Abstract
Expansion of the genetic code with nonstandard amino acids (nsAAs) has enabled biosynthesis of proteins with diverse new chemistries. However, this technology has been largely restricted to proteins containing a single or few nsAA instances. Here we describe an in vivo evolution approach in a genomically recoded Escherichia coli strain for the selection of orthogonal translation systems capable of multi-site nsAA incorporation. We evolved chromosomal aminoacyl-tRNA synthetases (aaRSs) with up to 25-fold increased protein production for p-acetyl-L-phenylalanine and p-azido-L-phenylalanine (pAzF). We also evolved aaRSs with tunable specificities for 14 nsAAs, including an enzyme that efficiently charges pAzF while excluding 237 other nsAAs. These variants enabled production of elastin-like-polypeptides with 30 nsAA residues at high yields (~50 mg/L) and high accuracy of incorporation (>95%). This approach to aaRS evolution should accelerate and expand our ability to produce functionalized proteins and sequence-defined polymers with diverse chemistries.
Expansion of the genetic code by incorporation of nsAAs into proteins has enabled template-based incorporation of >100 nsAAs containing diverse chemical groups, including post-translational modifications, photocaged amino acids, bioorthogonal reactive groups, and spectroscopic labels1–5. For example, site-specific incorporation of nsAAs at a single position has been used to engineer protein-drug conjugates6, cross-linking proteins7, and enzymes with altered or improved function8,9. Incorporation of nsAAs at multiple sites could further extend the function and properties of proteins and biomaterials by allowing synthesis of polypeptide polymers with programmable combinations of natural and nonstandard amino acids. However, multi-site nsAA incorporation has so far been limited by inefficiencies associated with the translation machinery and the cellular hosts in which the recombinant proteins are produced10.
There are two common strategies for expressing recombinant proteins containing nsAAs. The first substitutes a close synthetic analog for a natural amino acid in an auxotrophic strain3. This approach has been used to tag, identify, and study newly synthesized proteomes in a variety of cell types11,12, to produce nsAA-containing biopolymers with improved stability13 and with conductive chemical groups14, and to facilitate characterization of structural proteins15. However, the nsAA must be a close analog of the natural amino acid it replaces16, and the eliminated amino acid is excluded in the recombinant protein3 and in the entire proteome, causing growth defects that can reduce protein yields. Alternatively, nsAAs can be incorporated by codon reassignment or frameshift codons using orthogonal translation systems consisting of an aaRS capable of charging only a cognate tRNA that is not aminoacylated by endogenous aaRSs2,4. Typically, a UAG stop codon is assigned to the nsAA, and the orthogonal tRNA anticodon is mutated to CUA. Pairs of aaRSs and tRNAs from organisms such as Methanocaldococcus jannaschii and Methanosarcina species have been used to incorporate a wide variety of nsAAs4,17 into proteins in bacterial hosts4,18,19, but at only one or a few instances within a polypeptide chain5,10.
The first challenge for multi-site nsAA incorporation using codon reassignment is to overcome competition between the orthogonal nsAA-tRNACUA and essential translation machinery (e.g., release factor 1, RF1) for the UAG codon, which reduces full-length protein production and limits the number of nsAAs that can be incorporated into a single protein20–23. To address this, we recently recoded all instances of the UAG codon to the synonymous UAA codon in E. coli21,24. This genomically recoded organism permitted the deletion of RF1 and, therefore, elimination of translational termination at UAG codons. In this organism, UAG was changed from a stop to a sense codon, provided the appropriate translation machinery was present21,24. Nevertheless, a second challenge to multi-site nsAA incorporation by codon reassignment is that the evolved aaRSs show ~100- to 1,000-fold reduced activity5,19 compared with native enzymes, resulting in inefficient nsAA acylation19,25,26, low levels of nsAA-tRNA and low protein yields21,27,28, particularly with multi-site nsAA incorporation20. We hypothesize that current approaches rely on multi-copy plasmids for aaRS and tRNA overexpression to overcome enzyme inefficiency, which masks differences between modestly and highly active aaRSs capable of multi-site nsAA incorporation.
In this study, we describe an in vivo protein evolution approach to isolate more efficient aaRS variants for multi-site incorporation of diverse nsAAs. Specifically, we used multiplex automated genome engineering (MAGE)29,30 to generate libraries of chromosomally integrated aaRSs in a genomically recoded organism containing both positive- and negative-selection markers. Using this approach, we demonstrate the ability to isolate aaRS variants with increased efficiency and tunable nsAA specificities for a variety of nsAAs. We tested the selected variants on elastin-like polypeptide (ELP) fusion proteins that contain up to 30 UAG codons. ELPs are a family of unstructured protein-polymers composed of a VPGXG repeat, where X, the guest residue position, is permissive for any amino acid except proline31 and is therefore also permissive to nsAAs. We demonstrate incorporation of 30 nsAAs per protein with high yields (~50 mg/L) and >95% fidelity of nsAA incorporation at each UAG codon.
RESULTS
Genome-wide recoding improves multi-site nsAA incorporation
We first characterized the ability of a known orthogonal translation system32 to incorporate 3–30 nsAAs per protein in the genomically recoded organism. We previously demonstrated reduced natural suppression and elimination of protein truncation in this strain (at three UAGs)21. In this study, we constructed three fluorescent protein standards (Fig. 1a): a superfolder GFP33 containing three UAG codons at positions 39, 151 and 182 (GFP(3UAG)), and two ELP-GFP fusion proteins where the ELP contains 10 (ELP(10UAG)-GFP) or 30 (ELP(30UAG)-GFP) UAG codons at the guest residue positions. ELPs were fused to the N terminus of superfolder GFP, and control (wild-type, WT) proteins with tyrosine codons substituted for UAGs were similarly constructed (Supplementary Notes 1 and 2).
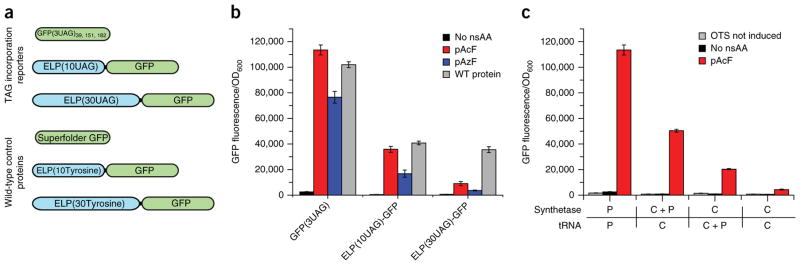
Figure 1.
Evaluation of multi-site nsAA incorporation and expression profiles on the activity of M. jannaschii derived pAcF orthogonal translation system (OTS). (a) Schematic illustration of reporter proteins for incorporation of 3, 10 and 30 nsAAs and equivalent control wild-type (WT) protein. (b) Incorporation of 3, 10 and 30 nsAAs (pAcF or pAzF) in a single protein by the plasmid-based M. jannaschii–derived pAcF OTS in the genomically recoded organism. (c) Production of superfolder GFP containing three UAG sites (GFP(3UAG)) by pAcFRS and tRNACUA expressed by plasmid or chromosomal integration. GFP(3UAG) is drastically reduced as a result of reduction in OTS transcript copy number, and partially rescued by supplementation of plasmids bearing either aaRS or tRNACUA. n = 3, error bars; mean ± s.d.
The genomically recoded organism21 was co-transformed with the reporter gene and an orthogonal translation system plasmid32 containing an aaRS:tRNA pair previously engineered for incorporation of p-acetyl-3-phenylalanine (pAcF), that is also able to incorporate p-azido-3-phenylalanine17 (pAzF). All fusion proteins resulted in quantifiable signals. GFP fluorescence assays indicated that multi-site pAcF incorporation in the recoded strain produced 110%, 87% and 25% of pAcF containing GFP(3UAG), ELP(10UAG)-GFP and ELP(30UAG)-GFP fluorescence, respectively, and 75%, 32% and 6% of pAzF containing GFP(3UAG), ELP(10UAG)-GFP and ELP(30UAG)-GFP compared to WT proteins (Fig. 1b). Similarly, the yield of purified ELP(30UAG)-GFP containing pAcF expressed in small batch cultures was 18% and 8% compared to expression of WT protein in the genomically recoded organism or parent (nonrecoded) strain, respectively (Table 1). The yield of pAzF containing ELP(30UAG)-GFP in the genomically recoded organism was too low to allow for purification. Although the genomically recoded organism improved yields of proteins containing multiple nsAAs, we hypothesized that yield would be enhanced by further evolution of the orthogonal translation systems.
Table 1.
Yield of purified ELP(30UAG)-GFP expressed in GRO by various OTSs in the presence of nsAAs
| OTS | Yield (mg/L) | nsAA | Strain |
|---|---|---|---|
| pAcFRSa | 3.04 ± 1.4 | pAcF | Non-recoded E. coli |
| WT-TyrRSb | 38.7 ± 4.3 | pAcF | Non-recoded E. coli |
| pAcFRS | 10.5 ± 5.5 | pAcF | GRO |
| pAcFRS | N.D.c | pAzF | GRO |
| pAzFRS | N.D.c | pAzF | GRO |
| pAcFRS.1.t1 | 52.6 ± 6.3d | pAcF | GRO |
| pAzFRS.2.t1 | 39.05 ± 3.4d | pAcF | GRO |
| pAzFRS.2.t1 | 41.9 ± 6 | pAzF | GRO |
| pAcFRS.2.t1 | 64.5 ± 9.7 | BuY | GRO |
| pAcFRS.2.t1 | 53 ± 5.4 | 4CF3F | GRO |
| pAzFRS.2.t1 | 48.2 ± 11.2 | 4ClF | GRO |
| WT-TyrRSb | 67.7 ± 6.2 | No nsAA | GRO |
| WT-TyrRSb | 58.7 ± 5 | pAcF | GRO |
| WT-TyrRSb | 61.4 ± 10.1 | pAzF | GRO |
Expression too low to allow sufficient purification for A280 measurement, therefore protein quantity was estimated based on fluorescence of semi-purified protein extracts.
WT ELP(30Tyrosine)-GFP proteins contain no UAGs and were expressed using E. coli native translation machinery. However, expression of ELP(30Tyrosine)-GFP was measured in the presence of pAcF or pAzF to assess for potential toxic effects of these nsAAs on protein expression.
N.D.: expression too low to allow purification of reporter protein by inverse transition cycling. Data are reported as mean ± s.d. calculated from purification of n = 3 independently grown and purified cultures.
P < 0.05 compared with pAcFRS progenitor with pAcF.
OTS, orthogonal translation system; GRO, genomically recoded organism.
Chromosomal integration highlights aaRS enzyme inefficiency
We constructed a genomically recoded organism strain containing a chromosomally integrated orthogonal translation system to enable MAGE-based evolution of the aaRS and to characterize its performance in this context. A DNA cassette based on the pAcF orthogonal translation system plasmid32 consisting of an inducible M. jannaschii-based pAcF aaRS (pAcFRS), a constitutive tRNACUA and a tolC selection marker (Supplementary Fig. 1)) was assembled and integrated in a known intergenic region (Supplementary Note 1) in the genomically recoded organism using λ Red recombination34. Subsequently, UAG codons were inserted by MAGE in four permissive sites in the tolC cassette, to enable negative selection (Supplementary Note 1).
We then characterized the effect of varying aaRS (i.e., pAcFRS) and tRNACUA concentration on GFP(3UAG) production in the genomically recoded organism. The reduction in copy number caused by genomic integration of the orthogonal translation system resulted in a ~20-fold decrease in the yield of GFP(3UAG) in the RF1-deficient genomically recoded organism, highlighting the impaired efficiency of this orthogonal translation system (Fig. 1c). Individually increasing either pAcFRS or tRNACUA concentration by supplementation with plasmids resulted in partial restoration of GFP(3UAG) expression (Fig. 1c), suggesting impaired binding of pAcFRS to pAcF and to its cognate tRNACUA, likely because the M. jannaschii TyrRS (MjTyrRS) natively recognizes the GUA anticodon32. These results suggest that elevated levels of pAcFRS and tRNACUA expression compensate for their reduced enzymatic activity.
Evolution of orthogonal translation systems in vivo
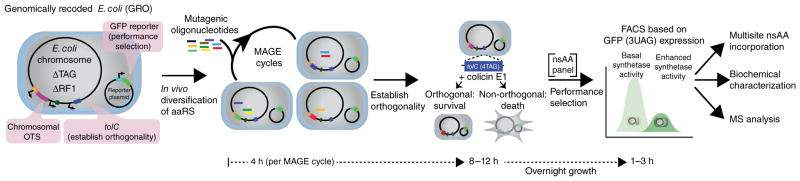
In prior work, we used MAGE to generate a genomic library of ribosome binding site sequences in which genetic diversity can be increased simply by increasing the number of MAGE cycles29. Here we sought to evolve protein function with a chromosomal orthogonal translation system in a genomically recoded organism by successive rounds of diversification with MAGE and negative and positive selection bypassing conventional intermediate plasmid extraction and transformation steps (Fig. 2). To generate an aaRS library, we designed a pool of synthetic single-stranded DNA (ssDNA) oligonucleotides to mutagenize the selected amino acid targets, and used several rounds of MAGE to create a diverse library. The resulting cell population was then subjected to a tolC-based negative-selection step35 wherein mutated aaRS variants capable of mischarging tRNACUA with natural amino acids permitted read-through of a tolC construct containing four UAG sites, rendering the organism sensitive to colicin E1 (Supplementary Fig. 2). Thus, the negative-selection marker is dormant unless colicin E1 is present, eliminating the need to replace or modify the cellular host for positive or negative selection. The remaining orthogonal library was subsequently screened for improved GFP(3UAG) production in the presence of either pAcF or pAzF. aaRS variants with improved performance were isolated by two rounds of fluorescence-activated cell sorting (FACS). Finally, biochemical and proteomic analyses were performed and the resulting isolated variants were evaluated for their ability to produce proteins containing up to 30 instances of pAcF or pAzF, as well as 236 other nsAAs (Supplementary Note 3). This workflow was designed for streamlined selection from diversified populations or further diversification of selected mutants to improve or tune the properties (e.g., activity, specificity) of selected aaRSs for a variety of nsAAs (Fig. 2).
Figure 2.
Evolution of chromosomally integrated aaRS variants. The genomically recoded organism (GRO) is engineered to contain a single chromosomal copy of the aaRS for diversification using MAGE, a negative-selection marker for removal of nonorthogonal translation systems (OTS) (capable of incorporation of natural amino acids), and a GFP marker for fluorescence-based identification and isolation of improved variants. Site-directed mutagenesis of chromosomally integrated translation components by MAGE generates a highly diversified population, which is subsequently subjected to tolC- and colicin E1–mediated negative selection in the absence of nsAAs. UAG suppression in GFP(3UAG) enables FACS of orthogonal aaRS libraries in the presence of the desired nsAA to identify improved variants. The selected aaRS variants are evaluated for multi-site nsAA incorporation, in vitro activity and protein purity.
Evolution of chromosomally integrated aaRSs variants
We used a reported crystal structure for the MjTyrRS36 to inform the diversification of 12 residues in the amino acid binding pocket surrounding the variable side chain of the nsAA (compared with typically six or fewer residues18,37,38, with few exceptions targeting nine residues39), and five residues at the aaRS-tRNACUA anticodon recognition interface (Fig. 3a). Synthetic degenerate ssDNA oligonucleotides were designed to randomize the residues in the nsAA binding pocket and aaRS-tRNACUA binding interface separately (Supplementary Table 1) to distinguish between improved nsAA binding and tRNACUA recognition.
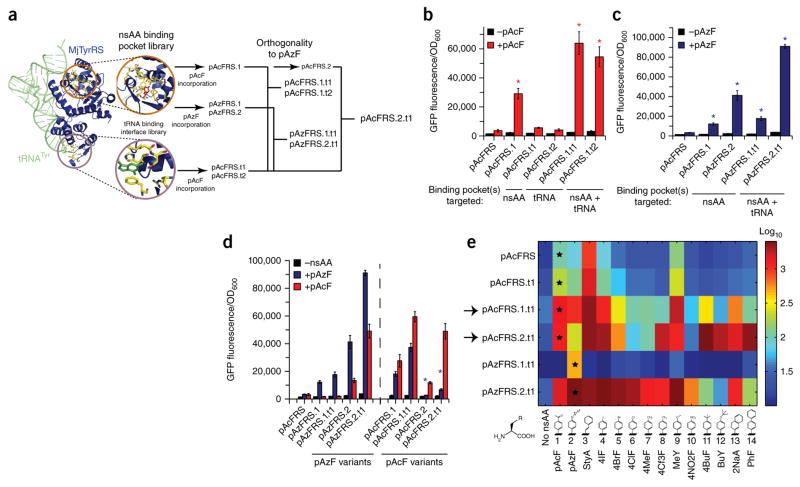
Figure 3.
Evolution and characterization of chromosomally integrated pAcFRS and pAzFRS variants with improved efficiencies. (a) Crystal structure of MjTyrRS in complex with tRNACUA and tyrosine. Insets highlight the amino acid binding pocket and the tRNACUA anticodon binding interface with a schematic representation of the libraries generated from pAcFRS (a mutant of the MjTyrRS) and aaRS variants isolated following each library diversification and selection steps. (b,c) pAcFRS (b) and pAzFRS (c) variants assayed by GFP(3UAG) fluorescence. n = 3; error bars, mean ± s.d. *P < 0.05 indicates comparison of each evolved aaRS with pAcFRS in the +pAcF or +pAzF condition. (d,e) Specificities of pAcFRS and pAzFRS variants for pAcF, pAzF (d), and 14 phenylalanine derivatives (e), as assayed by GFP(3UAG) fluorescence. Variant pAcFRS.2.t1 was evolved from variant pAcFRS.1.t1 for reduced pAzF incorporation (marked by black arrows). n = 3; error bars, mean ± s.d. *P < 0.05 indicates comparison of pAcFRS.2 with the progenitor pAcFRS.1.
We screened the diversified populations of variants after five and nine MAGE cycles (Supplementary Fig. 3 and Supplementary Note 4) by induction of GFP(3UAG) in the presence of pAcF or pAzF and performed two rounds of FACS to isolate cells with improved aaRS activity (Supplementary Fig. 4). The nsAA binding library produced variants with improved pAcFRS and pAzFRS incorporation efficiency (Fig. 3a and Supplementary Table 2). Individual colonies selected after FACS were sequenced revealed an aaRS variant for improved pAcF incorporation (pAcFRS.1: A167D mutation) capable of about an eightfold higher GFP(3UAG) production compared with the progenitor enzyme, pAcFRS32 (Fig. 3b). In addition, individual colony analysis of sorted populations revealed two top variants for pAzF incorporation (pAzFRS.1: (G158V, C159M, R162D, A167Y) and pAzFRS.2: (E107T, F108Y, Q109M) mutations) capable of producing ~3.5- and ~12-fold more GFP(3UAG) than the progenitor enzyme (Fig. 3c).
Similarly, screening of the library for aaRS-tRNACUA binding optimization (screened for enhanced GFP(3UAG) production with pAcF) revealed two mutants, pAcFRS.t1 and pAcFRS.t2 ((R257G) or (R257C, F261E) mutations, respectively), both exhibiting ~1.5 fold higher GFP(3UAG) production compared to the progenitor enzyme (Fig. 3b). We then combined mutations isolated for nsAA binding and tRNA binding via MAGE, which produced four variants. These chromosomally integrated variants harboring mutations for improved pAcF or pAzF and tRNACUA binding resulted in a synergistic ~17-fold (pAcFRS.1.t1), ~15-fold (pAcFRS.1.t2), ~5.5-fold (pAzFRS.1.t1) and ~25-fold (pAzFRS.2.t1) increase in GFP(3UAG) production compared with the progenitor enzyme (Fig. 3b,c). Correct incorporation of pAcF or pAzF into all three sites in GFP(3UAG) was confirmed by mass spectrometry (MS; Supplementary Table 3).
Evolution of aaRS variants with tunable nsAA specificities
As several previously described aaRS variants incorporate numerous nsAAs17,40 (termed polyspecificity17), we first determined the polyspecificity of each of our chromosomally integrated aaRS variants by assaying GFP(3UAG) production in the presence of pAcF or pAzF (Fig. 3d) as well as in the presence of 236 other nsAAs41. These assays revealed polyspecificity in each of our variants with the exception of pAzFRS.1 and pAzFRS.1.t1, which were exceptionally specific to pAzF (Fig. 3e and Supplementary Fig. 5b–m).
Based on previous studies of polyspecificity17,40, we hypothesized that a customized diversification-selection experiment designed to alter the nsAA binding pocket to reject a specific nsAA would create a pocket capable of accepting new, previously excluded, nsAAs. Therefore, an additional round of evolution was performed to increase the specificity of pAcFRS.1 toward pAcF while excluding pAzF. pAc-FRS.1 was subjected to five additional MAGE cycles with an oligonucleotide pool designed to preserve the (A167D) mutation, responsible for improved activity of pAcFRS.1, and randomize the remaining 11 sites in the nsAA binding pocket. This library was subjected to tolC negative selection in the presence of pAzF, and the remaining orthogonal library was screened for improved GFP(3UAG) in the presence of pAcF by FACS. Individual colony sequencing revealed that the sorted population was enriched in an aaRS mutant (pAcFRS.2 (L65V, A167D) mutations). Comparison of GFP(3UAG) expression in the presence of pAcF or pAzF confirmed an increase in selectivity for pAcFRS.2 and pAcFRS.2.t1 toward pAcF over pAzF (Fig. 3d,e marked by black arrow). Upon polyspecificity analysis of chromosomally integrated progenitor (pAcFRS), first-generation (pAcFRS.1.t1) and second-generation (pAcFRS.2.t1) aaRSs, we found that altering the binding pocket to exclude pAzF resulted in the selection of a variant that efficiently incorporates nsAAs (i.e., compounds 11, 12 and 14; Fig. 3e) not incorporated by other variants. Furthermore, each of the 14 different nsAAs could be incorporated at high efficiency by selecting the appropriate aaRS variant, informed by the aaRS-nsAA specificity heat map (Fig. 3e).
Efficient multi-site nsAA incorporation by evolved aaRSs
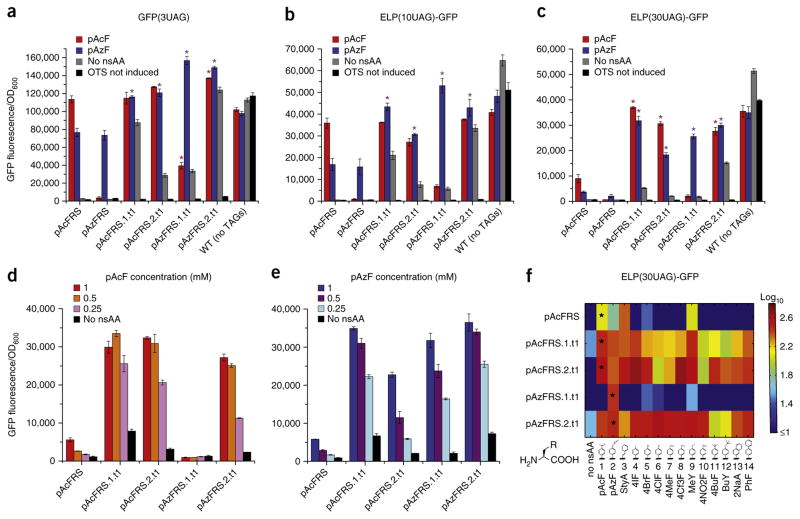
To evaluate whether the evolved aaRSs can improve multi-site nsAA incorporation, we co-transformed the genomically recoded organism with a plasmid carrying the reporter protein with three, ten or 30 UAGs, or WT equivalents and episomal versions of each orthogonal translation system variant (Supplementary Fig. 6). Plate-based fluorescence analysis indicated increased incorporation of either pAcF or pAzF by our evolved orthogonal translation systems for expression of GFP(3UAG) up to 1.1-fold (pAcF) or twofold (pAzF), ELP(10UAG)-GFP up to 1.1-fold (pAcF) or 3.2-fold (pAzF), and ELP(30UAG)-GFP up to 4-fold (pAcF) or 7-fold (pAzF) (Fig. 4a–c and Supplementary Fig. 7). Purification of ELP(30UAG)-GFP containing pAcF expressed by the genomically recoded organism in small-batch cultures revealed a fivefold increase in protein production of up to 54 mg/L (i.e., >90% of WT-protein expression under similar conditions) and high-yield expression of ELP(30UAG)-GFP containing pAzF (~35 mg/L, compared with very low yields generated by progenitor pAcFRS or pAzFRS, that could not be purified) (Table 1 and Supplementary Fig. 8). In addition, we evaluated the production of ELP(30UAG)-GFP in the presence of up to fourfold reduced concentrations of pAcF or pAzF. This analysis revealed that several of our enzyme variants are capable of efficient production of ELP(30UAG)-GFP with 0% or <20% loss in protein yield with twofold or fourfold reduced pAcF concentration (Fig. 4d), respectively, or with <5% or <30% loss in protein yield with twofold or fourfold reduced pAzF concentration (Fig. 4e), respectively. Notably, our evolved aaRSs outperformed the progenitor synthetase at all nsAA concentrations.
Figure 4.
Evaluation of multi-site nsAA incorporation by evolved aaRS variants expressed on multi-copy plasmids. Production of GFP(3UAG) (a), ELP(10UAG)-GFP (b) and ELP(30UAG)-GFP (c) by progenitor and evolved orthogonal translation systems expressed on multi-copy plasmids in the genomically recoded organism compared with WT (no UAG) proteins. Production of ELP(30UAG)-GFP by progenitor and evolved orthogonal translation systems expressed on multi-copy plasmids in the genomically recoded organism in the presence of variable pAcF (d) or pAzF (e) concentrations (n = 3, error bars, s.d. *P < 0.05 indicates comparison of evolved aaRS with pAcFRS in the +pAcF or +pAzF condition). (f) Efficiency and specificity of progenitor and evolved pAcFRS and pAzFRS variants for 14 phenylalanine derivatives, as assayed by ELP(30UAG)-GFP fluorescence. Data shown are the average of two independent experiments each with n = 3.
Biochemical in vitro characterization of the aaRS variants was carried out by ATP-PPi exchange and tRNACUA aminoacylation and confirmed increased activity and respective specificities of our evolved aaRS variants. Although improvement to the kcat/Km of the progenitor enzyme was modest in absolute terms (i.e., ~9.5-fold for variants pAcFRS.1.t1 and 8.7-fold for pAzFRS.2.t1 for tRNA aminoacylation and ~5.4-fold for variant pAcFRS.1.t1 and 3.3-fold for pAzFRS.1.t1 for ATP-ppi exchange, Supplementary Tables 4 and 5), the in vivo ELP-GFP suppression efficiency relative to the nsAA supply (Fig. 4d,e) suggested that the nsAA concentration inside the cell was close to the Km. Taken together, these data show that the evolved enzymes with increased pAcF and pAzF incorporation are robust enough to provide sufficient nsAA-tRNA for complete nsAA incorporation. This is likely because the pAcFRS and pAzFRS variants have to supply aa-tRNA to only ~1,500 UAG codons (30 UAG codons on ~50 plasmid copies), whereas TyrRS serves ~40,000 codons in E. coli42.
We then evaluated the ability of our plasmid-based orthogonal translation systems to produce ELP(30UAG)-GFP with a panel of 14 nsAAs. A fluorescence assay indicated increased production of ELP(30UAG)-GFP by select aaRS variants for every nsAA in this panel compared with the progenitor pAcFRS (Fig. 4f), and purification of ELP(30UAG)-GFP containing select nsAAs confirmed high yield (48–65 mg/L, compared with ~65 mg/L for WT protein, Table 1) expression. This analysis also revealed that whereas the pAzFRS.1.t1 maintains stringent specificity for pAzF, the specificity of variant pAcFRS.2.t1 for pAcF over pAzF decreases when expressed on a multicopy plasmid. These results demonstrate that the ELP-GFP fusion protein resolves previously encountered issues of misfolding and aggregation caused by multi-site nsAA incorporation in the GFP open reading frame20, while retaining the ability to assay chemically diverse nsAA incorporation by GFP fluorescence.
When expressed on multicopy plasmids in the absence of nsAAs, all of our evolved variants showed increased protein production compared with the progenitor enzyme (Fig. 4a–c, gray bars), which may suggest incorporation of natural amino acids. Accordingly, our plasmid-based, but not the chromosomal-based, variants failed the negative-selection step (Supplementary Fig. 9). However, time-course analysis of GFP(3UAG) or ELP(30UAG)-GFP expression revealed a reduced rate of protein production in the absence of the nsAA (Supplementary Figs. 10 and 11), and background GFP production was reduced with increasing numbers of UAGs (Fig. 4a–c, gray bars), indicating that incorporation of the nsAA is favored over natural amino acids.
Proteomics analyses confirms accurate nsAA incorporation
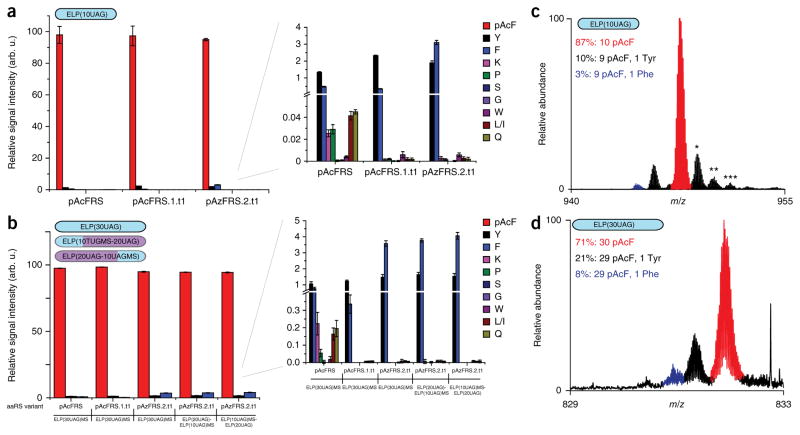
We pursued two complementary MS approaches to carefully examine and assay the fidelity of multi-site nsAA incorporation. To quantify the identities of the amino acid at the UAG codons, we used a multiple reaction monitoring (MRM)-based-MS assay21,43. We chose to characterize the most efficient variants (pAcFRS.1.t1 and pAzFRS.2.t1) for production of specially designed reporters, ELP(10UAG)-GFPMS and ELP(30UAG)-GFPMS, and to examine the effect of TAG codon position (i.e., at the N or C terminus) on nsAA incorporation accuracy. Shotgun liquid chromatography (LC)-tandem MS (MS/MS) analysis identified the most prevalent misincorporated amino acids at UAG codons (Supplementary Table 6), and MRM revealed that the pAc-FRS, pAcFRS.1.t1 and pAzFRS.2.t1 orthogonal translation systems incorporate pAcF at >95% of the peptides for all constructs examined (Fig. 5a,b and Supplementary Note 5).
Figure 5.
Quantitative MS evaluation of the purity of multi-site nsAA incorporation by evolved aaRS variants expressed on multi-copy plasmids. (a,b) Relative intensities of reporter peptides originating from ELP(10UAG) (a) and ELP(30UAG) (b) reporters containing pAcF, produced by progenitor and evolved orthogonal translation systems expressed on multi-copy plasmids in the genomically recoded organism. n = 4; error bars represent confidence interval calculated at the 95% confidence level. (c) Partial top-down mass spectrum of recombinant ELP(10UAG), after removal of the GFP UAG by trypsin digestion; the isotopically resolved 14+ charge state [M+14H]14+ is shown. Mass values are for the most abundant species, incorporating 10 pAcF residues (isotopic distribution in red: theoretical: 13,245.68 Da, experimental: 13,245.62 Da ± 4.5 p.p.m.). To the left of the main peak, species incorporating 9 pAcF and 1 Tyr residue (theoretical: 13,219.66 Da, experimental: 13,219.57 Da ± 6.8 p.p.m.) or 9 pAcF and 1 Phe residues (theoretical: 13,203.67 Da, experimental: 13,203.67 Da ± 7.6 p.p.m.) are colored black and blue, respectively. Species marked with *, ** and *** are M+O (+16 Da), M+2O (+32 Da) and M+3O (+48 Da), respectively, typical artifacts of analysis in electrospray MS. (d) Partial top-down mass spectrum of recombinant ELP(30UAG), after removal of the GFP UAG by trypsin digestion; the isotopically resolved 46+ charge state [M+46H]46+ is shown. Mass experimental vs. mass theoretical (1.8 p.p.m. error) values are for the most abundant species, incorporating 30 pAcF residues (isotopic distribution in red: theoretical: 38,198.30 Da, experimental: 38,198.37 Da ± 1.8 p.p.m.), and species incorporating 29 pAcF and 1 Tyr residues (theoretical: 38,172.28 Da, experimental: 38,172.35 Da ± 1.8 p.p.m.) or 29 pAcF and 1 Phe (theoretical: 38,156.29 Da, experimental: 38,156.30 Da ± 0.3 p.p.m.) residues are colored black and blue, respectively.
To examine the effect of UAG codon position with respect to the N terminus, we constructed reporters ELP(10UAG)MS-ELP(20UAG) and ELP(20UAG)-ELP(10UAG)MS (Fig. 5b and Supplementary Note 2). We found that N-terminal nsAA incorporation accuracy is independent of UAG codon position and of any local mRNA differences. Analysis of pAzF incorporation accuracy by pAzFRS.1.t1 was also performed with ELP(10UAG)-GFPMS and results were similar to that of pAcF (Supplementary Table 7 and Supplementary Figs. 12 and 13). We also observed low levels of K, P, S, G, W, L/I and Q with a striking decrease in misincorporation of K, P, L/I and Q content in proteins produced by pAcFRS.1.t1 and pAzFRS.2.t1, but not by pAzFRS.1.t1 (Fig. 5a,b and Supplementary Fig. 13).
To complement the nsAA quantitation at single sites from MRM, we used LC-MS of intact proteins to obtain quantitative analyses of the ELP(10UAG) and ELP(30UAG) polymers. Using pAcFRS.1.t1 aaRS, we observed that 87% of the ELP(10UAG) proteins had ten pAcF residues. Analysis of the ELP(30UAG) protein showed that 71% had 30 pAcF residues (Fig. 5c,d and Supplementary Fig. 14a,b). These values were used to calculate the single-site accuracy of pAcF incorporation (pure full-length product = (single-site accuracy)n, n equals #UAG codons) and showed 98.6% and 98.9% accuracy for ELP(10UAG) and ELP(30UAG), respectively, which is in quantitative agreement with MRM (97.3% and 98.2%, respectively). Similarly, pAzFRS.2.t1 produced full-length ELP(10UAG) and ELP(30UAG) pure proteins at ratios consistent with MRM analysis (Supplementary Fig. 14c,d).
We then assessed whether our approach affects the robustness of the host organism or host proteins. The fitness of the genomically recoded organism expressing the progenitor and evolved orthogonal translation systems was not impaired due to misincorporation of pAcF or pAzF by native synthetases at non-UAG codons (Supplementary Fig. 15), consistent with our prior findings21. Next, we conducted analysis of ELP(10UAG)-GFPMS expressed in the presence of the evolved pAcFRS and pAcF and examined the highly expressed GFP-derived peptides FEGDTLVNR and SAMPEGYVQER for pAcF incorporation at F and Y, as they are most structurally similar to pAcF and more likely to mischarge. The results of this analysis revealed only F and Y incorporation, suggesting that pAcF misincorporation is below the level of detection (<0.01%) if it exists at all. We therefore conclude that our system is orthogonal as designed and misincorporation is unlikely to occur at native proteins, which express at levels lower than ELP-GFP. Taken together, these results show that our selected aaRS variants are capable of orthogonal, efficient, multi-site nsAA incorporation in the genomically recoded organism strain along with high protein yields, purity and robust cell fitness.
DISCUSSION
We present an approach for the evolution of aaRS variants capable of multi-site nsAA incorporation in recoded organisms. MAGE has been used to generate combinatorial genomic libraries by targeting multiple and distal genetic loci29,30. Although libraries of chromosomally integrated genes can be generated with other approaches (e.g., recombineering), the low rate of recombination achieved with these methods and limited ability for multi-site combinatorial mutagenesis in vivo result in reduced library complexity29. Here, mutagenesis by MAGE enabled simultaneous targeting of an expanded number of residues in the nsAA binding pocket. These libraries facilitated selection of improved aaRSs for a variety of nsAAs—a direct result of the increased number of targeted residues as several of the mutated residues in these variants were not included in previous screens37,44, including a pAzFRS variant (pAzFRS.1) that is both more efficient and more specific for pAzF than previously reported pAzFRSs. Although we do not explore the full diversity of our library in a single evolution (i.e., diversification and selection) experiment (~1015 for 12 amino acid targets), we were able to perform successive rounds of diversification and selection, without plasmid extraction or transformation steps, by increasing the number of MAGE cycles, changing the mutagenic ssDNA pool, targeting different areas in the aaRS (i.e., the nsAA or tRNA binding site, which can be targeted consecutively or concurrently) or changing the negative-selection conditions (e.g., including pAzF). We expect that the modular nature of our approach will facilitate multiplexed, automated diversification of many aaRS-nsAA pairs, with broad application to other proteins and pathways.
The evolution of a chromosomally integrated protein is an additional advantage of our workflow. Consistent with our original hypothesis, expression of aaRS variants from multi-copy plasmids masked the differences between aaRSs of low, modest and high activity for a protein with only three nsAAs; these differences became evident for a protein with 30 nsAAs (Fig. 4c) or when the aaRS variants were expressed from the chromosome (Fig. 3b–e). Several of our selected aaRS variants are unique in that they support high levels of protein expression in the absence of an nsAA and death in the presence of the negative-selection marker when expressed from multi-copy plasmids. These observations indicate that our evolved variants would not survive the negative selections conventionally used with plasmid-based orthogonal translation system libraries and therefore could not have been isolated by such approaches. Despite this property, we show high fidelity of nsAA incorporation, which suggests that enhancement of aaRS efficiency may be achieved if aaRS levels are lowered during negative selection such as by reduction of arabinose concentration, reduced strength promoters, or low copy–number plasmids, or if selection stringencies are lowered as previously suggested39.
Using the genomically recoded organism and newly evolved aaRS variants, we achieved site-specific multi-site nsAA incorporation with high yields and high purity. Because incorporation accuracy was not biased with respect to position, it should not affect bulk polymer properties. Previous attempts to incorporate more than one instance of an nsAA per protein in strains with no or attenuated RF1 activity showed at best a 33% yield of WT protein when incorporating three instances of an nsAA into superfolder GFP (<20.5 mg/L)45 and 3% yield of WT when incorporating ten instances of an nsAA into GFP (0.4 mg/L)20. Although ELPs are a well-expressed family of proteins46, we expect that our approach will improve multi-site nsAA incorporation in a diverse set of natural and recombinant proteins and protein polymers.
In future work, similar orthogonal translation system libraries can be constructed for the Methanosarcina mazei PylRS28 and the O-phosphoseryl-tRNA18 synthetase or for co-evolution of multiple orthogonal translation system components (e.g., aaRS, tRNA, EF-Tu) to enable incorporation of chemically diverse, bulky, and highly charged amino acids5,18. In addition, increasing the targeted residue pool (>12 sites), integrated with computational protein design47 or selection strategies that link cell viability to the incorporation of an nsAAs into essential proteins48,49, will enable strategic targeting and partial randomization of specified residues to increase library coverage.
Multi-site nsAA incorporation will allow control over the chemical and physical properties of protein-based biomaterials. Indeed, limited to only one or a few instances of site-specific nsAA incorporation, most previous work has centered on tag-and-modify approaches or simple protein decorations. Our approach enables site-specific nsAA incorporation where multiple identical nsAAs provide the dominant physical and biophysical properties to biopolymers. As ELPs undergo a sharp soluble-to-insoluble phase transition at their transition temperature, which depends on the ELP composition46, ELPs similar to those in this study could be used as a scaffold for smart biomaterials responsive to light, electro-magnetic field and various analytes. Multi-site nsAA incorporation will also allow the design and production of post-translationally modified proteins (e.g., kinases18) for the study and treatment of disease or of new biologics (e.g., antibodies10) with multiple instances of novel chemical functionalities. As genomically recoded organisms with more free codon channels are constructed21,50, the selection of aaRS variants with tunable or exclusive nsAA specificities enabled by our evolution approach could provide orthogonal coding channels for incorporation of two or more nsAAs within a single protein or sequence-defined polymers.
ONLINE METHODS
See Supplementary Notes 1 and 2 for DNA or protein sequences and Supplementary Notes 5 and 6 for detailed materials and methods for the MRM based- and intact MS.
Assembly of orthogonal translation system integration cassette
To generate an orthogonal translation system integration cassette, previously published32 p-acetyl-3-phenylalanine aaRS (pAcFRS) gene downstream of the araBAD promoter, a constitutive tRNACUA under the control of the proK promoter and a tolC expression cassette were amplified using primers containing genomic homology regions or terminal sequence overlaps for Gibson Assembly51. The integration cassette was then assembled using Gibson Assembly Master Mix (New England BioLabs) according to the manufacturer’s instructions. The orthogonal translation system integration cassette was then amplified by PCR consisting of 2 μl of Gibson Assembly product, 10 pmol each of sense and antisense DNA primers, 50 μl Hot-Start HiFi Mastermix enzyme (Kapa Biosystems) and water for a final volume of 100 μl. The PCR reaction conditions were 95 °C for 2 min for initial denaturation, followed by 30 cycles at 98 °C for 30 s, 65 °C for 30 s and 72 °C for 5 min. The resulting PCR product was visualized on a 1% agarose gel stained with SYBR Safe DNA stain (Invitrogen) and the correct size band was excised and purified using a gel extraction kit (Qiagen). Genomic integration of orthogonal translation system cassette into the genomically recoded organism (E. coli C321.A, CP006698.1, GI:54981157) was performed with 100 ng of the DNA cassette as previously described52. Colonies were screened for correct integration by colony PCR and verified by Sanger sequencing.
MAGE evolution of orthogonal translation systems
Liquid cell cultures were inoculated from colonies grown at 30 °C to mid-logarithmic growth (optical density at 600 nm of ~0.7) in a shaking incubator. To induce expression of the lambda-red recombination proteins (Exo, Beta and Gam), cell cultures were shifted to 42 °C for 15 min and then immediately chilled on ice. In a 4 °C environment, 1 ml of cells was centrifuged at 16,000g for 30 s. Supernatant medium was removed and cells were resuspended in milli-Q water. The cells were spun down, the supernatant was removed and the cells were washed a second time. After a final 30 s spin, supernatant water was removed and oligos prepared at a concentration of 5–6 μM in DNase-free water were added to the cell pellet. The oligo-cell mixture was transferred to a pre-chilled 1 mm gap electroporation cuvette (Bio-Rad) and electroporated under the following parameters: 1.8 kV, 200 V and 25 mF. Luria-Bertani broth with minimal salts Luria-Bertani broth containing 5 g/L NaCl (LB-min) medium (3 ml) was immediately added to the electroporated cells. The cells were recovered from electroporation and grown at 30 °C for 3–3.5 h. Once cells reached mid-logarithmic growth they were used in additional MAGE cycles, isolated, or assayed for genotype and/or phenotype analysis.
Negative selection
Following the last MAGE cycle, cultures were immediately resuspended in 1ml of LB-min medium containing 0.2% arabinose and colicin E1. After 8 h of incubation at 34 °C, cells were transferred to 3 ml of LB-min medium, grown to an OD600 of 1.0 in a shaking incubator at 250r.p.m., and frozen in glycerol (20% w/w).
Sequencing of aaRS libraries
For the MAGE 5 and 9 libraries, we initially established >270 clonal isolates and performed targeted sequencing of the aaRS region to obtain a snapshot of the genetic complexity (Genewiz, Inc.). To further examine the complexity of our library, we conducted high throughput (HT) sequencing of the target region of the nsAA binding site (498 bp, Supplementary Note 4) in libraries created after five and nine MAGE cycles, used for the pAcF and pAzF evolution experiments. To create libraries for HT sequencing, genomic DNA of each of ~2 × 109 cells of diversified populations was extracted using a Qiagen Genomic DNA purification kit and 30 cycles of PCR were applied for targeted amplification of the 498 bp region of the mutant aaRS gene. Illumina libraries were prepared by the Yale Center for Genome Analysis (YCGA) with each strain given a unique barcode for pooling. Up to two libraries were pooled for sequencing using Illumina’s MiSeq and HiSeq technologies. To capture sequencing information across the entire 498-bp region, 2 × 250 bp paired-end reads were collected for each library. See Supplementary Note 2 for detailed presentation and analysis of sequencing data and estimates of aaRS library complexity.
Plasmid construction
Plasmids bearing GFP-based reporter genes were constructed by insertion of reporter protein genes to a previously described plasmid harboring the gene coding for wild-type GFP, a colE1 origin of replication and a kanamycin resistance marker21. The genes encoding for GFP(3UAG) and superfolder GFP were chemically synthesized (IDT), and inserted in place of the existing wild-type GFP gene using the flanking restriction sites EcoRI and HindIII. The gene encoding for ELP(10UAG) or ELP(10Tyr) flanked by BseRI restriction sites, were chemically synthesized (GeneArt, Life Technologies) and inserted sequentially (up to ELP(30UAG) and ELP(30Tyr)) into the BseRI restriction site located at the N terminus of the GFP gene as previously described53. A DNA cassette encoding for a leader protein sequence (‘MSKGP’) was then inserted at the N terminus of the ELP gene to optimize protein expression.
Plasmids bearing the orthogonal translation system components were constructed by insertion of aaRS genes to a previously described plasmid harboring a p15A origin of replication and a chloramphenicol resistance marker21,32. aaRS genes were PCR-amplified from chromosomal templates and inserted sequentially in place of the progenitor pAcFRS gene using the flanking restriction sites BglII and SalI (for copy 1) and NdeI and PstI (for copy 2).
Analysis of GFP expression by intact cell fluorescence measurements
Liquid cell cultures of strains harboring chromosomally integrated orthogonal translation systems and GFP reporter plasmids were inoculated from frozen stocks and grown to confluence overnight. For plate-based assays, strains harboring orthogonal translation systems and GFP reporter plasmids were inoculated from frozen stocks and grown to confluence overnight. Cultures were then inoculated at 1:20 dilution in LBmin media supplemented with 30 μg/ml kanamycin and allowed to grow at 34 °C to an OD600 of 0.5–0.8 in a shaking plate incubator at 650 r.p.m. (~3 h). Cultures and inducers were added individually to each well. aaRS expression was then induced by the addition of 0.2% arabinose, GFP expression was induced by the addition of 60 ng/μl anhydrotetracycline, and the appropriate nsAA was added at a concentration of 1 mM. Cells were incubated at 34 °C for an additional 16 h. Liquid cell cultures of strains harboring plasmid-based orthogonal translation systems and GFP or ELP-GFP reporter plasmids were inoculated from frozen stocks and grown to confluence overnight. Cultures were then inoculated at 1:20 dilution in 2× YT media supplemented with 30 μg/ml kanamycin and 20 μg/ml chloramphenicol. Cultures were inoculated by addition of 1:20 confluent cell culture (grown overnight) and aaRS expression was simultaneously induced by the addition of 0.2% arabinose, and the appropriate nsAA was added to a concentration of 1 mM. Cultures and inducers were added individually to each well. Cells were allowed to grow at 34 °C to an OD600 of 0.5–0.8 in a shaking plate incubator at 650 r.p.m. (~3 h), reporter protein expression was then induced by the addition of 60 ng/μl anhydrotetracycline, and cells were incubated at 34 °C for an additional 16 h.
For 384-well plate-based assays, fluorescence and OD600 were directly measured following expression. For 96-well plate–based assays, cells were centrifuged at 4,000g for 4 min. Supernatant medium was removed and cells were resuspended in PBS. This process was repeated twice with PBS. GFP fluorescence was measured on a Biotek spectrophotometric plate reader using excitation and emission wavelengths of 395 and 509 nm, respectively). Fluorescence signals were normalized by dividing the fluorescence counts by the OD600 reading. The nsAAs used in this study were purchased from Sigma-Aldrich (St. Louis, MO), ChemImpex (Wood Dale, IL), and Bachem (Torrance, CA). Solutions of nsAAs (50 mM) were made in water or 50 mM NaOH; these stock solutions were diluted 50- or 100-fold (to 1 mM final concentration) into medium used for bacterial growth.
Each isolated aaRS was treated as a separate experiment, and GFP levels were compared with the progenitor from which it was evolved by a one-tailed heteroscedastic t-test. Significance is reported where P was found to be <0.01. To verify aaRS activity, we independently re-introduced (by MAGE) the specific identified mutations into the aaRS in the genomically recoded organism strain and repeated the evaluation of aaRS activity in these strains. Dot-plot graphs of collected data points can be found in Supplementary Note 7.
Flow cytometry analysis and cell sorting
GFP expression was induced as above. Following ~16 h of induction, cells were washed twice in PBS and diluted 1:100 in PBS. Cell fluorescence analysis and sorting was performed using a FACS-Aria flow cytometer (BD-Biosciences) and FACS Diva software. Sorted fractions were recovered for 1 h in 0.5–1 ml LBmin media before small aliquots were plated on LBmin plates supplemented with 30 μg/ml kanamycin for individual colony analysis. The remaining mixed culture was grown to confluence in LBmin media with 30 μg/ml kanamycin and frozen at −80 °C to maintain diversity.
ELP expression and purification
Before batch expression, starter cultures (2 ml) of 2× YT media supplemented with 30 μg/ml kanamycin and 20 μg/ml chloramphenicol were inoculated with transformed cells from a fresh agar plate or from stocks stored at −80 °C, and incubated overnight at 34 °C while shaking at 250 r.p.m. Expression cultures (250 ml flasks containing 50 ml of 2× YT media, antibiotics, 0.2% arabinose and 1 mM of the nsAA) were inoculated with 0.5 ml of the starter culture and incubated at 34 °C for 4 h and then reporter protein expression was induced with 60 μg/ml anhydrotetracycline.
Cells were harvested 24 h after inoculation by centrifugation at 4,000g for 15 min at 4 °C. The cell pellet was resuspended by vortex in ~1.5 ml PBS buffer and stored at −80 °C or immediately purified. For purification, resuspended pellets were lysed by ultrasonic disruption (9 cycles of 10 s sonication separated by 40 s intervals). Poly(ethyleneimine) (0.2 ml of 10% solution) was added to each lysed suspension before centrifugation at 15,000g for 3 min to separate cell debris from the soluble cell lysate.
All ELP constructs were purified by a modified inverse transition cycling (ITC) protocol consisting of multiple “hot” and “cold” spins using sodium citrate to trigger the phase transition. Before purification, the soluble cell lysate was incubated for 1–2 min at ~65 °C to denature native E. coli proteins. For “hot” spins, the ELP phase transition was triggered by adding sodium citrate to the cell lysate or the product of a previous cycle of ITC at a final concentration of ~0.5 M. The solutions were then centrifuged at 14,000g for 3 min and the pellets were resuspended in PBS, followed by a 3–5 min “cold” spin performed without addition of sodium citrate to remove denatured contaminant. Additional rounds of ITC were carried out as needed until sufficient purification was achieved.
Protein concentration was calculated by measuring the OD280 of purified protein stocks according to the following extinction coefficients for ELP(30UAG)-GFP (Supplementary Fig. 16): Tyr (WT protein): 63,610, pAcF: 82,510, pAzF: 75,990, BuY: 22,450, 4CF3F: 19,027, 4ClF: 20,905.
Intact mass measurements
Intact mass measurements of GFP(3UAG) were performed by electrospray MS on an Agilent 6550 QTOF instrument after external calibration. Spectra deconvolution was performed with Agilent MassHunter Qualitative Analysis software v. B.06.00 Bioconfirm Intact mass module using the maximum entropy deconvolution algorithm. Intact mass measurements of ELP(10UAG)-GFP and ELP(30UAG)-GFP were obtained on a 12T LTQ-FT (Thermo) mass spectrometer fitted with a custom nano-spray ionization source and the data were analyzed using QualBrowser, part of the Xcalibur software packaged with the ThermoFisher LTQ-FT. Materials and methods for all intact mass measurements are detailed in the Supplementary Note 6.
aaRS expression and purification
The genes of pAcFRS variants were cloned into pET15a and then used to transform Rosetta cells for expression. For each variant, the expression strain was grown on 500 ml of LB media supplemented with 100 μg/ml ampicillin at 37 °C to an OD600 of 0.6–0.8 and the protein expression was induced by the addition of 0.5 mM isopropyl β-3-thiogalacto-pyranoside. Cells were incubated at 30 °C for an additional 3 h and harvested by centrifugation at 5,000g for 10 min at 4 °C. The cell paste was suspended in 15 ml of lysis buffer (50 mM Tris (pH 7.5), 300 mM NaCl, 20 mM imidazole) and lysed by sonication. The crude extract was centrifuged at 30,000g for 30 min at 4 °C. The soluble fraction was loaded onto a column containing 2 ml of Ni-NTA resin (Qiagen) previously equilibrated with 20 ml lysis buffer. The column was washed with 20 ml lysis buffer, and the bound protein was then eluted with 2 ml of 50 mM Tris (pH 7.5), 300 mM NaCl, 300 mM imidazole. Purified proteins were dialyzed with 50 mM HEPES-KOH (pH 7.5), 50 mM KCl, 1 mM DTT and 50% glycerol, and stored at − 80 °C for further studies.
ATP-PPi exchange assay
A 25 μl ATP-PPi exchange reaction contained the following components: 100 mM HEPES-KOH (pH 7.5), 30 mM KCl, 10 mM MgCl2, 2 mM DTT, 2 mM KF, 2 mM NaPPi, 5 mM ATP, 5 μM enzyme, 2 μCi/μl of (γ-32P)-labeled ATP (PerkinElmer) and varied concentrations of amino acids (0.25, 0.5, 1.25, 2.5, 5, 10 and 20 mM, respectively). The reactions were incubated at 37 °C. Time points were taken at 2 min, 5 min and 10 min by plotting 1-μl aliquots from the reaction immediately to the PEI-cellulose plates (Merck). For each reaction, 1 μl of blank reaction mixture containing no enzymes was set as background. The reaction mixtures were separated on the plates in 1 M urea and 1 M monopotassium phosphate. The plates were then scanned in a Molecular Dynamics Storm 860 phosphorimager (Amersham Biosciences). The ratio of ATP to PPi was determined to monitor reaction progress. The kinetic constants were derived from plotting initial velocity of a series of reactions that contained varied concentrations of amino acids. The data were analyzed by GraFit 5.0 (Erithacus Software).
tRNA transcription and purification
Template plasmid containing tRNA gene was purified with the plasmid maxi kit (Qiagen), and 100 μg of plasmid was digested with BstNI (New England BioLabs). The BstNI-digested template DNA was purified by phenol chloroform extraction, followed by ethanol precipitation and resolved in double distilled water. A His-tagged T7 RNA polymerase was purified over a column of Ni-NTA resin according to manufacturer’s instructions (Qiagen). The transcription reaction (40 mM Tris (pH 8); 4 mM each of UTP, CTP, GTP and ATP at pH 7.0; 22 mM MgCl2; 2 mM spermidine; 10 mM DTT; 6 μg pyrophosphatase (Roche Applied Science); 60 μg/ml BstNI digested DNA template, approximately 0.2 mg/ml T7 RNA polymerase) was performed in 10-ml reaction volumes for overnight at 37 °C. The tRNA was purified on 12% denaturing polyacrylamide gel containing 8 M urea and TBE buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA). UV shadowing illuminates the pure tRNA band, which is excised and extracted three times with 1 M sodium acetate pH 5.3 at 4 °C. The tRNA extractions were then ethanol precipitated, dissolved in RNase-free distilled water, pooled and finally desalted using a Biospin 30 column (Bio-Rad). The ratio of aminoacylated tRNA to total tRNA was determined to monitor reaction progress.
tRNA folding and 32P labeling
The tRNA was refolded by heating to 100 °C for 5 min and slow cooling to room temperature. At 65 °C, MgCl2 was added to a final concentration of 10 mM to aid folding. A His-tagged CCA-adding enzyme was purified over a column of Ni-NTA resin according to manufacturer’s instructions (Qiagen). 16 μM folded tRNA in 50 mM Tris (pH 8.0), 20 mM MgCl2, 5 mM DTT and 50 μM NaPPi was incubated at room temperature for 1 h with approximately 0.2 mg/ml CCA-adding enzyme and 1.6 μCi/μl of (α-32P)-labeled ATP (PerkinElmer). The sample was phenol/chloroform extracted and then passed over a Bio-spin 30 column (Bio-Rad) to remove excess ATP.
Aminoacylation assay
A 20 μl aminoacylation reaction contained the following components: 50 mM HEPES-KOH (pH 7.2), 25 mM KCl, 10 mM MgCl2, 5 mM DTT, 10 mM ATP, 25 μg/ml pyrophosphatase (Roche Applied Science), 2 mM amino acids. All plateau tRNA aminoacylation levels were determined at 37 °C according to the reaction conditions described above with 500 nM enzyme, 5 μM unlabeled tRNA plus 100 nM 32P-labeled tRNA. Time points were taken at 5 min, 20 min and 60 min by removing 2 μl aliquots from the reaction and immediately quenching the reaction into an ice-cold 3 μl quench solution (0.66 μg/μl nuclease P1 (Sigma) in 100 mM sodium citrate (pH 5.0)). For each reaction, 2 μl of blank reaction mixture containing no enzymes was added to the quench solution as the start time point. The nuclease P1 mixture was then incubated at room temperature for 30 min and 1 μl aliquots were spotted on PEI-cellulose plates (Merck) and developed in running buffer containing 5% acetic acid and 100 mM ammonium acetate. Radioactive spots for AMP and AA-AMP (representing free tRNA and aminoacyl-tRNA, respectively) were separated and then visualized and quantified by phosphorimaging by a Molecular Dynamics Storm 860 phosphorimager (Amersham Biosciences). The ratio of aminoacylated tRNA to total tRNA was determined to monitor reaction progress.
Supplementary Material
Acknowledgments
We thank K. Bilguvar and J. Knight (Yale Center for Genome Analysis) for conducting next-generation sequencing experiments; T. Wu (Yale West Campus Analytical Core facility) for conducting intact MS experiments; B. Gassaway for assistance with shotgun MS experiments. We are grateful to members of the Isaacs laboratory, J. Ling and G. Church for critical discussions and feedback. This work was supported by the Defense Advanced Research Projects Agency contracts N66001-12-C-4020 and N66001-12-C-4211 to (F.J.I., J.R., D.S., and M.C.J.), U.S. Department of Energy (DE-FG02-02ER63445 to F.J.I.) grants GM22854 to D.S. and GM67193 to N.L.K. from the National Institute for General Medical Sciences, T32GM007205 and 1F30CA196191 (A.D.H.), Army Research Office (W911NF- 11-1-0445 to M.C.J.), the David and Lucile Packard Foundation (M.C.J.), the Camille Dreyfus Teacher-Scholar Program (M.C.J.), DuPont, Inc. (F.J.I.) and the Arnold and Mabel Beckman Foundation (F.J.I.).
Footnotes
Accession codes. The synthetase variants evolved in this study are deposited at Genbank (KT996130–KT996140). The GFP and ELP-GFP reporter proteins used in this study are deposited at Genbank (KT996141–KT996147).
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.A. designed ELP constructs, and conducted and interpreted multi-site nsAA incorporation experiments. M.A., A.D.H. and F.J.I. designed, conducted and interpreted synthetase evolution experiments. H.-R.A. and J.R. conducted and interpreted MS experiments. I.N. and N.L.K. performed and interpreted top-down MS experiments. A.D.H, A.L.G. and F.J.I. analyzed NextGen sequencing experiments. C.F., D.W.M. and D.S. conducted and interpreted biochemical experiments. Y.-S.W. executed nsAA screens. S.H.H. tested ELP expression in vitro. M.A., A.D.H. and A.J.R. performed crystal structure analysis and target selection. N.J.M. constructed and characterized the tolC variant used for negative selection. F.J.I., J.R. and D.S. directed the studies and interpreted data. M.A. and F.J.I. wrote the paper with assistance from A.D.H., N.J.M., D.S., M.C.J., I.N., N.L.K. and J.R.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Seitchik JL, et al. Genetically encoded tetrazine amino acid directs rapid site-specific in vivo bioorthogonal ligation with trans-cyclooctenes. J Am Chem Soc. 2012;134:2898–2901. doi: 10.1021/ja2109745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 3.Link AJ, Mock ML, Tirrell DA. Non-canonical amino acids in protein engineering. Curr Opin Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 5.O’Donoghue P, Ling J, Wang YS, Söll D. Upgrading protein synthesis for synthetic biology. Nat Chem Biol. 2013;9:594–598. doi: 10.1038/nchembio.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian F, et al. A general approach to site-specific antibody drug conjugates. Proc Natl Acad Sci USA. 2014;111:1766–1771. doi: 10.1073/pnas.1321237111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furman JL, et al. A genetically encoded aza-Michael acceptor for covalent cross-linking of protein-receptor complexes. J Am Chem Soc. 2014;136:8411–8417. doi: 10.1021/ja502851h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang M, et al. Evolution of iron(II)-finger peptides by using a bipyridyl amino acid. ChemBioChem. 2014;15:822–825. doi: 10.1002/cbic.201300727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F, Niu W, Guo J, Schultz PG. Unnatural amino acid mutagenesis of fluorescent proteins. Angew Chem Int Edn Engl. 2012;51:10132–10135. doi: 10.1002/anie.201204668. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Liu CC. Biological applications of expanded genetic codes. ChemBioChem. 2014;15:2335–2341. doi: 10.1002/cbic.201402159. [DOI] [PubMed] [Google Scholar]
- 11.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuet KP, Tirrell DA. Chemical tools for temporally and spatially resolved mass spectrometry-based proteomics. Ann Biomed Eng. 2014;42:299–311. doi: 10.1007/s10439-013-0878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishi Y, et al. Different effects of 4-hydroxyproline and 4-fluoroproline on the stability of collagen triple helix. Biochemistry. 2005;44:6034–6042. doi: 10.1021/bi047887m. [DOI] [PubMed] [Google Scholar]
- 14.Kothakota S, Mason TL, Tirrell DA, Fournier MJ. Biosynthesis of a periodic protein containing 3-thienylalanine - a step toward genetically-engineered conducting polymers. J Am Chem Soc. 1995;117:536–537. [Google Scholar]
- 15.Bae JH, et al. Incorporation of beta-selenolo[3,2-b]pyrrolyl-alanine into proteins for phase determination in protein X-ray crystallography. J Mol Biol. 2001;309:925–936. doi: 10.1006/jmbi.2001.4699. [DOI] [PubMed] [Google Scholar]
- 16.Kirshenbaum K, Carrico IS, Tirrell DA. Biosynthesis of proteins incorporating a versatile set of phenylalanine analogs. ChemBioChem. 2002;3:235–237. doi: 10.1002/1439-7633(20020301)3:2/3<235::AID-CBIC235>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 17.Young DD, et al. An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry. 2011;50:1894–1900. doi: 10.1021/bi101929e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park HS, et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–1154. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umehara T, et al. N-acetyl lysyl-tRNA synthetases evolved by a CcdB-based selection possess N-acetyl lysine specificity in vitro and in vivo. FEBS Lett. 2012;586:729–733. doi: 10.1016/j.febslet.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 20.Johnson DB, et al. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinemann IU, et al. Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 2012;586:3716–3722. doi: 10.1016/j.febslet.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukai T, et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010;38:8188–8195. doi: 10.1093/nar/gkq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiltschi B, Wenger W, Nehring S, Budisa N. Expanding the genetic code of Saccharomyces cerevisiae with methionine analogs. Yeast. 2008;25:775–786. doi: 10.1002/yea.1632. [DOI] [PubMed] [Google Scholar]
- 26.Nehring S, Budisa N, Wiltschi B. Performance analysis of orthogonal pairs designed for an expanded eukaryotic genetic code. PLoS One. 2012;7:e31992. doi: 10.1371/journal.pone.0031992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odoi KA, Huang Y, Rezenom YH, Liu WR. Nonsense and sense suppression abilities of original and derivative Methanosarcina mazei pyrrolysyl-tRNA synthetase-tRNAPyl pairs in the Escherichia coli BL21(DE3) cell strain. PLoS One. 2013;8:e57035. doi: 10.1371/journal.pone.0057035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher RR, Li Z, Lewis AO, Isaacs FJ. Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA. Nat Protoc. 2014;9:2301–2316. doi: 10.1038/nprot.2014.082. [DOI] [PubMed] [Google Scholar]
- 31.MacEwan SR, Chilkoti A. Elastin-like polypeptides: biomedical applications of tunable biopolymers. Biopolymers. 2010;94:60–77. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]
- 32.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 33.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 34.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeVito JA. Recombineering with tolC as a selectable/counter-selectable marker: remodeling the rRNA operons of Escherichia coli. Nucleic Acids Res. 2008;36(1):e4. doi: 10.1093/nar/gkm1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi T, et al. Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion (vol. 102, pages 1366, 2005) Proc Natl Acad Sci USA. 2005;102:1366–1371. doi: 10.1073/pnas.0407039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz KC, et al. A genetically encoded infrared probe. J Am Chem Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooley RB, et al. Structural basis of improved second-generation 3-nitro-tyrosine tRNA synthetases. Biochemistry. 2014;53:1916–1924. doi: 10.1021/bi5001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stokes AL, et al. Enhancing the utility of unnatural amino acid synthetases by manipulating broad substrate specificity. Mol Biosyst. 2009;5:1032–1038. doi: 10.1039/b904032c. [DOI] [PubMed] [Google Scholar]
- 41.Ko JH, et al. Pyrrolysyl-tRNA synthetase variants reveal ancestral aminoacylation function. FEBS Lett. 2013;587:3243–3248. doi: 10.1016/j.febslet.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo LT, et al. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc Natl Acad Sci USA. 2014;111:16724–16729. doi: 10.1073/pnas.1419737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aerni HR, Shifman MA, Rogulina S, O’Donoghue P, Rinehart J. Revealing the amino acid composition of proteins within an expanded genetic code. Nucleic Acids Res. 2015;43:e8. doi: 10.1093/nar/gku1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chin JW, et al. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 45.Wu IL, et al. Multiple site-selective insertions of noncanonical amino acids into sequence-repetitive polypeptides. ChemBioChem. 2013;14:968–978. doi: 10.1002/cbic.201300069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer DE, Chilkoti A. Quantification of the effects of chain length and concentration on the thermal behavior of elastin-like polypeptides. Biomacromolecules. 2004;5:846–851. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- 47.Tinberg CE, et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501:212–216. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rovner AJ, et al. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mandell DJ, et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lajoie MJ, et al. Probing the limits of genetic recoding in essential genes. Science. 2013;342:361–363. doi: 10.1126/science.1241460. [DOI] [PubMed] [Google Scholar]
- 51.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 52.Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer DE, Chilkoti A. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: examples from the elastin-like polypeptide system. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.