Abstract
We tested the impact of individual PBP 5 mutations on expression of ampicillin resistance in Enterococcus faecium using a shuttle plasmid designed to facilitate expression of cloned pbp5 in ampicillin-susceptible E. faecium D344SRF. Substitutions that had been implicated in contributing to the resistance of clinical strains conferred only modest levels of resistance when they were present as single point mutations. The levels of resistance were amplified when some mutations were present in combination. In particular, a methionine-to-alanine change at position 485 (in close proximity to the active site) combined with the insertion of a serine at position 466 (located in a loop that forms the outer edge of the active site) was associated with the highest levels of resistance to all β-lactams. Affinity for penicillin generally correlated with β-lactam MICs for the mutants, but these associations were not strictly proportional.
Ampicillin resistance in Enterococcus faecium is due to expression of the low-affinity class B penicillin-binding protein 5 (PBP 5) (15). Early studies suggested that higher levels of ampicillin resistance in Enterococcus hirae (similar to those in E. faecium) were achieved by increasing levels of PBP 5 expression (5). However, higher levels of resistance in clinical isolates are only rarely associated with increased levels of PBP 5 expression (12, 16). More commonly, mutations that are presumed to lower the affinity for β-lactam antibiotics have been identified within pbp5 genes of highly resistant clinical isolates (1, 12, 16). It has been presumed that these mutations serve to lower the affinity of the PBP 5 molecule for β-lactam antibiotics, yielding higher MICs as a result.
The specific impacts of individual or multiple pbp5 mutations on the resistance level have been difficult to determine because most analyses have been performed with clinical isolates, in which factors other than the PBP 5 amino acid sequence may contribute to resistance. In a previous study (14) a single mutation (M485A) was introduced into a cloned pbp5, and the impact of this mutation was relatively minor. However, the plasmid used in those experiments contained only pbp5 and its promoter, and expression of resistance prior to introduction of the mutation was minimal (ampicillin MIC, 6 μg/ml) (14). The role of PBP 5 in expression of ampicillin resistance in E. faecium C68, a clinical isolate resistant to high levels of ampicillin (MICs, 256 to 512 μg/ml), was reported recently (11). pbp5 of C68 is located downstream of two open reading frames, designated ftsWEfm and psr. It was observed that expression of ampicillin resistance from a cloned version of the C68 pbp5 was higher when pbp5 was located downstream of ftsWEfm and psr (ampicillin MICs, 64 to 128 μg/ml) than when it was cloned with only its own promoter (ampicillin MICs, 8 to 16 μg/ml) (data not shown) (11). The role of the putative ftsWEfm gene product is unknown, although recent work suggests that its homologue in Escherichia coli may serve as a chaperone protein for PBP 3 (4). We reasoned that the more robust expression of resistance from the larger construct would facilitate analysis of the impact of specific pbp5 mutations on expression of β-lactam resistance in E. faecium. In this study, we used the larger construct to assess the contributions of different individual mutations to expression of ampicillin resistance in E. faecium. We also correlate these resistance levels with the affinity of the PBP 5 protein for radiolabeled penicillin. Finally, we offer some mechanistic rationales for our findings based on the structure of PBP 5.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The host strain for all experiments was E. faecium D344SRF (11), a rifampin- and fusidic acid-resistant derivative of clinical strain E. faecium D344R in which the spontaneous loss of pbp5 and its surrounding region resulted in an ampicillin-susceptible phenotype. It was used in these experiments to ensure that the PBP 5 proteins encoded by our shuttle plasmids would be the only PBP 5 proteins expressed by the test strains. Loss of the surrounding regions also precluded recombination between the plasmid constructs and the chromosome of D344SRF. E. faecium C68 is an ampicillin-resistant, VanB-type vancomycin-resistant clinical isolate. E. faecium D366 is an ampicillin-susceptible, VanB-type E. faecium clinical isolate originating from France. Both strains have been described previously (1, 12).
Construction of plasmid vectors.
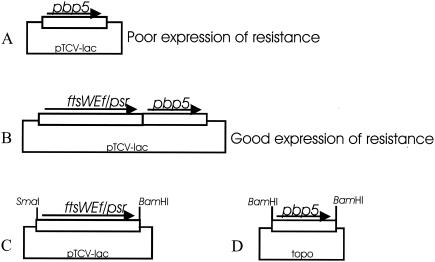
ftsWEfm and psr from E. faecium C68 (extending from upstream of the ftsWEfm promoters to downstream of the pbp5 promoter [Fig. 1 ]) were amplified by using primers containing SmaI (upstream) and BamHI (downstream) sites and were cloned into the multiple-cloning site of shuttle vector pTCV-lac (10), yielding pCWR620. Wild-type and mutant pbp5 genes were inserted into the BamHI site of pCWR620 for use in these experiments, as described in the legend to Fig. 1. The plasmids were transformed into E. coli HB101 containing pRK24 (mobilizing plasmid) (10). They were then transferred by conjugation (10) into E. faecium D344SRF with selection on brain heart infusion agar (BHI) plates containing kanamycin (1,500 μg/ml).
FIG. 1.
Cloning strategy to create mutants. pbp5 cloned by itself expresses resistance poorly (A). However, when it is cloned downstream of ftsWEf and psr from strain C68, the level of resistance expression is higher (B). So we first created a plasmid in which ftsWEf and psr were cloned into the shuttle vector pTCV-lac using restriction enzymes SmaI and BamHI, with the BamHI site of the insert located downstream of psr (C). We then separately amplified pbp5 from E. faecium D366 (susceptible strain) from the ribosome-binding site to downstream of the transcription terminator (with BamHI sites engineered on both ends) and cloned this into the commercially available E. coli plasmid vector pCR-XL-TOPO (Invitrogen) (D). Site-directed mutagenesis was performed with pbp5 in this plasmid. After sequence confirmation, the insert was removed by digestion with BamHI and inserted downstream of psr in the pTCV-lac construct, resulting in several variants (B) in which mutant pbp genes were located downstream of ftsWEf and psr. The proper orientation was confirmed by PCR amplification.
MIC determinations.
MICs were determined by an agar dilution method with BHI agar. Plates were inoculated by using a multipoint inoculator with an inoculum standardized by spectrophotometry to yield 104 CFU/spot. Since the impact of the PBP 5 mutations on the MICs of β-lactam antibiotics was often subtle, we used arithmetic increases in antibiotic concentrations rather than the standard doubling dilutions for our MIC determinations. The antibiotic concentrations tested were as follows: for ampicillin, 10 to 200 μg/ml in 10-μg/ml increments and then 300 and 400 μg/ml; for piperacillin, 70, 80, and 90 μg/ml and then 100 to 1,000 μg/ml in 100-μg/ml increments; for ceftriaxone and ticarcillin, from 1,000 to 4,000 μg/ml in 200-μg/ml increments and then 5,000, 6,000, 7,000, and 8,000 μg/ml. Growth was often slow in the first 24 h, so the plates were read at 24, 48, and 72 h. The results reported here reflect the determinations at 48 h. MICs were determined at least in duplicate, and for some more than 10 times (see the explanation in footnote b of Table 1). The results are given in Table 1 as the arithmetic means of the MIC determinations.
TABLE 1.
Mutagenized pbp5 genes and expression of β-lactam resistance in E. faecium
| Plasmida | Amino acid at the following position in PBP 5:
|
MIC (μg/ml)b
|
||||||
|---|---|---|---|---|---|---|---|---|
| 485 | 499 | 629 | Ser at position 466′ | Ampicillin | Piperacillin | Ticarcillin | Ceftriaxone | |
| pCWR624 (wild type) | M | I | E | − | 38 | 205 | 1,733 | 2,150 |
| pCWR633 | T | − | 34 | 162 | 1,480 | 1,760 | ||
| pCWR634 | V | − | 42 | 171 | 1,500 | 2,050 | ||
| pCWR635 | T | − | 48 | 226 | 2,183 | 2,450 | ||
| PCWR697 | A | 55 | 123 | 2,933 | 3,266 | |||
| pCWR651 | + | 46 | 250 | 2,233 | 2,383 | |||
| pCWR661 | T | + | 56 | 237 | 2,550 | 3,050 | ||
| pCWR662 | V | + | 65 | 293 | 4,750 | 3,233 | ||
| pCWR663 | T | + | 92 | 341 | 4,366 | 3,233 | ||
| pCWR698 | A | + | 110 | 197 | 5,666 | 4,133 | ||
| pCWR666c | A | T | V | + | 185 | 607 | 4,828 | 6,350 |
| C68 (clinical strain) | A | T | V | + | 250 | >1,000 | >8,000 | 8,000 |
All plasmid constructs were in strain D344SRF. Ampicillin MICs for D344SRF alone and D344SRF containing the vector plasmid pTCV-lac were repeatedly ≤2 μg/ml.
Values represent the arithmetic means of multiple MIC determinations. The number of replicate MICs varied between the different constructs. This variation resulted from the variation in timing of construct availability, since mutants with double mutations were derived from mutants with single mutations. For example, the wild-type gene construct (pCWR624) was available from the beginning of the experiments and was therefore included in all MIC tests (greater than 10), whereas pCWR698 was the most recently constructed and was tested only in duplicate. All available constructs were tested for each MIC determination, so all constructs except pCWR698 were tested on at least four occasions.
pCWR666 contains mutations in addition to those designated (F497G, R598Q, P667S) and contains the pbp5 gene from E. faecium C68.
We based our choices of sites for mutations on the report by Rybkine et al. (12), in which common mutations found in highly resistant strains were identified. Those investigators considered the methionine-to-alanine or methionine-to-threonine substitution at position 485, as well as the added serine found at position 466′ in strain H80761, to be particularly important. Both the M485A change and the added serine at position 466′ were present in the PBP 5 of E. faecium C68. We also elected to introduce I499T and E629V mutations that were present in C68 and other resistant clinical isolates described previously (4), but not in the PBP 5 from ampicillin-susceptible strain E. faecium D366.
Expression of large quantities of PBP 5 for binding studies.
In order to perform PBP 5 binding affinity analysis, large quantities of protein were isolated by cloning truncated versions of PBP 5 into expression vector pMC56/RBSII (2), a 2,275-bp cloning vector derived from pDS56/RBSII (the ampicillin resistance determinant was replaced with the chloramphenicol resistance determinant, and the origin of replication was reversed). Amplification products were generated from templates consisting of 5 ng of shuttle vector containing different pbp5 mutants (pCWR624 through pCWR666) in a 50-μl mixture. Amplification continued for 16 cycles with the Quikchange kit (Stratagene) by use of the conditions recommended for a site-directed mutagenesis reaction. Mutagenesis was not performed in these reactions; rather, the conditions were used to minimize the potential for the introduction of undesirable mutations during amplification. Oligonucleotides were designed to amplify the portion of pbp5 that excluded the first 36 amino acids that constitute the transmembrane portion of the protein and to include BamHI sites on each end of the amplified product: BamHI36a (5′-CAGGATCCTATCAAGAAACCCAAGCAGTAGAAGCTGG-3′) and pbp5 pastterm (5′-CATGGATCCAGTTGTGAAAGAGACATCGT-3′). PCR products were ligated to Zero Blunt topo (Invitrogen) and transformed into E. coli DH10B. The insert was excised by using BamHI, ligated to the BamHI site of PMC56/RBSII, and transformed into E. coli DH10B. The correct sequence was confirmed for each mutant in this host, after which the plasmids were transformed into E. coli M15(pREP4) cells for protein extraction. The final deduced protein contained a sequence of eight amino acids (Met Arg Gly Ser Tyr Gln Glu Thr) preceding the start of the truncated PBP 5 at amino acid 36 (Tyr).
PBP analysis.
PBP 5 was isolated from E. coli M15(pREP4) harboring plasmids encoding the PBP 5 proteins with their membrane anchors deleted. Twenty-milliliter cultures were grown in BHI agar at 37°C and exposed to 1 mM isopropyl-β-d-thiogalactopyranoside for 1 h after an optical density of 0.6 at 600 nm was achieved. After centrifugation and resuspension in 1 ml of phosphate buffer (pH 7, 50 mM) the cells were exposed to 30 s of sonication (Branson Sonic Power Co., Danbury, Conn.). After ultracentrifugation, 5 to 10 μl of the supernatant (25 μg of protein) was used to label the soluble PBP 5 with various amounts (1 to 256 μg/ml) of [14C]penicillin (2.11 GBq/mmol; Amersham Bioscience) at 35°C for 30 min. PBP 5 was separated by sodium dodecyl sulfate-polyacrylamide gel (10%) electrophoresis and was detected as described previously (15). The relative quantities of labeled PBP 5 were estimated after densitometry scanning of the gels with the GEL ANALYST perfect image master (version 4.01; Clara Vision, Orsay, France).
RESULTS AND DISCUSSION
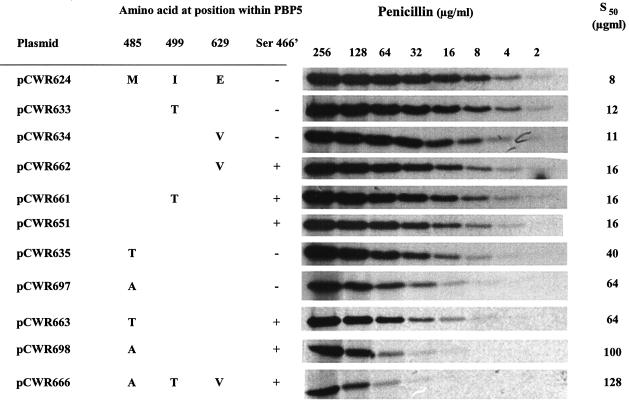
The results of MIC testing for ampicillin, piperacillin, ticarcillin, and ceftriaxone are shown in Table 1. These data indicate that single mutations yield little in terms of increased MICs of the different antimicrobial agents, with at most a 50% increase in MIC associated with any single mutation. Combinations of the different mutations with the addition of a serine at position 466′ yield higher MICs, in most cases doubling the MIC compared to those associated with the PBP 5 of parent strain D366. The combination of all three mutations, as embodied by pbp5 of E. faecium C68 (pCWR666 in Table 1), yields the highest MICs of all compounds tested, suggesting that the interaction of all the mutations together yields the lowest affinity of all. The impacts of the different mutations on the activities of different classes of antibiotics vary considerably. For example, the alanine substitution at position 485, when combined with the serine added at position 466′, appeared to have a greater impact on the activities of ticarcillin (3.2-fold), ampicillin (2.8-fold), and ceftriaxone (2-fold) than on that of piperacillin (no change) (Table 1). Conversely, the threonine substitution at position 485 in combination with the added serine appeared to have a greater impact on the activities of ampicillin (2.4-fold) and ticarcillin (2.5-fold) than on those of piperacillin (1.6 fold) and ceftriaxone (1.5 fold). The valine substitution at position 629 had little impact when it was present by itself, but it was associated with detectable increases in the MICs of all antimicrobials when it was combined with the added serine at position 466′.
The affinities of the different mutant PBP 5 proteins for penicillin correlated with the MICs. The PBPs associated with lower ampicillin MICs exhibited a greater affinity for [14C]penicillin than did the more resistant variants (Fig. 2). These data indicate that the primary amino acid sequence of PBP 5 exerts an impact on the affinity for penicillin and that reduced affinity translates into lower susceptibility in vitro. However, this correlation was not strictly proportional. As an example, saturation of PBP 5 by penicillin showed at least a 12-fold decrease in affinity between the wild-type PBP 5 (pCWR624) and the PBP 5 with M485A and the additional serine at position 466′ (pCWR698) (Fig. 2; Table 1). In contrast, the ampicillin MICs for the strains harboring pCWR698 increased only threefold. It therefore seems clear that the decreased affinity of PBP 5 is not the only factor involved in the expression of β-lactam resistance in E. faecium.
FIG. 2.
Affinities of different PBP 5 mutants for binding to penicillin. Different PBP 5 mutants with their membrane anchors deleted were produced in E. coli after truncated versions of pbp5 genes amplified from the different pCWR plasmids listed in the figure were cloned in pMC56/RBSII (Table 1). The amino acids at the indicated positions within PBP 5 are shown; + and −, presence and absence of Ser at position 466′, respectively; Penicillin (μg/ml), the concentrations of penicillin used to saturate the different PBP 5 proteins; S50, the concentration of penicillin needed to saturate 50% of PBP 5.
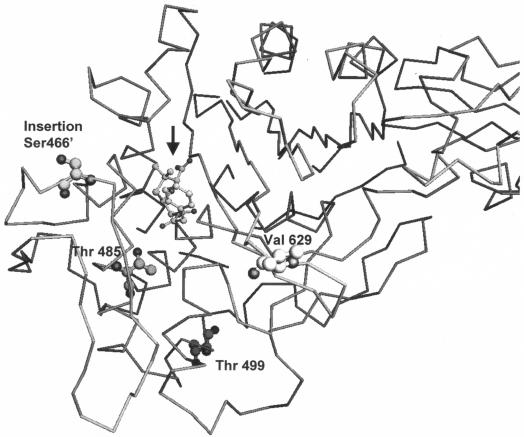
Examination of the positions of the various mutations in the X-ray crystal structure of PBP 5 (13; D. Kostrewa, A. D'Arcy, G. E. Dale, M. G. P. Page, and F. K. Winkler, unpublished data) reveals that most of the substitutions occur around the active-site region (Fig. 3). The exception is position 499, which is quite distant from the active site. This residue is located in the middle of helix 3 of the transpeptidase domain, on the surface of the protein almost opposite the active-site cleft. The amino acid side chain projects into the surrounding solvent, and therefore, the I499T substitution would not be expected to have any significant effect on the local structure or electric field. Indeed, the single substitution at this position has little effect on the susceptibilities to β-lactams.
FIG. 3.
Structure of the E. faecium PBP 5. The α-carbon atom trace of the transpeptidase domain of PBP 5 complexed with benzylpenicillin (arrow) (13; Kostrewa et al., unpublished), prepared using MOLOC (6), is shown. The benzylpenicillin moiety of the acyl enzyme is shown in a small ball-and-stick representation, and the side chains of the mutated residues are highlighted by larger ball-and-stick representations.
Position 485 is located inside the active site, very close to the active-site serine at position 422, the residue that is acylated by β-lactams (Fig. 3), and substitutions should be expected to have strong effects. The side chain of methionine packs in a hydrophobic pocket and interacts with the side chain of lysine 425, possibly helping to position it for activation of serine 422. The substitutions to threonine and alanine that occur in the resistant mutants both result in a smaller side chain that would not fill the hydrophobic pocket so well and could result in the lysine at position 425 not being so well positioned with respect to the serine at position 422 and, thus, in decreased reactivities toward β-lactams. Position 629 lies at the end of a turn between the β1 and β2 strands, which form one edge of the active site (Fig. 3). The amino acid side chain projects into the surrounding solvent and does not interact directly with the β-lactam antibiotic. The E629V substitution is an unfavorable one for the overall stability of the protein, as it introduces a hydrophobic residue in place of a hydrophilic one that would be easily hydrated. Hydrophobic residues are strongly disfavored in loops (8) but do occur more frequently in the end positions of turns (3). This loop does move somewhat during the acylation by benzylpenicillin (Kostrewa et al., unpublished), as does the corresponding structure in Streptococcus pneumoniae PBP 2x (7, 9). Thus, the replacement of glutamate by valine may have consequences for the dynamic conformation of the loop, and by restricting mobility it may decrease the ability of the protein to bind to β-lactams. The remote location would suggest that this should be, by itself, a relatively weak effect, which is confirmed by the modest changes in the measured affinities observed.
The last position to consider is the insertion of the serine at position 466′ after the serine at position 466. This residue is located in a loop that forms the other edge of the active site relative to that defined by the β1 strand. It is a rather flexible structure, as is the homologous loop in S. pneumoniae PBP 2x. The loop in the latter protein undergoes considerable restructuring during the reaction with β-lactams (9, 13), and this structure also appears to be the part of PBP 5 that is most affected by the acylation reaction (Kostrewa et al., unpublished). The consequences of the insertion on the structural dynamics of the loop are difficult to predict, but the potential for an influence on β-lactam binding is manifest.
The effects of the individual mutations are not simply additive: the serine insertion has no measurable effect on the modest change in affinity produced by the E629V substitution but does seem to amplify the effects of the Met485 substitutions and the I499T substitution.
In conclusion, this extensive in vitro mutagenesis study showed that many amino acid substitutions present in the resistant clinical isolates (11, 12) were associated with a decreased affinity of PBP 5, which translated into different levels of β-lactam resistance expression. No single mutation conferred a substantial increase in resistance to any of the four antibiotics tested. In particular, these results highlight the role of the additional serine at position 466′, which was encountered only in the most resistant clinical isolates and only in combination with other mutations. Further work will be required to define the other factors that contribute to the ultimate resistance levels expressed in clinical isolates.
Acknowledgments
We thank H. P. Weber for help in preparing the figure of the crystal structure of E. faecium PBP 5.
These studies were supported by a grant from the National Institute of Allergy and Infectious Diseases (grant R01 AI45626).
REFERENCES
- 1.Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chittock, R. S., S. Ward, A. S. Wilkinson, P. Caspers, B. Mensch, M. G. Page, and C. W. Wharton. 1999. Hydrogen bonding and protein perturbation in beta-lactam acyl-enzymes of Streptococcus pneumoniae penicillin-binding protein PBP2x. Biochem. J. 338(Pt 1):153-159. [PMC free article] [PubMed] [Google Scholar]
- 3.Chou, P. Y., and G. D. Fasman. 1977. Beta-turns in proteins. J. Mol. Biol. 115:135-175. [DOI] [PubMed] [Google Scholar]
- 4.Eberhardt, C., L. Kuerschner, and D. S. Weiss. 2003. Probing the catalytic activity of a cell division-specific transpeptidase in vivo with beta-lactams. J. Bacteriol. 185:3726-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontana, R., M. Aldegheri, M. Ligozzi, H. Lopez, A. Sucari, and G. Satta. 1994. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 38:1980-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerber, P. R., and K. Muller. 1995. MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J. Comput. Aided Mol. Des. 9:251-268. [DOI] [PubMed] [Google Scholar]
- 7.Gordon, E., N. Mouz, E. Duee, and O. Dideberg. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299:477-485. [DOI] [PubMed] [Google Scholar]
- 8.Leszczynski, J. F., and G. D. Rose. 1986. Loops in globular proteins: a novel category of secondary structure. Science 234:849-855. [DOI] [PubMed] [Google Scholar]
- 9.Pares, S., N. Mouz, Y. Petillot, R. Hakenbeck, and O. Dideberg. 1996. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 3:284-289. [DOI] [PubMed] [Google Scholar]
- 10.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galatosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 11.Rice, L. B., L. L. Carias, R. Hutton-Thomas, F. Sifaoui, L. Gutmann, and S. D. Rudin. 2001. Penicillin-binding protein 5 and expression of ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 45:1480-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rybkine, T., J.-L. Mainardi, W. Sougakoff, E. Collatz, and L. Gutmann. 1998. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of β-lactam resistance. J. Infect. Dis. 178:159-163. [DOI] [PubMed] [Google Scholar]
- 13.Sauvage, E., F. Kerff, E. Fonze, R. Herman, B. Schoot, J. P. Marquette, Y. Taburet, D. Prevost, J. Dumas, G. Leonard, P. Stefanic, J. Coyette, and P. Charlier. 2002. The 2.4-Å crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell. Mol. Life Sci. 59:1223-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sifaoui, F., M. Arthur, L. Rice, and L. Gutmann. 2001. Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrob. Agents Chemother. 45:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williamson, R., C. LaBouguenec, L. Gutmann, and T. Horaud. 1985. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to penicillin. J. Gen. Microbiol. 131:1933-1940. [DOI] [PubMed] [Google Scholar]
- 16.Zorzi, W., X. Y. Zhou, O. Dardenne, J. Lamotte, D. Raze, J. Pierre, L. Gutmann, and J. Coyette. 1996. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J. Bacteriol. 178:4948-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]