Abstract
Azole resistance in Candida albicans can be due to upregulation of multidrug transporters belonging to ABC (ATP-binding cassette) transporters (CDR1 and CDR2) or major facilitators (CaMDR1). Upregulation of these genes can also be achieved by exposure to fluphenazine, resulting in specific upregulation of CDR1 and CDR2 and by exposure to benomyl, resulting in specific CaMDR1 upregulation. In this study, these two different states of gene upregulation were used to determine coregulated genes that often share similar functions or similar regulatory regions. The transcript profiles of a laboratory strain exposed to these drugs were therefore determined and compared with those of two matched pairs of azole-susceptible and -resistant strains expressing CDR1 and CDR2 (CDR strains) or CaMDR1 (MDR isolates). The results obtained revealed that, among 42 commonly regulated genes (8.6% of all regulated genes) between fluphenazine-exposed cells and CDR isolates, the most upregulated were CDR1 and CDR2 as expected, but also IFU5, RTA3 (which encodes putative membrane proteins), HSP12 (which encodes heat shock protein), and IPF4065 (which is potentially involved in stress response). Interestingly, all but HSP12 and IPF4065 contain a putative cis-acting drug responsive element in their promoters. Among the 57 genes (11.5% of all regulated genes) commonly regulated between benomyl-exposed cells and MDR isolates, the most upregulated were CaMDR1 as expected but also genes with oxido-reductive functions such as IFD genes, IPF5987, GRP2 (all belonging to the aldo-keto reductase family), IPF7817 [NAD(P)H oxido-reductase], and IPF17186. Taken together, these results show that in vitro drug-induced gene expression only partially mimics expression profiles observed in azole-resistant clinical strains. Upregulated genes in both drug-exposed conditions and clinical strains are drug resistance genes but also genes that could be activated under cell damage conditions.
Candida albicans is an important opportunistic fungal pathogen in human. It can cause mucosal and systemic infections in immunocompromised patients (20). Among the different antifungal agents available, the class of azoles has been used extensively during the past 20 years. Due to the repeated use of this agent especially in human immunodeficiency virus-positive patients with recurrent oropharyngeal candidiasis, treatment failures were observed to be associated with emergence of azole-resistant C. albicans strains (22). We have been interested in understanding the molecular mechanisms of resistance to azole antifungal agents in C. albicans. Our work and those of others have already demonstrated the importance of multidrug transporters in this phenomenon, although other mechanisms such as target site alterations were also identified (20, 21, 24, 26-28, 30-32). The upregulation of multidrug transporter genes leads to enhanced efflux of azoles and therefore results in decreased drug accumulation and reduced inhibition of their target encoded by the ERG11 gene. At least two families of multidrug transporters, the ABC (ATP-binding cassette) transporter family and the major facilitator superfamily (MFS) have been shown to be involved in antifungal resistance. Cdr1p and Cdr2p (Candida drug resistance, ABC transporters) and CaMdr1p (C. albicans multidrug resistance 1, MFS transporter) were shown to be major mediators of azole resistance. It was previously demonstrated that each gene encoding these proteins can be upregulated in distinct clinical azole-resistant strains (27, 28). Multidrug transporter genes can also be induced transiently by treating cells with different drugs. For example, estradiol and fluphenazine exposure can induce CDR1 and CDR2 (10), whereas benomyl and H2O2 can induce CaMDR1 (17). The regulatory elements permitting the upregulation of these genes have been determined only for CDR1 and CDR2. A 22-bp drug-responsive element (DRE) is known to be required for induction of both CDR1 and CDR2 (10). Transient upregulation of these genes therefore mimics gene expression levels measured in clinical azole-resistant isolates. However, in vitro exposure of C. albicans to these drugs as well as natural history of clinical isolates may result in profound gene expression alterations additional to those already observed.
Several genome-wide studies performed in Saccharomyces cerevisiae (1, 3) have established that exposure to drugs can trigger the expression of genes that either belong to pathways affected by these drugs or are involved in resistance to the tested drugs. On the other hand, exposure of S. cerevisiae to specific agents can reveal clusters of coordinately regulated genes that possess common regulatory elements recognized by the same transcription factors (11, 33). A few similar genome-wide studies have been undertaken in clinical C. albicans isolates. Rogers et al. and Cowen et al. (8, 23) have analyzed transcript profiles of azole-resistant isolates, while de Backer et al. (9) have determined the effect of azole treatment on gene expression in C. albicans. In the present study we aimed to identify genes commonly regulated between cells transiently exposed to drugs and azole-resistant clinical strains. These two different conditions should enable the identification of genes commonly regulated in addition to those already known, such as CDR1, CDR2, and CaMDR1. As mentioned above for S. cerevisiae, coordinately regulated genes are helpful in the delimitation of common regulatory elements deduced from the analysis of their promoters. We therefore undertook genome-wide transcript profiling experiments with matched pairs of clinical isolates—one pair with a resistant strain upregulating CDR genes and another pair with a resistant strain upregulating CaMDR1—and with strains exposed to drugs known to induce multidrug transporter genes, namely, fluphenazine and benomyl.
The results showed that transcripts profiles of the azole-resistant strain upregulating the CDR genes revealed more genes regulated in common with profiles of fluphenazine-treated cells than with profiles of benomyl-treated cells. Moreover, transcript profiles of the azole-resistant strain upregulating CaMDR1 exhibited transcript profiles resembling more closely those found in benomyl-treated cells. As expected, some of the commonly regulated genes observed in both the selected azole-resistant isolate and drug-treated cells possess common regulatory elements. These genes shared functions in drug resistance but also in response to cell-damaging conditions.
MATERIALS AND METHODS
C. albicans strains and growth conditions.
Two pairs of C. albicans azole-susceptible and azole-resistant clinical isolates and the laboratory strain CAF2-1 were used in this study and are described in Table 1. Each pair of isolates consists of strains with matched Ca3 profiles (4, 5). Strains were cultivated at 30°C under constant shaking in liquid complete medium YEPD consisting of 1% Bacto Peptone (Difco Laboratories, Basel, Switzerland), 0.5% yeast extract (Difco) and 2% glucose (Fluka, Buchs, Switzerland).
TABLE 1.
C. albicans strains used in this study
| Strain name | Genotype or description | MIC (μg/ml) of fluconazole | Reference(s) |
|---|---|---|---|
| CAF2-1 | Δura3::imm434/URA3 | 0.5 | 16 |
| DSY294 | Azole-susceptible clinical strain C43a | 0.25 | 4, 28 |
| DSY296 | Azole-resistant clinical strain C56a | 128 | 4, 28 |
| DSY2285 | Azole-susceptible clinical strain 26a | 0.25 | 5, 6 |
| DSY2286 | Azole-resistant clinical strain 91a | 16 | 5, 6 |
C. albicans clinical isolates with original strain numbering.
Microarray design.
Eurogentec (Seraing, Belgium) C. albicans cDNA Microarray slides were used in this study. Briefly, 6,039 open reading frames from PCR amplicons 300 bp long were spotted in duplicate onto each aldehyde coated glass slide onto 32 blocks. The microarray covers nearly 98% of the total number of C. albicans genes and includes 27 control genes in each block, including, for example, negative controls (intergenic regions), cross hybridization controls (S. cerevisiae genes), and dynamic range controls by a serial dilution of TEF3 gene from 1 to 1/32.
RNA isolation and probe labeling.
Each C. albicans strain was grown overnight in 5 ml of YEPD medium under constant agitation at 30°C. For RNA extractions, cultures were diluted at a density of 0.75 × 107 cells per ml in 15 ml of fresh YEPD medium and were grown at 30°C with agitation for 2 h to reach a density of 1.5 × 107 cells per ml. At this point cultures of clinical isolates were centrifuged at 4°C, 5 min at 5,500 × g for subsequent RNA isolation. Strain CAF2-1 was exposed either for 20 min to 10 mg liter−1 of fluphenazine (Sigma-Aldrich, Buchs, Switzerland) or for 30 min to 25 mg liter−1 of benomyl (Riedel de Hahn, Seelze, Germany) at 30°C under agitation. This strain was cultivated in parallel under the same conditions except that no drugs were added. After drug exposure, cultures were centrifuged at 4°C and 5,500 × g for RNA isolation. Total RNA was extracted using glass beads as described previously (25). A least 200 μg of each RNA was purified according to the supplier (RNeasy kit; QIAGEN Inc., Chatsworth, Calif.). Concentration of purified RNA was measured spectrophotometrically at A260 and A280 and adjusted to 2.5 mg ml−1. RNA was stored at −80°C until use.
For RNA labeling, 25 μg of total RNA were mixed with 8 μl of 5× First Strand buffer (Invitrogen, Basel, Switzerland), 1 μl of 0.1 μM C. albicans-specific primer mix Plus (including T20VN and OligodT18-21) (Eurogentec), 3 μl of 6.67 mM (each) deoxynucleoside triphosphate (Roche, Basel, Switzerland), 1 μl of 1 mM dCTP (Roche), 1.5 μl of 1 mM cyanin 3(Cy3)-dCTP (Amersham Biosciences, Otelfingen, Switzerland) for RNA of non-drug-exposed or azole-susceptible clinical strains or 1.5 μl of 1 mM cyanin 5 (Cy5)-dCTP (Amersham Biosciences) for RNA of drug-exposed or azole-resistant clinical strains, 4 μl of 100 mM dithiothreitol (Invitrogen), 1 μl of RNasin (20 to 40 U/μl; Promega, Wallisellen, Switzerland), and diethyl pyrocarbonate-treated water to a volume of 40 μl. The mixture was heated at 65°C for 5 min and cooled at 42°C for 5 min. The reverse transcription (RT) reaction was performed for 1 h at 42°C with 1 μl of RNasin and 200 U of SuperScript II reverse transcriptase (Invitrogen). The reaction was stopped and RNA degraded by addition of 5 μl of EDTA (50 mM, pH 8.0) and 2 μl of NaOH (10 N) and incubation at 65°C for 20 min. The reaction was neutralized with 4 μl of acetic acid (5 M). The labeled probes (cDNA) were purified according to QiaQuick PCR purification kit protocol (QIAGEN Inc.). The elution step was performed twice with prewarmed H2O (42°C) and centrifuged at maximum speed for 1 min. The purified labeled probes were concentrated using microcon-30 filter (Amicon, Wallisellen, Switzerland) to a final volume of 5 μl.
Hybridization of the arrays.
Cy5- and Cy3-labeled cDNAs synthesized from RNA of cells from which transcription profiles have to be compared were pooled with 5 μl of heat-denatured salmon sperm DNA (10 mg ml−1), boiled at 95°C for 2 min, and chilled on ice. Probes were then mixed with 40 μl of hybridization buffer (DIG easy hyb; Roche), applied to a C. albicans microarray slide and covered with a Lifterslips (25 by 44 mm; Erie Scientific Co., Portsmouth, Maine) during all hybridization steps, and loaded onto the hybridization chamber (Corning Inc., Corning, N.Y.) which was kept wet by loading 10 μl of H2O in each existing well. After overnight hybridization at 42°C, coverslips were removed from slides by dipping in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% sodium dodecyl sulfate. Slides were washed for 5 min at room temperature with occasional agitation and rinsed with 0.2× SSC for 5 min at room temperature with occasional agitation. Slides were then spin dried (500 × g, 5 min) and stored protected from light until scanning.
Quantitative analysis of the microarrays.
Hybridized microarray slides were scanned with a ScanArray 4000 (Perkin-Elmer, Schwerzenbach, Switzerland) at a 10-μm resolution. The following wavelengths were used for photoexcitation: 532 nm for Cy3- and 635 nm for Cy5-labeled cDNA. The intensity of each laser light was determined by equalizing the Cy3 and the Cy5 fluorescent signals of the dynamic range controls (TEF3) at 3 different blocks for each slide. The resulting 16-bit TIFF files of each signal were quantified and converted into a text file with the ImaGene software (version 4.0; BioDiscovery Inc., El Segundo, Calif.).
Normalization control.
Experiments from each hybridization were normalized using a LOWESS analysis for print-tip variability and for slide-to-slide variability using the publicly available software from Lund University (http://www.braju.com/R/com.braju.sma/) run in R project environment (http://cran.r-project.org/). In order to determine a significant threshold, we performed a LOWESS normalization only on the TEF3 spots, TEF3 being an housekeeping gene. The significant threshold was considered above the most modulated TEF3 hybridization signals. A fourfold differential expression limit was therefore chosen in this study in the comparison of pairwise experiments. Normalized data can be downloaded from http://www.hospvd.ch/imulunder the file name Microarray data: array_data_sanglard.xls.
Cluster analysis.
Cluster analysis was performed using the CLUSTER program version 2.11 (12). Genes with a twofold differential expression under at least one condition were selected for the clustering of the global analysis (see Fig. 1 and 2). Genes with a fourfold differential expression under at least one condition were selected for clustering of pairwise microarray experiments (see Fig. 3 and 5). Signal intensities of each selected gene were compared between the different conditions using the complete linkage clustering program in the uncentered correlation mode of hierarchical clustering. Results were viewed using TREEVIEW (version 1.6) (12).
FIG. 1.
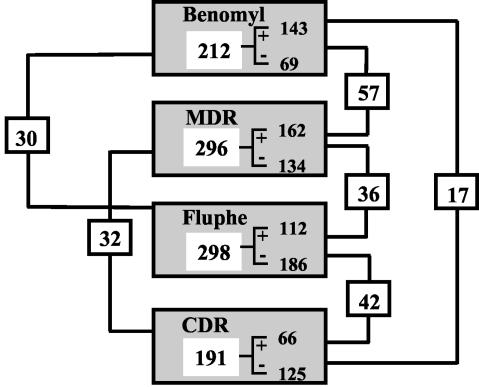
Numbers of C. albicans genes with at least twofold differences in levels of expression. For each experiment, the number of up- or downregulated genes is given. Pairwise comparison between each experiment yielded numbers of commonly regulated genes and these numbers are given linked to each compared experiments.
FIG. 2.
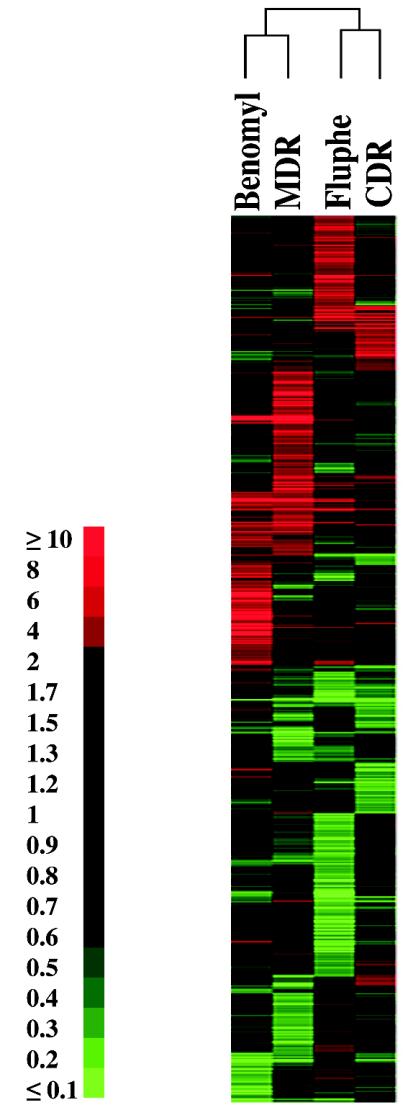
Cluster analysis of the four experiments undertaken in this study. The threshold value used in the analysis is at least a twofold increase in gene expression. The cluster was constructed as indicated in Materials and Methods. A quantitative color scale of gene expression ratio in expression values (n-fold) is given at the left side of the figure.
FIG. 3.
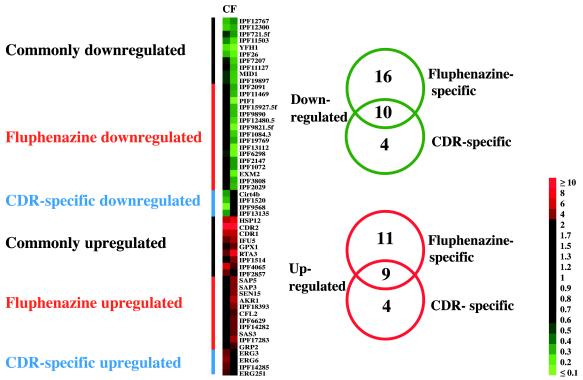
Cluster analysis of the fluphenazine and CDR experiments. The threshold used in this analysis is a fourfold difference in expression. Genes have been arranged in different categories according to their expression profiles. The number of genes in each category is shown at the right side of the figure. Abbreviations: C, CDR; F, fluphenazine.
FIG. 5.
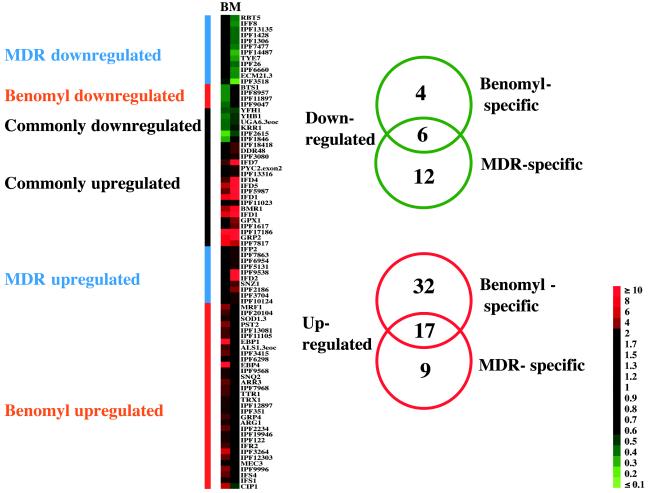
Cluster analysis of the benomyl and MDR experiments. The threshold used in this analysis is of fourfold difference of expression.
Northern blot analysis.
Northern blotting was carried out as described previously (28). DNA probes used in this study were generated by PCR using pairs of primers described in Table 2. 32P-DNA labeled probes were generated by random priming (14). Quantifications of Northern blot bands were performed by scanning the hybridized membranes in an Instant Imager (Packard Instrument Company, Meriden, Conn.). Signals were integrated by the software supplied by the manufacturer and normalized to the corresponding values of an internal standard. This standard was a TEF3 probe originating from a 0.7-kb EcoRI-PstI fragment from pDC1 described by Hube et al. (18). The signals obtained for the same gene in two conditions were expressed as a ratio of normalized signal intensities. The ratio obtained for the selected genes (see Fig. 4 and 6) could be thus compared with those calculated in microarray experiments.
TABLE 2.
Primers used for Northern blot and RT-PCR analysisa
| Primer name | Sequence |
|---|---|
| IPF1514 F | CTGCAAGTGCATTAAGACAT |
| IPF1514 R | ATACGAATCGGACATTTTTC |
| IPF2857 F | F-ATGTTGCGTCAAAGTTTATTC |
| IPF2857 R | CTACATCAAATCTTCTGGTC |
| IPF4065 F | ATGGCTAAAACTTCTCCAC |
| IPF4065 R | TTAATTAGCAGCGTCACCATTAC |
| IPF6629 F | ATGACTGACGGTAAATTCCC |
| IPF6629 R | TTAATTGTGGATTCTCTTTAAAA |
| IPF14285 F | F-GATCCACCCTTATCACCTAA |
| IPF14285 R | AGTAGCAGAATCTGGTGGTT |
| CDR1 F | GTTGTTTTGGGGAGACCCGGTGCT |
| CDR1 R | CTTACCAGCACCAGATGCTCCCAT |
| CDR2 F | ATTCAATTCACGGAAATCGGATA |
| CDR2 R | GTGCACAGTGCACACATTAACC |
| ERG3 F | ATCATCTGGTCTTCTGTAAGATTT |
| ERG3 R | AAATTCATTCTTTTCACCGATTGT |
| IPF2186 F | CAACGAGCAAACGTCAGAAA |
| IPF2186 R | CCCAAACCCAAGCAAATCTA |
| IPF3264 F | GACAACTCCCCAAGTTACAT |
| IPF3264 R | AAATCAGCACCTAAGCTTTC |
| IPF3415 F | AACTTGCTAAACAAGGTGGT |
| IPF3415 R | GAATCTAACTTCTCGATGGC |
| IPF3704 F | GCACACGTTTGTCATGGAAC |
| IPF3704 R | ACTTGTCGCCAAAGCCTAGA |
| IPF7817 F | TGCTAGAAGAGCTGTTGAAA |
| IPF7817 R | TAATCGTTACCACCAGATGA |
| IPF7863 F | ACCGAATTGCCACTTGATTC |
| IPF7863 R | TGTCGATAGCACCAGCAAAG |
| IPF9538 F | CTGGATGCAAATCAAGCAGA |
| IPF9538 R | TTTTGTCAAACGGCCATGTA |
| IPF17186 F | GTACTTTTGGATGGGATGAT |
| IPF17186 R | TTCAGCAACTTGTTTAATGG |
| CaMDR1 F | AAGTCGCACGCGTAAAATGCATTACAGATTTTTAAGAGAT |
| CaMDR1 R | TCAGCGACGCGTCTAATTAGCATACTTAGATCTTGCTCT |
| CIP1 F | ATTACAAGAAAAGAACCGGA |
| CIP1 R | GTCATCAGCATTGATACCAG |
| IFD1 F | CGTTGTTGAGAAAGGGTGGGCA |
| IFD1 R | CAATAGTATTCTCCATTTTACC |
| IFD4 F | GGTGCCTCATCTATGAAAACA |
| IFD4 R | CGTTAATCTTATCTGCGTCTCT |
| IFD7 F | AATGATGTTGTCGAAAAGGGA |
| IFD7 R | CAATAGCTTTGTCATTGTCGTT |
| ERG6 F | TATTCCCAATTGACTCATCA |
| ERG6 R | TACCATCACCGACTTCAATA |
| GPX1 F | ATGTCTCAATTTTACGAATTAGC |
| GPX1 R | CTTCAACAATTCTTCAATCTTGG |
| GRP2 F | AATCCAGTATATGTATTTGGTCC |
| GRP2 R | CTCAATCTTACTTTCAGCTTTCT |
| HSP12 F | ATTTCTACTAAAATCAACGAAGC |
| HSP12 R | GACAACTCCACTCACGTATTC |
| IFU5 F | AAACCCACCACAAGTTCCTG |
| IFU5 R | CTTGGGGCATTAGACCTTGA |
| RTA3 F | TACAGAATGGACTCCTACCT |
| RTA3 R | GCCGTACGATTTAATCGA |
| SAP3 F | CTGATTTATGGGTTCCTGAT |
| SAP3 R | CACCAAAAATAATTTGTCCA |
| SAP5 F | GCTTCTACTGGGCAAATTAT |
| SAP5 R | CTTCACTTTCACGAATACGA |
| DDR48 F | TTTCGGTTTCGGTAAAGACG |
| DDR48 R | TGTCAGTGTTTGAGGAGCCA |
| EBP1 F | GCCTCTAATAAAAGAACCGA |
| EBP1 R | CAAAGTTTCCTTTCCAAATC |
| EBP4 F | TGGAAGAAAATCAATGATGA |
| EBP4 R | CAATTTTTTCAGCACCAACTA |
| IFR2 F | TCAAGCCACTATCAAATACG |
| IFR2 R | AAGCCCTTGAAAAGTTGTAG |
| MRF1 F | GGCGGATATAACGATGCAGT |
| MRF1 R | ACTTTCGATTGTGGATTGGC |
| SNZ1 F | CCGGTTTAGCACAAATGCTT |
| SNZ1 R | CCCATCAATTCACCCAAATC |
| TRX1 F | TTCACGTTGTCACTGAAGTT |
| TRX1 R | GAGAAGCCAAAGCTTGTTTA |
| TTR1 F | CAACAATGGTTTCATCTCAA |
| TTR1 R | CAGCTTTGATTTTGTCATCT |
| IFD2 F | GCAACAAGAGATAATTTCCCA |
| IFD2 R | CAAGATGTAATAACTTAAAAG |
| IFD5 F | TGTTGTTGAAAAAGGGTTGACG |
| IFD5 R | CAATAGTTTTATCAGCTTCACG |
| TEF3 F | AGAAACCGTCCACTTGTTGG |
| TEF3 R | GCAAGAAAATGGTGGCAAAT |
| DFG5 F | ATAATTGCGGTGGTGGGTTA |
| DFG5 R | CACATGTGACTCCATCGGAC |
| PHR2 F | CTCCAGTTTGGTCTGGTGGT |
| PHR2 R | CGGTTCTGACGGTACCAGAT |
Primers were designed using the primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi/primer3_www.cgi) on the ORF sequence. Abbreviations: F, forward primer; R = reverse primer.
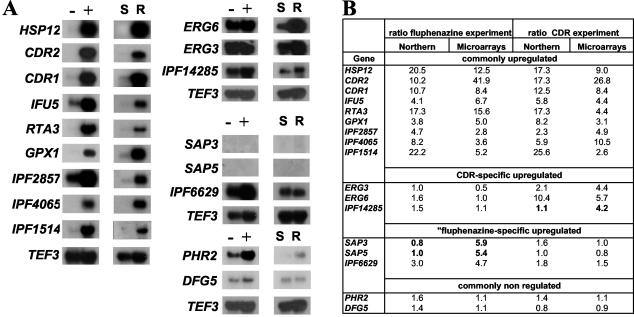
FIG. 4.
Verification of differential expression of selected genes upregulated in the fluphenazine and CDR experiments. (A) Northern blot analysis. The TEF3 gene was used as an RNA loading control. (B) Expression ratio obtained by microarray experiments and by Northern blot analysis. These ratios were obtained as described in Materials and Methods. Numbers in boldface type indicate discrepancies between both analysis. Symbols: (−) and (+), nonexposed or fluphenazine-exposed conditions, respectively; (S), azole-susceptible strain DSY294; (R), azole-resistant strain DSY296.
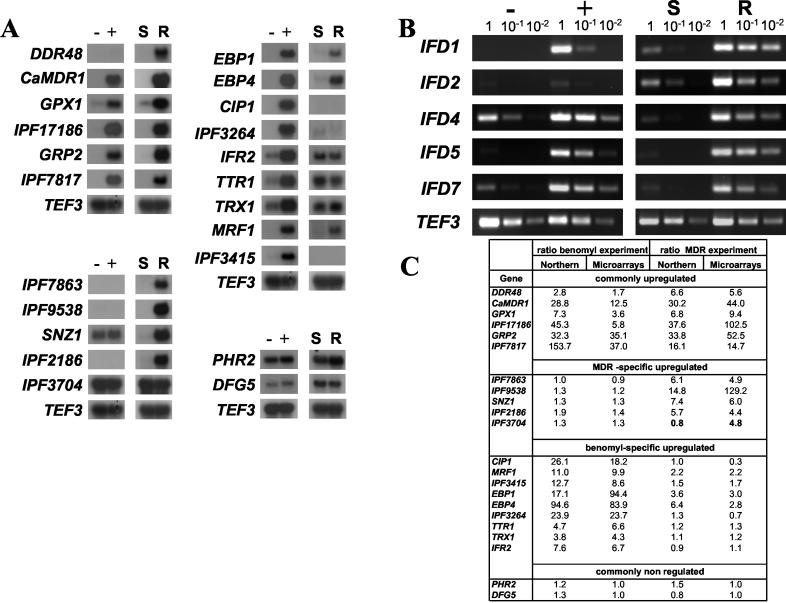
FIG. 6.
Verification of differential expression of selected genes upregulated in the benomyl and MDR experiments. (A) Northern blot analysis. TEF3 was used as internal control. (B) Evaluation of differential expression by RT-PCR on the IFD genes family upregulated in the benomyl and MDR experiments. (C) Expression ratio obtained by microarray experiments and by Northern blot analysis. These ratios were obtained as described in Materials and Methods. Numbers in boldface type indicate discrepancies between both analysis. Symbols: (−) and (+), nonexposed or benomyl-exposed conditions; (S), azole-sensitive strain DSY2285; (R), azole-resistant strain DSY2286.
RT-PCR analysis.
RT was performed on 2 μg of RNA using Superscript II (Invitrogen) with random primers (Invitrogen) following instructions of the manufacturer. PCR were performed by mixing 2 μl of serial dilutions of cDNAs in a total volume of 50 μl with 5 μl of 10× PCR buffer (Roche), 0.5 μl each of forward and reverse primer (100 μM), 5 U of Taq DNA polymerase (Roche), and 2 μl of 10 mM deoxynucleoside triphosphates (Roche) in a final volume of 50 μl. PCR was performed under the following conditions: one cycle for 4 min at 94°C; 30 cycles with 1 min at 94°C, 1 min at 54°C, and 2 min at 72°C; and one terminal cycle for 10 min at 72°C. Equivalent PCR product volumes were loaded on a 1% agarose gel and separated by electrophoresis in 1× TAE (40 mM Tris, 10 mM EDTA, 0.1% acetic acid [pH 8.5]). Primer sequences are described in Table 2.
RESULTS
Experimental design and global gene expression results.
Previous results showed that multidrug transporter upregulation in C. albicans can be observed in several distinct situations, either by drug exposure or as the result of development of azole resistance in clinical isolates (10). It can be expected that other genes will be commonly regulated in these isolates under these conditions. Since these genes often share the same transcriptional activators or utilize similar gene activation pathway, it is therefore of interest to determine groups of commonly regulated genes in conditions known for upregulation of multidrug transporter genes. Transcript profiling with available C. albicans microarrays comprising almost the entire genome represents an appropriate tool to address this problem.
In vitro treatment of C. albicans with drugs such as fluphenazine or benomyl leads to transient upregulation of the ABC transporters CDR1 and CDR2 and of the major facilitator gene CaMDR1, respectively (10, 17). Therefore, transcript profiles of cells treated with fluphenazine or benomyl were first compared with profiles of cells in nontreated conditions. These experiments are referred to herein as the fluphenazine and benomyl experiments, respectively. Next, transcript profiles of azole-susceptible and azole-resistant C. albicans clinical isolates were compared with each other. The azole-susceptible strain DSY294 (MIC of fluconazole, 0.25 μg/ml) was first compared with its matched azole-resistant isolate DSY296 (MIC of fluconazole, 128 μg/ml), which is a strain known to upregulate CDR1 and CDR2 (28). This experiment was named CDR. The azole-susceptible isolate DSY2285 (MIC of fluconazole, 0.25 μg/ml) was then compared with its matched azole-resistant isolate DSY2286 (MIC of fluconazole, 16 μg/ml), which is a strain upregulating CaMDR1 (6). We called this the MDR experiment.
Figure 1 summarizes the number of genes differentially expressed in these different experiments. The threshold value for the selected genes was a twofold difference of gene expression between the experiments, which is a value generally accepted in genome-wide expression profiling. These genes were further divided to give the number of genes being either up- or downregulated in each experiment. Given that approximately 90 to 95% of the genes spotted on the microarrays gave validated data, meaning that reading of spot intensities for individual genes could be made in all hybridized arrays, the numbers given in Fig. 1 indicate that 3 to 4% of the total number of genes in the C. albicans genome exhibited transcriptional changes in these conditions. However, only a portion of the differentially expressed genes in these experiments could be commonly regulated between the different experiments. A cluster analysis using these data was performed in order to group these genes according to their expression patterns. The results of this analysis are represented in Fig. 2 and indicate that transcript profiles of the MDR and benomyl experiments and of the CDR and fluphenazine experiments are grouped together. These results are consistent with previous observations indicating that fluphenazine induces CDR genes and that a similar event, i.e., constitutive upregulation of CDR genes, can be measured in the azole-resistant isolate DSY296 used in the CDR experiment. A similar relationship can be made for benomyl that induces CaMDR1 and constitutive upregulation of the same gene existing in the azole-resistant isolate DSY2286 used in the MDR experiment. These results prompted us to compare in details genes differentially expressed between, first, the fluphenazine and CDR experiments and, second, the benomyl and the MDR experiments.
Comparative analysis of both the fluphenazine and the CDR microarray experiments. (i) Classification of expression clusters.
Since it appeared from the global analysis of the microarray data that the fluphenazine and CDR experiments were more related to each other than to the other experiments, these two sets of microarray data were compared using a cluster analysis with a threshold of fourfold differential expression in at least one experiment. This threshold value was used since it is situated above the upper limit of TEF3 expression variation and is therefore yielding genes with more relevant differential expression as outlined in Materials and Methods. Six different groups of differentially expressed genes were observed after cluster analysis among which three contained upregulated genes and the others contained downregulated genes (Fig. 3, left panel). The three clusters of each category were further distributed into commonly regulated genes, fluphenazine-specific or CDR-specific genes. In the group of upregulated genes listed in Fig. 3 (right panel) we identified nine commonly regulated genes, 11 fluphenazine-specific genes and 4 CDR-specific genes. The functions of these genes, grouped in functional categories such as transport, response to stress, ergosterol biosynthesis, or others, are listed in Tables 3 to 5. As summarized in Table 3, CDR1 and CDR2 show the largest change in expression (from 8.4- to 41.9-fold in both the CDR and fluphenazine experiments), while other genes such as GPX1 and IPF1514 are only upregulated (5.0- and 5.2-fold, respectively) above the 4-fold threshold value only in the fluphenazine experiment. These genes, although their increase in upregulation in the CDR experiment is 3.1- and 2.6-fold, respectively, still satisfy their categorization in commonly regulated genes, because of the algorithm used in the cluster analysis. This phenomenon can be observed for a few other genes listed in Tables 3 to 8. Among the downregulated genes (at least fourfold), 10 commonly regulated genes, 16 fluphenazine-specific genes, and 4 CDR-specific genes could be discriminated.
TABLE 3.
Genes upregulated in the fluphenazine and CDR experiments
| Function group and name | Functiona | DRE sequenceb | Fold increase (−) in expt
|
|
|---|---|---|---|---|
| CDR | Fluphenazine | |||
| Transport | ||||
| CDR2 | C. albicans drug resistance protein 2 | CGGAAATCGGATA (−221) | 26.8 | 41.9 |
| CDR1 | C. albicans drug resistance protein 1 | CGGATATCGGATA (−460) | 8.4 | 8.4 |
| Response to stress | ||||
| HSP12 | Heat shock protein (by homology); similar to S. cerevisiae HSP12 | 9.0 | 12.6 | |
| GPX1 | Glutathione peroxidase (by homology); similar to S. cerevisiae HYR1 | 3.1 | 5.0 | |
| IPF4065 | Similar to S. cerevisiae YHR087w (involved in stress response) | 10.5 | 3.7 | |
| Other | ||||
| RTA3 | Probable transmembrane protein; similar to S. cerevisiae YOR049c (involved in lipid transport activity) | CGGAAcTCGGAAA (−595) | 4.4 | 15.7 |
| IPF1514 | Unknown | 2.6 | 5.2 | |
| IFU5 | Unknown; similar to S. cerevisiae YFL010C | CGGAAATCGGATA (−248) | 4.3 | 6.7 |
| IPF2857 | Unknown | 4.9 | 2.8 | |
Gene functions were obtained using the CandidaDB and MycoPathPD databases available from the Pasteur Institute and from Incyte.
Bases indicated in lowercase type are mismatches compared to the following generic DRE sequence: CGGWWWTCGGWWW. Position of the DRE relative to the start codon is indicated in parentheses.
TABLE 5.
Genes specifically induced in the CDR experimenta
| Function group and name | Function | Fold increase (−) in expt
|
|
|---|---|---|---|
| CDR | Fluphenazine | ||
| Ergosterol bio- synthesis | |||
| ERG3 | C-5,6 sterol desaturase | 4.4 | 0.5 |
| ERG6 | C-24 sterol transmethylase | 5.7 | 1.0 |
| ERG251 | C-4 sterol methyl oxidase (by homology); similar to S. cerevisiae ERG25 | 4.1 | 1.1 |
| Other (IPF14285) | Unknown | 4.3 | 1.1 |
See Table 3 footnotes for details.
TABLE 8.
Genes specifically induced in the benomyl experiment
| Function group and name | Function | Cap1p binding site position(s) | Fold increase (−) in expt
|
|
|---|---|---|---|---|
| MDR | Benomyl | |||
| Other | ||||
| IPF2234 | Unknown | −339 | 1.1 | 7.4 |
| IPF122 | Unknown | −859, −520 | 1.1 | 5.1 |
| IPF13081 | Unknown | 1.4 | 5.7 | |
| IPF9568 | Unknown | 1.4 | 4.6 | |
| IPF6298 | Unknown | 1.4 | 4.0 | |
| IPF351 | Unknown; similar to S. cerevisiae YMR090w | −78 | 1.2 | 4.1 |
| IFS4 | Pirin protein (by homology) | −536, −87, −294, −273 | 0.5 | 7.2 |
| IFS1 | Pirin protein (by homology); similar to S. cerevisiae YDR014w | −418, −274 | 0.6 | 4.1 |
| CIP1 | Cadmium-induced protein c (by homology) | −559 | 0.3 | 18.1 |
| IPF19946 | Nucleotide-binding protein (by homology); similar to S. cerevisiae NBP35 | 1.1 | 4.7 | |
| ARG1 | Argininosuccinate synthetase (by homology); similar to S. cerevisiae YOL058w | 1.1 | 4.2 | |
| MRF1 | Mitochondrial respiratory function protein (by homology); similar to S. cerevisiae YBR026c | 2.2 | 9.9 | |
| IPF3415 | Mitochondrial inner membrane protease (by homology); similar to S. cerevisiae YIM1 | −168 | 1.7 | 8.6 |
| EBP1 | NADPH dehydrogenase, estrogen binding protein | 3.0 | 94.4 | |
| EBP4 | NADPH dehydrogenase, estrogen binding protein | 2.8 | 83.8 | |
| IPF20104 | Alcohol dehydrogenase (by homology); similar to S. cerevisiae ADH6 | −179 | 1.7 | 5.5 |
| SNQ2 | Similar to S. cerevisiae YNL334C (transaminase activity, cellular response to starvation) | 1.0 | 5.1 | |
| ARR3 | Arsenite transporter activity (by homology); similar to S. cerevisiae YPR201w | −328 | 1.0 | 7.5 |
| Cell wall maintenance | ||||
| IPF9996 | Similar to S. cerevisiae YDR371w (chitinase activity) | −119 | 0.7 | 9.3 |
| ALS1 | Similar to S. cerevisiae YJR151c (cell wall organization and biogenesis) | −424 | 1.5 | 5.8 |
| Response to stress | ||||
| IPF12897 | Putative oxidoreductase (by homology); similar to S. cerevisiae YJR096w | 1.2 | 4.2 | |
| IPF11105 | Probable quinone oxidoreductase; similar to S. cerevisiae YBR046c | −580 | 1.4 | 7.9 |
| PST2 | 1,4-Benzoquinone reductase (by homology), similar to S. cerevisiae YDR032c | −290 | 1.5 | 9.8 |
| IPF3264 | Similar to S. cerevisiae YML131w (oxidoreductase activity, response to stress) | −306 | 0.7 | 23.7 |
| SOD1 | Cu,Zn-superoxide dismutase; similar to S. cerevisiae YJR104C (oxidative stress response) | −752 | 1.8 | 4.1 |
| TTR1 | Glutaredoxin (by homology); similar to S. cerevisiae YDR513w (response to pH, oxidative stress and heat) | −137 | 1.3 | 6.6 |
| TRX1 | Thioredoxin (by homology); similar to S. cerevisiae YLR043c (response to oxidative stress) | −216, −257, −467, −652, −920 | 1.2 | 4.3 |
| GRP4 | Putative reductase (by homology); similar to S. cerevisiae YOL151w (induced by osmotic stress) | −389 | 1.2 | 6.5 |
| IFR2 | Similar to S. cerevisiae YNL134c (response to heat shock) | 1.1 | 6.7 | |
| IPF7968 | Similar to S. cerevisiae YNL229c (transcription corepressor activity, salinity response) | −161 | 1.0 | 6.0 |
| IPF12303 | Similar to S. cerevisiae YNL229c (transcription corepressor activity, salinity response) | −38, −176 | 0.8 | 8.1 |
| MEC3 | Similar to S. cerevisiae YLR288c (response to stress) | 0.8 | 4.0 | |
See Table 6 footnotes for details.
(ii) Validation by Northern analysis.
Some of the upregulated genes and especially those commonly regulated or those with low intensity signals (SAP3 and SAP5) were analyzed by Northern blot to confirm the microarray data (Fig. 4A). Radioactive signals obtained by Northern blotting were quantified and compared with fluorescence signals of the microarray analysis (Fig. 4B). Comparison of signal ratios obtained by microarrays or by Northern blot analysis showed a good correlation between the two methods (Fig. 4B). Eight out of the nine genes previously defined as commonly upregulated genes exhibit mRNA signals with ratio above the fourfold cutoff value that was used in the microarray analysis. The remaining gene, IPF2857, possessed a signal ratio in clinical strains of only 2.3 by Northern analysis compared to 4.9 obtained by microarray analysis. Given that Northern blot analysis might be less sensitive than microarray analysis, this gene could be therefore classified in the commonly upregulated genes. Three out of 11 genes belonging to the fluphenazine-specific upregulated genes were verified by Northern analysis. SAP3 and SAP5 were chosen because they showed low intensity signals in the microarray analysis (Fig. 4B). IPF6629 was chosen because it contains a DRE-like sequence (see below) in its promoter. SAP3 and SAP5 mRNA signals were almost absent under the conditions tested, with ratios of 0.8 and 1.0, respectively. In contrast, IPF6629 was significantly upregulated in Northern blot with a ratio of 3.0. None of these three genes showed a difference in gene expression between the azole-susceptible and azole-resistant strains DSY294 and DSY296, which is consistent with the microarray experiments. Three genes out of the four present in the group of CDR-specific upregulated genes were analyzed by Northern blot. ERG genes (ERG3 and ERG6) were confirmed as CDR-specific, since their expression ratios in clinical strains were 2.1 and 10.4, respectively. Both genes were not induced by fluphenazine (ratios of 1.0 and 1.6, respectively, by Northern blotting). In contrast IPF14285 was not confirmed as a CDR-specific upregulated gene, since its expression ratio was only 1.1. In Northern blot this gene does not appear upregulated in fluphenazine nor in CDR experiments (Fig. 4A). The expression of PHR2 and DFG5 was taken as a control, since the expression of these genes was changed neither in the fluphenazine nor in the CDR experiments. The absence of regulation of these two genes was confirmed by Northern blot (Fig. 4B). In conclusion, signals revealed by Northern blot analysis were correlating well with those obtained by microarray analysis, except for SAP3 and SAP5 that were not upregulated by fluphenazine treatment and for IPF14285 that was not upregulated in the azole-resistant strain DSY296.
(iii) Functional aspects of differentially expressed genes.
Among the nine commonly upregulated genes, four genes (CDR1, CDR2, IFU5, and RTA3) contain in their promoter a sequence motif similar to the DRE (CGGWWWTCGGWWW) that was previously described to be important for CDR2 and CDR1 upregulation (10) (Table 3). Although the inspection of genome data identified a total of 10 genes with a perfect match to this DRE and 177 with a single mismatch DRE in their promoters, this DRE sequence is not present in the promoters of CDR-specific upregulated genes and only present in one gene (IPF6629) among the nine fluphenazine-specific upregulated genes (Tables 4 and 5). We therefore conclude that the DRE is strongly linked to a pathway controlling the expression of genes involved in azole resistance. Since the exact function of RTA3 and IFU5 is not yet determined in C. albicans, it is not possible to attribute a common functional role to the four commonly upregulated and DRE-containing genes described above. The product of these four genes share, however, transmembrane segments and thus a membrane localization, therefore suggesting that the common regulation of these genes in the experimental conditions tested in this study has an impact in membrane functions.
TABLE 4.
Genes specifically induced in the fluphenazine experimenta
| Function group and name | Function | DRE sequence | Fold increase (−) in expt
|
|
|---|---|---|---|---|
| CDR | Fluphenazine | |||
| Virulence factor | ||||
| SAP5 | Secreted aspartyl proteinase 5; virulence factor | 0.8 | 5.4 | |
| SAP3 | Secreted aspartyl proteinase 3; virulence factor | 1.0 | 5.9 | |
| Response to stress | ||||
| CFL2 | Similar to S. cerevisiae YNR060w (oxidoreductase activity) | 0.7 | 4.1 | |
| IPF6629 | Similar to S. cerevisiae YLR109w (alkyl hydroperoxide reductase) | gGGTATTCGGTTA (−600) | 1.5 | 4.7 |
| SAS3 | Silencing protein (by homology); similar to S. cerevisiae YBL052c (involved in stress response) | 1.2 | 4.5 | |
| IPF17283 | Similar to S. cerevisiae YCR061w (salinity response, response to heat) | 2.4 | 6.7 | |
| GRP2 | Reductase (by homology); similar to S. cerevisiae YOL151w (induced by osmotic stress) | 1.7 | 4.2 | |
| Other | ||||
| SEN15 | tRNA splicing endonuclease delta subunit (by homology) | 0.9 | 5.4 | |
| IPF18393 | Similar to S. cerevisiae YDR469w (DNA repair activity) | 0.6 | 5.3 | |
| IPF14282 | Mucin proteins (by homology) | 1.4 | 4.4 | |
| AKR1 | Ankyrin repeat-containing protein (by homology); similar to S. cerevisiae YDR264c | 0.9 | 7.7 | |
See Table 3 footnotes for details.
Regarding the functions of other genes, we observed that among the nine fluphenazine-specific genes, five were potentially involved in different stress responses such as oxidative stress (CFL2 and IPF6629), osmotic stress (GRP2 and IPF17282), or more general stress responses (SAS3) (Table 4). In contrast the CDR-specific genes were belonging to the ergosterol biosynthetic pathway (ERG3, ERG6, and ERG251) (Table 5). Downregulated genes in the two experiments belonged essentially to genes with unknown functions in the three clusters (Fig. 3). Downregulated genes with known functions do not share a common functional category (data not shown). These genes have potential functions in stress response (IPF2091) or in transport (IPF12300, IPF9821, and MID1).
Comparative analysis of the benomyl versus MDR microarray experiments. (i) Classification of expression clusters.
Microarray data from the related benomyl and MDR experiments were also compared by cluster analysis (Fig. 5). Genes that were differentially expressed by at least a fourfold difference were classified into six clusters. As previously observed for the CDR and fluphenazine experiments up- and downregulated genes were distributed into three clusters containing the commonly regulated genes, the benomyl-specific and the MDR-specific genes. As depicted in the right panel of Fig. 5, among the 22 downregulated genes, 6, 12, and 4 were commonly regulated genes, MDR-specific and benomyl-specific genes, respectively. Similarly, upregulated genes contained 17, 9, and 32 commonly regulated genes, MDR-specific and benomyl-specific genes, respectively. The functions of these genes, grouped in functional categories such as transport, response to stress, cell wall maintenance, or others, are listed in Tables 6 to 8.
TABLE 6.
Genes upregulated in the benomyl and MDR experiments
| Function group and name | Functiona | Cap1p binding site position(s)b | Fold increase (−) in expt
|
|
|---|---|---|---|---|
| MDR | Benomyl | |||
| Other | ||||
| IPF3080 | Unknown | −213, −708 | 4.1 | 1.6 |
| IPF1617 | Unknown | −207 | 7.3 | 3.2 |
| IPF11023 | Unknown | 4.3 | 2.6 | |
| Transport (CaMDR1) | Multidrug transporter activity; similar to S. cerevisiae FLR1 | −532 | 44.0 | 12.6 |
| Response to stress | ||||
| IPF18418 | Unknown; similar to S. cerevisiae YHR029c (induced by cell-damaging conditions) | 5.4 | 1.6 | |
| IPF13316 | Unknown; similar to S. cerevisiae YNL010w (induced by cell-damaging conditions) | 4.1 | 1.7 | |
| IPF5987 | Unknown; similar to S. cerevisiae YPR127w (protein with similarity to S. pombe pyridoxal reductase, aldo-keto reductase family) | 70.2 | 7.9 | |
| IPF17186 | Unknown; similar to S. cerevisiae YDR533c (induced by cell-damaging conditions) | −720 | 102.5 | 56.8 |
| PYC2 | Pyruvate carboxylase 2; similar to S. cerevisiae YBR218c | 5.1 | 1.9 | |
| GPX1 | Glutathione peroxidase; similar to S. cerevisiae HYR1 | 9.4 | 3.6 | |
| GRP2 | Reductase; similar to S. cerevisiae YOL151w (induced by osmotic stress) | −444 | 52.5 | 35.1 |
| IPF7817 | Putative NADH-dependent flavin oxidoreductase by homology; similar to S. cerevisiae OYE3 | −743 | 14.7 | 37.0 |
| DDR48 | Stress protein; similar to S. cerevisiae YMR173w (involved in the production of or recovery from mutation) | 5.6 | 1.7 | |
| IFD1 | Benzyl alcohol dehydrogenase activity (by homology); similar to S. cerevisiae YPL088w (aldo-keto reductase family) | 748.3 | 32.6 | |
| IFD4 | Benzyl alcohol dehydrogenase activity (by homology); similar to S. cerevisiae YPL088w (aldo-keto reductase family) | 129.6 | 6.7 | |
| IFD5 | Benzyl alcohol dehydrogenase activity (by homology); similar to S. cerevisiae YPL088w (aldo-keto reductase family) | 511.2 | 16.1 | |
| IFD7 | Benzyl alcohol dehydrogenase activity (by homology); similar to S. cerevisiae YPL088w (aldo-keto reductase family) | 158.9 | 5.4 | |
Gene functions were obtained using the CandidaDB and MycoPathPD databases available from the Pasteur Institute and from Incyte.
The sequence of the Cap1p-binding site considered is TTA(C/G)TAA (15).
(ii) Validation by Northern blot analysis and RT-PCR.
To validate the data obtained with microarrays, Northern blot analysis were performed with the most upregulated genes from each group and also with four genes (IFR2, IPF3415, TTR1, and TRX1) tested in another published microarray analysis (13). In general, Northern analysis confirmed the microarray data. The transcription of 6 chosen genes among the 17 commonly upregulated ones (DDR48, BMR1/CaMDR1, GPX1, IPF17186, GRP2, and IPF7817) was increased as expected from the MDR and benomyl microarray experiments. Among 5 genes from the group of 9 MDR-specific upregulated genes (IPF7863, IPF9538, IPF2186, IPF3704, and SNZ1), all except one (IPF3704) appeared effectively only upregulated in the clinical resistant strain DSY2286. As shown in Fig. 6, IPF3704 had an expression ratio of only 0.8. Nine genes selected from the group of the 32 benomyl upregulated genes were also verified by Northern analysis. CIP1, MRF1, EBP1, EBP4, and IPF3264 were chosen because they corresponded to the most upregulated genes. The four others (TTR1, TRX1, IFR2, and IPF3415) were genes already shown as upregulated under oxidative stress conditions as shown by Enjalbert et al. (13). As shown in Fig. 6, all these genes were induced by benomyl treatment. Nevertheless, three of them (MRF1, EBP1, and EBP4) were also upregulated in the clinical resistant strain DSY2286 with expression ratios of 2.2, 3.6, and 6.4 in clinical strains, respectively. Therefore, these three genes could not be considered as benomyl-specific upregulated genes, but rather as commonly regulated genes. As previously mentioned above in the cluster analysis of the fluphenazine and CDR experiments, some genes can be grouped in specific expression categories by the clustering program algorithm, even though their upregulation ratios would group them in other categories. We observed in our microarray analysis that several genes of the IFD family (genes coding for putative benzyl alcohol dehydrogenases) were upregulated in both experimental conditions, either by benomyl treatment or in clinical strains. Since these genes are possessing high degree of similarity, it is not possible to distinguish individual members by Northern blot analysis. The expression of these genes was therefore verified by semiquantitative RT-PCR using primers specific for each gene. As shown in Fig. 6B, PCR signals demonstrated that IFD1, IFD4, IFD5, and IFD7 were upregulated by at least 100-fold after benomyl exposure and in the clinical azole-resistant strain DSY2286, while IFD2 remained only strongly upregulated in DSY2286. These results were consistent with the microarrays data (in the MDR experiment, IFD1, IFD5, IFD7, IFD4, and IFD2 were upregulated by 683-, 511-, 158-, 129-, and 98-fold, respectively; in the benomyl experiment, IFD1, IFD5, IFD7, and IFD4 were upregulated by 31-, 18-, 7-, and 8-fold, respectively). Taken together, the results of Northern and RT-PCR analysis correlated well with those obtained with microarray analysis except for four genes (MRF1, IPF3704, EBP1, and EBP4).
(iii) Functional aspects of differentially expressed genes.
Among the genes upregulated in a specific category, we investigated if their promoters could share putative common regulatory sequence(s). CaMDR1 belongs to the commonly upregulated genes and its promoter contains one putative Cap1p binding site, TTAG/CTAA (15), localized at bp −532 (with respect to the ATG start codon). Cap1p was shown earlier to be involved in multidrug resistance and oxidative stress response in C. albicans (2, 34). The promoters of genes containing this motif were therefore scanned using algorithms available at http://genolist.pasteur.fr/CandidaDB/. This motif is highly distributed in the C. albicans genome, since 1,637 open reading frames contain at least one perfect Cap1p binding site sequence in their promoters. Among the 29 benomyl-specific genes, 21 contained this sequence in their promoter (Table 8). Since none of the MDR specific genes and only a few of the commonly upregulated genes contained such a site in their promoters (Tables 6 and 7), this element could be rather considered as a benomyl-specific regulatory element. The transcription factor Cap1p is known to be involved in oxidative stress response (34). It is interesting that among the 29 benomyl-specific upregulated genes, 7 (IPF2897, IPF11105, PST2, IPF3264, SOD1, TTR1, and TRX1) are potentially implicated in this type of stress response. Five other genes included in this cluster play a role in other stress responses such as osmotic (GRP4) or heat shock stress responses (IFR2) (Table 8). Among the 17 commonly regulated genes, 9 are also involved essentially in oxidative stress response (PYC2, GPX1, GRP2, IPF7817, IFD1, IFD4, IFD5, and IFD7), and 1 of them (DDR48) corresponds to a protein potentially involved in the production of mutation or recovery from mutation. Interestingly, one of the members of the IFD gene family (IFD4), the product of which could have benzyl alcohol dehydrogenase activity, has been shown to be identical to the gene CSH1 coding for a surface hydrophobicity-associated protein (29). IFD proteins, if all located at the cell surface, could therefore play an important role at the interface between the cell and its environment.
TABLE 7.
Genes specifically induced in the MDR experiment
| Name | Functiona | Fold increase (−) in expt
|
|
|---|---|---|---|
| MDR | Benomyl | ||
| IFP2 | Unknown | 4.6 | 0.7 |
| IPF6954 | Unknown | 4.3 | 0.9 |
| IPF5131 | Unknown | 4.3 | 0.9 |
| IPF2186 | Unknown; similar to S. cerevisiae YGR110w | 10.2 | 1.4 |
| IPF3704 | Similar to S. cerevisiae YNL218w (DNA-dependent ATPase activity) | 4.8 | 1.3 |
| IPF7863 | Similar to S. cerevisiae YNL335w (nitrile hydratase activity) | 4.9 | 0.9 |
| IPF9538 | Similar to S. cerevisiae YNL202w (2,4-dienoyl-coenzyme A reductase NADPH activity) | 129.2 | 1.1 |
| IFD2 | Benzyl alcohol dehydrogenase activity (by homology); similar to S. cerevisiae YPL088w | 95.0 | 1.1 |
| SNZ1 | Stationary-phase protein (by homology); similar to S. cerevisiae YMR096w (putative pyridoxine [vitamin B6] biosynthetic enzyme) | 6.0 | 1.3 |
See Table 6, footnote a, for details.
DISCUSSION
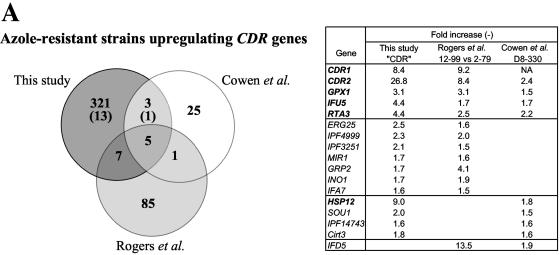
In this work, we compared transcript profiling between clinical strains with known azole resistance mechanisms and laboratory strains exposed to drugs known to induce genes involved in azole resistance. Previous experiments performed by our group and others have established that specific multidrug transporter genes can be induced by distinct agents, thus resulting in similar gene expression patterns than those observed in the azole-resistant isolates. Until now, only a specific set of genes has been analyzed in response to these drugs. Gupta et al. (17) showed that benomyl, methotrexate, and H2O2 could enhance the expression of CaMDR1 in C. albicans. On the other hand, fluphenazine, estradiol, or terbinafine can induce the expression of CDR1 and CDR2 in the same yeast (10). However, these drugs can induce the expression of additional genes and therefore address the validity of the comparison between transient induction of multidrug transporters and their constitutive upregulation in azole-resistant clinical strains. This study clarifies this question and establishes the number of genes commonly regulated in each situation. A few gene profiling experiments have been carried out with azole-resistant strains until now. Rogers and coworkers (23) have used a well-known series of sequential isolates first analyzed by White (30, 31). In these isolates, an ordered expression of both types of multidrug transporter genes (i.e., CDR and CaMDR1 genes) occurred over time. Another genome-wide study was performed by Cowen et al. (7) with sequential isolates that developed azole resistance by upregulation of CDR genes or CaMDR1. Although the type of microarray, the gene annotation, and the signal intensities were different between these different studies, we attempted to compare the transcript profiles obtained in these studies with our analysis. We compared first the transcript profiles of the MDR experiment with those obtained by isolate 2-80 (isolate that upregulates CaMDR1) versus isolate 2-79 (azole-susceptible isolate) (study published by Rogers et al. [23]) and with those of isolate D12-330 versus an azole-susceptible isolate (study published by Cowen et al. [7]). Only genes upregulated by more than 1.5-fold were taken into consideration. As shown in Fig. 7, the present study yielded a higher number of upregulated genes than measured in the two other studies. These discrepancies reflect not only differences in experimental conditions but also that the origin of isolates in the three studies is different. Figure 7 reveals that only a few genes are commonly regulated in strains upregulating CDR genes. Among these commonly regulated genes, we found CDR1, CDR2, RTA3, IFU5, and GPX1. It is remarkable that these genes are also among those that are also commonly regulated between the fluphenazine and the CDR experiment of this study. Beside being involved in drug resistance (for CDR1 and CDR2), these genes have in common that their S. cerevisiae homologues respond to conditions of cell damages that were performed in microarray experiments, as deduced from transcript profile information available at http://www.yeastgenome.org/. This suggests that the common regulation of these genes is not accidental, but constitutes a group of genes necessary for the adaptation of C. albicans to drug exposure. One would expect that these genes are controlled by a common regulatory mechanism. Since four of these five commonly regulated genes contained a DRE in their promoter, these results strongly suggest the existence of a common transcriptional pathway important for their regulation. The simplest hypothesis would be that a yet-unidentified transcription factor can bind to the DRE and be activated either after drug treatment or by a mutation(s) existing in azole-resistant strains, thus resulting in transcriptional activation of genes containing the DRE. Current experiments undertaken in our laboratory favor this hypothesis: a transcriptional activator of CDR1 and CDR2 has been isolated and alleles of this gene cloned from azole-resistant isolates are able to activate the transcription of CDR1 and CDR2 in an azole-susceptible laboratory strain (A. T. Coste, unpublished data).
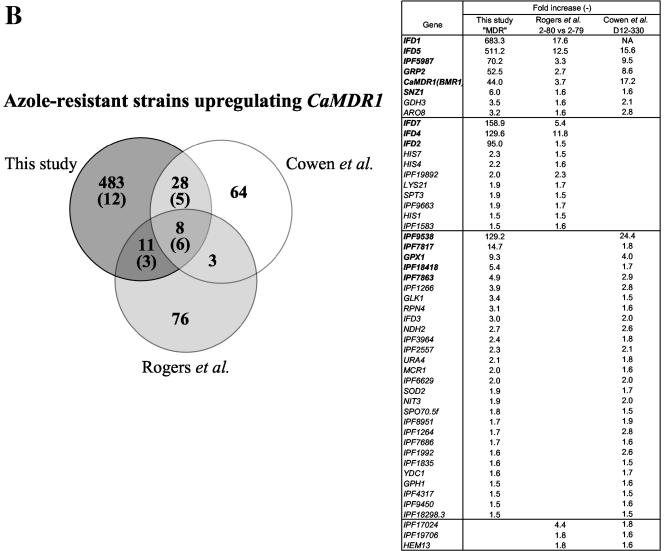
FIG. 7.
Comparison of the microarray data of (A) CDR and (B) MDR experiments with previously published microarray analysis performed with other azole-resistant strains (7, 23). Gene expression values obtained from these studies for selected genes with a threshold upregulation of 1.5 are given in the accompanying table and commonly regulated genes can be observed in a Venn diagram. Numbers in boldface type in the accompanying table and in brackets in the Venn diagram indicate the number of commonly regulated genes taking the present data set with a fourfold threshold. NA, not available.
When comparing the set of genes commonly regulated in isolates upregulating CaMDR1 by microarray experiments of this study and of those published by Cowen et al. (7) and Rogers et al. (23), only eight genes were upregulated in the three studies. We observed a higher number of genes commonly regulated (28) between the MDR experiment of our study and the set of strains used by Cowen et al. (7) than in the set of strain used by Rogers et al. (23). This probably reflects differences between strains used in these different studies. The genes commonly regulated in the three studies were corresponding to CaMDR1 (BMR1), GRP2, IFD5, IPF5987, and SNZ1. Besides the antifungal drug resistance function of CaMDR1, the other genes have oxido-reductive functions (GRP2, IFD5) or are potentially involved in pyridoxine (vitamin B6) synthesis (IPF5987 and SNZ1). It is, however, interesting that the S. cerevisiae homologues of all these five genes respond to cell damage conditions as can be determined from transcript profiling experiments available at http://www.yeastgenome.org/. Therefore, the common regulation of these genes constitute also a necessary process in the development of drug resistance mediated by the “CaMDR1” pathway. It is currently difficult to predict the identity of one or several regulatory elements responsible for the upregulation of these genes, since no detailed studies have been carried out to dissect the functional elements of the promoter of these genes. As mentioned above, some of these genes contained in their promoters a putative Cap1p binding site. Therefore, the role of this transcription factor in the regulation of these genes needs to be determined.
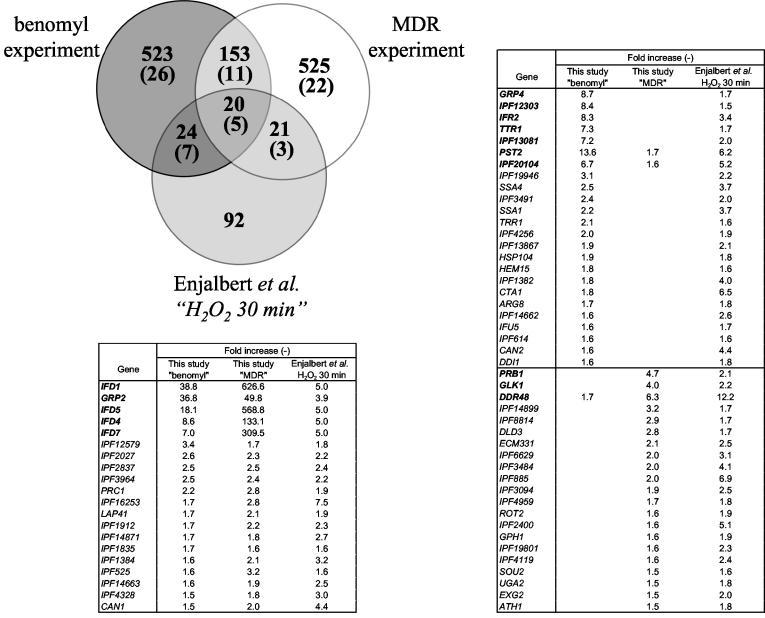
Since CaMDR1 can be also induced by H2O2 (17), it was also of interest to compare our results that were obtained using benomyl-exposed cells with another study published by Enjalbert et al. (13) investigating the genome-wide transcriptional response of C. albicans to several agents including H2O2 (Fig. 8). Interestingly, some of the H2O2 upregulated genes in the study of Enjalbert et al. (13) (IFD1, GRP2, IFD5, IFD4, IFD7, GRP4, IPF12303, IFR2, TTR1, IPF13081, PST2, and IPF20104) are those that were found induced also by benomyl in this study. Most of these genes are implicated in response to stress or have oxido-reductive functions (Table 8). Among these, GRP2, GRP4, IPF12303, TTR1, PST2, and IPF20104 contained a putative Cap1p binding site in their promoters (Tables 6 to 8). Therefore, a functional linkage involving Cap1p probably exists between benomyl and H2O2 exposure. It is possible that this transcription factor could be activated by exposure to these drugs. In S. cerevisiae, the functional homologue of Cap1p, Yap1p, is localized in the cytoplasm and it is only after its activation by oxidative stress that this factor migrates to the nucleus and activates the transcription of genes with Yap1p-binding sites in their promoters (19). A similar situation is existing in C. albicans, as shown by studies carried out by Zhang et al. (34).
FIG. 8.
Comparison of the microarray data of benomyl and MDR experiments with previously published microarray analysis on stress response after 30 min of H2O2 exposure (13). Gene expression values obtained from these studies for selected genes with a threshold upregulation of 1.5 are given in the accompanying table and commonly regulated genes can be observed in a Venn diagram. Numbers in boldface type in the accompanying table and in brackets in the Venn diagram indicate the number of commonly regulated genes taking the present data set with a fourfold threshold.
Molecular mechanisms responsible for azole resistance have been investigated in details in C. albicans. While the analysis of azole resistance has demonstrated that the major azole resistance mechanisms are involving drug efflux and target alterations, other yet unknown mechanisms are still possibly existing. Transcript profile analysis in clinical isolates may help to identify these alternative pathways. In this work, clinical isolates with resistance mechanisms involving alteration of gene expression have been investigated. As expected, the upregulation of efflux transporter genes could be verified by this type of analysis. Even though the overlap of transcript profiling between drug-exposed cells and azole-resistant isolates help us to determine clusters of genes needed for resistance development, the analysis of additional isolates with still-unknown resistance mechanisms will be an interesting option for the discovery of new resistance mechanisms.
Acknowledgments
This work is supported by EC grant QLK2-CT-2001-02377 and grant 3200B0-100747/1 from the Swiss Research Foundation.
We thank Philippe Hauser for critical reading of the manuscript.
REFERENCES
- 1.Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998-35015. [DOI] [PubMed] [Google Scholar]
- 2.Alarco, A. M., and M. Raymond. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bammert, G. F., and J. M. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boerlin, P., F. Boerlin-Petzold, J. M. Goudet, C. Durussel, J. L. Pagani, J. P. Chave, and J. Bille. 1996. Typing Candida albicans oral isolates from human immunodeficiency virus-infected patients by multilocus enzyme electrophoresis and DNA fingerprinting. J. Clin. Microbiol. 34:1235-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calabrese, D. 2002. Incidence of resistance mechanisms to antifungal agents in human yeast pathogens. University of Lausanne, Lausanne, Switzerland.
- 6.Calabrese, D., J. Bille, and D. Sanglard. 2000. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 146:2743-2754. [DOI] [PubMed] [Google Scholar]
- 7.Cowen, L. E., A. Nantel, M. S. Whiteway, D. Y. Thomas, D. C. Tessier, L. M. Kohn, and J. B. Anderson. 2002. Population genomics of drug resistance in Candida albicans. Proc. Natl. Acad. Sci. USA 99:9284-9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowen, L. E., D. Sanglard, D. Calabrese, C. Sirjusingh, J. B. Anderson, and L. M. Kohn. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Backer, M. D., T. Ilyina, X. J. Ma, S. Vandoninck, W. H. Luyten, and H. Vanden Bossche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 11.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 12.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enjalbert, B., A. Nantel, and M. Whiteway. 2003. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg, A., and B. Vogelstein. 1984. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137:266-267. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes, L., C. Rodrigues-Pousada, and K. Struhl. 1997. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17:6982-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta, V., A. Kohli, S. Krishnamurthy, N. Puri, S. A. Aalamgeer, S. Panwar, and R. Prasad. 1998. Identification of polymorphic mutant alleles of CaMDR1, a major facilitator of Candida albicans which confers multidrug resistance, and its in vitro transcriptional activation. Curr. Genet. 34:192-199. [DOI] [PubMed] [Google Scholar]
- 18.Hube, B., M. Monod, D. A. Schofield, A. J. Brown, and N. A. Gow. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 19.Kuge, S., M. Arita, A. Murayama, K. Maeta, S. Izawa, Y. Inoue, and A. Nomoto. 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21:6139-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen, M. H., J. E. Peacock, Jr., A. J. Morris, D. C. Tanner, M. L. Nguyen, D. R. Snydman, M. M. Wagener, M. G. Rinaldi, and V. L. Yu. 1996. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am. J. Med. 100:617-623. [DOI] [PubMed] [Google Scholar]
- 21.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rex, J. H., M. G. Rinaldi, and M. A. Pfaller. 1995. Resistance of Candida species to fluconazole. Antimicrob. Agents Chemother. 39:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers, P. D., and K. S. Barker. 2003. Genome-wide expression profile analysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob. Agents Chemother. 47:1220-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanglard, D., F. Ischer, and J. Bille. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45:1174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanglard, D., F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contributing to the resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC-transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 28.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singleton, D. R., J. Masuoka, and K. C. Hazen. 2001. Cloning and analysis of a Candida albicans gene that affects cell surface hydrophobicity. J. Bacteriol. 183:3582-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White, T. C. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14-alpha-demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wirsching, S., S. Michel, G. Kohler, and J. Morschhauser. 2000. Activation of the multiple drug resistance gene MDR1 in fluconazole-resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J. Bacteriol. 182:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimoto, H., K. Saltsman, A. P. Gasch, H. X. Li, N. Ogawa, D. Botstein, P. O. Brown, and M. S. Cyert. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277:31079-31088. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, X., M. De Micheli, S. T. Coleman, D. Sanglard, and W. S. Moye-Rowley. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36:618-629. [DOI] [PubMed] [Google Scholar]