Abstract
Depression symptoms following immune response to a challenge have been reported after the recovery from sickness. A RNA-Seq study of the dysregulation of the microglia transcriptome in a model of inflammation-associated depressive behavior was undertaken. The transcriptome of microglia from mice at day 7 after Bacille Calmette Guérin (BCG) challenge was compared to that from unchallenged Control mice and to the transcriptome from peripheral macrophages from the same mice. Among the 562 and 3,851 genes differentially expressed between BCG-challenged and Control mice in microglia and macrophages respectively, 353 genes overlapped between these cells types. Among the most differentially expressed genes in the microglia, serum amyloid A3 (Saa3) and cell adhesion molecule 3 (Cadm3) were over-expressed and coiled-coil domain containing 162 (Ccdc162) and titin-cap (Tcap) were under-expressed in BCG-challenged relative to Control. Many of the differentially expressed genes between BCG-challenged and Control mice were associated with neurological disorders encompassing depression symptoms. Across cell types, S100 calcium binding protein A9 (S100A9), interleukin 1 beta (Il1b) and kynurenine 3-monooxygenase (Kmo) were differentially expressed between challenged and control mice. Immune response, chemotaxis, and chemokine activity were among the functional categories enriched by the differentially expressed genes. Functional categories enriched among the 9,117 genes differentially expressed between cell types included leukocyte regulation and activation, chemokine and cytokine activities, MAP kinase activity, and apoptosis. More than 200 genes exhibited alternative splicing events between cell types including WNK lysine deficient protein kinase 1 (Wnk1) and microtubule-actin crosslinking factor 1(Macf1). Network visualization revealed the capability of microglia to exhibit transcriptome dysregulation in response to immune challenge still after resolution of sickness symptoms, albeit lower than that observed in macrophages. The persistent transcriptome dysregulation in the microglia shared patterns with neurological disorders indicating that the associated persistent depressive symptoms share a common transcriptome basis.
Introduction
Studies of behavioral and molecular changes in response to a challenge have exposed the relationship between brain inflammation and incidence of depression-like symptoms [1,2]. Peripheral infections can alter inflammatory cytokines elicited by microglia, the innate immune cells located in the brain. This alteration of cell signaling dysregulates pathways such as tryptophan metabolism that has been associated with depression-like behaviors. After peripheral challenge with Bacille Calmette-Guérin (BCG) mice display depressive-like behaviors 7 days to 1 month post challenge, well-past sickness recovery. Mice challenged with BCG exhibit sickness symptoms reflected by weight loss early in the first 2 days after challenge compared to control mice challenged with saline followed by recovery of weight by day 5. Recovery from sickness was confirmed by non-significant differences in horizontal locomotor activity and rearing at day 6 post challenge. Despite the recovery from sickness symptoms, depression-like behaviors including significant increase in the duration of immobility measured using the tail suspension test and the Porsolt forced swim test at day 6 and decrease in sucrose ingestion in the sucrose preference test at day 7 were recorded in mice challenged with BCG relative to control mice [3–5].
Brain microglia and peripheral macrophages are immune cells yet their response to immune challenge and impact on surrounding cells are different [6]. Transcriptome analysis have revealed common and unique profiles among these cell types [7]. This suggest that differences between the microglia and macrophage transcriptome could be directly associated with depression-like symptoms. Characterization of the differences between microglia and macrophage transcript isoform abundance, alternative splicing, gene differential expression, and networks after sickness recovery from BCG challenge is essential to understand the role of microglia on depression-like behaviors.
The objective of this study was to uncover the gene expression dysregulation in microglia from mice challenged with BCG that exhibit depression-like symptoms despite having recovered from the associated sickness. This work builds upon our prior study that confirmed in the same mouse populations comparable changes in body weights and other sickness indicators but significant differences in depression-like behaviors between BCG-challenged and Control groups at day 7 post-challenge [5]. Analyses supporting the objective of the present study include: 1) uncovering differential gene expression and functional categories between BCG-challenged and Control groups within cell types; 2) uncovering differential gene expression and functional categories between microglia and peripheral macrophages within BCG-challenged group; 3) detection of alternative splicing between microglia and peripheral macrophages in the BCG-challenged group; and 4) network visualization to uncover potential synergistic or antagonistic relationships between BCG-challenge and cell type groups.
Materials and Methods
Experiments
All animal care and experimental procedures adhered to NIH guidelines and were approved by the University of Illinois Institutional Animal Care and Use Committee. Measures were taken to minimize the number of animals used and the pain and suffering of the mice.
Microglia and peritoneal macrophages (hereby termed macrophages) were collected from approximately 22 weeks old male C57Bl/6J mice. Mice housing, management, and BCG challenge followed previously described protocols [5,8]. Briefly, mice were housed individually in standard polypropylene cages including corn cob litter. Housing was under a 12:12 h reversed light/dark cycle (lights on 10:00 PM-10:00 AM) with controlled environmental temperature (23°C) and humidity (45%). Mice were offered water and food (Teklad 8640 chow, Harlan Laboratories, Indianapolis, IN, USA) ad libitum and handled daily for one week prior to the trial to ensure adaptation. Mice were acclimated to the light cycle and facility for at least 3 weeks prior to the injection with BCG or saline. All mice were individually handled for a few minutes daily prior to the challenge.
The immune challenge involved live attenuated mycobacteria TICE BCG (50 mg wet weight of lyophilized culture containing 1x108 colony forming units or CFU/vial; Organon Teknika Corp. LLC, USA Inc., USA). Each vial’s reconstitution prior to inoculation followed provider instructions using preservative-free saline. No peritoneal mycobacteria infection was reported in mice by day 20 after intraperitoneal infection with BCG; whereas dissemination to other organs, including the spleen, bone marrow, and lung was observed [9]. There are no reports of BCG infection of microglia or brain and the peritoneum is a source of non-infected macrophages rather than macrophages from other tissues that could be infected with BCG.
Individual mice were challenged once with either 10 mg/mouse (BCG-challenged group, n = 12) or sterile saline solution (Control group, n = 12) administered via intraperitoneal injection [5]. No mouse became severely ill or died at any time prior to the experimental endpoint. Mice were euthanized by CO2 asphyxiation by trained personnel 7 days after BCG challenge and all efforts were made to minimize suffering. The endpoint was selected based on prior work that demonstrated the recovery from sickness yet persistence of depressive-like symptoms 7 days after challenge [5].
Macrophages were collected from peritoneal tissue using the proven protocols [8,10]. Abdomens were disinfected, skin was retracted, and the peritoneal cavity was flushed with Hank's Balanced Salt Solution (cold harvest medium). The peritoneal fluid was centrifuged and the resulting cell pellet was resuspended and plated. The medium was aspired after 2 h incubation to remove non-adherent cells. The surviving adherent cells constituted the non-thioglycollate elicited peritoneal macrophages and cells from individual mice were stored in Trizol at -80°C until RNA extraction [11].
After peritoneum collection (~5 minutes), mice were perfused, and the brains were excised and minced. Microglia were collected following established protocols [12]. In brief, brains were trypsinized [13], dissociated using cell screens (40 μm), centrifuged, resuspended in 30% Percoll (GE Healthcare, Princeton, NJ), then centrifuged for myelin removal. Brain cells were labeled with anti-CD11b (integrin alpha M antibody) magnetized Miltenyi MicroBeads (Miltenyi Biotec, Germany). Cells were separated in a magnetic field with MS columns (Miltenyi Biotec, Germany). The resulting Cd11b+ fraction collected was centrifuged, resuspended, and stored at -80°C. Flow cytometry validation of cell isolation encompassed cell staining with primary fluorescent antibodies for two primary markers for macrophages and microglia: CD11b and CD45 (protein tyrosine phosphatase, receptor type, C antibody) [14,15]. Fc receptors were blocked by incubation with anti-CD16/CD32 antibody before incubation with eBioscience anti-CD11b and anti-CD45 antibodies (eBioscience Inc., San Diego, CA). Surface receptor expression was identified using a Biosciences LSR II Flow Cytometry Analyzer with BD FACSDiva software (BD Biosciences, San Jose, CA). Antibody gating was determined using isotype-stained controls. Cells were ~93% Cd11b+ and ~91% Cd45+, confirming microglia enrichment.
RNA extraction from microglia followed the Tripsin method using a total RNA Kit (Omega Biotek, Norcross, GA) and a DNase step to remove DNA contamination [11]. The Agilent 2100 Bioanalyzer with RNA Pico chip (Agilent Technologies, Palo Alto, CA) was used to assess RNA Integrity Numbers. RNA Integrity Numbers were > 9 in 90% of the samples and > 7 in 100% of the 48 samples.
Identification of differential transcript isoform and gene expression
RNA libraries from individual mouse were sequenced using an Illumina HiSeq 2000 (Illumina, San Diego, CA) and 100nt long paired-end reads were obtained. Read quality control was implemented using FastQC [8,16]. Quality control analysis indicated that the Phred nucleotide quality score was > 30 across the length of the reads such that the read sequences were not trimmed.
Reads were mapped to the Genome Reference Consortium GRCm38 mouse assembly using Tophat2 (v 2.0.8) [17] with the Illumina iGenomes package (mm10; http://support.illumina.com/sequencing/sequencing_software/igenome.html). Reads were combined into transcripts and differential expression was tested using Cufflinks (v2.1.1) [18]. The specifications beyond the default settings used to obtain transcript abundance levels were: a) Upper Quartile Normalization was used to normalize the median transcript counts across libraries; b) multi-read correction for reads mapping to multiple sites; and c) fragment bias correction [19]. Four comparisons were evaluated: two pairwise contrasts between BGG-challenge groups within cell types, and two pairwise contrasts between cells within BCG-challenge groups. Results from genes with at least 10 mapped reads were considered. Multiple test adjustment used the Benjamini-Hoechberg false discovery rate (FDR) approach [20,21]. Genes exhibiting an FDR-adjusted P-value < 0.05 were considered differentially expressed.
Identification of alternative splicing events
Alternative splicing events characterizing differences between groups (i.e., cell types or challenge level) were identified using a quantitative approach [22]. Quantitative characterization considered genes detected in both groups, represented by at least two transcript isoforms, and with at least one transcript isoform differentially expressed between groups (FDR-adjusted P-value < 0.05) between groups in one direction (over- or under-expressed) and the rest of the isoforms not differentially expressed or differentially expressed in the other direction.
Identification of functional categories over-represented among gene profiles
Functional analyses of the transcript isoforms differentially expressed between BCG-challenge groups within cell types, between cell types within BCG-challenged groups, and of transcript isoforms expressed exclusively in one cell type were performed using hypergeometric testing and Gene Set Enrichment Analysis (GSEA). These analyses allowed the identification of Gene Ontology (http://www.geneontology.org/) biological processes, molecular functions, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (http://www.genome.jp/org/). The hypergeometric test was implemented in the Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov) [23,24]. Gene Ontology (GO) results were reported using the DAVID Functional Annotation Tool (FAT) classes to facilitate interpretation. Category enrichment was measured using Expression Analysis Systematic Explorer (EASE) scores computed based on a one-tailed jackknifed Fisher exact test [25,26]. The GO categories were clustered and the statistical significance of each cluster was assessed using an Enrichment Score (-log10 geometric mean of the cluster members EASE scores [27,28]. Functional annotation clusters were considered significant at Enrichment Score > 2 (comparable to P-value < 0.001) using the Mus musculus genome as background. Gene Set Enrichment Analysis was implemented using the GSEA-P software package [29,30]. This approach offered functional insights complementary to DAVID based on the consideration of the expression profile of all the genes analyzed and annotations to the Molecular Signature Database (MSigDB) [31].
Gene network visualization
Interpretation of findings and discovery of differences in gene co-expression associated with BCG challenge or cell type were enhanced using network visualization. Networks were visualized using the BisoGenet plug-in [27,28,32] within the Cytoscape environment [24,33]. BisoGenet enabled the visualization of associations between genes detected in the present study using information from the SysBiomics repository. Networks depicting genes as nodes and gene associations as edges and including at most one non-measured gene connecting observed genes were considered. The resulting gene networks were augmented with information on differential gene expression results from the comparison between BCG-challenged and cell type groups obtained in this study. The node size represented the differential expression P-value and the node color denoted over- or under-expression between challenge levels or cell types.
Results and Discussion
Global transcriptome profiles across immune challenge groups and cell types
Among more than 15,000 genes tested, 562 and 3,851 were differentially expressed (FDR-adjusted P-value < 0.05) between the BCG-challenged and Control groups in microglia and macrophages, respectively (Table 1). Differential abundance of transcript isoforms between BCG-challenged groups followed similar patterns albeit at lower absolute count number. Significant gene expression differences between cell types were more prevalent and extreme than between BCG-challenged groups. Approximately 9,117 genes (9,780 transcript isoforms) were differentially expressed between microglia and macrophages in the BCG-challenged group. Among these, 6,590 genes (8,144 transcripts) overlapped between cell types across BCG-challenged groups. The prevalent differential expression between cell types is consistent with prior transcriptomic comparisons and has been linked to differences in cell type origin and local environment [7,34,35].
Table 1. Number of genes and transcript isoforms analyzed and differentially expressed between BCG-challenged (BCG) and Control mice within cell type and between microglia and macrophages within challenge group and overlapping counts.
| Comparison | Genes | Transcript isoforms | ||||
|---|---|---|---|---|---|---|
| Analyzed | DE1 or Unique Annotated | DE or Unique Not annotated | Analyzed | DE or Unique Annotated | DE or Unique Not annotated | |
| Microglia Control vs. BCG | 15,293 | 562 | 18 | 39,005 | 317 | 6 |
| Macrophages Control vs. BCG | 13,245 | 3,851 | 286 | 32,743 | 2,381 | 160 |
| Overlap | 353 | 245 | ||||
| Control Microglia vs Macrophages | 15,287 | 9,860 | 0 | 29,110 | 11,113 | 0 |
| BCG Microglia vs. Macrophages | 15,828 | 9,117 | 890 | 42,035 | 9,780 | 735 |
| Overlap | 6,590 | 8,144 | ||||
| Unique to Macrophages Control | 23 | 42 | ||||
| Unique to Microglia Control | 236 | 421 | ||||
| Unique to Macrophages BCG | 3 | 20 | ||||
| Unique to Microglia BCG | 214 | 300 | ||||
DE1: differentially expressed genes or transcript isoforms (FDR-adjusted P-value < 0.05) or uniquely detected in either cell type.
Differences in the microglia transcriptome between BCG-challenged and Control mice
Differential gene and transcript isoform expression in microglia between BCG-challenged and Control mice are presented in Table 1. Among the differentially expressed genes in the microglia, 518 genes were over-expressed whereas only 44 genes were under-expressed in the BCG-challenged relative to Control mice. The predominance of over-expressed genes (92%) in response to BCG-challenge is consistent with previous reports of transcriptome changes in the microglia in response to lipopolysaccharide (LPS) challenge [36]. Differentially expressed genes between BCG-challenged and Control in microglia ((|log2(fold change)| > 3, FDR-adjusted P-value < 2.0 x 10−3) are listed in Table 2 together with supporting literature. An extended list of differentially expressed genes in microglia is provided in S1 Table.
Table 2. Most extreme differentially expressed genes between BCG-challenged and Control mice within microglia (|log2(fold change)| > 3, FDR-adjusted P-value < 2.0 x 10−3) and macrophages (|log2(fold change)| > 6, FDR-adjusted P-value < 3.0 x 10−4) and supporting literature review.
| Gene Symbol | Gene name NCBI | Log2(BCG/ Control) | References |
|---|---|---|---|
| Microglia; BCG-challenged vs Control mice | |||
| Saa3 | serum amyloid A 3 | 5.90 | [37–40] |
| Cadm3 | cell adhesion molecule 3 | 5.14 | [37] |
| Steap4 | STEAP family member 4 | 4.89 | [41,42] |
| Sele | selectin E | 4.80 | [43,44] |
| Cxcr1 | chemokine (C-X-C Motif) receptor 1 | 4.03 | [39,45,46] |
| Ifitm1 | interferon induced transmembrane protein 1 | 3.72 | [38,39,44,47] |
| Irg1 | immunoresponsive 1 homolog: involved in the inhibition of the inflammatory response. | 3.66 | [46,48,49] |
| Lrg1 | leucine-rich alpha-2-glycoprotein 1 | 3.57 | [40,50] |
| Prok2 | prokineticin 2 | 3.48 | [50] |
| Cfb | complement factor B | 3.39 | [38] |
| Slfn4 | schlafen 4 | 3.33 | [40,48] |
| Nxpe5 | neurexophilin and PC-esterase domain family, member 5 | 3.29 | [51] |
| Ly6i | lymphocyte antigen 6 complex, locus I | 3.29 | [38,49] |
| Oas3 | 2'-5'-oligoadenylate synthetase 3, 100kDa | 3.27 | [38,52] |
| Ifi205 | interferon activated gene 205 | 3.15 | [38,53] |
| Stfa1 | stefin A1 | 3.12 | [54] |
| Plac8 | placenta-specific 8 | 3.09 | [38,44] |
| Macrophages; BCG-challenged vs Control mice | |||
| S100a9 | S100 calcium binding protein A9 (calgranulin B) | 10.11 | [55] |
| Mrgpra2a | MAS-related GPR, member A2A | 10.10 | |
| Ly6i | lymphocyte antigen 6 complex, locus I | 9.42 | |
| Asprv1 | aspartic peptidase, retroviral-like 1 | 9.15 | |
| Ly6c2 | lymphocyte antigen 6 complex, locus C2 | 7.92 | [56] |
| Nos2 | nitric oxide synthase 2, inducible | 7.69 | [57] |
| Il1f9 | interleukin 1 family, member 9 | 7.66 | [58] |
| Ccl8 | chemokine (C-C motif) ligand 8 | 7.45 | [41] |
| 2010002M12Rik | interferon induced protein with tetratricopeptide repeats 1B like 2 | 7.32 | |
| Spon1 | spondin 1, (f-spondin) extracellular matrix protein | 7.23 | [59] |
| S100a8 | S100 calcium binding protein A8 (calgranulin A) | 7.15 | [60] |
| Cxcr2 | chemokine (C-X-C motif) receptor 2 | 7.02 | |
| Ifng | interferon gamma | 6.45 | [61] |
| Cxcl9 | chemokine (C-X-C motif) ligand 9 | 6.40 | |
| Gpr141 | G protein-coupled receptor 141 | 6.15 | |
| 2010005H15Rik | RIKEN cDNA 2010005H15 gene | 6.14 | |
| Iigp1 | interferon inducible GTPase 1 | 6.08 | |
Studies of behavioral and molecular changes in response to BCG challenge have exposed the relationship between brain inflammation and incidence of depression-like symptoms [1,2]. These studies have reported increased levels of inflammatory cytokines expression in the microglia. Similarly, the present study detected over-expression of genes related to inflammatory response 7 days after challenge (Table 2, S1 Table) including Serum amyloid A3 (Saa3), S100 calcium binding protein A8 (S100a8), S100 calcium binding protein A9 (S100a9), Fc-gamma receptors (Fcgr4 and Fcgr2b), prostaglandin-endoperoxide synthase 2 (Ptgs2), interleukin 1beta (Il1b), and interleukins receptors (Il1r1 and Il4ra).
Consistent with our findings, an immune challenge event can elicit over-expression of Saa3 in the brain. Also, the levels of Saa3 in microglia increase in response to mouse hepatitis virus-JHM [62] and a strain of Creutzfeldt-Jakob disease [63]. The expression of Saa3 could be induced by S100A8 and S100A9 and these were over-expressed as well. Altered expression of S100A9 has been reported in neurological diseases associated with inflammation and depression such as cerebral ischemia, traumatic brain injury, and Alzheimer’s disease [64,65].
Fc receptors are expressed in immune cells and connect humoral and cell-mediated response to pathogen infection [66]. Polymorphism in these receptors were associated with susceptibility to Guillain-Barré syndrome and multiple sclerosis, disorders that exhibit depression symptoms [67]. Increased levels of Fcgr4 and Fcgr2b expression, similar to that observed in this study, induce vascular damage and exacerbate neurodegenerative conditions in humans [68]. Activation of microglia through Fc gamma receptors results in phagocytosis and polarization to an M2b phenotype [68] and similarly, Fc receptor overexpression was correlated with over-expressed pro-inflammatory cytokines in the present and previous studies [69,70]. Similarly, Ptgs2 was over-expressed in the microglia in our study and has been related to inflammatory response in the brain and to neurodegeneration processes [71]. This gene has been proposed as a therapeutic target for neurodegenerative diseases that encompass depressive behaviors such as Parkinson, Alzheimer, and Huntington’s disease [72].
Over-expression of pro-inflammatory cytokines such as interleukin Il1b has been linked to neurodegenerative disorders such as Alzheimer’s disease [65,73]. Il1b has been linked to depressive behaviors [74,75] and therapies blocking Il1b expression in a mouse model of Alzheimer’s disease decreased the synthesis of S100 proteins, decreased fibrillar deposition, and protected mice from cognitive deficits [76]. Our transcriptome findings in the microglia are consistent with reports that mice express depression-like behaviors 7 days after of inoculation with BCG [5,77].
Differences in the macrophage transcriptome between BCG-challenged and Control mice
The comparison of gene expression profiles in macrophages between BCG-challenged and Control mice enabled us to uncover changes common to macrophage and microglia immune cells and changes unique to microglia cells. Differential gene and transcript isoform expression between BCG-challenged and Control mice in macrophages are listed in Table 1. Among the 3,851 differentially expressed genes in macrophages, 2,151 were over-expressed and 1,700 were under-expressed in the BCG-challenged relative to Control mice. The predominance of over-expressed genes (75%) in response to BCG-challenge is consistent with previous reports of transcriptome changes in the macrophages in response to BCG challenge [78] and consistent with the microglia changes previously described. Among these genes S100A9, S100A8, Il1b, interferon gamma (Ifng), nitric oxide synthase 2 inducible (Nos2), and interleukin 1 family member 9 (Il1f9) were identified. Like in microglia, many differentially expressed genes in macrophages were related with inflammatory response. Differentially expressed genes between BCG-challenged and Control in macrophages (|log2(fold change)| > 6, FDR-adjusted P-value < 3.0 x 10−4) are listed in Table 2 together with supporting literature. An extended list of differentially expressed genes in macrophages is provided in S1 Table.
The increase in expression of Ifng and Nos2 is a component in the macrophage response to infectious or inflammatory diseases represented in our study by the BCG challenge [61,79]. High levels of Ifng were found in patients diagnosed with major depression without pathogen stimuli [80–84]. Additionally, an increase in Ifng expression up-regulates Caspase 1 (Casp1), an enzyme that converts pro-Il1b into active mature Il1b [85] and that has a role in acute [86] and major depression [74,75]. Our results corroborated that the up-regulation of Ifng and Il1b (Table 2, S1 Table) is correlated with the development of depression-like behavior in BCG-challenged mice [5,77].
Transcriptome differences between BCG-challenged and Control mice shared between cell types
Among the genes differentially expressed between BCG-challenged and Control mice, 353 genes overlapped between microglia and macrophages (Table 1). Shared profiles were identified in genes belonging to families involved in immune response including: S100A8, S100A9, Il1r1, and Il1b. Kynurenine 3-monooxygenase (Kmo) was over-expressed (FDR-adjusted P-value < 0.05) in BCG-challenged relative to Control mice, both in microglia and macrophages (S1 Table). Kmo catalyzes the conversion of kynurenine to 3-hydroxykynurenine and modulation of KMO activity has been implicated in several neurodegenerative diseases [87]. Over-expression of Il1b and Kmo was observed in the microglia of mice that also exhibited social withdrawal after LPS injection [88]. Also, over-expression of Kmo was reported in the rat whole brain after a systemic challenge with LPS [89,90] and in human brain cells after Il1b treatment [91]. On the other hand, a study of mouse whole brain after BCG-challenge reported over-expression of indoleamine 2,3-dioxygenase 1 (Ido1) but no differential expression of Kmo [92]. Both, Kmo and Ido1 reduce the level of circulating tryptophan and production of serotonin and increase the levels of tryptophan metabolites that have cytotoxic effect [1,2]. In the present study, Kmo was over-expressed but Ido1 and the ortholog Ido2 were not differentially expressed in the microglia between BCG-challenged and Control mice. Our study supports the emerging notion that Kmo could be induced by immune challenge [93,94] and, as potential target of inflammatory cytokines in the kynurenine pathway, Kmo could be associated with behavioral disorders.
Functional analysis of microglia and macrophages transcriptomic differences between BCG-challenged and Control mice
Analysis of enriched functional categories based on the comparison of genes expression profiles between BCG-challenged and Control mice in microglia and macrophages offered insights into the biological processes and pathways associated with BCG-challenge. Table 3 summarizes the clusters of functional categories surpassing a DAVID Enrichment Score > 4. S2 Table presents the complete list of categories that exhibited an Enrichment Score > 2 and corresponding gene count.
Table 3. Most significant clusters (DAVID Enrichment Score ES > 4) of enriched Gene Ontology (GO) biological processes (BP) and molecular functions (MF) among the genes differentially abundant between BCG-challenged and Control mice within cell type.
| Cell type and cluster1 | Terms2 | ES |
|---|---|---|
| Microglia | ||
| BP | ~defense response ~inflammatory response ~response to wounding | 12.96 |
| BP | ~chemotaxis ~taxis ~locomotory behavior | 5.63 |
| MF | ~carbohydrate binding ~polysaccharide binding ~pattern binding | 5.12 |
| MF | ~peptidase inhibitor activity ~endopeptidase inhibitor activity ~enzyme inhibitor activity | 4.88 |
| BP | ~immune effector process ~adaptive immune response ~adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 4.75 |
| BP | ~cell adhesion ~biological adhesion ~cell-cell adhesion | 4.37 |
| Macrophages | ||
| BP | ~defense response ~response to wounding ~inflammatory response | 13.32 |
| MF | ~carbohydrate binding ~polysaccharide binding ~pattern binding | 8.18 |
| BP,MF | ~chemotaxis ~taxis ~chemokine activity | 7.79 |
| BP | ~cell activation ~leukocyte activation ~lymphocyte activation | 6.17 |
| BP | ~vasculature development ~blood vessel development ~blood vessel morphogenesis | 6.12 |
| BP | ~regulation of cytokine production ~positive regulation of cytokine production ~positive regulation of multicellular organismal process | 5.97 |
| BP | ~positive regulation of immune system process ~positive regulation of response to stimulus ~positive regulation of immune response | 5.86 |
| BP | ~immune effector process ~leukocyte mediated immunity ~adaptive immune response | 5.02 |
| BP | ~apoptosis ~programmed cell death ~cell death | 4.77 |
| BP | ~regulation of phosphorylation ~regulation of phosphorus metabolic process ~regulation of phosphate metabolic process | 4.34 |
| BP,MF | ~GTPase regulator activity ~nucleoside-triphosphatase regulator activity ~GTPase activator activity | 4.20 |
| BP | ~positive regulation of immune system process ~regulation of leukocyte activation ~regulation of cell activation | 4.01 |
1 Each row corresponds to a cluster of Functional Annotation Tool (FAT) GO categories.
2 The three GO terms exhibiting most significant enrichment P-value in each cluster are listed, separated by “~”. Additional information in each cluster is provided in S2 Table.
Enriched functional categories shared by both cell types included immunological response, chemotaxis, cell migration and apoptosis among others biological processes and molecular functions (Table 3, S2 Table). These clusters with highest enrichment overlap between microglia and macrophages, corroborating the persistence of systemic response 7 days after BCG-challenge. The enrichment of apoptosis-related processes observed in this and previous studies [95] is associated with the over-expression of Nos2 in both cell types (Table 2). Likewise, the enrichment of monocyte and leukocyte migration processes (Table 3) reported here and by others [96] is in correspondence with the over-expression of S100A9 and S100A8 (Table 2) in both cell types. Higher leukocyte migration, activity of the pro-inflammatory cytokines [97], and apoptosis [98] were all observed in patients with major depression disorders. Additionally, the observed enrichment of protein metabolism including inhibition of enzymes and peptidases is compatible to changes in protein metabolism and downregulation of proteolysis reported in patients with bipolar depression [99,100] and schizophrenia [101].
Complementary functional analysis of the microglia and macrophages profiles using GSEA (Tables 4 and 5, respectively) confirmed the previous gene list enrichment and further enabled the discrimination of enrichment among over- and under-expressed genes in BCG-challenged relative to Control mice. S3 and S4 Tables include additional categories enriched at P-value < 0.05 and with a minimum of 10 genes in microglia and macrophages, respectively. A remarkable GSEA finding in microglia was the downregulation of inositol metabolism because this result is consistent with changes in this metabolite in schizophrenia, bipolar, and depression disorders [101,102].
Table 4. Gene Set Enrichment Analysis (GSEA) categories enriched among transcript isoforms over-expressed (FDR-adjusted P-value < 5.0 x 10−4 > 10 transcript isoforms) and under-expressed (P-value < 0.05 > 10 transcript isoforms) in BCG-challenged relative to Control mice in microglia.
| Categories | NG1 | P-value | FDR 2 |
|---|---|---|---|
| Over-expressed in BCG-challenged vs Control mice | |||
| Defense response | 147 | <0.1E-05 | <0.1E-05 |
| Inflammatory response | 79 | <0.1E-05 | <0.1E-05 |
| Response to other organism | 41 | <0.1E-05 | <0.1E-05 |
| Immune response | 152 | <0.1E-05 | <0.1E-05 |
| Response to virus | 31 | <0.1E-05 | <0.1E-05 |
| Response to external stimulus | 183 | <0.1E-05 | <0.1E-05 |
| Immune system process | 219 | <0.1E-05 | <0.1E-05 |
| Response to wounding | 113 | <0.1E-05 | <0.1E-05 |
| KEGG cytokine cytokine receptor interaction | 146 | <0.1E-05 | <0.1E-05 |
| Multi organism process | 77 | <0.1E-05 | <0.1E-05 |
| Response to biotic stimulus | 69 | <0.1E-05 | <0.1E-05 |
| Locomotory behavior | 57 | <0.1E-05 | 7.98E-05 |
| Cation homeostasis | 58 | <0.1E-05 | 8.65E-05 |
| Cellular cation homeostasis | 56 | <0.1E-05 | 1.49E-04 |
| G protein coupled receptor binding | 30 | <0.1E-05 | 2.80E-04 |
| Ion homeostasis | 65 | <0.1E-05 | 3.30E-04 |
| Under-expressed in BCG-challenged vs Control mice | |||
| Nervous system development | 228 | <0.1E-05 | 1.00E+00 |
| Polysaccharide binding | 19 | 2.85E-03 | 7.91E-01 |
| Glycosaminoglycan binding | 18 | 5.46E-03 | 5.51E-01 |
| KEGG inositol phosphate metabolism | 42 | 8.10E-03 | 7.12E-01 |
| Central nervous system development | 73 | 1.24E-02 | 9.92E-01 |
| Protein secretion | 16 | 1.43E-02 | 6.71E-01 |
| Transmission of nerve impulse | 85 | 2.27E-02 | 6.72E-01 |
| KEGG Parkinson’s disease | 94 | 3.03E-02 | 7.31E-01 |
| Cellular protein complex assembly | 30 | 4.28E-02 | 1.00E+00 |
| Synaptic transmission | 75 | 4.76E-02 | 8.86E-01 |
1 NG: number of genes.
2 FDR: adjusted P-value.
Table 5. Gene Set Enrichment Analysis (GSEA) categories enriched among transcript isoforms over-expressed (FDR-adjusted P-value < 1.5 x 10−2 > 10 transcript isoforms) and under- expressed (FDR-adjusted P-value < 5 x 10−4 > 10 transcript isoforms) in BCG-challenged relative to Control mice in macrophages.
| Categories | NG1 | P-value | FDR2 |
|---|---|---|---|
| Over-expressed in BCG-challenged relative to Control mice | |||
| KEGG ECM receptor interaction | 45 | <0.1E-05 | 9.96E-04 |
| Defense response | 144 | <0.1E-05 | 5.88E-03 |
| Inflammatory response | 73 | <0.1E-05 | 1.28E-02 |
| Serine type peptidase activity | 18 | <0.1E-05 | 1.31E-02 |
| Serine hydrolase activity | 19 | <0.1E-05 | 1.32E-02 |
| KEGG proteasome | 43 | <0.1E-05 | 1.48E-02 |
| Under-expressed in BCG-challenged relative to Control mice | |||
| KEGG ribosome | 78 | <0.1E-05 | <0.1E-05 |
| Structural constituent of ribosome | 75 | <0.1E-05 | <0.1E-05 |
| Gated channel activity | 27 | <0.1E-05 | <0.1E-05 |
| Cation channel activity | 29 | <0.1E-05 | 2.39E-04 |
1 NG: number of genes.
2 FDR: adjusted P-value.
Gene networks of microglia and macrophages transcriptomic differences between BCG-challenged and Control mice
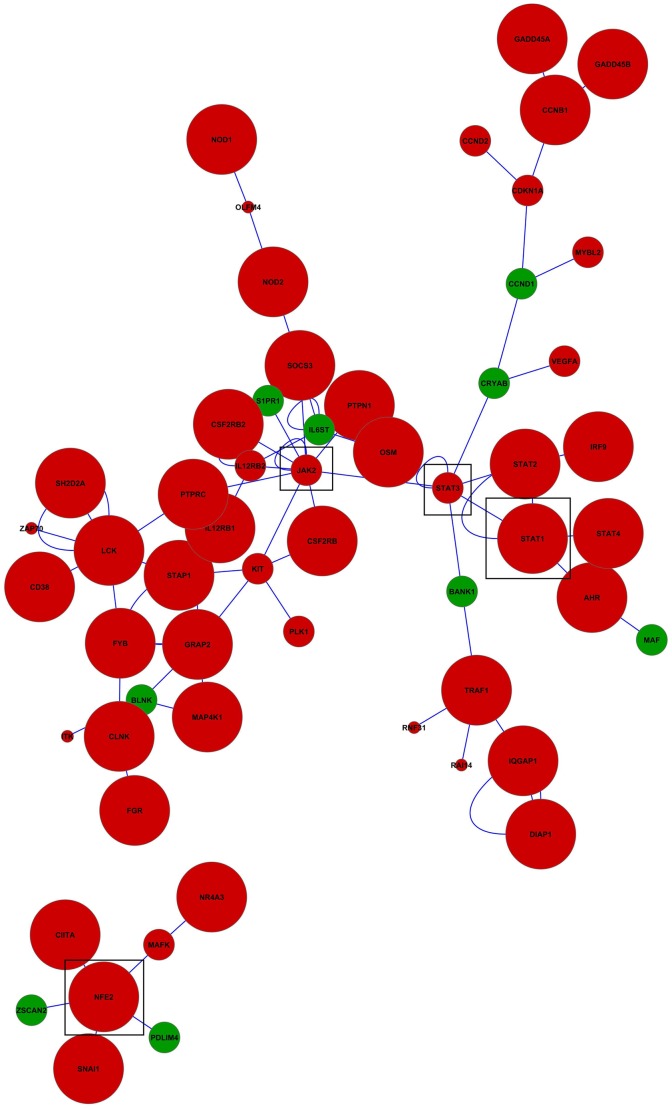
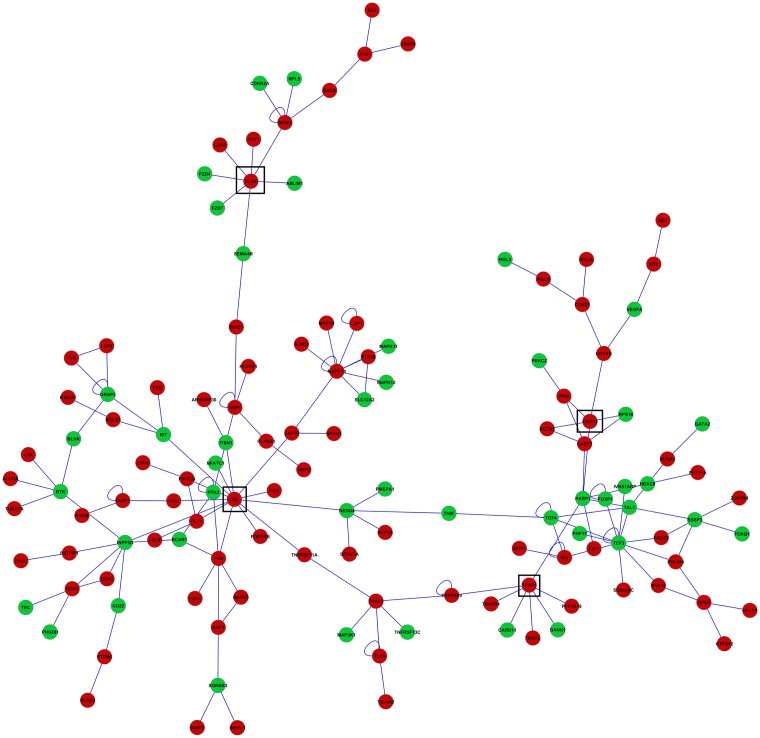
Visualization of networks of genes differentially expressed between BCG-challenged and Control mice in microglia (Fig 1) and macrophages (Fig 2) augmented our understanding of the molecular relationships among genes. Networks including more than 5 connected genes are discussed. The predominance of over-expressed genes relative to under-expressed genes in both networks is in agreement with their predominance among the most differentially expressed genes in Table 2 and with the enriched categories detected using GSEA (Tables 4 and 5). These networks also exhibit a predominance of genes related to immune response.
Fig 1. Network of genes differentially expressed between BCG-challenged and Control mice in microglia.
Red (green) nodes denote genes over- (under-) expressed in BCG-challenged relative to Control mice. Node size represents the P-value were larger nodes indicates more extreme significance (FDR-adjusted P-value < 0.001 larger nodes; P-value < 0.05 intermediate nodes; P-value < 0.1 small nodes). Edges denote known relationships between genes in the SysBiomics repository. Framed genes (squares) are discussed in the manuscript.
Fig 2. Network of genes differentially expressed between BCG-challenged and Control mice in macrophages.
Red (green) nodes denote genes over- (under-) expressed in BCG-challenged relative to Control mice. All genes were differentially expressed at FDR-adjusted P-value < 0.001. Edges denote known relationships between genes in the SysBiomics repository. Framed genes (squares) are discussed in the manuscript.
The microglia network (Fig 1) includes 57 gene nodes and 84% of the genes were over-expressed in BCG-challenged relative to Control (most at FDR-adjusted P-value < 0.001). In this network, Janus kinase 2 (Jak2), signal transducer and activator of transcription 3 (Stat3), signal transducer and activator of transcription 1 (Stat1), and nuclear factor erythroid derived 2 (Nfe2) were well-connected gene nodes linking to 11, 7, 5, and 5 other gene nodes, respectively. These genes were over-expressed in BCG-challenge relative to Control mice. Stat1 was affiliated to the enriched category “response to bacterium” in functional cluster 1 (Table 3) whereas nuclear factor erythroid derived 2 (Nfe2) was also over-expressed in BCG-challenge relative to the Control mice, although the corresponding categories were enriched below the set threshold.
The macrophages network (Fig 2) included 129 gene nodes and 66% of the genes were over-expressed in BCG-challenged relative to Control (FDR-adjusted P-value < 0.001). Casitas B-lineage lymphoma (Cbl) was the most connected gene node, linked to 13 other nodes. This gene was over-expressed in BCG-challenged relative to Control mice yet was not affiliated to the functional categories enriched beyond the set threshold. Discs large homolog 4 (Dlg4) and tumor necrosis factor receptor-associated factor 1 (Traf1) were over-expressed genes in BCG-challenged relative to Control mice and each gene was connected to 7 gene nodes. Dlg4 was affiliated to enriched categories including behavior, endocytosis, and structural molecule activity across 3 functional clusters whereas Traf1 was affiliated to enriched categories related to apoptosis across 2 functional clusters (Table 3). Caspase 3 (Casp3) was linked to 5 other gene nodes and was also over-expressed in BCG-challenged relative to Control mice. Casp3 was affiliated to several enriched categories including apoptosis, kinase activities, and cells activation across 6 functional clusters (Table 3).
Transcriptome differences between microglia and macrophages from BCG-challenged mice
The study of transcript isoforms and genes differentially expressed between microglia and macrophages 7 days after BCG challenge offered additional insights into the roles of both cell types after immune challenge (Table 1). Over 9,780 transcript isoforms corresponding to 9,117 genes were differentially expressed between microglia and macrophages in BCG-challenged mice. The genes most differentially expressed between microglia and macrophages from BCG-challenged mice (log2(fold change) > |6|, FDR-adjusted P-value < 2.0 x 10−4) are listed in Table 6 together with supporting literature review. An extended list of differentially expressed genes between microglia and macrophages is provided in S5 Table.
Table 6. Most extreme differentially expressed genes (FDR-adjusted P-value < 2.0 x 10−4) between microglia and macrophages in BCG-challenged mice and supporting literature review.
| Gene Symbol | NCBI Gene Name | Log2(Macrophages/Microglia) | Reference |
|---|---|---|---|
| Col8a1 | collagen, type VIII, alpha 1 | -13.70 | [103] |
| Kcne2 | potassium voltage-gated channel, Isk-related subfamily, gene 2 | -13.63 | [104] |
| Slc38a5 | solute carrier family 38, member 5 | -11.61 | |
| Lrrc3 | predicted gene 884 | -11.49 | [105] |
| Nav3 | neuron navigator 3 | -11.27 | [106] |
| Gm694 | predicted gene 694 | -11.07 | |
| Capn3 | calpain 3 | -11.06 | [107] |
| Gpr34 | G protein-coupled receptor 34 | -10.49 | [39,105,106] |
| Lrrn1 | leucine rich repeat protein 1, neuronal | -10.30 | [108] |
| Wwox | WW domain-containing oxidoreductase | -10.24 | |
| Vstm4 | V-set and transmembrane domain containing 4 | -10.07 | |
| Ecscr | endothelial cell surface expressed chemotaxis and apoptosis regulator | -9.95 | [39] |
| Cenpf | centromere protein F | -9.93 | |
| Gm10790 | predicted gene 10790 | -9.93 | |
| Tspan7 | tetraspanin 7 | -9.79 | [105] |
| Epn3 | epsin 3 | -9.76 | |
| Tmem100 | transmembrane protein 100 | -9.73 | [105] |
| Leprel1 | prolyl 3-hydroxylase 2 | -9.72 | [105] |
| Ebf3 | early B cell factor 3 | -9.71 | [105] |
| Rnu11 | U11 small nuclear RNA | -9.65 | |
| Wdr52 | cilia and flagella associated protein 44 | -9.60 | |
| Olfml3 | olfactomedin-like 3 | -9.60 | [39,105] |
| Trpm3 | transient receptor potential cation channel, subfamily M, member 3 | -9.58 | |
| Fat3 | FAT tumor suppressor homolog 3 (Drosophila) | -9.56 | |
| Adcy2 | adenylate cyclase 2 | -9.52 | |
| Kcnj13 | potassium inwardly-rectifying channel, subfamily J, member 13 | -9.48 | |
| Gpr56 | adhesion G protein-coupled receptor G1 | -9.46 | [39,105] |
| Sema4g | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G | -9.45 | |
| Ecm1 | extracellular matrix protein 1 | 6.89 | [39] |
| F10 | coagulation factor X | 6.90 | |
| Spib | Spi-B transcription factor (Spi-1/PU.1 related) | 6.90 | |
| Cxcr5 | chemokine (C-X-C motif) receptor 5 | 6.98 | [39] |
| Cxcl3 | chemokine (C-X-C motif) ligand 3 | 7.07 | [39] |
| Flrt3 | fibronectin leucine rich transmembrane protein 3 | 7.14 | [39] |
| Selplg | P-selectin glycoprotein ligand 1 | 7.16 | [39] |
| Cd19 | CD19 antigen | 7.17 | [39] |
| Retnla | resistin like alpha | 7.25 | [39] |
| Tnfrsf8 | tumor necrosis factor receptor superfamily, member 8 | 7.42 | |
| Cd79a | CD79A antigen (immunoglobulin-associated alpha) | 7.49 | |
| Mcoln2 | mucolipin 2 | 7.54 | |
| Vsig4 | V-set and immunoglobulin domain containing 4 | 7.55 | |
| Fcrla | Fc receptor-like A | 7.64 | |
| Ms4a1 | membrane-spanning 4-domains, subfamily A, member 1 | 7.68 | |
| Cd163l1 | CD163 molecule-like 1 | 7.76 | |
| Gjb5 | gap junction protein, beta 5 | 7.86 | |
| Vmn2r24 | vomeronasal 2, receptor 24 | 7.86 | |
| Scn4a | sodium channel, voltage-gated, type IV, alpha | 7.88 | |
| Il9r | interleukin 9 receptor | 7.95 | |
| Blk | B lymphoid kinase | 8.01 | |
| F7 | coagulation factor VII | 8.11 | [40] |
| Fcrl5 | Fc receptor-like 5 | 8.14 | |
| Serpinb2 | serine (or cysteine) peptidase inhibitor, clade B, member 2 | 8.21 | [39] |
| Pou2af1 | POU domain, class 2, associating factor 1 | 8.44 | |
| Ptges | prostaglandin E synthase | 8.66 | [109] |
| Cxcl13 | chemokine (C-X-C motif) ligand 13 | 9.30 | [39] |
| Nos2 | nitric oxide synthase 2, inducible | 9.33 | [79] |
| Cd5l | CD5 antigen-like | 9.34 | [39] |
| Arg1 | arginase, liver | 10.05 | [110] |
The list of genes over-expressed in microglia relative to macrophages from BCG-challenged mice confirms the similar molecular mechanisms shared between neurological disorders and response to immune challenge that also share depressive behaviors. Among the differentially expressed genes, collagen type VIII alpha 1 (Col8a1) and potassium voltage-gated channel Isk-related subfamily gene 2 (Kcne2) were over-expressed meanwhile arginase liver (Arg1) and Nos2 were under-expressed in microglia relative to macrophages from BCG-challenged mice. Col8a1 is up-regulated during repair processes in the mouse brain and collagen subunits have been linked to axonal guidance, synaptogenesis and Schwann cell differentiation [111,112]. Genomic rearrangements involving this gene have been linked to Tourette syndrome, a neuropsychiatric disorder that can encompass depression symptoms [113]. In addition to Col8a1, other genes over-expressed in microglia relative to macrophages from BCG-challenged mice have been linked to neuropsychiatric disorders that encompass depression symptoms (Table 6) including: leucine rich repeat protein 1 neuronal (Lrrn1), tetraspanin 7 (Tspan7), early B cell factor 3 (Ebf3), neuron navigator 3 (Nav3), and WW domain-containing oxidoreductase (Wwox). Over-expression of Lrrn1 was linked to autism spectrum disorder and Tourette syndrome [114]. Tspan7 was associated with Huntington's chorea, fragile X syndrome, and myotonic dystrophy [115]. Changes in the expression of Ebf3 was observed in obsessive-compulsive disorder [116]. Nav3 and WW domain-containing oxidoreductase (Wwox) have been implicated in Alzheimer's disease [117,118]. Nav3 was also associated with amyotrophic lateral sclerosis, Parkinson׳s disease [118], and was up-regulated in microglia compare to macrophages in response to hypoxia after stroke [106].
Kcne2 was among the genes over-expressed in microglia compared to macrophages in BCG-challenged mice. Expression of K+ channel genes follows nitric oxide changes after LPS stimulation indicating that K+ channels are involved in microglia activation [104]. Also, sialic acid binding Ig-like lectin H (Siglech) is considered a microglia signature gene [105] and G protein-coupled receptor 34 (Gpr34) is highly expressed in microglia and regulates the function, morphology, and phagocytosis of microglia during neuroinflammation [119,120].
Among the genes under-expressed in microglia relative to macrophages from BCG-challenged mice, the enzymes encoded by Arg1 and Nos2 compete for the same substrate: L-arginine [121,122]. The over-expression of Nos2 in macrophages is controlled by cytokines in response to a pathogen or inflammatory diseases [79] and also up-regulation of Nos2 and Arg1 were found in mouse and rat macrophages treated with LPS [123]. MicroRNA 155 (Mir155) was also over-expressed in macrophages relative to microglia (S5 Table). This microRNA is a crucial regulator of apoptosis and cell fate decisions in BCG-challenged macrophages and over-expression triggers mitogen-activated protein kinases (MAPK) cascades [124]. The genes under-expressed in microglia relative to macrophages after BCG challenge offer insights into the molecular mechanisms that are differentially regulated due to their potential negative impact in the brain.
Functional analysis of transcriptomic differences between microglia and macrophages from BCG-challenged mice
Enriched functional categories offered additional insights into the roles of microglia and macrophages after sickness recovery from a BCG challenge. Table 7 summarizes the DAVID clusters of functional categories surpassing Enrichment Score > 4 and S6 Table presents the complete list of categories surpassing Enrichment Score > 2 and corresponding gene counts. Functional categories encompassing genes differentially expressed between cell types included leukocyte regulation and activation, chemokine and cytokine activities, MAP kinase activity, and apoptosis (Table 7). These results are consistent with reports of enriched categories in LPS-challenged microglia [125] and BCG-challenged macrophages [124]. Enrichment of apoptosis (Table 7) is associated with the over-expression of Nos2 and Arg1 in macrophages because these genes regulate the production of peroxynitrites that in turn induce apoptosis [126,127].
Table 7. Most significant clusters (DAVID Enrichment Score ES > 4) of enriched Gene Ontology (GO) biological processes (BP) and molecular functions (MF) among the transcript isoforms differentially abundant between microglia and macrophages from BCG-challenged mice.
| GO cluster1 | Terms2 | ES |
|---|---|---|
| BP | ~response to wounding~inflammatory response~defense response | 8.72 |
| BP | ~regulation of cell activation~regulation of leukocyte activation~positive regulation of immune system process | 6.72 |
| BP | ~cell migration~localization of cell~cell motility | 6.55 |
| BP | ~regulation of cytokine production~regulation of cytokine biosynthetic process~positive regulation of cytokine biosynthetic process | 6.44 |
| MF,BP | ~purine nucleotide binding~ribonucleotide binding~purine ribonucleotide binding | 6.14 |
| MF,BP | ~taxis~chemotaxis~chemokine activity | 6.04 |
| BP | ~cell death~death~programmed cell death | 5.92 |
| BP | ~regulation of apoptosis~regulation of programmed cell death~regulation of cell death | 5.53 |
| BP | ~regulation of cell activation~regulation of leukocyte activation~regulation of lymphocyte activation | 5.26 |
| BP | ~vasculature development~blood vessel development~blood vessel morphogenesis | 4.59 |
| BP | ~hemopoietic or lymphoid organ development~immune system development~hemopoiesis | 4.59 |
| BP | ~regulation of cytokine production~regulation of interferon-gamma production~positive regulation of cytokine production | 4.26 |
| MF | ~GTP binding~guanyl ribonucleotide binding~guanyl nucleotide binding | 4.07 |
1 Each row corresponds to a cluster of Functional Annotation Tool (FAT) GO categories.
2 The three GO terms exhibiting most significant enrichment P-value in each cluster are listed, separated by “~”. Additional information in each cluster is provided in S6 Table
Enrichment of categories within over- or under-expressed genes in microglia relative to macrophages detected using GSEA offered confirmatory and complementary information. Table 8 lists GSEA findings at FDR-adjusted P-value < 0.01 and including > 10 transcript isoforms and S7 Table includes a more extensive list of categories enriched at FDR-adjusted P-value < 0.05 and including > 10 transcript isoforms per category. The enrichment of categories associated with immune response, ribosome and cytokine activity among genes under-expressed in microglia relative to macrophages could be related to the potential detrimental neurological effect of microglia activation [128]. Among the enriched categories including genes over-expressed in the microglia, dysregulation of the tight junction pathway has been related to anxiety behaviors and altered signaling in the brain [129]. Similarly enriched categories included brain, central and nervous system development categories are consistent with reports of functional signatures in microglia [105].
Table 8. Gene Set Enrichment Analysis (GSEA) categories enriched (FDR-adjusted P-value < 0.01, > 10 transcript isoforms) among transcript isoforms under-expressed in microglia relative to peripheral macrophages in BCG-challenged mice.
| Category | NG1 | P-value | FDR2 |
|---|---|---|---|
| Adaptive immune response | 21 | <0.1E-05 | <0.1E-05 |
| Adaptive immune response | 20 | <0.1E-05 | <0.1E-05 |
| Positive regulation of immune response | 20 | <0.1E-05 | <0.1E-05 |
| Regulation of immune response | 22 | <0.1E-05 | <0.1E-05 |
| Regulation of immune system process | 46 | <0.1E-05 | 1.05E-03 |
| Chemokine receptor binding | 28 | <0.1E-05 | 1.18E-03 |
| Positive regulation of immune system process | 37 | <0.1E-05 | 1.34E-03 |
| Positive regulation of response to stimulus | 27 | <0.1E-05 | 1.57E-03 |
| KEGG primary immunodeficiency | 30 | <0.1E-05 | 1.88E-03 |
| KEGG ribosome | 81 | <0.1E-05 | 1.90E-03 |
| Regulation of lymphocyte activation | 28 | <0.1E-05 | 2.07E-03 |
| Cellular defense response | 38 | <0.1E-05 | 2.28E-03 |
| Lymphocyte activation | 47 | <0.1E-05 | 3.78E-03 |
| G protein coupled receptor binding | 35 | <0.1E-05 | 4.34E-03 |
| Chemokine activity | 27 | <0.1E-05 | 4.62E-03 |
| Positive regulation of multicellular organismal process | 47 | <0.1E-05 | 4.63E-03 |
| Cytokine activity | 61 | <0.1E-05 | 6.65E-03 |
| B cell activation | 18 | <0.1E-05 | 7.59E-03 |
| Locomotory behavior | 61 | <0.1E-05 | 8.08E-03 |
| Leukocyte activation | 51 | <0.1E-05 | 8.95E-03 |
| KEGG hematopoietic cell lineage | 52 | <0.1E-05 | 9.40E-03 |
1 NG: number of genes.
2 FDR: adjusted P-value.
Alternative splicing between microglia and macrophages from BCG-challenged mice
Significant evidence of alternative splicing was detected in the comparison between cell types relative to challenge levels. Table 9 summarizes genes with at least nine transcript isoforms and at least two differentially expressed (FDR-adjusted P-value < 0.05) between microglia and macrophages. S8 Table lists all genes that match the broader alternative splicing definition of at least one transcript isoform differentially expressed among multiple isoforms.
Table 9. Genes exhibiting an alternative splicing event between microglia and macrophages in BCG-challenged mice including at least nine transcript isoforms and at least two over- or under-expressed (FDR-adjusted P-value < 0.05) transcript isoforms between cell types.
| Gene | NCBI Gene Name | Differential Expression | ||
|---|---|---|---|---|
| Under1 | Over2 | Non3 | ||
| Macf1 | microtubule-actin crosslinking factor 1 | 6 | 10 | 15 |
| Rbm5 | RNA binding motif protein 5 | 6 | 9 | 14 |
| Bag6 | BCL2-associated athanogene 6 | 7 | 7 | 13 |
| Arhgef1 | Rho guanine nucleotide exchange factor (GEF) 1 | 7 | 7 | 11 |
| Rbm39 | RNA binding motif protein 39 | 6 | 7 | 12 |
| Gapvd1 | GTPase activating protein and VPS9 domains 1 | 7 | 5 | 11 |
| Josd2 | Josephin domain containing 2 | 4 | 7 | 10 |
| Lrrc16a | leucine rich repeat containing 16A | 6 | 5 | 9 |
| Rnf220 | ring finger protein 220 | 2 | 8 | 9 |
| Gripap1 | GRIP1 associated protein 1 | 6 | 4 | 9 |
| Anapc1 | anaphase promoting complex subunit 1 | 5 | 5 | 9 |
| Arhgap4 | Rho GTPase activating protein 4 | 2 | 8 | 9 |
| Hnrnpl | heterogeneous nuclear ribonucleoprotein L | 4 | 6 | 9 |
| Il15ra | interleukin 15 receptor, alpha chain | 5 | 5 | 9 |
| Srsf5 | serine/arginine-rich splicing factor 5 | 3 | 7 | 8 |
| Ythdc1 | YTH domain containing 1 | 3 | 6 | 8 |
| Brwd1 | bromodomain and WD repeat domain containing 1 | 4 | 5 | 8 |
| Rtn4 | reticulon 4 | 4 | 5 | 8 |
| Sfrs18 | PNN interacting serine/arginine-rich | 4 | 5 | 8 |
| Wdr13 | WD repeat domain 13 | 5 | 4 | 8 |
| Wnk1 | WNK lysine deficient protein kinase 1 | 5 | 4 | 7 |
| Ctage5 | CTAGE family, member 5 | 3 | 6 | 6 |
| Hivep3 | human immunodeficiency virus type I enhancer binding protein 3 | 4 | 5 | 5 |
1Over: transcript isoforms over-expressed in microglia.
2Under: transcript isoforms under-expressed in microglia.
3Non: not differentially expressed transcript isoforms (FDR-adjusted P-value < 0.05).
Alternative splicing events differentiating cell types were identified in 387 genes including 1,607 transcript isoforms. Confirming our findings, at least 72 of these genes have recorded alternative splicing events in the microglia as described in the database of alternative splicing of brain cells [130]. The alternative splicing cassette characterized by the inclusion or exclusion of an exon was the most frequent type of splicing event with 68% of occurrences. Alternative 3′ and alternative 5′ splicing events characterized by alternative usage of a splicing site on the 3′ and 5′ end of an exon, respectively were the second most common and the less common events amounting to 25% and 7% of the transcript isoforms, respectively. Intron retention (the inclusion or exclusion of a segment previously annotated to be an intron); tandem cassette (the inclusion or exclusion of two or more tandem exons); and mutually exclusive exons (the inclusion of one exon in one transcript and inclusion of a different exon in another transcript) were identified in 19%, 13%, and 11% of the 387 genes investigated, respectively. More than one alternative splicing mode was observed in some genes including WNK lysine deficient protein kinase 1 (Wnk1), tripartite motif-containing 33 (Trim33), and SWI/SNF related matrix associated actin dependent regulator of chromatin subfamily c member 2 (Smarcc2). Among the genes exhibiting alternative splicing events in the microglia, several genes have been associated with neurological disorders. Schizophrenia has been associated with microtubule-actin crosslinking factor 1 (Macf1) [131,132], Wnk1 [131], and Fused in sarcoma (Fus) [133]. Our results confirm alternative splicing events that are shared between neurological disorders and microglia after immune challenge and both linked to depressive behaviors.
Gene networks of transcriptomic differences between microglia and macrophages from BCG-challenged mice
Additional understanding of the relationship among genes in microglia and macrophages from BCG-challenged mice was gained from networks visualization. A network was depicted considering the 2,487 most extreme differentially expressed genes between microglia and macrophages (log2(fold change) > |2|, FDR-adjusted P-value < 2.0 x 10−4). From these, sub-networks connecting more than 5 genes are presented to facilitate visualization and interpretation. A gene network was constructed that included 146 genes with 39% of the genes over-expressed in microglia and the rest under-expressed (Fig 3). The predominance of genes under-expressed in microglia relative to macrophages suggests that gene dysregulation leading to negative neurological effects is more contained and quickly resolved in microglia than in macrophages after a BCG challenge.
Fig 3. Network of genes differentially expressed between microglia and macrophages in BCG-challenged mice.
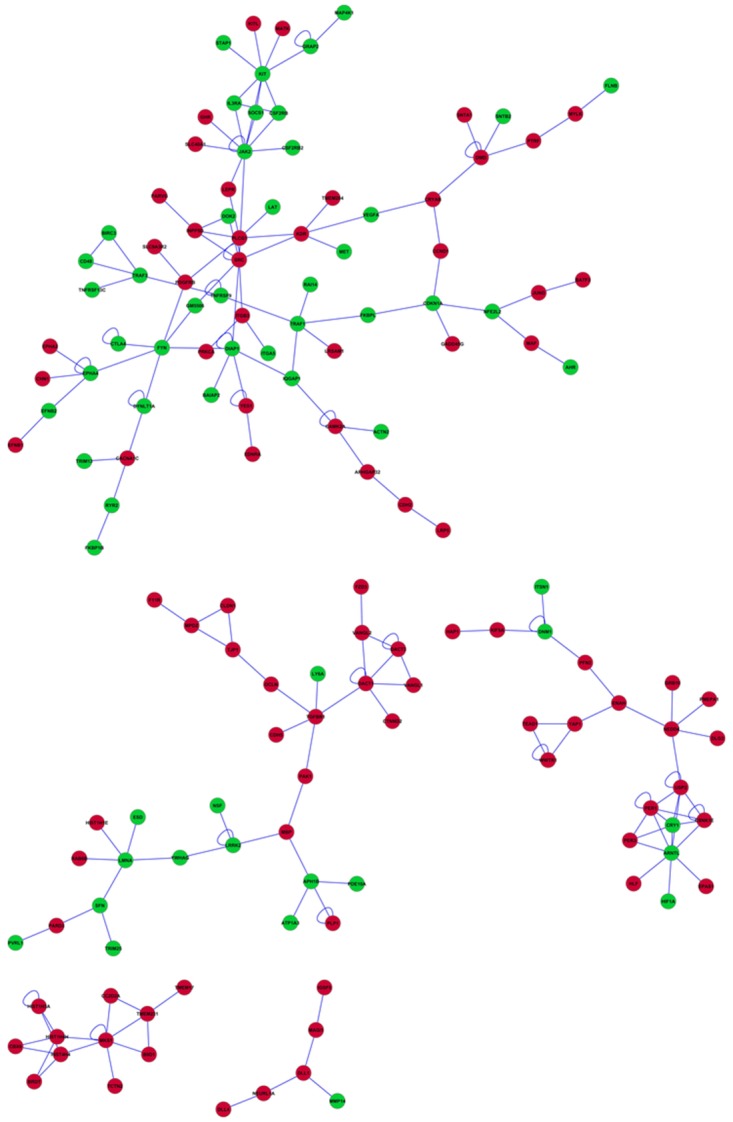
Red (green) nodes denote genes over- (under-) expressed in BCG-challenged relative to Control mice. All genes were differentially expressed at FDR-adjusted P-value < 0.0001. Edges denote known relationships between genes in the SysBiomics repository. Framed genes (squares) are discussed in the manuscript.
Janus kinase (Jak2) was the most connected gene in the network, linked to 11 other genes (Fig 3). This gene was under-expressed in microglia relative to macrophages and was affiliated to the enriched categories of protein kinase activity, nucleotide binding, apoptosis, cell differentiation, and regulation of phosphorylation in four functional clusters (Table 7). Kit oncogene (Kit) was under-expressed in microglia relative to macrophages, was linked to eight other genes, and was affiliated to the enriched categories cell migration, chemotaxis, nucleotide binding, phosphorylation, protein kinase activity, apoptosis, leukocyte proliferation and differentiation distributed across 11 functional clusters (Table 7). Diaphanous homolog 1 (Diap1) was also under-expressed in microglia, was connected to seven gene nodes, and was affiliated to the functional categories actin cytoskeleton and protein organization that were enriched below the threshold.
The most connected genes over-expressed in microglia relative to macrophages (Fig 3) included Rous sarcoma oncogene (Src), ubiquitin specific peptidase 2 (Usp2), and transforming growth factor, beta receptor I (Tgfbr1) and these genes were linked to 9, 7, and 5 other genes, respectively. Src is affiliated to enriched categories including cell migration, protein kinase activity, nucleotide binding, apoptosis, and cell adhesion in six clusters (Table 7). Ups2 is affiliated to enriched categories including phosphate and phosphorus metabolic process in one cluster and Tgfbr1 is affiliated to enriched categories including cell migration, nucleotide binding, phosphorylation, protein kinase activity, angiogenesis and apoptosis in seven clusters (Table 7). The genes most connected in the network enriched apoptosis, cell migration, chemotaxis, and cell adhesion categories. This finding is in agreement with reports of the functional response of macrophages [124,134] and microglia [135] after a bacterial challenge.
This research studied the transcriptome of whole-brain microglia. However, microglia from different brain regions is likely to contribute unequally to depressive behaviors. Likewise, some brain regions are more responsive to periphery immune challenges than others. Additional studies that can parse the differential effect of periphery BCG challenge across brain regions and the corresponding changes in the transcriptome will offer more precise insights into the association between the transcriptome changes in response to immune challenge within brain regions and corresponding depressive-like behaviors.
Conclusions
The impact of immune challenge on the microglia after recovery from sickness and the implications on depressive behaviors was studied. The transcriptome of microglia 7 days after BCG challenge was compared to the corresponding transcriptome from unchallenged Control mice and to macrophages from the same mice. The number of genes differentially expressed between BCG-challenged and Control mice suggests the capacity of microglia to restrain or quickly resolve transcriptomic dysregulation relative to macrophages. The differential expression of Kmo in microglia between BCG-challenged and Control mice suggests that Kmo is a potential target of pro-inflammatory cytokines in the kynurenine pathway and a potential factor of depressive-like symptoms that remain after sickness symptoms subside. The over-expression of a number of genes, including Ifng and Il1b, in microglia relative to macrophages offered further evidence of transcriptome conditions associated with depression-like symptoms. Functional analysis highlighted the enrichment of categories including immune response and chemotaxis by genes over-expressed in BCG-challenged compared to Control mice in both cell types. Network visualization uncovered the key role of Jak2, Stat3, Stat1, and Nfe2 as hub genes, dysregulated and connected to other dysregulated genes in the microglia 7 days after BCG challenge.
The large number of differentially expressed genes between cell types from BCG-challenged mice speaks to the response of these cells to immune challenge. Also, a number of genes exhibiting differential splicing events between microglia and macrophages in this study have been linked to neurological disorders. Network visualization depicted the capability of microglia to exhibit transcriptome dysregulation after sickness recovery from immune challenge, albeit lower than macrophages. The present study provides ample evidence that the microglia transcriptome dysregulation after BCG challenge is shared with neurological disorders that also exhibit depressive behaviors.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
Data are available from the NCBI/GEO repository GSE76069 and GSE70941 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sxghgcmorrulhwr&acc=GSE76069; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ulczkewahfmnbov&acc=GSE70941).
Funding Statement
This study is supported by the National Institutes of Health R21 MH 096030 (SRZ), http://www.nih.gov; National Institutes of Health R01 MH 090127 (RM), http://www.nih.gov; National Institutes of Health R01 SUB UT 00000712 (KWK), http://www.nih.gov; National Institutes of Health R01 MH083767 (KWK), http://www.nih.gov; US Department of Agriculture ILLU-538-632 (SRZ), http://www.usda.gov; US Department of Agriculture ILLU-538-909 (SRZ), http://www.usda.gov; 5P30DA018310 NIH National Institute of Drug Abuse to Bruce Southey (BRS).
References
- 1.Dantzer R, O'Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9: 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelley KW, O'Connor JC, Lawson MA, Dantzer R, Rodriguez-Zas S, McCusker RH. Aging leads to prolonged duration of inflammation-induced depression-like behavior caused by Bacillus Calmette-Guerin. Brain Behav Immun. 2013;32: 63–9. 10.1016/j.bbi.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreau M, Andre C, O'Connor JC, Dumich SA, Woods JA, Kelley KW, et al. Inoculation of Bacillus Calmette-Guerin to mice induces an acute episode of sickness behavior followed by chronic depressive-like behavior. Brain Behav Immun. 2008;22: 1087–1095. 10.1016/j.bbi.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Connor JC, Lawson MA, Andre C, Moreau M, Lestage J, Castanon N, et al. Lipopolysaccharide-induced depressive-like behavior is mediated by indoleamine 2,3-dioxygenase activation in mice. Mol Psychiatry. 2009;14: 511–522. 10.1038/sj.mp.4002148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Zas SL, Nixon SE, Lawson MA, Mccusker RH, Southey BR, O’Connor JC, et al. Advancing the understanding of behaviors associated with Bacille Calmette Guérin infection using multivariate analysis. Brain Behav Immun. 2015;44: 176–186. 10.1016/j.bbi.2014.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci. 2011;14: 1227–35. 10.1038/nn.2923 [DOI] [PubMed] [Google Scholar]
- 7.Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang L, Means TK, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16: 1896–905. 10.1038/nn.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nixon SE, González-Peña D, Lawson MA, McCusker RH, Hernandez AG, O'Connor JC, et al. Analytical workflow profiling gene expression in murine macrophages. J Bioinform Comput Biol. 2015; 13: 1550010 10.1142/S0219720015500109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ufimtseva E. Mycobacterium-host cell relationships in granulomatous lesions in a mouse model of latent tuberculous infection. BioMed Res Int. 2015; 948131 10.1155/2015/948131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol 2008;Chapter 14: Unit 14.1 10.1002/0471142735.im1401s83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelsthorpe AR, Gelsthorpe K, Sokol RJ. Rapid isolation of total RNA from small samples of hypocelluar dense connective tissues. BioTechniques. 1997;22: 1082–1086. [DOI] [PubMed] [Google Scholar]
- 12.Nikodemova M, Watters JJ. Efficient isolation of live microglia with preserved phenotypes from adult mouse brain. J Neuroinflammation. 2012; 9: 147 10.1186/1742-2094-9-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Pena D, Nixon S, Lawson M, McCusker R, Dantzer R, Kelley K, et al. Impact of the increased in the biological samples and alternatives vs. defaults setting in the transcripts analysis. 6th International Conference on Bioinformatics and Computational Biology (BICoB-2014). 2014; Las Vegas, NV: Bioinformatics Applications 1 16–19p.
- 14.Fenn AM, Hall JCE, Gensel JC, Popovich PG, Godbout JP. IL-4 signaling drives a unique arginase+/Il-1beta+ microglia phenotype and recruits macrophages to the inflammatory CNS: consequences of age-related deficits in IL-4Ralpha after traumatic spinal cord injury 2014;34: 8904–8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford AL, Goodsall AL, Hickey WF, Sedgwick JD. Normal adult ramified microglia separated from other central nervous system macrophages by flow cytometric sorting. Phenotypic differences defined and direct ex vivo antigen presentation to myelin basic protein-reactive CD4+ T cells compared. J Immunol. 1995;154: 4309–21. [PubMed] [Google Scholar]
- 16.Institute B. FastQC A Quality Control tool for High Throughput Sequence Data. 2013. Available: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 17.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31: 46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5: 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;59: 289–300. [Google Scholar]
- 21.Caetano-Anolles K, Mishra S, Rodriguez-Zas SL. Synergistic and antagonistic interplay between Myostatin gene expression and physical activity levels on gene expression patterns in triceps brachii muscles of C57/BL6 mice. PLoS One. 2015;10: e0116828 10.1371/journal.pone.0116828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadeque A, Serao NV, Southey BR, Delfino KR, Rodriguez-Zas SL. Identification and characterization of alternative exon usage linked glioblastoma multiforme survival. BMC Med Genomics. 2012;5: 59 10.1186/1755-8794-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 24.Delfino KR, Rodriguez-Zas SL. Transcription factor-microRNA-target gene networks associated with ovarian cancer survival and recurrence. PLoS One. 2013;8: e58608 10.1371/journal.pone.0058608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serao NV, Delfino KR, Southey BR, Beever JE, Rodriguez-Zas SL. Cell cycle and aging, morphogenesis, and response to stimuli genes are individualized biomarkers of glioblastoma progression and survival. BMC Med Genomics. 2011;4: 49 10.1186/1755-8794-4-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delfino KR, Serao NV, Southey BR, Rodriguez-Zas SL. Therapy-, gender- and race-specific microRNA markers, target genes and networks related to glioblastoma recurrence and survival. Cancer Genomics Proteomics. 2011;8: 173–183. [PMC free article] [PubMed] [Google Scholar]
- 27.Serao NV, Gonzalez-Pena D, Beever JE, Faulkner DB, Southey BR, Rodriguez-Zas SL. Single nucleotide polymorphisms and haplotypes associated with feed efficiency in beef cattle. BMC Genet. 2013;14: 94 10.1186/1471-2156-14-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serao NV, Gonzalez-Pena D, Beever JE, Bollero GA, Southey BR, Faulkner DB, et al. Bivariate genome-wide association analysis of the growth and intake components of feed efficiency. PLoS One. 2013. October 29; 8:e78530 10.1371/journal.pone.0078530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamayo P, Steinhardt G, Liberzon A, Mesirov JP. The limitations of simple gene set enrichment analysis assuming gene independence. Stat Methods Med Res. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27: 1739–1740. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin A, Ochagavia ME, Rabasa LC, Miranda J, Fernandez-de-Cossio J, Bringas R. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11: 91–91. 10.1186/1471-2105-11-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13: 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13: 1118–28. 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ginhoux F, Greter M, Leboeuf M, Nandi S. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010; 330:841–5. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das A, Chai JC, Kim SH, Park KS, Lee YS, Jung KH, et al. Dual RNA sequencing reveals the expression of unique transcriptomic signatures in lipopolysaccharide-induced BV-2 microglial cells. PLoS One. 2015;10: e0121117 10.1371/journal.pone.0121117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith RP. Transcriptional Regulation in the Peripheral Nervous System and the Role of STAT3 in Axon Regeneration. University of Miami Scholarly Repository; 2008. [Google Scholar]
- 38.Szulzewsky F, Pelz A, Feng X, Synowitz M, Markovic D, Langmann T, Holtman IR, Wang X, Eggen BJ, Boddeke HW, Hambardzumyan D, Wolf SA, Kettenmann H. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1.PLoS One. 2015;10:e0116644 10.1371/journal.pone.0116644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang L, Means TK, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16: 1896–905. 10.1038/nn.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13: 1118–1128. 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M, Hou X, Zhang P, Hao Y, Yang Y, Wu X, et al. Microarray gene expression profiling analysis combined with bioinformatics in multiple sclerosis. Mol Biol Rep. 2013;40: 3731–7. 10.1007/s11033-012-2449-3 [DOI] [PubMed] [Google Scholar]
- 42.Ebert S, Walczak Y, Rema C, Langmann T. Microglial activation and transcriptomic changes in the blue light-exposed mouse retina. Adv Exp Med Biol. 2012;723: 619–32. 10.1007/978-1-4614-0631-0_79 [DOI] [PubMed] [Google Scholar]
- 43.Mabbott NA, Kenneth Baillie J, Kobayashi A, Donaldson DS, Ohmori H, Yoon S, et al. Expression of mesenchyme-specific gene signatures by follicular dendritic cells: insights from the meta-analysis of microarray data from multiple mouse cell populations. Immunology. 2011;133: 482–98. 10.1111/j.1365-2567.2011.03461.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Israelsson C, Bengtsson H, Kylberg A, Kullander K, Lewen A, Hillered L, et al. Distinct cellular patterns of upregulated chemokine expression supporting a prominent inflammatory role in traumatic brain injury. J Neurotrauma. 2008;25: 959–74. 10.1089/neu.2008.0562 [DOI] [PubMed] [Google Scholar]
- 45.Krause D, Jobst A, Kirchberg F, Kieper S, Hartl K, Kastner R, et al. Prenatal immunologic predictors of postpartum depressive symptoms: a prospective study for potential diagnostic markers. Eur Arch Psychiatry Clin Neurosci. 2014;264: 615–624. 10.1007/s00406-014-0494-8 [DOI] [PubMed] [Google Scholar]
- 46.Li H, Gang Z, Yuling H, Luokun X, Jie X, Hao L, et al. Different neurotropic pathogens elicit neurotoxic CCR9- or neurosupportive CXCR3-expressing microglia. 2006;177: 3644–56. [DOI] [PubMed] [Google Scholar]
- 47.Hwang Y, Kim J, Shin JY, Kim JI, Seo JS, Webster MJ, et al. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl Psychiatry. 2013;3: e321 10.1038/tp.2013.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takasaki I, Taniguchi K, Komatsu F, Sasaki A, Andoh T, Nojima H, et al. Contribution of spinal galectin-3 to acute herpetic allodynia in mice. Pain. 2012;153: 585–92. 10.1016/j.pain.2011.11.022 [DOI] [PubMed] [Google Scholar]
- 49.Martinez AN, Mehra S, Kaushal D. Role of interleukin 6 in innate immunity to Mycobacterium tuberculosis infection. J Infect Dis. 2013;207: 1253–61. 10.1093/infdis/jit037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamashita N, Tsukahara H, Tsuge M, Nagaoka Y, Yashiro M, Saito Y, et al. Pathogenic mechanisms of influenza A(H1N1)pdm09 infection elucidated on gene expression profiling. J Neurovirol. 2013;55: 572–7. [DOI] [PubMed] [Google Scholar]
- 51.Brodziak F, Meharg C, Blaut M, Loh G. Differences in mucosal gene expression in the colon of two inbred mouse strains after colonization with commensal gut bacteria. PLoS One. 2013;8: e72317 10.1371/journal.pone.0072317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen M, Muckersie E, Forrester JV, Xu H. Immune activation in retinal aging: a gene expression study. Invest Ophthalmol Vis Sci. 2010;51: 5888–96. 10.1167/iovs.09-5103 [DOI] [PubMed] [Google Scholar]
- 53.Suga A, Sadamoto K, Fujii M, Mandai M, Takahashi M. Proliferation potential of Muller glia after retinal damage varies between mouse strains. PLoS One. 2014;9: e94556–e94556. 10.1371/journal.pone.0094556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laziu D, Alborzi F, Marcuzzi GP, Angel P, Hess J, Pfister H, et al. Enhanced StefinA and Sprr2 expression during papilloma formation in HPV8 transgenic mice. J Dermatol Sci. 2011;62: 84–90. 10.1016/j.jdermsci.2011.02.006 [DOI] [PubMed] [Google Scholar]
- 55.Gebhardt C, Nemeth J, Angel P, Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. 2006;72: 1622–31. [DOI] [PubMed] [Google Scholar]
- 56.Stier MT, Spindler KR. Polymorphisms in Ly6 genes in Msq1 encoding susceptibility to mouse adenovirus type 1. Mamm Genome. 2012;23: 250–8. 10.1007/s00335-011-9368-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shiloh MU, MacMicking JD, Nicholson S, Brause JE, Potter S, Marino M, et al. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity. 1999;10: 29–38. [DOI] [PubMed] [Google Scholar]
- 58.Foster AM, Baliwag J, Chen CS, Guzman AM, Stoll SW, Gudjonsson JE, et al. IL-36 promotes myeloid cell infiltration, activation, and inflammatory activity in skin. J Immunol. 2014;192: 6053–61. 10.4049/jimmunol.1301481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmer GD, Attur MG, Yang Q, Liu J, Moon P, Beier F, et al. F-spondin deficient mice have a high bone mass phenotype. PLoS One. 2014;9: e98388 10.1371/journal.pone.0098388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okada K, Arai S, Nakase H, Kohno H, Nakamura F, Takeda M, et al. Autocrine pathways involving S100A8 and/or S100A9 that are postulated to regulate the immunological functions of macrophages in rats. Biochem Biophys Res Commun. 2015;456: 415–20. 10.1016/j.bbrc.2014.11.099 [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Wakeham J, Harkness R, Xing Z. Macrophages are a significant source of type 1 cytokines during mycobacterial infection. J Clin Invest. 1999;103: 1023–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rempel JD, Quina LA, Blakely-Gonzales P, Buchmeier MJ, Gruol DL. Viral induction of central nervous system innate immune responses. J Virol. 2005;79: 4369–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker CA, Manuelidis L. Unique inflammatory RNA profiles of microglia in Creutzfeldt-Jakob disease. Proc Natl Acad Sci U S A. 2003;100: 675–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ha T, Chang K, Kim Ja, Kim H, Kim S, Chong YH, et al. S100a9 knockdown decreases the memory impairment and the neuropathology in Tg2576 mice, AD animal model. PLoS One. 2010;5: e8840 10.1371/journal.pone.0008840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang W, Tan M, Yu J, Tan L. Role of pro-inflammatory cytokines released from microglia in Alzheimer's disease. PLoS One. 2015;3: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Sorge NM, van der Pol W, Jansen MD, Geleijns KP, Kalmijn S, Hughes RA, et al. Severity of Guillain–Barré syndrome is associated with Fcγ Receptor III polymorphisms. J Neuroimmunol. 2005;162: 157–164. [DOI] [PubMed] [Google Scholar]
- 67.Vedeler CA, Myhr KM, Nyland H. Fc receptors for immunoglobulin G—a role in the pathogenesis of Guillain-Barre syndrome and multiple sclerosis. J Neuroimmunol. 2001;118: 187–93. [DOI] [PubMed] [Google Scholar]
- 68.Fuller JP, Stavenhagen JB, Teeling JL. New roles for Fc receptors in neurodegeneration-the impact on Immunotherapy for Alzheimer's Disease. Neurosci. 2014;8: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8: 958–969. 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mosser DM. The many faces of macrophage activation. J Leukoc Biol. 2003;73: 209–212. [DOI] [PubMed] [Google Scholar]
- 71.Kawano T, Anrather J, Zhou P, Park L, Wang G, Frys KA, et al. Prostaglandin E2 EP1 receptors: downstream effectors of COX-2 neurotoxicity. Nat Med. 2006;12: 225–9. [DOI] [PubMed] [Google Scholar]
- 72.Norden DM, Godbout JP. Review: microglia of the aged brain: primed to be activated and resistant to regulation. Neuropathol Appl Neurobiol. 2013;39: 19–34. 10.1111/j.1365-2990.2012.01306.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dinarello CA, Simon A, van der Meer J.W.. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov. 2012;11: 633–52. 10.1038/nrd3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goshen I, Kreisel T, Ben-Menachem-Zidon O, Licht T, Weidenfeld J, Ben-Hur T, et al. Brain interleukin-1 mediates chronic stress-induced depression in mice via adrenocortical activation and hippocampal neurogenesis suppression. Mol Psychiatry. 2007;13: 717–728. [DOI] [PubMed] [Google Scholar]
- 75.Norman GJ, Karelina K, Zhang N, Walton JC, Morris JS, Devries AC. Stress and IL-1beta contribute to the development of depressive-like behavior following peripheral nerve injury. Mol Psychiatry. 2010;15: 404–14. 10.1038/mp.2009.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kitazawa M, Cheng D, Tsukamoto MR, Koike MA, Wes PD, Vasilevko V, et al. Blocking IL-1 signaling rescues cognition, attenuates tau pathology, and restores neuronal beta-catenin pathway function in an Alzheimer's disease model. J Immunol. 2011;187: 6539–49. 10.4049/jimmunol.1100620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O'Connor J, Andre C, Wang Y, Lawson MA, Szegedi SS, Lestage J, et al. Interferon-gamma and tumor necrosis factor-alpha mediate the upregulation of indoleamine 2,3-dioxygenase and the induction of depressive-like behavior in mice in response to bacillus Calmette-Guerin. J Neurosci. 2009;29: 4200–9. 10.1523/JNEUROSCI.5032-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pan F, Zhao Y, Zhu S, Sun C, Lei L, Feng X, et al. Different transcriptional profiles of RAW264.7 infected with Mycobacterium tuberculosis H37Rv and BCG identified via deep sequencing. PLoS One. 2012;7: e51988 10.1371/journal.pone.0051988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15: 323–50. [DOI] [PubMed] [Google Scholar]
- 80.Schindler H, Lutz MB, Rollinghoff M, Bogdan C. The production of IFN-gamma by IL-12/IL-18-activated macrophages requires STAT4 signaling and is inhibited by IL-4. J Immunol. 2001;166: 3075–82. [DOI] [PubMed] [Google Scholar]
- 81.Myint A, Leonard B, Steinbusch H, Kim Y. Th1, Th2, and Th3 cytokine alterations in major depression. J Affect Disord. 2005;88: 167–173. [DOI] [PubMed] [Google Scholar]
- 82.Pavon L, Sandoval-Lopez G, Eugenia Hernandez M, Lori-a F, Estrada I, Perez M, et al. Th2 cytokine response in Major Depressive Disorder patients before treatment. J Neuroimmunol. 2006;172: 156–65. [DOI] [PubMed] [Google Scholar]
- 83.Hernandez ME, Mendieta D, Martinez-Fong D, Lori-a F, Moreno J, Estrada I, et al. Variations in circulating cytokine levels during 52 week course of treatment with SSRI for major depressive disorder. Eur Neuropsychopharmacol. 2008;18: 917–24. 10.1016/j.euroneuro.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 84.Simon NM, McNamara K, Chow CW, Maser RS, Papakostas GI, Pollack MH, et al. A detailed examination of cytokine abnormalities in Major Depressive Disorder. Eur Neuropsychopharmacol. 2008;18: 230–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li P, Allen H, Banerjee S, Seshadri T. Characterization of mice deficient in interleukin-1 beta converting enzyme. J Cell Biochem. 1997;64: 27–32. [DOI] [PubMed] [Google Scholar]
- 86.Levine J, Barak Y, Chengappa KN, Rapoport A, Rebey M, Barak V. Cerebrospinal cytokine levels in patients with acute depression. Neuropsychobiology. 1999;40: 171–6. [DOI] [PubMed] [Google Scholar]
- 87.Winkler D, Beconi M, Toledo-Sherman LM, Prime M, Ebneth A, Dominguez C, et al. Development of LC/MS/MS, high-throughput enzymatic and cellular assays for the characterization of compounds that inhibit kynurenine monooxygenase (KMO). J Biomol Screen. 2013;18: 879–889. 10.1177/1087057113489731 [DOI] [PubMed] [Google Scholar]
- 88.Corona AW, Huang Y, O’Connor JC, Dantzer R, Kelley KW, Popovich PG, et al. Fractalkine receptor (CX3CR1) deficiency sensitizes mice to the behavioral changes induced by lipopolysaccharide. J Neuroinflamm. 2010;7: 1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Connor TJ, Starr N, O'Sullivan JB, Harkin A. Induction of indolamine 2, 3-dioxygenase and kynurenine 3-monooxygenase in rat brain following a systemic inflammatory challenge: A role for IFN-γ? Neurosci Lett. 2008;441: 29–34. 10.1016/j.neulet.2008.06.007 [DOI] [PubMed] [Google Scholar]
- 90.Molteni R, Macchi F, Zecchillo C, Dell'Agli M, Colombo E, Calabrese F, et al. Modulation of the inflammatory response in rats chronically treated with the antidepressant agomelatine. Eur Neuropsychopharmacol. 2013;23: 1645–1655. 10.1016/j.euroneuro.2013.03.008 [DOI] [PubMed] [Google Scholar]
- 91.Zunszain PA, Anacker C, Cattaneo A, Choudhury S, Musaelyan K, Myint AM, et al. Interleukin-1β: a new regulator of the kynurenine pathway affecting human hippocampal neurogenesis. Neuropsychopharmacology. 2012;37: 939–949. 10.1038/npp.2011.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O'Connor JC, Lawson MA, Andre C, Briley EM, Szegedi SS, Lestage J, et al. Induction of IDO by Bacille Calmette-Guerin is responsible for development of murine depressive-like behavior. J Immunol. 2009;182: 3202–3212. 10.4049/jimmunol.0802722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parrott JM, O’Connor JC. Kynurenine 3-monooxygenase: an influential mediator of neuropathology. Front Psychiatry. 2015;6: 116 10.3389/fpsyt.2015.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Campbell BM, Charych E, Lee AW, Moller T. Kynurenines in CNS disease: regulation by inflammatory cytokines. Front Neurosci. 2014;8: 12 10.3389/fnins.2014.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xaus J, Comalada M, Valledor AF, Lloberas J, Lopez-Soriano F, Argiles JM, et al. LPS induces apoptosis in macrophages mostly through the autocrine production of TNF-alpha. Blood. 2000;95: 3823–31. [PubMed] [Google Scholar]
- 96.Eue I, Pietz B, Storck J, Klempt M, Sorg C. Transendothelial migration of 27E10+ human monocytes. Int Immunol. 2000;12: 1593–604. [DOI] [PubMed] [Google Scholar]
- 97.Maes M. Evidence for an immune response in major depression: a review and hypothesis. Prog Neuropsychopharmacol Biol Psychiatry. 1995;19: 11–38. [DOI] [PubMed] [Google Scholar]
- 98.Eilat E, Mendlovic S, Doron A, Zakuth V, Spirer Z. Increased apoptosis in patients with major depression: A preliminary study. J Immunol. 1999;163: 533–534. [PubMed] [Google Scholar]
- 99.Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61: 300–8. [DOI] [PubMed] [Google Scholar]
- 100.Ryan MM, Lockstone HE, Huffaker SJ, Wayland MT, Webster MJ, Bahn S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol Psychiatry. 2006;11: 965–978. [DOI] [PubMed] [Google Scholar]
- 101.Bousman CA, Chana G, Glatt SJ, Chandler SD, Lucero GR, Tatro E, et al. Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: convergent pathway analysis findings from two independent samples. Am J Med Genet B Neuropsychiatr Genet. 2010;153B: 494–502. 10.1002/ajmg.b.31006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chiappelli J, Rowland LM, Wijtenburg SA, Muellerklein F, Tagamets M, McMahon RP, et al. Evaluation of myo-inositol as a potential biomarker for depression in Schizophrenia. Neuropsychopharmacology. 2015;40: 2157–2164. 10.1038/npp.2015.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Buga A, Scholz CJ, Kumar S, Herndon JG, Alexandru D, Cojocaru GR, et al. Identification of new therapeutic targets by genome-wide analysis of gene expression in the ipsilateral cortex of aged rats after stroke. PLoS One. 2012;7: e50985–e50985. 10.1371/journal.pone.0050985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jou I, Pyo H, Chung S, Jung SY, Gwag BJ, Joe EH. Expression of Kv1.5 K+ channels in activated microglia in vivo. Glia. 1998;24: 408–14. [PubMed] [Google Scholar]
- 105.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17: 131–43. 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fumagalli S, Perego C, Pischiutta F, Zanier ER, De Simoni M. The ischemic environment drives microglia and macrophage function. Front Neurol. 2015;6: 81–81. 10.3389/fneur.2015.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Trager N, Haque A, Ray SK, Das A, Banik NL. Role of Calpain in immunobiology of neurodegenerative diseases In: Anonymous role of proteases in cellular dysfunction. Springer; 2014. pp. 3–15. [Google Scholar]
- 108.Solga AC, Pong WW, Walker J, Wylie T, Magrini V, Apicelli AJ, et al. RNA-sequencing reveals oligodendrocyte and neuronal transcripts in microglia relevant to central nervous system disease. Glia. 2015;63: 531–548. 10.1002/glia.22754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Milano S, Arcoleo F, Dieli M, D'Agostino R, D'Agostino P, De Nucci G, et al. Prostaglandin E 2 regulates inducible nitric oxide synthase in the murine macrophage cell line J774. Prostaglandins. 1995;49: 105–115. [DOI] [PubMed] [Google Scholar]
- 110.Weisser SB, McLarren KW, Voglmaier N, van Netten-Thomas C,J., Antov A, Flavell RA, et al. Alternative activation of macrophages by IL-4 requires SHIP degradation. Eur J Immunol. 2011;41: 1742–53. 10.1002/eji.201041105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fox MA. Novel roles for collagens in wiring the vertebrate nervous system. Curr Opin Cell Biol. 2008;20: 508–513. 10.1016/j.ceb.2008.05.003 [DOI] [PubMed] [Google Scholar]
- 112.Hubert T, Grimal S, Carroll P, Fichard-Carroll A. Collagens in the developing and diseased nervous system. Cell Mol Life Sci. 2008;66: 1223–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nag A, Bochukova EG, Kremeyer B, Campbell DD, Muller H, Valencia-Duarte AV, et al. CNV analysis in Tourette syndrome implicates large genomic rearrangements in COL8A1 and NRXN1. PLoS One. 2013;8: e59061 10.1371/journal.pone.0059061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Clarke RA, Lee S, Eapen V. Pathogenetic model for Tourette syndrome delineates overlap with related neurodevelopmental disorders including Autism. Transl Psychiatry. 2012;2: e158 10.1038/tp.2012.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Virtaneva KI, Emi N, Marken JS, Aruffo A, Jones C, Spurr NK, et al. Chromosomal localization of three human genes coding for A15, L6, and S5.7 (TAPA1): all members of the transmembrane 4 superfamily of proteins. Immunogenetics. 1994;39: 329–34. [DOI] [PubMed] [Google Scholar]
- 116.Stewart SE, Yu D, Scharf JM, Neale BM, Fagerness JA, Mathews CA, et al. Genome-wide association study of obsessive-compulsive disorder. Mol Psychiatry. 2013;18: 788–98. 10.1038/mp.2012.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Teng C, Yang Y, Chen Y, Kuo Y, Sze C. Role of WWOX/WOX1 in Alzheimer's disease pathology and in cell death signaling. Front Biosci (Elite Ed). 2013;5: 72–85. [DOI] [PubMed] [Google Scholar]
- 118.Zong Y, Yu P, Cheng H, Wang H, Wang X, Liang C, et al. miR-29c regulates NAV3 protein expression in a transgenic mouse model of Alzheimer×3s disease. Brain Res. 2015;1624: 95–102. 10.1016/j.brainres.2015.07.022 [DOI] [PubMed] [Google Scholar]
- 119.Bedard A, Tremblay P, Chernomoretz A, Vallieres L. Identification of genes preferentially expressed by microglia and upregulated during cuprizone-induced inflammation. Glia. 2007;55: 777–789. [DOI] [PubMed] [Google Scholar]
- 120.Preissler J, Grosche A, Lede V, Le Duc D, Kragel K, Matyash V, et al. Altered microglial phagocytosis in GPR34-deficient mice. Glia. 2015;63: 206–215. 10.1002/glia.22744 [DOI] [PubMed] [Google Scholar]
- 121.Wu G, Morris SM. Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336 (Pt 1): 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2: 907–16. [DOI] [PubMed] [Google Scholar]
- 123.Sonoki T, Nagasaki A, Gotoh T, Takiguchi M, Takeya M, Matsuzaki H, et al. Coinduction of nitric-oxide synthase and arginase I in cultured rat peritoneal macrophages and rat tissues in vivo by lipopolysaccharide. J Biol Chem. 1997;272: 3689–93. [DOI] [PubMed] [Google Scholar]
- 124.Ghorpade DS, Leyland R, Kurowska-Stolarska M, Patil SA, Balaji KN. MicroRNA-155 is required for Mycobacterium bovis BCG-mediated apoptosis of macrophages. Mol Cell Biol. 2012;32: 2239–53. 10.1128/MCB.06597-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Svensson C, Part K, Kannis-Beres K, Kaldmae M, Fernaeus SZ, Land T. Pro-survival effects of JNK and p38 MAPK pathways in LPS-induced activation of BV-2 cells. Biochem Biophys Res Commun. 2011;406: 488–492. 10.1016/j.bbrc.2011.02.083 [DOI] [PubMed] [Google Scholar]
- 126.Bronte V, Serafini P, Mazzoni A, Segal DM, Zanovello P. L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol. 2003;24: 302–6. [DOI] [PubMed] [Google Scholar]
- 127.Virag L, Szabo E, Gergely P, Szabo C. Peroxynitrite-induced cytotoxicity: mechanism and opportunities for intervention. Toxicol Lett. 2003;140–141: 113–24. [DOI] [PubMed] [Google Scholar]
- 128.Starossom SC, Mascanfroni ID, Imitola J, Cao L, Raddassi K, Hernandez SF, et al. Galectin-1 deactivates classically activated microglia and protects from inflammation-induced neurodegeneration. Immunity. 2012;37: 249–63. 10.1016/j.immuni.2012.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gormanns P, Mueller NS, Ditzen C, Wolf S, Holsboer F, Turck CW. Phenome-transcriptome correlation unravels anxiety and depression related pathways. J Psychiatr Res. 2011;45: 973–979. 10.1016/j.jpsychires.2010.12.010 [DOI] [PubMed] [Google Scholar]
- 130.Zhang G, Bogdanova N, Gao T, Song JJ, Cragg MS, Glennie MJ, et al. Fcy receptor-mediated inflammation inhibits axon regeneration. PLoS One. 2014;9: e88703–e88703. 10.1371/journal.pone.0088703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17: 142–53. 10.1038/mp.2011.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506: 185–90. 10.1038/nature12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Merner ND, Girard SL, Catoire H, Bourassa CV, Belzil V, Riviare J, et al. Exome sequencing identifies FUS mutations as a cause of essential tremor. Am J Hum Genet. 2012;91: 313–9. 10.1016/j.ajhg.2012.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hmama Z, Pena-Diaz S, Joseph S, Av-Gay Y. Immunoevasion and immunosuppression of the macrophage by Mycobacterium tuberculosis. Immunol Rev. 2015;264: 220–32. 10.1111/imr.12268 [DOI] [PubMed] [Google Scholar]
- 135.Liu B, Wang K, Gao HM, Mandavilli B, Wang JY, Hong JS. Molecular consequences of activated microglia in the brain: overactivation induces apoptosis. J Neurochem. 2001;77: 182–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Data are available from the NCBI/GEO repository GSE76069 and GSE70941 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sxghgcmorrulhwr&acc=GSE76069; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ulczkewahfmnbov&acc=GSE70941).