Abstract
High-level resistance to ertapenem was produced by β-lactamases of groups 1, 2f, and 3 in a strain of Klebsiella pneumoniae deficient in Omp35 and Omp36. From a wild-type strain producing ACT-1 β-lactamase, ertapenem-resistant mutants for which the ertapenem MICs were up to 128 μg/ml and expression of outer membrane proteins was diminished could be selected.
Ertapenem is a potent carbapenem antibiotic for most clinical isolates of Klebsiella pneumoniae, with a typical MIC at which 90% of the isolates tested are inhibited of 0.03 to 0.06 μg/ml (6, 9), but occasional strains for which the MICs are ≥16 μg/ml have been detected (6, 7). In one such strain resistance was dependent on the presence of the plasmid-mediated extended-spectrum β-lactamase (ESBL) SHV-2 and additional host events presumably affecting ertapenem permeativity (7). Further studies were undertaken to elucidate the contribution of β-lactamase and host mutation to such exceptional resistance.
The K. pneumoniae strain for which the ertapenem MIC was 16 μg/ml was treated with ethidium bromide to cure the resident plasmid. The ertapenem MIC for the resulting strain, C2, was still elevated at l μg/ml, and the strain was found to be defective in expression of outer membrane porins OmpK35 and OmpK36 (10). To evaluate the influence of different β-lactamases on the ertapenem susceptibility of this strain, plasmids were introduced by mating with R+ derivatives of Escherichia coli J53 Azir (met pro; azide resistant) (8), with selection on medium lacking the growth requirements of the donor and containing an antibiotic to which the plasmid provided resistance, if possible a non-β-lactam so as to avoid inadvertent selection of additional mutations. A few nonconjugative plasmids were introduced by electroporation. MICs were determined by agar dilution on Mueller-Hinton medium with an inoculum of 104 organisms per spot according to NCCLS protocols (12). E. coli ATCC 25922 was used for quality control. Antibiotics were obtained from Sigma (St. Louis, Mo.) (cefotaxime) and the pharmaceutical companies AstraZeneca (meropenem), Bristol-Meyers Squibb (cefepime), GlaxoSmithKline (ceftazidime), and Merck & Co. (cefoxitin, ertapenem, and imipenem).
In Table 1 the K. pneumoniae C2 derivatives are listed according to the β-lactamase classification scheme of Bush et al. (4). The highest ertapenem MICs (≥128 μg/ml) were achieved by β-lactamase group 1 enzymes ACT-1, DHA-1, and FOX-1 and by group 2f enzyme KPC-1. KPC-1 is a known carbapenemase (14) and was encoded by a multicopy plasmid, while group 1 enzymes have been reported to express carbapenem resistance in strains lacking outer membrane porins (3, 10). Other group 1 enzymes provided a lesser degree of ertapenem resistance, with FOX-3 and FOX-5 β-lactamases conferring MICs of only 8 μg/ml. Group 1 enzymes providing ertapenem resistance also increased resistance to imipenem and meropenem but with diminishing effect: the highest imipenem MIC was 64 μg/ml, and the highest meropenem MIC was 16 μg/ml. With group 2be (ESBL) enzymes, ertapenem MICs of ≥16 μg/ml were conferred by several TEM-type ESBLs, but the maximum MIC with SHV- or CTX-M-type ESBLs was only 8 μg/ml. Susceptibility to imipenem and meropenem was less affected than that to ertapenem. Most group 2c and 2d enzymes had no effect on ertapenem susceptibility, but OXA-2 β-lactamase was exceptional in providing an ertapenem MIC of 32 μg/ml with little if any effect on imipenem or meropenem susceptibility. As expected, the group 3 metallo-β-lactamase VIM-2 elevated the ertapenem MIC for strain C2 to 64 μg/ml, with concomitantly increased resistance to the other carbapenems.
TABLE 1.
Susceptibilities of porin-deficient K. pneumoniae strain C2 containing various plasmid-mediated β-lactamases
| Enzyme group | β-Lacta- mase | Plasmid | MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cefepime | Cefotaxime | Cefoxitin | Ceftazidime | Ertapenem | Imipenem | Meropenem | |||
| None | 0.5 | 0.5 | 64 | 0.5 | 1 | 1 | 0.25 | ||
| 1 | ACC-1 | pSLK54 | 8 | 128 | ≥1,024 | ≥256 | 32 | 8 | 4 |
| ACT-1 | pMG251 | 2 | 128 | ≥1,024 | 64 | ≥128 | 64 | 16 | |
| CMY-2 | pMG250 | 4 | 128 | ≥1,024 | 128 | 32 | 32 | 8 | |
| DHA-1 | pMG247 | 4 | 256 | ≥1,024 | ≥256 | 128 | 64 | 8 | |
| FOX-1 | pGL3a | 16 | ≥256 | ≥1,024 | ≥256 | 128 | 64 | 16 | |
| FOX-3 | p1734 | 2 | 32 | ≥1,024 | 128 | 8 | 2 | 2 | |
| FOX-5 | pMG252 | 4 | 64 | ≥1,024 | 128 | 8 | 2 | 1 | |
| LAT-1 | pHP15 | 8 | ≥256 | ≥1,024 | ≥256 | 64 | 32 | 16 | |
| MIR-1 | pMG233 | 2 | 64 | ≥1,024 | 32 | 32 | 32 | 8 | |
| MOX-1 | pRMOX-1 | 4 | 32 | ≥1,024 | 64 | 32 | 8 | 8 | |
| MOX-2 | pKOL | 8 | 64 | ≥1,024 | 128 | 16 | 2 | 2 | |
| 2b | TEM-1 | R1 | 0.5 | 0.5 | 64 | 0.5 | 2 | 1 | 0.5 |
| TEM-2 | RP1 | 4 | 0.5 | 64 | 1 | 8 | 2 | 2 | |
| 2be | TEM-3 | pCFF04 | 32 | 128 | 64 | 128 | 8 | 2 | 1 |
| TEM-4 | pUD16 | 32 | 128 | 64 | 32 | 16 | 2 | 4 | |
| TEM-5 | pCFF14 | 16 | 16 | 64 | 256 | 16 | 4 | 1 | |
| TEM-6 | pMG226 | 16 | 4 | 16 | ≥256 | 32 | 1 | 1 | |
| TEM-7 | pIF100 | 32 | 2 | 64 | 64 | 16 | 2 | 2 | |
| TEM-8 | Plasmid from E. coli CF804 | 32 | 16 | 64 | ≥256 | 16 | 2 | 0.5 | |
| TEM-9 | pMG228 | 32 | 16 | 64 | ≥256 | 32 | 4 | 0.5 | |
| TEM-10 | pMG223 | 16 | 4 | 64 | ≥256 | 16 | 2 | 0.5 | |
| TEM-11 | pMG244 | 16 | 2 | 64 | 64 | 16 | 2 | 4 | |
| TEM-12 | pMG224 | 16 | 2 | 64 | 64 | 16 | 2 | 0.5 | |
| TEM-15 | pMG274 | 16 | 64 | 64 | ≥256 | 8 | 2 | 2 | |
| TEM-16 | Plasmid from K. pneumoniae CF1304 | 16 | 4 | 64 | ≥256 | 8 | 1 | 0.5 | |
| TEM-19 | pMG289 | 8 | 64 | 32 | 64 | 16 | 4 | 1 | |
| TEM-20 | pUD30 | 64 | 32 | 64 | 16 | 4 | 2 | 2 | |
| TEM-21 | pUD31 | 16 | 128 | 64 | 256 | 16 | 4 | 0.5 | |
| TEM-22 | Plasmid from E. coli HB101 TEM-22 | 8 | 64 | 64 | ≥256 | 16 | 2 | 0.25 | |
| TEM-24 | Plasmid from K. pneumoniae CF1104 | 16 | 32 | 64 | ≥256 | 8 | 0.5 | 0.5 | |
| TEM-25 | Plasmid from E. coli CF1609 | 16 | 32 | 64 | 32 | 16 | 4 | 0.25 | |
| TEM-26 | pMG225 | 32 | 4 | 64 | ≥256 | 8 | 1 | 0.25 | |
| TEM-52 | pMG276 | 8 | 32 | 64 | 64 | 8 | 4 | 0.5 | |
| TEM-61 | pMG290 | 8 | 4 | 16 | ≥256 | 16 | 4 | 2 | |
| TEM-71 | pMG259 | 1 | 0.5 | 64 | 256 | 8 | 4 | 0.125 | |
| TEM-88 | pMG272 | 8 | 32 | 64 | 64 | 16 | 2 | 0.5 | |
| 2b | SHV-1 | R1010 | 0.5 | 0.5 | 64 | 0.5 | 2 | 0.5 | 0.5 |
| 2be | SHV-2 | pMG258 | 16 | 4 | 64 | ≥256 | 8 | 1 | 0.25 |
| SHV-3 | pUD18 | 32 | 64 | 64 | ≥256 | 4 | 2 | 1 | |
| SHV-4 | pUD21 | 8 | 16 | 64 | 256 | 4 | 0.5 | 0.25 | |
| SHV-5 | pAFF2 | 16 | 32 | 64 | ≥256 | 8 | 2 | 2 | |
| SHV-6 | pSLH47 | 1 | 0.5 | 64 | 32 | 8 | 0.5 | 1 | |
| SHV-12 | pMG242 | 4 | 4 | 32 | ≥256 | 4 | 2 | 2 | |
| SHV-18 | pMG266 | 4 | 8 | 64 | 64 | 8 | 1 | 2 | |
| CTX-M-5 | pCLL3417a | 128 | ≥256 | 64 | 4 | 8 | 2 | 1 | |
| CTX-M-14 | pMG267 | 32 | 32 | 64 | 8 | 8 | 2 | 2 | |
| 2b | HMS-1 | R997 | 0.5 | 0.5 | 64 | 1 | 2 | 0.5 | 1 |
| 2c | PSE-1 | pMG217 | 0.5 | 0.5 | 64 | 4 | 2 | 1 | 0.5 |
| PSE-4 | pUZ8::Tn1405 | 0.5 | 0.5 | 64 | 0.5 | 2 | 1 | 2 | |
| CARB-3 | pUZ8::Tn1408 | 1 | 0.5 | 64 | 0.5 | 1 | 1 | 1 | |
| CARB-4 | pUZ8::Tn1413 | 0.5 | 0.25 | 32 | 0.5 | 1 | 1 | 2 | |
| SAR-1 | pUK657 | 0.5 | 0.5 | 64 | 0.5 | 2 | 1 | 0.5 | |
| 2d | OXA-1 | RGN238 | 4 | 0.5 | 64 | 0.5 | 2 | 1 | 2 |
| OXA-2 | R46 | 0.5 | 0.5 | 64 | 16 | 32 | 2 | 1 | |
| OXA-3 | R55 | 0.25 | 0.25 | 16 | 2 | 1 | 0.5 | 2 | |
| OXA-4 | pMG203 | 1 | 0.5 | 64 | 0.5 | 1 | 1 | 0.25 | |
| OXA-5 | pMG54 | 0.5 | 0.5 | 64 | 0.5 | 1 | 1 | 0.25 | |
| OXA-7 | pMG202 | 1 | 1 | 32 | 1 | 1 | 1 | 2 | |
| OXA-10 | pUZ8::Tn1404 | 0.5 | 0.5 | 64 | 0.5 | 2 | 1 | 2 | |
| LCR-1 | pUZ8::Tn1412 | 0.5 | 0.5 | 64 | 0.5 | 2 | 0.5 | 1 | |
| 2f | KPC-1 | pBR322-catI-blaKPC-1a | 128 | 128 | 128 | 64 | ≥128 | ≥128 | ≥128 |
| 3 | VIM-2 | pNOR2001a | 4 | 64 | 128 | 32 | 64 | 64 | 128 |
Multicopy recombinant plasmid.
Cefepime MICs of ≥32 μg/ml were produced in strain C2 with some TEM- and SHV-type ESBLs, by CTX-M-5 and M-14, and by KPC-1 β-lactamase. MICs of cefotaxime and ceftazidime were ≥32 μg/ml with group 1 enzymes as well as some TEM- and SHV-type ESBLs, KPC-1 and VIM-2.
Carbapenem resistance decreased markedly when the plasmid host had a wild-type complement of porins. The native ertapenem MIC for a susceptible clinical isolate of K. pneumoniae (strain 002) was 0.015 μg/ml, which increased to 0.5 μg/ml when plasmid pMG251, encoding ACT-1 β-lactamase (1), was introduced and to only 4 μg/ml when plasmid-mediated KPC-1 was present. From 002(pMG251) making ACT-1 β-lactamase, spontaneous mutants could be selected on Mueller-Hinton agar containing 2 μg of ertapenem per ml at a frequency of 6 × 10−8, for which the ertapenem MIC was 4 μg/ml. Such a first-step mutant gave rise to colonies on medium with 64 μg of ertapenem per ml at a frequency of 8 × 10−9. The ertapenem MIC for a second-step mutant was 128 μg/ml. β-Lactamase production by the mutants was unchanged. Table 2 shows the susceptibilities of these strains to other β-lactams. Resistance to imipenem and meropenem increased along with that to ertapenem, while susceptibility to ceftazidime and cefepime was less affected.
TABLE 2.
Susceptibilities of K. pneumoniae 002(pMG251) and its derivatives to β-lactams
| Strain | MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|
| Cefepime | Cefotaxime | Cefoxitin | Ceftazidime | Ertapenem | Imipenem | Meropenem | |
| 002(pMG251) | 0.125 | 2 | 512 | 8 | 0.5 | 2 | 0.125 |
| 002(pMG251) Ertapenemr MIC 4a | 1 | 32 | ≥1,024 | 16 | 4 | 16 | 1 |
| 002(pMG251) Ertapenemr MIC 4 carrying pQE7Kb | 0.5 | 16 | ≥1,024 | 8 | 2 | 2 | 0.5 |
| 002(pMG251) Ertapenemr MIC 4 carrying pSHA25Kc | 0.125 | 2 | 512 | 8 | 0.25 | 2 | 0.125 |
| 002(pMG251) Ertapenemr MIC 128d | 2 | 32 | ≥1,024 | ≥32 | 128 | ≥128 | 16 |
| 002(pMG251) Ertapenemr MIC 128 carrying pQE7K | 1 | 32 | ≥1,024 | ≥32 | 16 | 8 | 2 |
| 002(pMG251) Ertapenemr MIC 128 carrying pSHA25K | 0.06 | 1 | 512 | 8 | 0.25 | 2 | 0.125 |
Ertapenemr MIC 4, resistant derivative for which the ertapenem MIC was 4 μg/ml.
Codes for OmpK36.
Codes for OmpK37.
Ertapenemr MIC 128, resistant derivative for which the ertapenem MIC was 128 μg/ml.
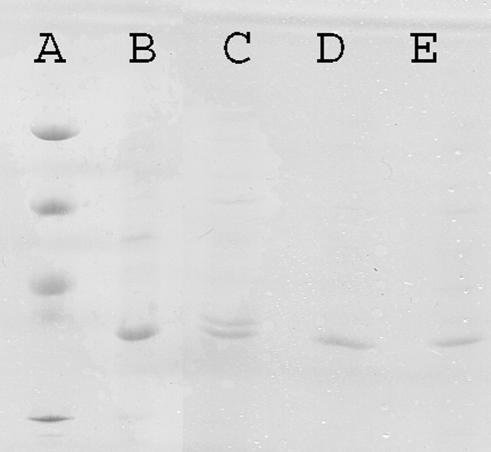
Outer membrane proteins of strain 002(pMG251) and its two ertapenem-resistant derivatives were prepared (10) as sodium lauryl sarcosinate (2%)-insoluble material from cell envelopes obtained by sonication of bacteria after growth in nutrient broth (Difco) and were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis by using the PhastSystem with PhastGel 12.5 medium (Pharmacia Biotech). Two protein bands were seen with strain 002 at about 35 and 36 kDa, probably corresponding to an OmpA-like protein and a porin, respectively. The upper band was lost in both of the ertapenem-resistant mutants (Fig. 1).
FIG. 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of outer membrane proteins with protein standards of 97, 66, 49, and 30 kDa (lane A), K. pneumoniae C2 (lane B), K. pneumoniae 002(pMG251) (lane C), the K. pneumoniae 002(pMG251) ertapenem-resistant derivative for which the MIC was 4 μg/ml (lane D), and the K. pneumoniae 002(pMG251) ertapenem-resistant derivative for which the MIC was 128 μg/ml (lane E).
To elucidate the events responsible for ertapenem resistance further, plasmids pSHA25K, encoding OmpK36, and pQE7K, encoding OmpK37 (5), were introduced into the ertapenem-resistant K. pneumoniae 002(pMG251) derivatives by electroporation. Both plasmids determine kanamycin resistance, as does pMG251, but neomycin could be used to select for plasmid acquisition. Transfer of plasmid pSHA16K, encoding OmpK35 (5), was also attempted, but despite being based on the same vector as the other Omp constructs, pSHA16K proved to be incompatible with pMG251 as either the entering or resident plasmid.
Acquisition of either pSHA25K or pQE7K increased β-lactam susceptibility for both the first- and second-step ertapenem-resistant mutants. Plasmid pSHA25K had the greater effect (Table 2). Porin loss was thus clearly involved in mutations to enhanced ertapenem resistance, but since both high- and low-level resistance mutants had susceptibility restored, the sequence of events responsible for the two resistance levels has not been established. Enhanced β-lactam efflux might be involved, but compounds reported to inhibit quinolone efflux in K. pneumoniae or other organisms, such as 25 μg of reserpine (Sigma) per ml (13), 100 μM carbonyl cyanide m-chlorophenylhydrazone (Sigma) (11), or 80 μg of Phe-Arg β-naphthylamide (Sigma) per ml (2), failed to block ertapenem resistance in the K. pneumoniae 002(pMG251) derivatives.
Ertapenem resistance in K. pneumoniae thus depended on production of particular β-lactamases and defects in permeability. In most strains loss of susceptibility was more marked for ertapenem than for imipenem or meropenem. Additional clinical isolates with this resistance phenotype have recently been reported (D. L. Paterson, R. A. Bonomo, J. Kolano, S. Patel-Brown, K. M. Hujer, L. B. Rice, and V. L. Yu, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1886, 2002; J. P. Quinn, A. M. Hujer, C. R. Bethel, P. Schreckenberger, and R. A. Bomomo, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-671, 2003).
Acknowledgments
This study was supported in part by a grant from Merck & Co., Inc.
We thank A. Doménech for providing plasmids pQE7K, pSHA16K, and pSHA25K.
REFERENCES
- 1.Alvarez, M., J. H. Tran, N. Chow, and G. A. Jacoby. 2004. Epidemiology of conjugative plasmid-mediated AmpC β-lactamases in the United States. Antimicrob. Agents Chemother. 48:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baucheron, S., H. Imberechts, E. Chaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar typhimurium phage type DT204. Microb. Drug Resist. 8:281-289. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doménech-Sánchez, A., S. Hernández-Allés, L. Martínez-Martínez, V. J. Benedí, and S. Albertí. 1999. Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in β-lactam antibiotic resistance. J. Bacteriol. 181:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs, P. C., A. L. Barry, and S. D. Brown. 2001. In vitro activities of ertapenem (MK-0826) against clinical bacterial isolates from 11 North American medical centers. Antimicrob. Agents Chemother. 45:1915-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby, G., P. Han, and J. Tran. 1997. Comparative in vitro activities of carbapenem L-749,345 and other antimicrobials against multiresistant gram-negative clinical pathogens. Antimicrob. Agents Chemother. 41:1830-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacoby, G. A., and P. Han. 1996. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J. Clin. Microbiol. 34:908-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livermore, D. M., M. W. Carter, S. Bagel, B. Wiedemann, F. Baquero, E. Loza, H. P. Endtz, N. van den Braak, C. J. Fernandes, L. Fernandes, N. Frimodt-Moller, L. S. Rasmussen, H. Giamarellou, E. Giamarellos-Bourboulis, V. Jarlier, J. Nguyen, C. E. Nord, M. J. Struelens, C. Nonhoff, J. Turnidge, J. Bell, R. Zbinden, S. Pfister, L. Mixson, and D. L. Shungu. 2001. In vitro activities of ertapenem (MK-0826) against recent clinical bacteria collected in Europe and Australia. Antimicrob. Agents Chemother. 45:1860-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Martínez, L., A. Pascual, S. Hernández-Allés, D. Alvarez-Díaz, A. I. Suárez, J. Tran, V. J. Benedí, and G. A. Jacoby. 1999. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazzariol, A., J. Zuliani, G. Cornaglia, G. M. Rossolini, and R. Fontana. 2002. AcrAB efflux system: expression and contribution to fluoroquinolone resistance in Klebsiella spp. Antimicrob. Agents Chemother. 46:3984-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. NCCLS, Wayne, Pa.
- 13.Ribera, A., A. Jurado, J. Ruiz, F. Marco, O. Del Valle, J. Mensa, J. Chaves, G. Hernández, M. T. Jiménez de Anta, and J. Vila. 2002. In vitro activity of clinafloxacin in comparison with other quinolones against Stenotrophomonas maltophilia clinical isolates in the presence and absence of reserpine. Diagn. Microbiol. Infect. Dis. 42:123-128. [DOI] [PubMed] [Google Scholar]
- 14.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]