Abstract
Objective
To validate independent associations between branched-chain amino acids (BCAA) and other metabolites with coronary artery disease (CAD).
Methods
We conducted mass-spectrometry-based profiling of 63 metabolites in fasting plasma from 1983 sequential patients undergoing cardiac catheterization. Significant CAD was defined as CAD index ≥ 32 (at least one vessel with ≥95% stenosis; N = 995) and no CAD as CAD index ≤ 23 and no previous cardiac events (N = 610). Individuals (N = 378) with CAD severity between these extremes were excluded. Principal components analysis (PCA) reduced large numbers of correlated metabolites into uncorrelated factors. Association between metabolite factors and significant CAD vs. no CAD was tested using logistic regression; and between metabolite factors and severity of CAD was tested using linear regression.
Results
Of twelve PCA-derived metabolite factors, two were associated with CAD in multivariable models: factor 10, composed of BCAA (adjusted odds ratio, OR, 1.20; 95% CI 1.05–1.35, p = 0.005) and factor 7, composed of short-chain acylcarnitines, which include byproducts of BCAA metabolism (adjusted OR 1.30; 95% CI 1.14–1.48, p = 0.001). After adjustment for glycated albumin (marker of insulin resistance [IR]) both factors 7 (p = 0.0001) and 10 (p = 0.004) remained associated with CAD. Severity of CAD as a continuous variable (including patients with non-obstructive disease) was associated with metabolite factors 2, 3, 6, 7, 8 and 9; only factors 7 and 10 were associated in multivariable models.
Conclusions
We validated the independent association of metabolites involved in BCAA metabolism with CAD extremes. These metabolites may be reporting on novel mechanisms of CAD pathogenesis that are independent of IR and diabetes.
Keywords: Metabolism, Coronary artery disease, Branched chain amino acids
1. Introduction
CAD is the leading cause of death in developed countries and is projected to become the largest cause of mortality in some developing countries by the year 2015 [1,2]. CAD is a complex, heritable disease and CAD risk models remain incomplete. As such, metabolomics (the study of small-molecule metabolites that are byproducts of cellular metabolism) could be instrumental in better understanding the development of CAD and identifying factors that discriminate the severity of CAD lesions noninvasively.
We previously used targeted, quantitative metabolic profiling in a case–control study of CAD [3]. In that study, we found that a PCA derived metabolite factor composed of BCAA and related metabolites, and another factor composed of some amino acids involved in the urea cycle pathway, were associated with extremes of CAD burden after adjusting for known clinical risk factors. We sought to validate these findings using a similarly-selected nested-case–control population with extremes of CAD. These individuals were selected from within a larger, sequential cohort of individuals referred for cardiac catheterization at Duke University Medical Center (DUMC). This analysis was carried out as a component of the Measurement to Understand Reclassification of Disease of Cabarrus and Kannapolis Horizon 1 Cardiovascular Disease Study (MURDOCK CV) [4]. To better understand the relationship of metabolite factors with CAD burden, we also conducted an exploratory analysis to evaluate the relationship of CAD severity as a continuous variable with metabolite factor levels in the entire cohort.
2. Methods
2.1. Study population
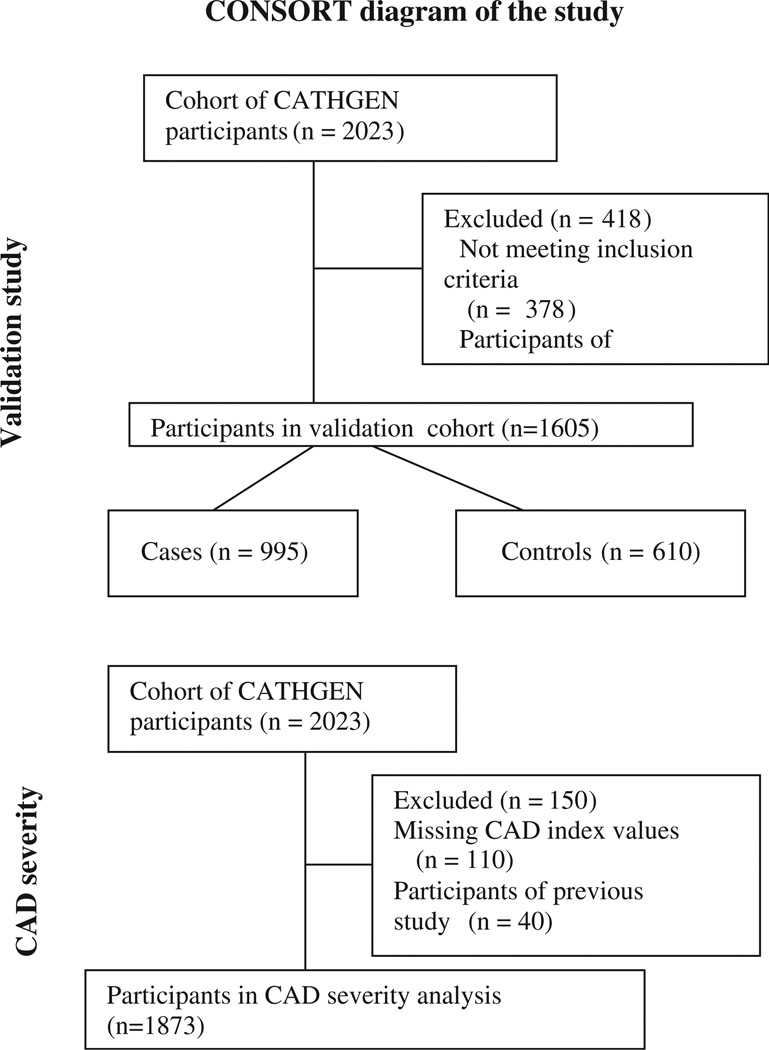
The CATHGEN biorepository consists of consenting, consecutive patients who underwent cardiac catheterization at DUMC between 2001 and 2010. All subjects were fasting for at least 6 h and arterial blood was collected through the femoral sheath at the time of catheterization. The blood samples were immediately chilled to 4 °C, centrifuged (within 30 min of collection), plasma separated, aliquoted, and frozen at −80 °C. As part of the MURDOCK CV study, 2023 sequential CATHGEN participants recruited between 2004 and 2007 were selected for study [4]. Of these, 40 individuals were participants in our previous case–control study and were excluded from the analyses, leaving 1983 individuals (Fig. 1). The Duke Databank for Cardiovascular Disease (DDCD), provided data on demographics, medical history, angiographic findings and longitudinal follow-up.
Fig. 1.
Participants included in sub groups.
Given our primary goal of validation, cases and controls were defined in a similar manner to our previous case–control study [3]. Specifically, cases with significant CAD were defined as individuals with a CAD index ≥ 32 (i.e. at least one epicardial coronary artery with 95% stenosis), where CAD index is a numeric summary of the angiographic extent of CAD [5]. Controls were defined as having a CAD index ≤ 23 (no individual epicardial coronary artery with >50% stenosis) and no history of prior or subsequent myocardial infarction, percutaneous coronary intervention, coronary artery bypass, heart transplant or peripheral vascular disease resulting in 995 cases with significant obstructive CAD and 610 control patients without significant obstructive CAD or history of adverse cardiovascular outcomes for primary validation analyses (Fig. 1). The 378 patients with intermediate degrees of CAD who did not meet criteria for either case or control group were excluded from the primary validation analysis. Exploratory analyses included the entire cohort to assess whether the severity of CAD as a continuous independent variable was associated with metabolite factor levels (dependent variable).
The Duke University Institutional Review Board (IRB) approved both the CATHGEN biorepository and the MURDOCK CV study. Written informed consent was obtained from all subjects for participation in the CATHGEN biorepository. The MURDOCK CV study was approved with a waiver of informed consent and HIPAA authorization.
2.2. Laboratory methods
Quantitative, targeted mass-spectrometry based metabolite profiling [6], was used to measure levels of 45 acylcarnitines, 15 amino acids and three conventional metabolites (ketones, nonesterified fatty acids and β-hydroxybutyrate) as a part of the main MURDOCK CV study [4]. Conventional metabolites were assayed on a Beckman–Coulter DxC600 clinical chemistry analyzer with reagents from Wako (Richmond, VA). Methodology and coefficients of variation for each assay have been reported [6,7]. Mass-spectroscopy (MS) metabolite measurement for acylcarnitines and amino acids used previously reported methods [8,9]. The laboratory (Sarah W. Stedman Nutrition and Metabolism Center, Duke Institute of Molecular Physiology metabolomics/biomarker core laboratory) was blinded to clinical data at the time of profiling. Glycated albumin was measured using the method described by T. Kouzuma et al. [10]. Serum was added to albumin digestion reagent, incubated and absorbance measured at 546 nm. Then ketoamine oxidase reagent was added, incubated, and absorbance measured again at 546 nm. The difference between measurements was calculated, and the concentration of glycated albumin was determined by comparison to a calibration curve. Total albumin in the sample was determined by the addition of serum to bromocresol green, incubation and absorbance measurement at 660 nm; followed by concentration determined by comparison to a calibration curve. The percentage of glycated albumin was determined by a simple ratio of these two measurements.
2.3. Statistical analysis
Metabolites with >25% of values reported as “0” (i.e. below the lower limits of quantification) were not further analyzed (C6 and C7-DC acylcarnitines). To reduce the large number of correlated metabolites residing in overlapping pathways into a smaller number of uncorrelated factors, PCA was used [3,6,7,11]. PCA factors with an eigenvalue ≥1.0 were retained based on the commonly used Kaiser criterion [12]. Varimax rotation was used to produce interpretable factors [13]. Individual metabolites with an absolute value for factor load of ≥0.4 were reported as composing a given factor. Scoring coefficients were constructed and used to calculate factors cores for each individual (weighted sum of the standardized metabolites within that factor, weighted on the factor loading for each metabolite). In exploratory analyses, partial least squares (PLS) analyses were also used as an alternative method for data reduction. To validate the association of metabolite factors and individual metabolites univariable and multivariable logistic regression models, adjusting for traditional CAD risk factors (age, sex, race, BMI, dyslipidemia, hypertension, diabetes, family history of CAD, and smoking) were constructed. Given the known role of one of our key metabolite factors (BCAA) in IR and diabetes, and that the diabetes variable included in the multivariable model is a clinical diagnosis that may underestimate the presence of IR; we further adjusted for glycated albumin in the multivariable logistic regression models.
To evaluate the association of metabolite factor levels with CAD severity (CAD index) as a continuous variable in the entire cohort of 1983 patients, univariable and multivariable linear regression models were constructed with the PCA factor levels as continuous dependent variables. The relationship of CAD index with PCA factors (factors 6, 7 and 10) as observed from box-and whisker plots was not perfectly linear; hence we explored using CAD index with and without spline transformation (had only a modest impact on the linearity of the relationship). We also explored using CAD index as a dependent and independent variable in linear regression models. Regardless of analytic technique, we found similar results. In this full cohort, we also assessed the association of number of diseased vessels on angiography as another measure of the severity of CAD with metabolite factor levels using linear regression.
Our primary objective was focused on validation of our previous findings; the analyses of metabolite factors with CAD severity was a priori defined as exploratory.
All statistical analyses were performed using SAS version 9.1 (Cary NC). A p value <0.05 was considered statistically significant. No adjustments were made for multiple comparisons as our analyses are focused on replicating previous findings and statistical significance is aimed at the beta for the PCA factors.
3. Results
The baseline clinical characteristics of the overall MURDOCK CV study population were previously presented [4]. Table 1 presents clinical characteristics of the CAD cases and controls used in this study. As expected, CAD cases had higher rates of clinical risk factors than controls, with the exception of low-density lipoprotein (LDL) cholesterol [14].
Table 1.
Baseline clinical characteristics of the case–control study population.
| Cases | Controls | p* | |
|---|---|---|---|
| (N = 995) | (N = 610) | ||
| Age, mean (SD) | 64.5 (10.9) | 57.0 (11.7) | <0.0001 |
| Sex, % male | 71.4 | 45.6 | <0.0001 |
| Race, % white | 78.6 | 63.4 | <0.0001 |
| Hypertension, % | 72.3 | 61.2 | <0.0001 |
| Diabetes, % | 34.1 | 26.6 | 0.0016 |
| Family history of CAD, % | 41.2 | 28.2 | <0.0001 |
| Current smoking, % | 52.2 | 36.4 | <0.0001 |
| Body mass index, mean (SD) | 29.5 (6.5) | 30.9 (8.2) | 0.0005 |
| CAD index, mean (SD) | 52.7 (18.5) | 4.1 (8.2) | <0.0001 |
| No. of coronary arteries with >75% stenosis | <0.0001 | ||
| 0 | 0 | 555 | |
| 1 | 285 | 0 | |
| 2 | 310 | 0 | |
| 3 | 398 | 0 | |
| History of MI, % | 37.9 | 0 | |
| History of dyslipidemia, % | 68.0 | 42.6 | <0.0001 |
| Total cholesterol, mean (SD) | 148.2 (39.9) | 159.0 (37.1) | <0.0001 |
| LDL cholesterol, mean (SD) | 90.1 (32.8) | 97.9 (31.4) | <0.0001 |
| HDL cholesterol, mean (SD) | 32.1 (11.7) | 36.6 (12.6) | <0.0001 |
| Triglycerides, mean (SD) | 134.9 (100.6) | 118.0 (77.5) | <0.0001 |
p-Value for difference between cases and controls.
3.1. Derivation of metabolite factors
PCA retained twelve metabolite factors (Table 2). Although these factors represented metabolites grouping in biologically plausible pathways, the factors were slightly different from the twelve PCA factors generated in our previous study [3]. Specifically the BCAA factor from our previous study (composed of phenylalanine, tyrosine, leucine/isoleucine, methionine, valine, C5 acylcarnitine, and alanine), grouped into three different factors in the PCA analysis for the current study, namely factor 6 (valine, leucine/isoleucine, methionine, phenylalanine, and tyrosine), factor 10 (valine, leucine/isoleucine, and glutamate/glutamine), and factor 7 (C3 and C5 acylcarnitines, byproducts of mitochondrial BCAA catabolism) (Table 2). The components of factor 9 containing amino acids reporting on the urea cycle (arginine, histidine, citrulline, and Ci4-DC: C4-DC) from the previous study did not group into any one factor in our PCA analysis; thus, we analyzed the individual metabolites from this factor in the current study.
Table 2.
Comparison of the metabolite factors generated by PCA with factors generated from previous cohort study by Shah et al. [3].
| Factor | Sequential cohort (N = 1983) | Cohort study (N = 388) | ||||
|---|---|---|---|---|---|---|
| Description | Component metabolitesa | Eigenvalue | Varianceb | Description | Component metabolitesa | |
| 1 | Medium chain acylcarnitines | C8, C10:1, C10, C12:1, C12, C14:2, C14:1, C14, C14:1-OH/C12:1-DC, C16:2, C16:1 | 14.19 | 23.3 | Medium chain acylcarnitines | C8, C10:1, C12, C10, C12:1, C10-OH:C8DC, C6-DC, C8:1-DC, C14:1, C14:2, C8:1-OH/C6:1-DC, C2 acylcarnitines |
| 2 | Short chain dicarboxyl acylcarnitines | Cit, C5OH/C3-DC, Ci4-DC/C4-DC, C5-DC, C6-DC, C10:3, C10:2, C10-OH/C8-DC, C12-OH/C10-DC, C6:1-DC/C8:1-OH, C8:1-DC | 6.45 | 10.5 | Long chain acylcarnitines | C18:1, C18:2, C18, C16, C16:1, C20:4, C14:1, C14:2, C16:2, C14:1-OH 3 |
| 3 | Long chain dicarboxyl acylcarnitines | C12-OH/C10-DC, C14-OH/C12-DC, C16-OH/C14-DC, C18:1-OH/C16:1-DC, C18-OH/C16-DC, C20, C20:1-OH/C18:1-DC, C20-OH/C18-DC | 5.12 | 8.4 | Long chain dicarboxyl/hydroxyl acylcarnitines | C18-OH/C16-DC, C20-OH/C18-DC, C20:1-OH/C18:1-DC, C16-OH/C14-DC, C18:1-OH/C16:1-DC, C14-OH/C12-DC, C12-OH:C10-DC, C14:1-OH, C20 |
| 4 | Ketone related | Ala, C2, C4-OH, Hbut, Ket | 4.06 | 6.7 | BCAA Related | Phe, Tyr, Leu/Ile, Met, Val, C5, Ala |
| 5 | Long chain acylcarnitines | C16, C18:2, C18:1, C18, C20:4, C16:1-OH/C14:1-DC | 2.66 | 4.4 | Ketone related | Ket, Hbut, Ala (−), C2, C4:OH, C14:1 |
| 6 | Branched chain amino acids & related metabolites | Val, Leu/Ile, Met, Phe, Tyr | 2.54 | 4.1 | Various | C8:1, C10:3 |
| 7 | Short chain acylcarnitines | C3, C4/Ci4, C5’s | 1.86 | 3.1 | Amino acids | Ser, Gly, FFA (−) |
| 8 | Amino acid factor | Gly, Ser, Pro, His, Orn, Arg, Cit | 1.60 | 2.6 | Dicarboxyls | C5-DC, C8:1-OH/C6:1-DC, Cit, C8:1-DC, C6-DC |
| 9 | Medium chain acylcarnitines | C8:1, C10:3, C10:2, C10:1 | 1.36 | 2.2 | Urea cycle related | Arg, His, Cit, Ci4-DC:C4DC (−) |
| 10 | Branched chain amino acids & related metabolites | Val, Leu/Ile, Glx | 1.30 | 2.1 | Short chain acylcarnitines | C3, C4:Ci4, C5 |
| 11 | Misc | Asx, C5:1, C22 | 1.24 | 2.1 | Various | C5:1, C18:2-OH (−), C22 (−) |
| 12 | Non-esterified fatty acids | Pro, C22, NEFA, Ala | 1.19 | 1.6 | Various | Asx, C22 |
The 12 metabolite factors generated by PCA, their description, individual components, eigen-values and variance explained by each PCA factor.
Only metabolites with |factor load| ≥ 0.4 for a specific that factor are listed, in order of magnitude of load for that factor; and a negative factor load for that factor is annotated with a (−).
Proportion of variance explained by that factor.
3.2. Association of PCA metabolite factors with significant CAD
In univariable analyses, factors 2, 3, 6, 7, 8, 9, and 10 were significantly associated with CAD (Table 3). Of these, factor 7 (short chain acylcarnitines, OR 1.30 [95% CI 1.14–1.48], P = 0.0001) and factor 10 (BCAA, OR 1.20 [95% CI 1.05–1.35], p = 0.005) remained significant in multivariable models (Table 3). After further adjustment for glycated albumin, factor 7 (OR 1.31 [95% CI 1.14–1.50], p = 0.0001) and factor 10 (OR 1.21 [95% CI 1.06–1.37], p = 0.004) remained associated with CAD. While the primary goal of this study was validation of our previous PCA-derived results, we also performed exploratory analyses using partial least squares (PLS) for data reduction. PLS analyses showed that 13% of the “outcome” of CAD could be explained by 15 metabolite factors. The top two PLS-derived factors combined explained 7% of the outcome, with one composed of several metabolites including Ci4-DC/C4-DC and citrulline (two of the “urea cycle factor” metabolites from the original paper), and the second one composed of the BCAA, aromatic amino acids (methionine, phenylalanine), glutamate/glutamine and C3 and C5 acylcarnitines, all of which are metabolites which composed the “BCAA factor” identified from our previous work. Thus, these PLS analyses corroborated our PCA results and also validated the findings from our previous work.
Table 3.
Univariable and multivariable associations of PCA factors with CAD case status.
| Factor | Name | Univariable | Multivariablea | ||
|---|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | ||
| 1 | Medium chain acylcarnitines | 1.00 (0.90, 1.10) | 0.93 | 0.95 (0.84, 1.07) | 0.36 |
| 2 | Short chain dicarboxyl acylcarnitines | 1.13 (1.01, 1.27) | 0.04 | 1.05 (0.94, 1.18) | 0.37 |
| 3 | Long chain dicarboxyl acylcarnitines | 1.40 (1.18, 1.67) | 0.0002 | 1.14 (0.97, 1.35) | 0.11 |
| 4 | Ketone related | 0.99 (0.89, 1.10) | 0.84 | 0.95 (0.84, 1.06) | 0.35 |
| 5 | Long chain acylcarnitines | 1.06 (0.95, 1.18) | 0.32 | 0.96 (0.85, 1.08) | 0.46 |
| 6 | Branched chain amino acids & related metabolites | 1.12 (1.02, 1.24) | 0.02 | 0.96 (0.85, 1.08) | 0.51 |
| 7 | Short chain acylcarnitines | 1.42 (1.26, 1.59) | 0.0001 | 1.30 (1.14, 1.48) | 0.0001 |
| 8 | Amino acid factor | 1.14 (1.03, 1.27) | 0.01 | 1.09(0.97, 1.23) | 0.15 |
| 9 | Medium chain acylcarnitines | 1.13 (1.02, 1.25) | 0.02 | 1.05 (0.93, 1.19) | 0.46 |
| 10 | Branched chain amino acids & related metabolites | 1.13 (1.02, 1.25) | 0.02 | 1.20 (1.05, 1.35) | 0.005 |
| 11 | Misc | 0.91 (0.82, 1.01) | 0.06 | 0.95 (0.85, 1.07) | 0.41 |
| 12 | Non-esterified fatty acids | 1.09 (0.98, 1.20) | 0.12 | 1.08 (0.96, 1.21) | 0.21 |
Adjusted for age, race, sex, diabetes, hypertension, smoking, dyslipidemia, family history of CAD, and BMI.
Values in bold represent factors that remained significantly associated with CAD in multivariable models.
3.3. Relationship of individual metabolites of factor 9 with significant CAD
When we examined the individual associations of the factor 9 from our previous work (arginine, histidine, citrulline and Ci4-DC/:C4DC) with CAD, we found that the levels of citrulline and Ci4-DC/:C4DC were higher in severe CAD cases than controls in univariable analysis, but not in multivariable models (Appendix 1).
3.4. Association of PCA factors with severity of CAD as a continuous measure
A higher CAD index was significantly associated with levels of factors 2, 3, 6, 7, 8, and 9 in univariable models (Table 4). After multivariable adjustment, severity of CAD was only significantly associated with levels of factor 7 (short chain acylcarnitines, p = 0.0003) and factor 10 (BCAA, p = 0.02; not significant in univariable modeling) (Table 4).
Table 4.
Univariable and multivariable associations of CADindex with PCA factor levels.
| Factor | Name | Univariable | Multivariablea | ||
|---|---|---|---|---|---|
| β, SE | p value | β, SE | p value | ||
| 1 | Medium chain acylcarnitines | −0.0001, 0.001 | 0.95 | −0.001, 0.001 | 0.28 |
| 2 | Short chain dicarboxyl acylcarnitines | 0.002, 0.001 | 0.008 | 0.001, 0.001 | 0.24 |
| 3 | Long chain dicarboxyl acylcarnitines | 0.003, 0.001 | 0.0002 | 0.002, 0.001 | 0.11 |
| Ketone related | 0.001, 0.001 | 0.20 | 0.0005, 0.001 | 0.60 | |
| 5 | Long chain acylcarnitines | 0.001, 0.001 | 0.15 | −0.0002, 0.001 | 0.86 |
| 6 | Branched chain amino acids & related metabolites | 0.002, 0.001 | 0.05 | −0.0008, 0.001 | 0.37 |
| 7 | Short chain acylcarnitines | 0.005, 0.001 | <0.0001 | 0.003, 0.001 | 0.0003 |
| 8 | Amino acid factor | 0.002, 0.001 | 0.02 | 0.002, 0.001 | 0.08 |
| 9 | Medium chain acylcarnitines | 0.002, 0.001 | 0.006 | 0.001, 0.001 | 0.21 |
| 10 | Branched chain amino acids & related metabolites | 0.002, 0.001 | 0.08 | 0.002, 0.001 | 0.02 |
| 11 | Misc | −0.001, 0.0009 | 0.18 | −0.0000, 0.001 | 0.99 |
| 12 | Non-esterified fatty acids | 0.002, 0.0009 | 0.09 | 0.001, 0.001 | 0.12 |
Adjusted for age, race, sex, diabetes, hypertension, smoking, dyslipidemia, family history of CAD, and BMI.
Values in bold represent factors that remained significantly associated with CAD in multivariable models.
Further, from univariable linear regression models we found that severity of CAD as measured by the number of diseased vessels was significantly associated with levels of factors 2, 3, 7, 9 and 10. After covariate adjustment, the severity of CAD as measured by number of diseased vessels was significantly associated with factor 7 (short-chain acylcarnitines, p = 0.0003) and factor 10 (BCAA, p = 0.01) (Appendix 2).
4. Discussion
In this study we validated results from our previous smaller case–control study using patients from a large sequential cohort. We found metabolites within the BCAA catabolic pathway are independently associated with significant CAD and expanded our previous findings by including a more detailed assessment of glucose control (glycated albumin). These results lend support to the hypothesis that these metabolites may mediate CAD development independently of their relationship with IR and diabetes. We also showed that our observations hold up when evaluating the association of metabolite factors with the continuous range of CAD severity, not just extremes of phenotype.
4.1. Association of BCAA with CAD, independent of IR or diabetes
BCAA and related metabolites have been strongly associated with IR in multiple studies [6,15–17]. Elevated BCAA levels were observed in diabetes patients compared with controls in a German cohort [18] and BCAA predicted incident diabetes in a prospective cohort study [19]. Elevations in BCAA levels were significantly associated with BMI and with IR after adjusting for BMI, sex, and pubertal stage in children [20]. In the Cardiovascular Risk in Young Finns Study, BCAA were demonstrated to be associated with IR in young, normoglycemic adults [21].
The diabetes variable included in our previous study was a clinical diagnosis that may have underestimated the prevalence of IR and diabetes. We further adjusted for glycated albumin to assess the independent association between BCAA and CAD in this study. Glycated albumin is a non-traditional glycemic marker shown to be more useful than hemoglobin A1C for monitoring blood glucose control in patients with poorly-controlled type 2 diabetes; being similar to hemoglobin A1C but integrates over a shorter period of time (14 days vs. 90 days) [22]. Both factors 7 and 10 remained significantly associated with CAD case status after additionally adjusting for glycated albumin, suggesting that the BCAA factor is associated with CAD independent of its relationship with IR, a known risk factor for development of CAD.
4.2. BCAA and IR
BCAA are essential amino acids derived from the diet that play an important role in the synthesis of peptides, sterols, ketones, and glucose [23]. In particular, leucine and isoleucine are involved in cell signaling, influencing cellular metabolism, protein synthesis and cell growth [6,24,25]. Isoleucine acts a nutrient regulator of glucose metabolism and isoleucine signaling is involved in the acceleration of metabolism of glucose [26]. Supplementation of BCAA in rats promoted IR [6] and similar observations were made in humans [6], primarily via chronic stimulation of the mTOR pathway and possibly by the activation of JNK in rats [6]. Branched chain keto acids presumably have a direct effect on mitochondrial function and cell viability, the exact mechanism of which is yet unknown [27].
4.3. BCAA and IR, independent of overweight/obesity
Studying 74 obese and 67 lean individuals, BCAA were associated with IR after adjusting for obesity [6]. Gastric bypass surgery resulted in better glycemic control and lower BCAA levels in morbidly obese and type 2 diabetes patients than equivalent weight loss from dietary interventions [28]. BCAA predicted the degree of improvement in IR following weight loss [28].
4.4. Validation of previous study
Of note, the BCAA factor identified in our previous study was composed of the BCAA valine and leucine/isoleucine, the aromatic amino acids phenylalanine and tyrosine (which share a transporter with the BCAA), C5 acylcarnitine, methionine, and alanine. In our current study, possibly due to the larger sample size facilitating resolution of clustered metabolites, this factor was split into three components, each with biological plausibility [1]: factor 6 (combination of BCAA and aromatic acids, which may be reporting on the pathway of shared transport) [2]; factor 7 (C3 and C5 acylcarnitines, which include byproducts of mitochondrial BCAA catabolism); and [3] factor 10 (BCAA, and glutamate/glutamine, the byproduct of the initial step of mitochondrial BCAA metabolism). We found statistical association only of factors 7 and 10 with case status, both of which report on mitochondrial BCAA catabolism, whereas factor 6 may be reporting on the shared transport mechanism with aromatic amino acids. Nevertheless, the concordance of findings for these metabolite factors with CAD in our validation cohort provides support for the role of these metabolites in cardiovascular disease. We also note that while the primary goal of the current study was validation of the prior results, the full multivariable results for factors 7 and 10 adjusted for cardiovascular risk factors and glycated albumin would survive Bonferroni correction at the level of 12 factors (p ≤ 0.004), supporting the robustness of these metabolites for CAD discrimination.
In our previous work, we had also found that a “urea cycle factor” (this factor had a strong load of arginine and may be reporting on amino acid and ammonia metabolism in the urea cycle) was independently associated with CAD severity; however, these metabolites did not cluster into one factor in the current study. Analysis of the individual metabolites revealed that citrulline and Ci4-DC:C4DC acylcarnitine were significantly associated with CAD severity in unadjusted analysis, but not after adjusting for clinical covariates.
In exploratory analyses and to capitalize on the full sequential cohort, we assessed whether the complete range of severity of CAD was associated with metabolite factor levels. Although there were statistically significant associations of CAD index with factors 7 and 10, it was in the absence of a clear linear relationship between increasing CAD severity and metabolite factor levels. Spline transformations of the data did not improve the linearity of the relationship, nor did it change the effect estimate considerably. Further, for factors 7 and 10 when evaluating number of diseased vessels reflecting CAD severity, we found significant associations. However, means of factor levels were primarily different between subjects with no diseased vessels (lower factor means) and any diseased vessels. The means of factor levels did not increase progressively with an increase in the number of diseased vessels. Thus, we conclude that the relationships across the whole range of severity of CAD as measured by CAD index or number of diseased vessels with metabolite factor levels is not strong as the association of metabolite levels with extremes of the CAD phenotype used in our case–control analysis.
4.5. Strengths and limitations
We recognize there are limitations to our study. The study population was a convenience sample of individuals referred for cardiac catheterization for concern of CAD. This population represents people who have a high burden of cardiovascular risk factors. However, the detailed angiographic phenotype is a definite strength. Though we did not account for dietary and environmental variations in metabolite levels in our study, all samples were collected from patients in a fasting state, and all metabolite profiling was done in a single batch to avoid batch effect confounders. We did not adjust for medication use or lipids in our analyses, as similar adjustments in our previous work showed minimal influence on associations of factors with CAD [3]. This is the largest study to date of metabolic profiling in CAD. The use of a high-throughput molecular technology (a quantitative, targeted metabolomic platform) has allowed us to identify metabolic signatures that independently discriminate extremes of CAD severity and validate our results from exploratory analyses in smaller sample sets. These findings could help in understanding the mechanisms of CAD development.
5. Conclusions
We have validated our previous findings showing BCAAs and related metabolites are independently associated with extremes of CAD severity, even after adjusting for diabetes and measures of IR. More studies are needed to elucidate the underlying biologic pathways that mediate the association of BCAAs with CAD severity.
Supplementary Material
Acknowledgments
Funding sources
The MURDOCK study was funded through a gift to Duke from the David H. Murdock Institute for Business and Culture; RO1HL095987 (Shah); UL1RR024128-01 (National Center for Research Resources [NCRR]; PO1 DK058398 (Newgard).
We thank the CATHGEN participants and Z. Elaine Dowdy for study coordination.
Footnotes
Disclosures
Shah, Kraus, Newgard and Newby hold a patent on an unrelated finding.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.atherosclerosis.2013.10.036.
Contributor Information
Sayanti Bhattacharya, Email: sayanti.bhattacharya@duke.edu.
Christopher B. Granger, Email: christopher.granger@duke.edu.
Damian Craig, Email: damian.craig@duke.edu.
Carol Haynes, Email: carol.haynes@duke.edu.
James Bain, Email: james.bain@duke.edu.
Robert D. Stevens, Email: rdjestev@duke.edu.
Elizabeth R. Hauser, Email: elizabeth.hauser@duke.edu.
Christopher B. Newgard, Email: newga002@mc.duke.edu.
William E. Kraus, Email: william.kraus@duke.edu.
L. Kristin Newby, Email: kristin.newby@duke.edu.
Svati H. Shah, Email: svati.shah@duke.edu.
References
- 1.Gupta R, Gupta VP. Meta-analysis of coronary heart disease prevalence in India. Indian Heart J. 1996;48(3):241–245. [PubMed] [Google Scholar]
- 2.Yusuf S, Reddy S, Ounpuu S, Anand S. Global burden of cardiovascular diseases: part II: variations in cardiovascular disease by specific ethnic groups and geographic regions and prevention strategies. Circulation. 2001;104(23):2855–2864. doi: 10.1161/hc4701.099488. [DOI] [PubMed] [Google Scholar]
- 3.Shah SH, Bain JR, Muehlbauer MJ, et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet. 2010;3(2):207–214. doi: 10.1161/CIRCGENETICS.109.852814. [DOI] [PubMed] [Google Scholar]
- 4.Shah SH, Granger CB, Hauser ER, et al. Reclassification of cardiovascular risk using integrated clinical and molecular biosignatures: design of and rationale for the Measurement to Understand the Reclassification of Disease of Cabarrus and Kannapolis (MURDOCK) Horizon 1 Cardiovascular Disease Study. Am Heart J. 2010;160(3):371–379. e2. doi: 10.1016/j.ahj.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 5.Smith LR, Harrell FE, Jr, Rankin JS, et al. Determinants of early versus late cardiac death in patients undergoing coronary artery bypass graft surgery. Circulation. 1991;84(Suppl. 5):III245–III253. [PubMed] [Google Scholar]
- 6.Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to IR. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah SH, Hauser ER, Bain JR, et al. High heritability of metabolomic profiles in families burdened with premature cardiovascular disease. Mol Syst Biol. 2009;5:258. doi: 10.1038/msb.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An J, Muoio DM, Shiota M, et al. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal IR. Nat Med. 2004;10(3):268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- 9.Chace DH, Hillman SL, Millington DS, Kahler SG, Roe CR, Naylor EW. Rapid diagnosis of maple syrup urine disease in blood spots from newborns by tandem mass spectrometry. Clin Chem. 1995;41(1):62–68. [PubMed] [Google Scholar]
- 10.Kouzuma T, Usami T, Yamakoshi M, Takahashi M, Imamura S. An enzymatic method for the measurement of glycated albumin in biological samples. Clin Chim Acta. 2002;324(1–2):61–71. doi: 10.1016/s0009-8981(02)00207-3. [DOI] [PubMed] [Google Scholar]
- 11.Huffman KM, Shah SH, Stevens RD, et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care. 2009;32(9):1678–1683. doi: 10.2337/dc08-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaiser HF. The application of electronic computers to factor analysis. Educ Psychol Meas. 1960;20:141–151. [Google Scholar]
- 13.Kaiser HF. The varimax criterion for analytic rotation in factor analysis. Psychometrika. 1958;23(3):187–200. [Google Scholar]
- 14.Wang TY, Newby LK, Chen AY, et al. Hypercholesterolemia paradox in relation to mortality in acute coronary syndrome. Clin Cardiol. 2009;32(9):E22–E28. doi: 10.1002/clc.20518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koves TR, Ussher JR, Noland RC, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle IR. Cell Metab. 2008;7(1):45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Shaham O, Wei R, Wang TJ, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng S, Rhee EP, Larson MG, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125(18):2222–2231. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suhre K, Meisinger C, Doring A, et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PloS One. 2010;5(11):e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCormack SE, Shaham O, McCarthy MA, et al. Circulating branched-chain amino acid concentrations are associated with obesity and future IR in children and adolescents. Pediatr Obes. 2013;8(1):52–61. doi: 10.1111/j.2047-6310.2012.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wurtz P, Soininen P, Kangas AJ, et al. Branched-chain and aromatic amino acids are predictors of IR in young adults. Diabetes Care. 2013;36(3):648–655. doi: 10.2337/dc12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee EY, Lee BW, Kim D, et al. Glycated albumin is a useful glycation index for monitoring fluctuating and poorly controlled type 2 diabetic patients. Acta Diabetol. 2011;48(2):167–172. doi: 10.1007/s00592-010-0242-0. [DOI] [PubMed] [Google Scholar]
- 23.Baquet A, Lavoinne A, Hue L. Comparison of the effects of various amino acids on glycogen synthesis, lipogenesis and ketogenesis in isolated rat hepatocytes. Biochem J. 1991;273(Pt 1):57–62. doi: 10.1042/bj2730057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha AK, Xu XJ, Lawson E, et al. Downregulation of AMPK accompanies leucine- and glucose-induced increases in protein synthesis and IR in rat skeletal muscle. Diabetes. 2010;59(10):2426–2434. doi: 10.2337/db09-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chotechuang N, Azzout-Marniche D, Bos C, et al. mTOR, AMPK, and GCN2 coordinate the adaptation of hepatic energy metabolic pathways in response to protein intake in the rat. Am J Physiol Endocrinol Metab. 2009;297(6):E1313–E1323. doi: 10.1152/ajpendo.91000.2008. [DOI] [PubMed] [Google Scholar]
- 26.Yoshizawa F. New therapeutic strategy for amino acid medicine: notable functions of branched chain amino acids as biological regulators. J Pharmacol Sci. 2012;118(2):149–155. doi: 10.1254/jphs.11r05fm. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Zhou M, Sun H, Wang Y. Branched-chain amino acid metabolism in heart disease: an epiphenomenon or a real culprit? Cardiovasc Res. 2011;90(2):220–223. doi: 10.1093/cvr/cvr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah SH, Crosslin DR, Haynes CS, et al. Branched-chain amino acid levels are associated with improvement in IR with weight loss. Diabetologia. 2012;55(2):321–330. doi: 10.1007/s00125-011-2356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.