Abstract
Hepatic stellate cells (HSCs) inhibit T cells, a process that could help the liver to maintain its immunoprivileged status. HSCs secrete latent TGFβ1, but the detailed mechanisms by which latent TGFβ1 is activated and whether it plays any role in HSC-mediated T-cell suppression remain unclear. Glycoprotein A repetitions predominant (GARP) is a surface marker of activated regulatory T cells (Tregs). GARP binds latent TGFβ1 for its activation, which is critical for Tregs to suppress effector T cells; however, it is still unclear whether GARP is present on HSCs and whether it has any impact on HSC function. In this study, we found that TGFβ1+/− HSCs, which produce reduced levels of TGFβ1, showed decreased potency in inhibiting T cells. We also found that pharmaceutical or genetic inhibition of the TGFβ1 signaling pathway reduced the T-cell-inhibiting activity of HSCs. In addition, using isolated primary HSCs, we demonstrated that GARP was constitutively expressed on HSCs. Blocking GARP function or knocking down GARP expression significantly impaired the potency of HSCs to suppress the proliferation of and IFNγ production from activated T cells, suggesting that GARP is important for HSCs to inhibit T cells. These results demonstrate the unexpected presence of GARP on HSCs and its significance in regard to the ability of HSCs to activate latent TGFβ1 and thereby inhibit T cells. Our study reveals a new mechanism for HSC-mediated immune regulation and potentially for other conditions, such as liver fibrosis, that involve HSC-secreted TGFβ1.
Keywords: Liver, immune regulation, GARP, fibrosis
Introduction
Transforming growth factor-beta1 (TGFβ1) plays critical roles in many physiological and pathological conditions, including cell differentiation and apoptosis, inflammation regulation, and tissue repair and fibrosis. TGFβ1 is first made by cells in a latent form in association with a dimeric protein called latency-associated peptide (LAP). The secreted latent TGFβ1 complex is then processed by different mechanisms to release active TGFβ1, which can then execute its many biological functions.
Glycoprotein-A repetitions predominant (GARP) has been identified as a cell-surface marker for activated foxp3+ regulatory T cells (Tregs) (1, 2). GARP is essential for the surface anchoring of latent TGFβ1 and its activation, both of which are required by Tregs to inhibit effector T cells and to induce tolerance via cell-to-cell contact (3–5). The distribution of GARP is thought to be restricted on platelets and activated Tregs (1). Little is known about whether GARP is expressed on other cells, and if it is, whether GARP plays any role in regulating the function of these cells.
Hepatic stellate cells (HSCs) account for about one third of all nonparenchymal cells in the liver (6). In normal liver, HSCs are involved in liver development and regeneration, and these cells also play a key role in the storage and transport of retinoids in the body. Under pathological conditions such as liver injury, HSCs are activated and heavily involved in liver fibrosis. It has been demonstrated that activated HSCs produce matrix proteins, including collagens, as well as different kinds of matrix metalloproteinases and tissue inhibitors of metalloproteinases (7). In addition, activated HSCs are an important major source of TGFβ1, which contributes to liver fibrosis through both autocrine and paracrine mechanisms, including stimulating the productions of matrix proteins, matrix metalloproteinases, and tissue inhibitors of metalloproteinases from HSCs (8).
Aside from the established role of HSCs in liver fibrosis, it has also been demonstrated by several groups, including ours, that HSCs have strong immunoregulatory activities, which could help to maintain the immunoprivileged status of liver (9–13). We have reported that HSCs directly inhibit T cells primarily through cell-to-cell contact and that programmed death-ligand 1 (PD-L1) on the surface of HSCs is an important molecule involved in this process (9, 13). In addition, we showed that HSCs indirectly regulate adaptive immune responses by inducing the propagation of myeloid-derived suppressor cells, which suppress both T- and B-cell responses (10, 14). However, despite the observation that activated HSCs are a major source of latent TGFβ1 in the liver (15), whether the TGFβ1 produced by HSCs contributes to the immunoregulatory activity of HSCs remains unclear, as do the mechanisms by which the HSC-produced latent TGFβ1 is activated.
In this study, we examined the role of TGFβ1 in HSC-mediated T-cell inhibition and the expression and function of GARP on primary HSCs. Our results suggest that GARP is constitutively expressed on HSCs and is required by HSCs to activate locally produced latent TGFβ1, thus inhibiting the proliferation of and inflammatory cytokine production from activated T cells.
Methods and Reagents
Mice
All mice used were on C57BL/6 background. Wild-type (WT), TGFβ1+/− knockout (KO) (16) and SMAD3 KOs(17) were purchased from The Jackson Laboratory (Bar Harbor, ME). PD-L1−/− mice (18) were a kind gift from Lieping Chen, MD, PhD (Department of Immunobiology and Yale Cancer Center, Yale University School of Medicine, New Haven, CT). All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committees at Cleveland Clinic and Case Western Reserve University.
Isolation of HSCs
Mouse HSCs were isolated from WT, TGFβ1+/− and PD-L1−/− mouse livers following protocols established in the lab as described in detail previously (13). In brief, HSCs were isolated from perfused mouse livers and cultured in RPMI-1640 medium supplemented with 20% fetal bovine serum in 5% CO2 in air at 37° C for 14–21 days. Cell viability was usually >90%, as determined by trypan blue exclusion, and cell purity was usually >90%, as determined by their staining positive for α-smooth muscle actin (immune staining) and negative for CD45 (flow), as previously described (10). Human primary HSCs were purchased from ScienCell Research Laboratories (Carlsbad, CA).
GARP expression assessments
To assess GARP expression at transcript levels, RT-PCR was performed to amplify GARP cDNA fragments from total RNAs purified from isolated HSCs. We used the following RT-PCR primers: P1: CAGCGTCGAGAGCAAGTG; P2: GCTTGGATGTCCAGTGAGA (for mouse GARP); P3: AAGTGCCCTGTAAGATGGTG; and P4: CAAGTTCAGGTAGATGAGTCTCG (for human GARP). Primers were designed to span different exons of GARP genes to ensure the amplification of cDNAs instead of contaminated genomic DNA, and RNAs without reserve transcription were used as negative controls. To confirm that the amplified products are GARP transcripts, the PCR bands were isolated from the gel using a Gel DNA extraction kit (Qiagen, CA) following manufacturer provided protocols and sequenced by the Sequencing Core Facility at Cleveland Clinic. For quantitative RT-PCRs (qRT-PCRs), GARP transcripts were quantitated using SYBR® Green Real-Time PCR Master Mixes (ABI Biosystems/Life Technologies, Carlsbad, CA) with the GARP primers P5: CAGCGTCGAGAGCAAGTG and P6: GCTTGGATGTCCAGTGAGAG and the GAPDH primers P7: AGGTCGGTGTGAACGGATTTG and P8: TGTAGACCATGTAGTTGAGGTCAG on an ABI 7500 real-time PCR machine, following manufacturer-provided protocols. To assess GARP expression at protein levels, Isolated HSCs were treated with Versene solution (Life technology, CA) to make single cell suspensions, then incubated with respective anti-GARP antibodies [2 µg/mL of an anti-mouse-GARP IgG, clone F011-5 or anti-human-GARP IgG, clone 7B11 (BioLegend, San Diego, CA)], or the same concentration of isotype control, followed by flow cytometry analysis on a flow cytometer (BD FACScalibur™, Franklin Lakes, New Jersey).
Weston blot of GARP
HSCs were treated with or without 100U IFN-γ for 48 hr. For negative controls, CD8+ T cells were isolated from WT C57BL/6 mouse splenocytes using a mouse CD8+ T cell isolation kit (Miltenyi Biotec, San Diego, CA), and for positive controls, mouse splenocytes after B cell depletion using a mouse B cell isolation kit (StemCell, Vancouver, Canada) were used. Cell lysates were prepared using NP-40 buffer (Sigma, MI) and total protein concentrations were determined by using a BCA protein concentration assay kit (Pierce, IL) following manufacturer provided protocols. Blotted membrane was incubated with 1 µg/ml of the sheep anti-GARP Ab (R&D, Minneapolis, MN) at 4°C overnight, then incubated with 2 µg /ml of the HRP-conjugated Rabbit anti-sheep IgG (Southern Biotech, Birmingham, AL) for 30min at room temperature. After washing, ECL western blotting regent (GE Healthcare Bio-Sciences, Pittsburg, PA) was applied for the detection. The same membrane was stripped and probed again using an anti-mouse actin antibody to serve as a loading control.
GARP knockdown by lentivirus-expressed shRNAs
Lentivirus-expressing short hairpin RNAs (shRNAs) designed to target different sites of the GARP transcript were ordered from Sigma (St. Louis, MO). To knock down GARP gene expression on HSCs, different combinations of the shRNA-expressing lentivirus or control empty virus were used to transfect HSCs following previously described protocols [18]. In brief, 0.5 × 106 HSCs were co-cultured with 8 µg/ml hexadimethrine bromide (Sigma Aldrich, St. Louis, MO) in the presence of 5–10 µl of lentiviral particles to the cells with a multiplicity of infections (MOI) of 0.5–1. Cells were incubated at 37°C in a humidified incubator in an atmosphere of 5% CO2 for 20 hrs, then washed and cultured in fresh media. The cells were cultured for another 4 days, then levels of GARP protein on them were assessed by flow cytometry.
T-cell inhibition assays
T cells from WT or SMAD3 KO mice were enriched using nylon wool columns and labeled with Carboxyfluorescein succinimidyl ester (CFSE), then activated with anti-CD3/CD28 monoclonal antibodies. Different numbers of WT or TGFβ1+/− HSCs were mixed with the activated T cells, in the presence of either a polyclonal sheep anti-GARP antibody or control IgGs (R&D Systems, Minneapolis, MN). HSC-mediated T-cell inhibition was assessed by measuring the proliferation of both CD4+ and CD8+ T cells and the production of IFNγ following protocols described before.
Immunofluorescence assays
Cells were plated in 4-well chamber slides (Thermo Scientific, Waltham, MA), washed with phosphate-buffered saline (PBS) twice, then fixed for 10 min in fresh 4% paraformaldehyde-PBS. After further washing in PBS, the cells were blocked with 5% bovine serum albumin (Sigma, St Louis, MO) for 2 h in PBS. Cells were then washed three more times in PBS with 1% bovine serum albumin and incubated with primary antibody and control IgGs (anti-GARP polyclone antibody [R&D Systems, Minneapolis, MN], anti-mouse LAP, clone TW7-16B4 [BioLegend, San Diego, CA], or anti-TGFβ polyclonal antibody [Santa Cruz Biotechnology, Dallas, TX]) overnight at 4°C. After washing, cells were incubated with respective secondary antibodies at room temperature for 1 hr. Slides were mounted with FluorSave™ reagent (EMD Millipore, Darmstadt, Germany) and kept in the dark until viewing.
Enzyme-linked immunosorbent assays
LAP, TGFβ1, and IFNγ levels in the culture supernatants were measured by standard enzyme-linked immunosorbent assays (ELISAs) following manufacturer-provided protocols.
Statistical Analysis
Each experiment was repeated at least twice or more with similar results. Results from a representative experiment were shown in each case. All data are shown as mean ± SD, student’s t-test was used to analyze the data using Microsoft Excel and GraphPad Prism 6 (GraphPad Software, San Diego, CA). Statistical significance was set at p < 0.05.
Results
TGFβ1 is important for HSCs to inhibit T cells
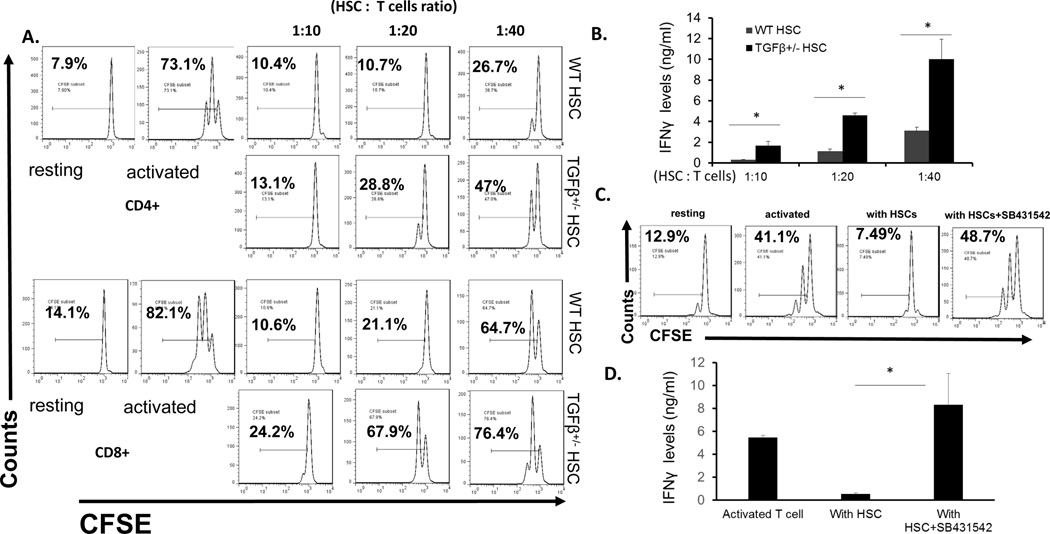
HSCs are known to be an important source of TGFβ1 (19, 20), but whether HSC-produced TGFβ1 contributes to the immunoregulatory activity of HSCs remains unclear. To clarify this issue, we isolated HSCs from WT or TGFβ1+/− mice (TGFβ1−/− mice die prematurely) (16). Consistent with previous reports (21–23), we found that TGFβ1 levels were significantly reduced in sera from TGFβ1+/− mice (data not shown). Upon comparing the T-cell inhibitory activity of the WT and TGFβ1+/− HSCs, we found that TGFβ1+/− HSCs had reduced potency in inhibiting T cells compared with WT HSCs (Fig. 1A, B), suggesting that TGFβ1 is needed for HSCs to inhibit T cells. In addition, we cultured WT HSCs with activated T cells in the presence or absence of the TGFβ signaling inhibitor SB-431542 (24), then assessed the proliferation of these T cells. These experiments showed that inhibiting TGFβ signaling by the inhibitor markedly augmented the proliferation of and IFNγ production from the activated T cells, even in the presence of HSCs (Fig. 1C.D), suggesting a significant role of TGFβ signaling in HSC-mediated T-cell inhibition.
Figure. 1. TGFβ1 is required for HSC to efficiently inhibit T cells.
WT and TGFβ1+/− HSCs were co-cultured with CFSE labeled, and anti-CD3/CD28 mAbs-activated T cells at different ratios (HSC: T cells). 72 h later, proliferations of the activated CD4+ and CD8+ T cells were assessed by flow cytometry (A) and IFNγ levels in the culture supernatants were measured by ELISA (B). Representative results of 3 different experiments. In addition, WT HSCs were co-cultured with CFSE labeled, and anti-CD3/CD28 mAbs-activated T cells at 1:15 ratio in the absence or presence of 2µM of the TGFβ signaling inhibitor SB431542, and proliferations of the activated T cells (C) and their production of IFNγ (D) were assessed in 72 h by flow cytometry and ELISA, respectively. Representative results of 2 different experiments.
The TGFβ1 signaling pathway in T cells is important for HSCs to inhibit T cells
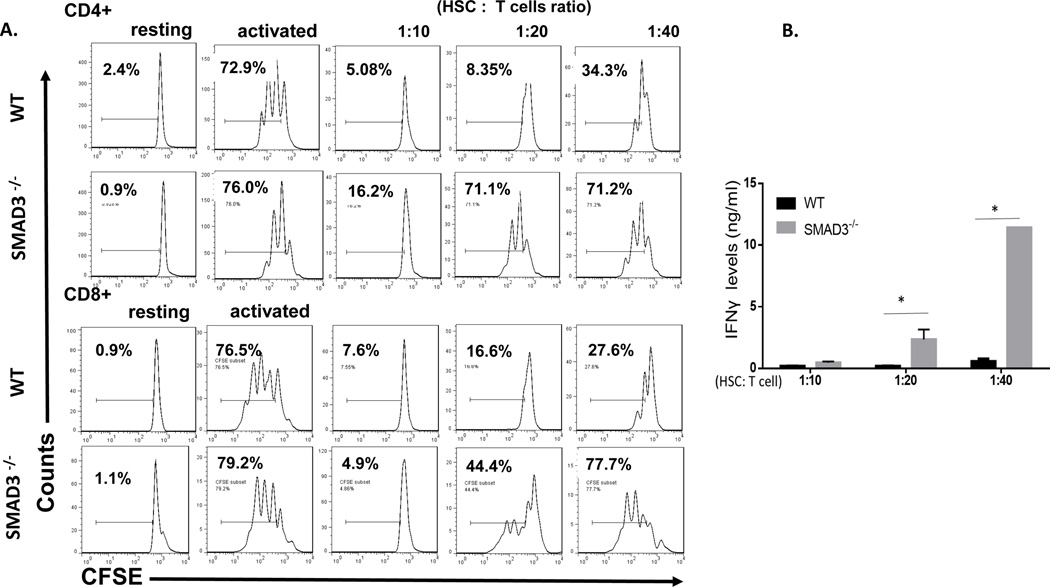
HSC-produced latent TGFβ1, after activation, could directly initiate signaling pathways in T cells to exhibit their T-cell inhibitory activity, or it could also act in an autocrine fashion for HSCs to indirectly inhibit T cells, e.g., by upregulating HSC expression of PD-L1. To address this issue, in light of previous reports that SMAD3 is a critical intracellular signal transducer and transcriptional modulator for TGFβ (17), we cultured HSCs with different numbers of activated WT and SMAD3−/− T cells, then assessed the proliferation and cytokine production of these T cells. These experiments showed that, compared with WT T cells, which were potently suppressed by the HSCs, SMAD3−/− T cells showed largely increased proliferation and IFNγ production (Fig. 2), indicating that HSC-produced TGFβ1 directly acted on these T cells to inhibit their proliferation and cytokine production and that the SMAD3 pathway of TGFβ1 signaling is important for the HSC-produced TGFβ1 to inhibit T cells. All the above results, taken together, have now revealed a previously unknown role of the HSC-produced TGFβ1 underlying the mechanism by which HSCs inhibit T cells.
Figure. 2. HSC-produced TGFβ1 directly inhibit T cells through the SMAD3 pathway.
WT HSCs were co-cultured with CFSE labeled, and anti-CD3/CD28 mAbs-activated WT or SMAD3−/− T cells at different ratios (HSC: T cells). 72 hr later, proliferation of the activated T cells was assessed by flow cytometry (A) and production of IFNγ by the activated T cells in the culture supernatants was measured by ELISA (B). *p<0.05 as calculated by student’s t-test. Representative results of 4 different experiments.
GARP is present on both human and mouse HSCs
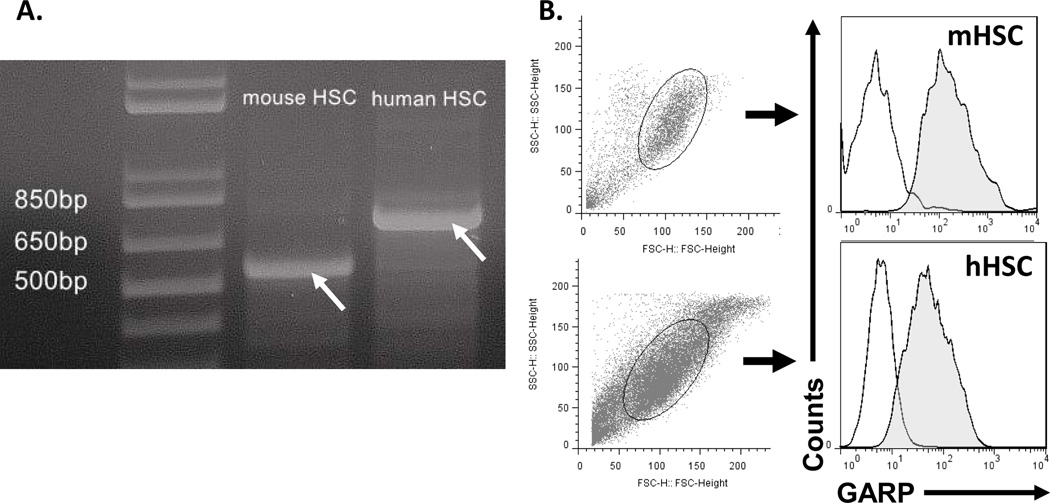
Previous studies have shown that GARP is critical for Tregs to activate latent TGFβ1 to inhibit effector T cells (1, 2). Thus we next explored whether GARP is also expressed on HSCs and whether it is important for HSCs to activate latent TGFβ1 for T cell inhibition. To determine whether HSCs constitutively express GARP, we first isolated total RNAs from mouse HSCs and carried out RT-PCRs to detect GARP transcripts. We found that cDNA fragment with the predicted size could be amplified by RT-PCR from mouse HSC total RNA (Fig. 3A), and further DNA sequencing results confirmed that these amplicons are indeed GARP transcripts (data not shown), suggesting that the GARP gene is constitutively transcribed in mouse HSCs. Using isolated primary human HSCs, we were also able to detect GARP transcripts in these cells by RT-PCR (Fig. 3A) followed by sequencing (data not shown). To determine the presence of GARP proteins on the surface of HSCs, we analyzed both human and mouse primary HSCs by flow cytometry and found that, consistent with the RT-PCR results, GARP proteins were detectable on the cell surface (Fig. 3B). All these data show that GARP is constitutively expressed and is present on the cell surface of both human and mouse HSCs. In the following experiments, we focused on mouse HSCs as our subject of research.
Figure 3. HSCs constitutively express GARP.
Total RNAs were isolated from mouse primary HSCs and human primary HSCs, then used for RT-PCR to amply the transcripts of GARP (arrows) using respective primers (A), these amplicons were purified and sequenced to confirm that they are indeed GARP transcripts (data not shown). Both mouse and human HSCs were also stained with respective anti-GARP mAb or isotype controls, then analyzed by flow cytometry (B) Empty areas, isotype control, shaded areas, GARP-specific Ab stainings. Representative results of 4 different experiments.
GARP expression levels are elevated on HSCs activated by IFNγ
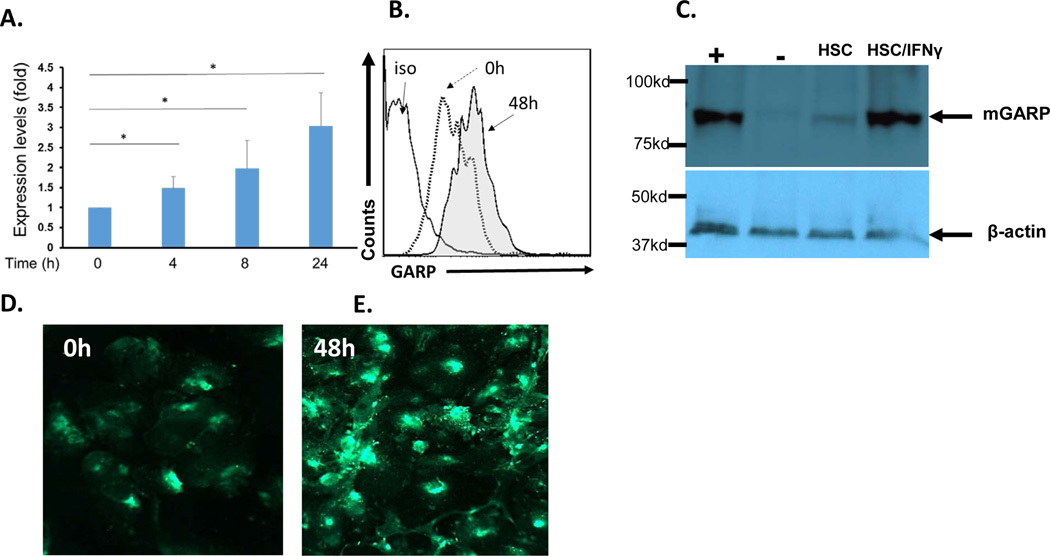
IFNγ is an important inflammatory cytokine that markedly upregulates PD-L1 expression on HSCs, thereby augmenting the T-cell inhibitory activity of HSCs (25). To test the effect of IFNγ on GARP expression, we first stimulated HSCs with 100 U of IFNγ, then isolated total RNAs at baseline (0) and after 4, 8, and 24 hrs to quantitated GARP transcripts by quantitative RT-PCR. These experiments showed that IFNγ stimulation upregulated the transcription of GARP in HSCs (Fig. 4A). We then incubated HSCs with different concentrations of IFNγ for 48 hrs, then assessed GARP protein levels on HSCs both by flow cytometric analysis, western blots and immunofluorescence staining. These experiments showed that consistent with increased levels of transcription as detected in early quantitative RT-PCR assays, GARP protein levels were elevated on the surface of HSCs after incubation with IFNγ (Fig. 4B,C D & E), suggesting that inflammatory cytokines such as IFNγ upregulate the expression of GARP on HSCs.
Figure 4. GARP expression is upregulated by IFNγ.
Mouse HSCs were incubated with 100U/ml of recombinant murine IFNγ, then collected at different time points to isolate total RNA for quantative RT-PCR to assess levels of GARP transcripts (A). The IFNγ stimulation experiments were repeated and this time, some of the HSCs were cultured on chamber slides. 48 hr after IFNγ stimulation, GARP expression on HSCs was assessed by flow cytometry (B), western blot (C) and immunofluorescence staining (D, E). Representative results of 4 different experiments except panel A, which are the combined results of 3 experiments, and panel C, which was done twice. *p < 0.05 as calculated by student’s t-test, +, positive control (non-B cell splenocytes); −, negative control (CD8+ T cells); HSC, HSCs without IFNγ stimulation; and HSC/IFNγ, HSCs with IFNγ stimulation for 48 hr; arrows, GARP band (~ 85KDa) and actin band (~40KDa). Western blot images were scanned and cropped to show the bands.
Blocking GARP function impairs the T-cell inhibitory activity of HSCs
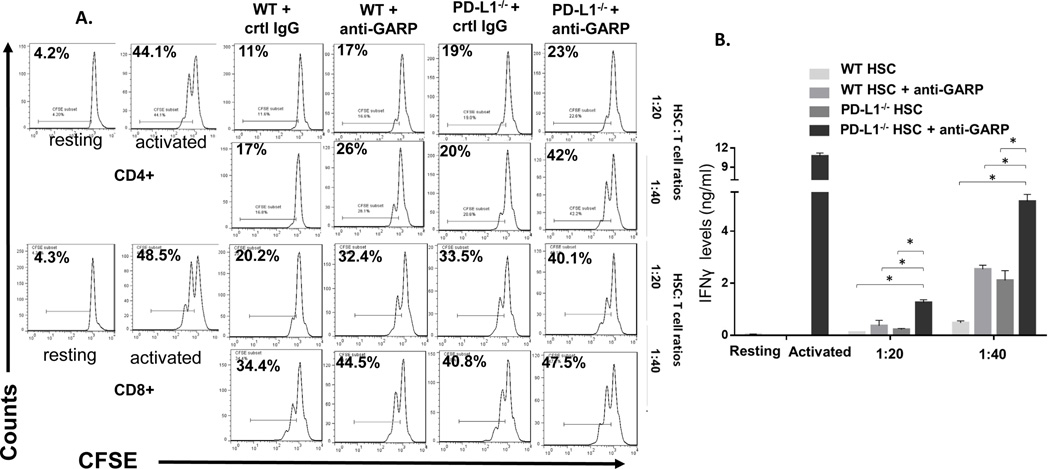
An important feature of HSCs is that they potently inhibit T cells through PD-L1, which could help to maintain the immunoprivileged status of liver (10). We next examined the role of the newly discovered GARP in HSC-mediated T-cell inhibition and its relevance with regard to PD-L1 on HSCs. In brief, we incubated WT or PD-L1−/− HSCs with different numbers of CFSE-labeled- and anti-CD3/anti-CD28 monoclonal antibody-activated T cells in the presence of GARP-blocking IgGs or control IgGs for 72 hrs, then analyzed the proliferation of activated T cells by flow cytometry. In addition to the T-cell proliferation assays, we also measured levels of IFNγ produced by the activated T cells in the culture supernatants by ELISA. These experiments showed that, consistent with previous reports (9, 10), in the presence of control IgGs, HSCs markedly inhibited T cells, as measured by both their proliferation and IFNγ production, and that PD-L1−/− HSCs exhibited reduced T-cell inhibitory activity (Fig. 5). In the presence of GARP-blocking IgGs, however, the ability of the WT HSCs to inhibit the proliferation of the activated T cells was also significantly reduced, as was their ability to suppress IFNγ production from these T cells. Interestingly, PD-L1−/− HSCs in the presence of GARP-blocking IgGs showed the weakest T-cell inhibitory activity (Fig. 5). These results suggest that in addition to PD-L1, GARP is another cell surface molecule required by HSCs to inhibit T cells.
Figure 5. GARP is required for HSC to efficiently inhibit T cells in addition to PD-L1.
WT or PD-L1−/− HSCs were co-cultured with CFSE-labeled, anti-CD3/CD28 mAb activated T cells at different ratios in the presence of control IgG or 10 µg/ml polyclonal anti-GARP IgGs. 72 h later, proliferation of the CD4+ and CD8+ T cells were assessed by flow cytometry (A). The experiment was repeated using WT HSC, PD-L1−/− HSCs and PD-L1−/− HSCs plus the GARP blocking IgGs, and IFNγ levels in the culture supernatants were measured by ELISA (B). * p<0.05 as calculated by student’s t-test, representative results of 2 different experiments.
Knocking down GARP expression impairs the T-cell inhibitory activity of HSCs
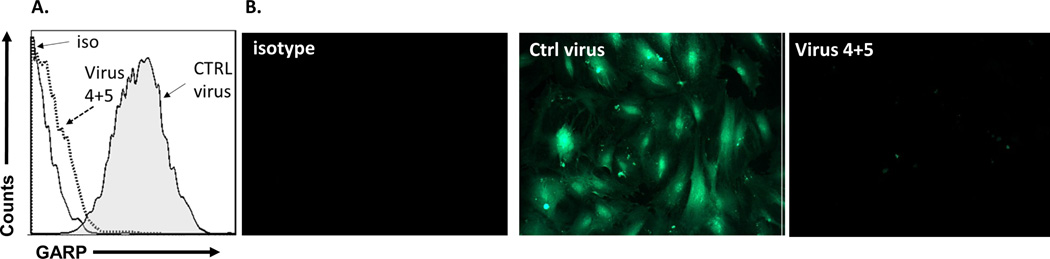
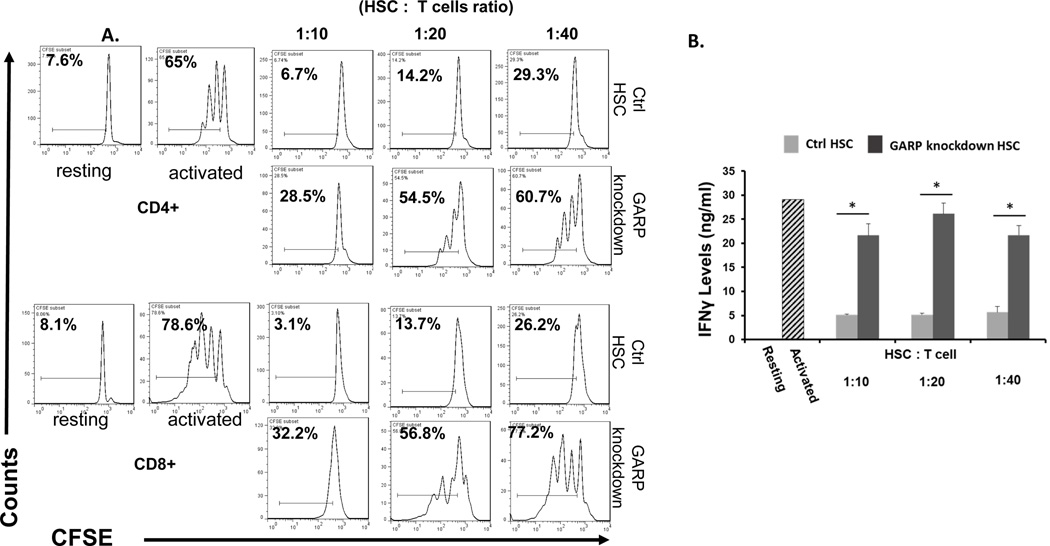
To further confirm the significance of GARP in HSCs regarding their T-cell inhibitory activity, but from a different perspective, we used a lentivirus-delivered shRNA approach to knock down GARP expression on HSCs. We screened combinations of lentivirus-expressed shRNAs targeting different sites of the GARP transcript by first infecting the cells with the recombinant virus, then assessing GARP levels on the infected cells 96 hrs later by flow cytometry and immunofluorescence staining. Using this method, we identified one combination of viral particles that were the most effective to knock down GARP expression on HSCs. It knocked down >90% of GARP expression in the infected HSCs (Fig. 6 A,B). The shRNA coding sequence in these two lentiviral particles is CCGGGATGCTACTCAGGACCTAATCCTCGAGGATTAGGTCCTGAGTAGCATCTTTTTG or CCGGATGCCAGCGGTGGAGCAATTACTCGAGTAATTGCTCCACCGCTGGCATTTTTTG, respectively. We then infected HSCs with these identified lentivirus-expressed shRNAs and 96 hrs later, after confirming the successful knockdown of GARP expression on the shRNA-infected HSCs, we compared their efficacy at inhibiting the proliferation of activated T cells and IFNγ production from the activated T cells with HSCs infected with the same number of control empty lentivirus at the same time. These experiments showed that HSCs with knocked-down GARP were less efficient at inhibiting activated T cells than the control HSCs (Fig. 7), confirming that GARP is important for HSCs to inhibit T cells.
Figure 6. Knocking down of GARP expression by lentivial-shRNAs.
Lentiviral particles containing shRNAs targeting different sites of GARP transcripts were used to transfect HSCS, then GARP protein levels on HSCs were assessed 72 hr later by flow cytometry (A) and immunofluorescence staining (B). One of the combinations, virus 4+5 was identified to be able to significantly knock down GARP expression on HSCs. Solid line, isotype control; dotted line, HSCs transfected with virus 4+5, shaded area, HSCs transfected with control virus. Representative results of 4 different experiments.
Figure 7. HSCs with GARP knocked down have reduced ability to inhibit T cells.
WT HSCs were transfected with the identified combination of shRNA virus (4+5) or control virus. 72 hr later, after GARP knock down was verified by flow cytometry in HSCs transfected with 4+5 viral particles, these HSCs were co-cultured with CFSE-labeled, anti-CD3/CD28 mAb activated T cells at different ratios for another 72 hrs. Proliferation of the CD4+ and CD8+ T cells were assessed by flow cytometry (A) and levels of IFNγ in the supernatants were measured by standard ELISA (B). Resting, T cells without activation (system negative control), Activated, T cells incubated with the anti-CD3/CD28 beads in the absence of HSCs (system positive control). Representative results of 3 different experiments.
Latent TGFβ1 and TGFβ1 co-localize with GARP on HSCs
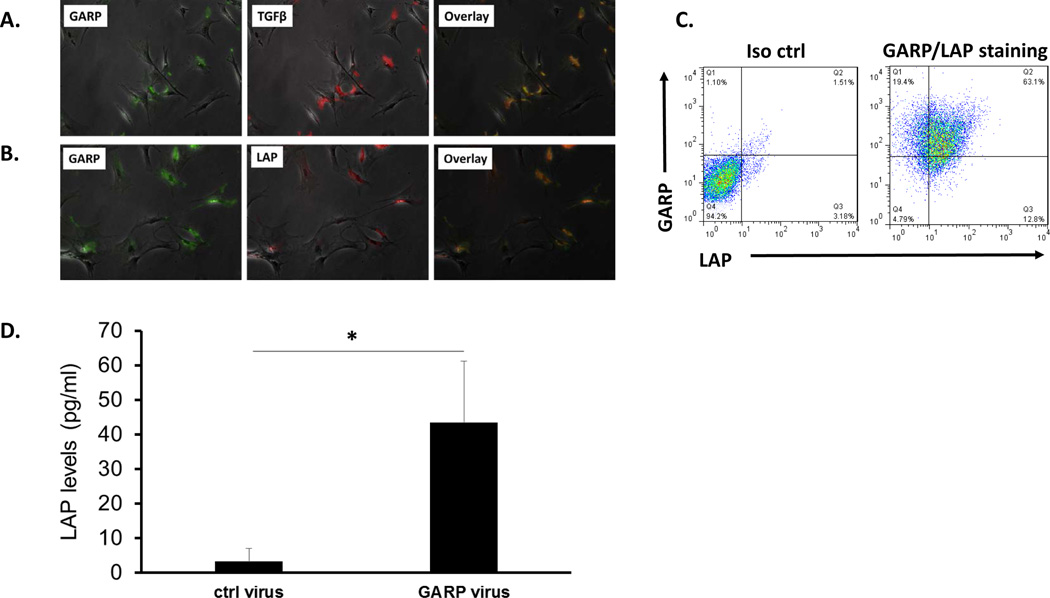
We have previously reported that cell-to-cell contact is required for HSCs to inhibit T cells (13), and we confirmed these results again in this study too (data not shown). With the new discovery that HSC-produced TGFβ1 is also required for HSC to efficiently inhibit T cells, this suggests that the GARP-activated, HSC-produced TGFβ1 works in a fashion similar to that reported in Tregs (3, 4), in which activated TGFβ1 is either released in close proximity during interactions of HSCs with effector T cells or bound to the cell surface of HSCs to interact with TGFβ1 receptors on T cells. To test this hypothesis, we first stained HSCs for GARP, latent TGFβ1 (LAP), and TGFβ1 using their respective antibodies, then examined the cells under a fluorescence microscope. These experiments revealed that, as demonstrated by previous flow cytometric studies, GARP is present on the surface of HSCs. Interestingly, there was also cytoplasmic staining of GARP inside HSCs. Both the latent TGFβ1 (LAP) and TGFβ1 itself were present on the surface of HSCs, and their signals co-localized with GARP staining, suggesting that both latent TGFβ1 (LAP) and TGFβ1 bind to GARP on HSCs (Fig. 8A, B). Flow cytometric analysis confirmed that majority of the HSCs are double positive for both GARP and LAP proteins (Fig. 8C). In addition, we measured levels of latent TGFβ1 (LAP) by ELISA in the culture supernatants of HSCs with or without the knocked-down GARP. These experiments showed that latent TGFβ1 (LAP) levels in the culture supernatants from HSCs with knocked-down GARP were significantly higher than those from control HSCs, suggesting that GARP is an important anchor of latent TGFβ1 (LAP) on HSCs (Fig. 8D).
Figure 8. Co-localization of latent TGFβ1 (LAP) and TGFβ1 with GARP on HSCs.
HSCs were cultured on chamber slides, and stained with respective antibodies against GARP, LAP and TGF β1. Slides were examined by a fluorescent microscope and representative pictures are taken and overlapped to show the co-localization of LAP and TGFβ1with GARP on HSCs (A, B). HSCs were also double-stained with a PE-labeled anti-GARP mAb (clone F011-5) and an APC labeled anti-LAP mAb (clone TW7-16B4), or respective isotype controls, and cells were analyzed by a flow cytometer (C). In another experiment, LAP levels in culture supernatants conditioned by HSCs with GARP knockdown (transfected with virus 4+5) or by control HSCs (transfected with empty virus) were measured by ELISA. (D). * p<0.05 as calculated by student’s t-test
Discussion
Using TGFβ1+/− HSCs and TGFβ signaling inhibitors, we demonstrated that HSC-derived TGFβ1 is required for HSCs to efficiently inhibit T cells. Using SMAD3−/− T cells, we showed that HSC-derived TGFβ1 acts directly on T cells, not through other autocrine mechanisms to indirectly inhibit T cells. Using RT-PCR, western blot and flow cytometric analysis, we found that GARP is constitutively expressed on both human and mouse primary HSCs. Using GARP-blocking antibodies to neutralize GARP function and GARP-specific shRNAs to knock down GARP expression on HSCs, we demonstrated that GARP is essential to HSCs to efficiently inhibit T cells. In addition, we found from immunofluorescence-staining studies that LAP and TGFβ1 co-localize with GARP on HSCs and that HSCs with knocked-down GARP expression exhibit higher levels of latent TGFβ1 (LAP) in the culture supernatants. These results, taken together, demonstrate that TGFβ1 is another major mechanism by which HSCs inhibit T cells. GARP is constitutively expressed on HSCs, and GARP binds HSC-produced latent TGFβ1, which is important for the activation of the latent TGFβ1 by other mechanisms for the inhibition of T cells. Unfortunately, because systemically knocking out GARP in mice is lethal (http://www2.brc.riken.jp/lab/animal/detail.php?reg_no=RBRC05951), and there is no HSC-specific GARP KO mouse available, we are not able to test these in vitro results in vivo.
Liver is a unique organ that is under chronic challenge from inflammatory processes. Liver is continuously exposed to various bacteria and food antigens, which interact with circulating T cells (26). The constitutive presence of foreign antigens in the liver requires tight control of local immune responses. In addition, liver allografts are spontaneously accepted in mice (27), and in humans, patients who have undergone liver allograft transplantation require significantly lower doses of immunosuppressive drugs to prevent rejection than patients who have received other solid-organ allografts (e.g., kidney or heart), and approximately 20%–40% patients who received liver allografts can gradually be weaned off immunosuppressive drugs without graft rejection (28–31), suggesting an immunoprivileged status of liver. Interestingly, although the whole liver may have an immunoprivileged status, transplanted allogeneic hepatocytes are highly immunogenic, both in rodents and humans (32, 33); this characteristic suggests that nonparenchymal cells are responsible for liver’s immunoprivileged status. Even though the precise mechanisms remain elusive, previous studies suggest a significant role of HSCs in maintaining liver homeostasis by regulating T cells. Our results, showing that HSCs inhibit T cells through GARP-associated activation of the locally produced latent TGFβ1, provide more evidence to support this concept.
Despite the observation that activated HSCs are an important source of TGFβ1 in the liver (8, 15), whether HSC-produced TGFβ1 is involved in HSC-mediated immune regulation has been unclear. In the present study, we found that TGFβ1+/− HSCs, which produce significantly lower levels of TGFβ1 than normal HSCs, also showed markedly impaired T-cell inhibitory function when assessed by both the activated T-cell proliferation and inflammatory cytokine production assays, suggesting that HSC-produced TGFβ1 is important for HSCs to direct inhibit T cells. In addition, we found that using either of two agents – the inhibitor SB4531542 to pharmaceutically inhibit TGFβ1 signaling or SMAD3−/− T cells, in which the TGFβ1 signaling pathway is genetically impaired – leads to significantly reduced T-cell inhibition by HSCs. These results further support the conclusion that TGFβ1 plays a critical role allowing HSCs to directly inhibit T cells and revealed a previously unrecognized mechanism by which HSCs regulate the immune system.
TGFβ1 is secreted by cells in a latent form, and the release of active TGFβ1 from the latent TGFβ1 complex is a critical regulatory step for TGFβ1 function and signaling (34). It has been demonstrated that cells secret latent TGFβ1 in which the active TGFβ1 is associate with LAP(4, 35). GARP was identified as a cell-surface marker and an important receptor for latent TGFβ1 on the surface of Treg cells (1–3). GARP functions by tethering and binding to latent TGFβ1 produced by Tregs, which helps αvβ8 integrin to activate TGFβ1 for Tregs to execute their regulatory activities (36). However, precisely how the Treg produced TGFβ1 inhibit effector T cells is still not clear. It is speculated that the activated TGFβ1 either keeps binding to GARP on the Treg cell surface to engage TGFβ1 receptors on effector T cells or is released in close proximity to the effector T cells during their interaction with Tregs for the inhibition (4). We observed that TGFβ1 and LAP co-localize with GARP on HSCs. We also showed that for HSCs to be able to inhibit T cells requires direct cell-to-cell contact; this finding suggests that the HSC-produced TGFβ1 might inhibit T cells in a similar fashion to that of Tregs.
GARP expression was only reported on platelets and Treg cells (1, 2). Results from our studies clearly showed that GARP is constitutively expressed on HSCs, which indicates that GARP should have a broader distribution other than solely on Tregs and platelets and could be a common mechanism involves in the presentation and activation of latent TGFβ1. Indeed, during the preparation of this article, we noticed a recent report showing that GARP is expressed on mesenchymal stem cells derived from bone marrow and adipose tissue and is a key factor allowing MSCs to inhibit T cells by regulating the bioavailability of TGFβ1 (37).
In contrast to their beneficial role in controlling immune responses in the liver, HSCs have a harmful effect of facilitating liver fibrosis (38). It has been reported that HSCs are an important source of latent TGFβ1 after liver injury and that activated TGFβ1 stimulates HSCs to produce matrix proteins and other enzymes that significantly contribute to liver fibrosis (39). However, the detailed mechanisms by which the HSC-produced latent TGFβ1 is activated are poorly understood. Our results suggest that the GARP-associated activation of latent TGFβ1 on HSCs is important not only for HSC inhibition of T cells, but also could be integrally involved in HSC-augmented liver fibrosis.
In summary, despite the prevailing belief that GARP expresses only on Tregs and platelets, our studies demonstrate that GARP is in fact constitutively expressed on HSCs and upregulated by the inflammatory cytokine IFNγ. GARP is required to anchor and activate HSC-produced latent TGFβ1, which allows HSCs to inhibit T cells. These results showed a wider-than-expected distribution of GARP on cells with immunosuppressive functions; thus our work has uncovered a novel mechanism by which HSCs control immune responses in the liver and has revealed a new pathway by which HSC-produced latent TGFβ1 is presented and activated, which could also be implicated in the pathogenesis of liver fibrosis.
Acknowledgments
This work was supported in part by Muscular Dystrophy Association Research Grant 234458 (FL) and NIH grant AR061564 (FL)
References
- 1.Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM. GARP (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated FOXP3+ regulatory T cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13445–13450. doi: 10.1073/pnas.0901944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang R, Kozhaya L, Mercer F, Khaitan A, Fujii H, Unutmaz D. Expression of GARP selectively identifies activated human FOXP3+ regulatory T cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13439–13444. doi: 10.1073/pnas.0901965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stockis J, Colau D, Coulie PG, Lucas S. Membrane protein GARP is a receptor for latent TGF-beta on the surface of activated human Treg. European journal of immunology. 2009;39:3315–3322. doi: 10.1002/eji.200939684. [DOI] [PubMed] [Google Scholar]
- 4.Wang R, Zhu J, Dong X, Shi M, Lu C, Springer TA. GARP regulates the bioavailability and activation of TGFbeta. Molecular biology of the cell. 2012;23:1129–1139. doi: 10.1091/mbc.E11-12-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards JP, Fujii H, Zhou AX, Creemers J, Unutmaz D, Shevach EM. Regulation of the expression of GARP/latent TGF-beta1 complexes on mouse T cells and their role in regulatory T cell and Th17 differentiation. Journal of immunology. 2013;190:5506–5515. doi: 10.4049/jimmunol.1300199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giampieri MP, Jezequel AM, Orlandi F. The lipocytes in normal human liver. A quantitative study. Digestion. 1981;22:165–169. doi: 10.1159/000198640. [DOI] [PubMed] [Google Scholar]
- 7.Lee UE, Friedman SL. Mechanisms of hepatic fibrogenesis. Best practice & research. Clinical gastroenterology. 2011;25:195–206. doi: 10.1016/j.bpg.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiological reviews. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charles R, Chou HS, Wang L, Fung JJ, Lu L, Qian S. Human hepatic stellate cells inhibit T-cell response through B7-H1 pathway. Transplantation. 2013;96:17–24. doi: 10.1097/TP.0b013e318294caae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou HS, Hsieh CC, Yang HR, Wang L, Arakawa Y, Brown K, Wu Q, Lin F, Peters M, Fung JJ, Lu L, Qian S. Hepatic stellate cells regulate immune response by way of induction of myeloid suppressor cells in mice. Hepatology. 2011;53:1007–1019. doi: 10.1002/hep.24162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang HR, Chou HS, Gu X, Wang L, Brown KE, Fung JJ, Lu L, Qian S. Mechanistic insights into immunomodulation by hepatic stellate cells in mice: a critical role of interferon-gamma signaling. Hepatology. 2009;50:1981–1991. doi: 10.1002/hep.23202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee WC, Yu MC, Chiang YJ, Wang HC, Lu L, Qian S. Liver stellate cells suppress dendritic cells through IL-10. Transplantation proceedings. 2005;37:10–11. doi: 10.1016/j.transproceed.2004.12.277. [DOI] [PubMed] [Google Scholar]
- 13.Yu MC, Chen CH, Liang X, Wang L, Gandhi CR, Fung JJ, Lu L, Qian S. Inhibition of T-cell responses by hepatic stellate cells via B7-H1-mediated T-cell apoptosis in mice. Hepatology. 2004;40:1312–1321. doi: 10.1002/hep.20488. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Tu Z, Qian S, Fung JJ, Markowitz SD, Kusner LL, Kaminski HJ, Lu L, Lin F. Myeloid-derived suppressor cells as a potential therapy for experimental autoimmune myasthenia gravis. Journal of immunology. 2014;193:2127–2134. doi: 10.4049/jimmunol.1400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bissell DM, Wang SS, Jarnagin WR, Roll FJ. Cell-specific expression of transforming growth factor-beta in rat liver. Evidence for autocrine regulation of hepatocyte proliferation. The Journal of clinical investigation. 1995;96:447–455. doi: 10.1172/JCI118055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, Karlsson S. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Richardson JA, Parada LF, Graff JM. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]
- 18.Dong H, Zhu G, Tamada K, Flies DB, van Deursen JM, Chen L. B7-H1 determines accumulation and deletion of intrahepatic CD8(+) T lymphocytes. Immunity. 2004;20:327–336. doi: 10.1016/s1074-7613(04)00050-0. [DOI] [PubMed] [Google Scholar]
- 19.Tahashi Y, Matsuzaki K, Date M, Yoshida K, Furukawa F, Sugano Y, Matsushita M, Himeno Y, Inagaki Y, Inoue K. Differential regulation of TGF-beta signal in hepatic stellate cells between acute and chronic rat liver injury. Hepatology. 2002;35:49–61. doi: 10.1053/jhep.2002.30083. [DOI] [PubMed] [Google Scholar]
- 20.Weng H, Mertens PR, Gressner AM, Dooley S. IFN-gamma abrogates profibrogenic TGF-beta signaling in liver by targeting expression of inhibitory and receptor Smads. Journal of hepatology. 2007;46:295–303. doi: 10.1016/j.jhep.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 21.Von Pfeil A, Hakenjos L, Herskind C, Dittmann K, Weller M, Rodemann HP. Irradiated homozygous TGF-beta1 knockout fibroblasts show enhanced clonogenic survival as compared with TGF-beta1 wild-type fibroblasts. International journal of radiation biology. 2002;78:331–339. doi: 10.1080/095530002753676200. [DOI] [PubMed] [Google Scholar]
- 22.Krause TJ, Katz D, Wheeler CJ, Ebner S, McKinnon RD. Increased levels of surgical adhesions in TGFbeta1 heterozygous mice. Journal of investigative surgery : the official journal of the Academy of Surgical Research. 1999;12:31–38. doi: 10.1080/089419399272746. [DOI] [PubMed] [Google Scholar]
- 23.Brooks WW, Conrad CH. Myocardial fibrosis in transforming growth factor beta(1)heterozygous mice. Journal of molecular and cellular cardiology. 2000;32:187–195. doi: 10.1006/jmcc.1999.1065. [DOI] [PubMed] [Google Scholar]
- 24.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Molecular pharmacology. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 25.Gu X, Wang Y, Xiang J, Chen Z, Wang L, Lu L, Qian S. Interferon-gamma triggers hepatic stellate cell-mediated immune regulation through MEK/ERK signaling pathway. Clinical & developmental immunology. 2013;2013:389807. doi: 10.1155/2013/389807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crispe IN, Giannandrea M, Klein I, John B, Sampson B, Wuensch S. Cellular and molecular mechanisms of liver tolerance. Immunological reviews. 2006;213:101–118. doi: 10.1111/j.1600-065X.2006.00435.x. [DOI] [PubMed] [Google Scholar]
- 27.Qian S, Demetris AJ, Murase N, Rao AS, Fung JJ, Starzl TE. Murine liver allograft transplantation: tolerance and donor cell chimerism. Hepatology. 1994;19:916–924. doi: 10.1002/hep.1840190418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazariegos GV, Reyes J, Marino IR, Demetris AJ, Flynn B, Irish W, McMichael J, Fung JJ, Starzl TE. Weaning of immunosuppression in liver transplant recipients. Transplantation. 1997;63:243–249. doi: 10.1097/00007890-199701270-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devlin J, Doherty D, Thomson L, Wong T, Donaldson P, Portmann B, Williams R. Defining the outcome of immunosuppression withdrawal after liver transplantation. Hepatology. 1998;27:926–933. doi: 10.1002/hep.510270406. [DOI] [PubMed] [Google Scholar]
- 30.Takatsuki M, Uemoto S, Inomata Y, Egawa H, Kiuchi T, Fujita S, Hayashi M, Kanematsu T, Tanaka K. Weaning of immunosuppression in living donor liver transplant recipients. Transplantation. 2001;72:449–454. doi: 10.1097/00007890-200108150-00016. [DOI] [PubMed] [Google Scholar]
- 31.de la Garza RG, Sarobe P, Merino J, Lasarte JJ, D'Avola D, Belsue V, Delgado JA, Silva L, Inarrairaegui M, Sangro B, Sola JJ, Pardo F, Quiroga J, Herrero JI. Trial of complete weaning from immunosuppression for liver transplant recipients: factors predictive of tolerance. Liver transplantation : official publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society. 2013;19:937–944. doi: 10.1002/lt.23686. [DOI] [PubMed] [Google Scholar]
- 32.Dhawan A, Puppi J, Hughes RD, Mitry RR. Human hepatocyte transplantation: current experience and future challenges. Nature reviews. Gastroenterology & hepatology. 2010;7:288–298. doi: 10.1038/nrgastro.2010.44. [DOI] [PubMed] [Google Scholar]
- 33.Bumgardner GL, Orosz CG. Unusual patterns of alloimmunity evoked by allogeneic liver parenchymal cells. Immunological reviews. 2000;174:260–279. doi: 10.1034/j.1600-0528.2002.017409.x. [DOI] [PubMed] [Google Scholar]
- 34.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. Journal of cell science. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 35.Massague J, Chen YG. Controlling TGF-beta signaling. Genes & development. 2000;14:627–644. [PubMed] [Google Scholar]
- 36.Edwards JP, Thornton AM, Shevach EM. Release of Active TGF-beta1 from the Latent TGF-beta1/GARP Complex on T Regulatory Cells Is Mediated by Integrin beta8. Journal of immunology. 2014;193:2843–2849. doi: 10.4049/jimmunol.1401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrillo-Galvez AB, Cobo M, Cuevas-Ocana S, Gutierrez-Guerrero A, Sanchez-Gilabert A, Bongarzone P, Garcia-Perez A, Munoz P, Benabdellah K, Toscano MG, Martin F, Anderson P. Mesenchymal stromal cells express GARP/LRRC32 on their surface: effects on their biology and immunomodulatory capacity. Stem cells. 2015;33:183–195. doi: 10.1002/stem.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nature communications. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bataller R, Brenner DA. Hepatic stellate cells as a target for the treatment of liver fibrosis. Seminars in liver disease. 2001;21:437–451. doi: 10.1055/s-2001-17558. [DOI] [PubMed] [Google Scholar]