of kinase (providing approximately 7.5 ng/μL kinase in the final reaction wells) and adjust up or down depending on the results of an initial PSPL assay.
We use a dedicated multiwell plate incubator to keep reaction plates at 30 °C. If a plate incubator is not available, an alternative is to line a Tupperware container with wet paper towels to make a humidified chamber that can be placed inside a standard 30 °C incubator.
One way to visualize the phosphorylation site motif is to display the data as a heat map (see Fig. 4B). Heat maps can be generated using Microsoft Excel. First log transform the normalized data so that the average value at a single position is 0. Then use the conditional formatting feature (three-color scale) to shade the negative, zero, and positive values with different colors.
References
- 1.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 2.Pinna LA, Ruzzene M. How do protein kinases recognize their substrates? Biochim Biophys Acta. 1996;1314:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 3.Kennelly PJ, Krebs EG. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 4.Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 5.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 6.Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 7.Shabb JB. Physiological substrates of cAMP-dependent protein kinase. Chem Rev. 2001;101:2381–2411. doi: 10.1021/cr000236l. [DOI] [PubMed] [Google Scholar]
- 8.Zhu G, Fujii K, Belkina N, Liu Y, James M, Herrero J, et al. Exceptional disfavor for proline at the P + 1 position among AGC and CAMK kinases establishes reciprocal specificity between them and the proline-directed kinases. J Biol Chem. 2005;280:10743–10748. doi: 10.1074/jbc.M413159200. [DOI] [PubMed] [Google Scholar]
- 9.Alexander J, Lim D, Joughin BA, Hegemann B, Hutchins JR, Ehrenberger T, et al. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal. 2011;4:ra42. doi: 10.1126/scisignal.2001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii K, Zhu G, Liu Y, Hallam J, Chen L, Herrero J, et al. Kinase peptide specificity: improved determination and relevance to protein phosphorylation. Proc Natl Acad Sci U S A. 2004;101:13744–13749. doi: 10.1073/pnas.0401881101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipchik AM, Killins RL, Geahlen RL, Parker LL. A peptide-based biosensor assay to detect intracellular Syk kinase activation and inhibition. Biochemistry. 2012;51:7515–7524. doi: 10.1021/bi300970h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutti JE, Porter MA, Cheely AW, Cantley LC, Wang X, Kireev D, et al. Development of a high-throughput assay for identifying inhibitors of TBK1 and IKKepsilon. PLoS One. 2012;7:e41494. doi: 10.1371/journal.pone.0041494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Allen MD. FRET-based biosensors for protein kinases: illuminating the kinome. Mol Biosyst. 2007;3:759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 15.Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linding R, Juhl Jensen L, Pasculescu A, Olhovsky M, Colwill K, Bork P, et al. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam HY, Kim PM, Mok J, Tonikian R, Sidhu SS, Turk BE, et al. MOTIPS: automated motif analysis for predicting targets of modular protein domains. BMC Bioinformatics. 2010;11:243. doi: 10.1186/1471-2105-11-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Songyang Z, Carraway KL, 3rd, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 19.Tegge WJ, Frank R. Analysis of protein kinase substrate specificity by the use of peptide libraries on cellulose paper (SPOT-method) Methods Mol Biol. 1998;87:99–106. doi: 10.1385/0-89603-392-9:99. [DOI] [PubMed] [Google Scholar]
- 20.Kettenbach AN, Wang T, Faherty BK, Madden DR, Knapp S, Bailey-Kellogg C, et al. Rapid determination of multiple linear kinase substrate motifs by mass spectrometry. Chem Biol. 2012;19:608–618. doi: 10.1016/j.chembiol.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglass J, Gunaratne R, Bradford D, Saeed F, Hoffert JD, Steinbach PJ, et al. Identifying protein kinase target preferences using mass spectrometry. Am J Physiol Cell Physiol. 2012;303:C715–727. doi: 10.1152/ajpcell.00166.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trinh TB, Xiao Q, Pei D. Profiling the substrate specificity of protein kinases by on-bead screening of peptide libraries. Biochemistry. 2013;52:5645–5655. doi: 10.1021/bi4008947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mok J, Kim PM, Lam HY, Piccirillo S, Zhou X, Jeschke GR, et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal. 2010;3:ra12. doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, et al. A rapid method for determining protein kinase phosphorylation specificity. Nature Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 25.Kemp BE, Graves DJ, Benjamini E, Krebs EG. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J Biol Chem. 1977;252:4888–4894. [PubMed] [Google Scholar]
- 26.Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 27.Dale S, Wilson WA, Edelman AM, Hardie DG. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- 28.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez FA, Raden DL, Davis RJ. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
- 30.Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ. Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem. 1990;265:14264–14269. [PubMed] [Google Scholar]
- 31.Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]