Abstract
The role of mitochondrial complex I in aging has been studied in both C. elegans and Drosophila, where RNAi knock down of specific complex I subunits has been shown to extend lifespan. More recently, studies in Drosophila have shown that an increase in mitochondrial activity, including complex I-like activity, can also slow aging. In this review, we discuss this apparent paradox. Improved maintenance of mitochondrial activity, mitochondrial homeostasis, may be responsible for lifespan extension in both cases. Decreased electron transport chain activity caused by reducing complex I subunit expression prompts an increase in stress response signaling that leads to enhanced mitochondrial homeostasis during aging. Increased complex I activity, as well as mitochondrial biogenesis, is expected to both directly counteract the decline in mitochondrial health that occurs during aging and may also increase cellular NAD+ levels, which have been linked to mitochondrial homeostatic mechanisms through activation of sirtuins. We suggest that manipulations that increase or decrease complex I activity both converge on improved mitochondrial homeostasis during aging, resulting in prolonged lifespan.
Keywords: Electron transport chain, Sirtuin, Mitochondrial homeostasis, Hormesis
Introduction
Metabolic rate has been closely associated with aging for as long as mechanisms of aging have been examined. Scientists as early as Aristotle characterized qualitative differences between the living and the dead, and deduced that the rate of aging must depend on the rate of change that organisms undergo before dying (Ross 1952). This intuitive idea that aging is determined by the rate of conversion of materials essential for life persisted through the turn of the previous century, compelling August Weismann to caution that, “The organism must not be looked upon as a heap of combustible material, which is completely reduced to ashes in a certain time the length of which is determined by size, and by the rate at which it burns…” (Weismann 1889).
Consequently, it comes as no surprise that initial forays into mechanistic studies of aging looked to metabolic rate as a fundamental cause of aging. Though these “rate-of-living” theories (Rubner 1908; Pearl 1928) have fallen out of favor, they set the stage for Denham Harman to delineate one of the most influential theories of aging that was the basis for many current studies of aging and longevity (Harman 1956). The Free Radical Theory of Aging (FRTA) theorizes that aging is not inherently caused by metabolism, but by accumulation of damage from reactive oxygen species (ROS) that are byproducts of metabolism. In recent years, however, the FRTA has faced a number of challenges and difficulties (Gems and Doonan 2009; Lapointe and Hekimi 2010). Indeed, it has been shown that while high levels of ROS are detrimental, moderately increased ROS levels can be beneficial. By a process known as hormesis, moderate increases in ROS have been shown to trigger stress responses that stimulate repair and protection pathways (Yun and Finkel 2014).
Aerobic metabolic processes in eukaryotes are compartmentalized and segregated in the mitochondria which functions both as the largest source of chemical energy in the form of ATP, and ROS (Wallace 2005). The major entryway of high energy electrons into the electron transport chain (ETC) is mitochondrial complex I (NADH-ubiquinone oxidoreductase) which serves multiple functions as a pace-setter for the ETC, a major producer of the electrochemical gradient across the inner mitochondrial membrane, and a major source of ROS production (Barja 1999; Papa et al. 2012). As expected from an enzyme that integrates many different functions, complex I is a complicated and highly organized holoenzyme comprised of more than 40 different subunits in mammals (Carroll et al. 2006; Gabaldón et al. 2005), with a growing list of accessory and assembly factors (Diaz et al. 2011; Cho et al. 2012). The assembly of complex I holoenzyme is an intricate multi-step process that involves the formation of multiple subassemblies (Mimaki et al. 2012). The fully assembled holoenzyme is thought to be L-shaped, embedded in the mitochondrial inner membrane by a hydrophobic membrane arm with a hydrophilic peripheral arm protruding into the mitochondrial matrix (Clason et al. 2010; Efremov et al. 2010).
In this review, we focus on studies showing that genetic manipulations affecting mitochondrial complex I can modulate lifespan in invertebrates. These studies include RNAi-based manipulations showing that knock down of certain complex I subunits can prolong both worm and fly lifespan. In addition, we discuss the more recent reports showing that up-regulation of mitochondrial activity, including complex I activity, can also prolong lifespan in flies.
Decreasing complex I and lifespan
The first evidence that knock-down of complex I subunits could positively impact metazoan lifespan came from genome-wide RNAi screens in C. elegans (Dillin et al. 2002; Lee et al. 2003). Much of the work in characterizing the adult lifespan effects of decreased complex I subunit expression in C. elegans involved RNAi knock down of nuo-2 (Dillin et al. 2002; Rea et al. 2007), the C. elegans homolog of mammalian NADH dehydrogenase iron-sulfur protein 3 (ndufs3) and D2030.4, the homolog of mammalian NADH dehydrogenase 1 beta subcomplex subunit 7 (ndufb7) (Lee et al. 2003). In addition to extending lifespan, reduced nuo-2 or D2030.4 expression led to an approximately 50 % reduction in ATP levels, and in the case of D2030.4, decreased oxygen consumption and morphological defects in mitochondria (Dillin et al. 2002; Lee et al. 2003). Worms under these RNAi knock down regimes lived significantly longer, but developmental time, size, and activity were all adversely affected by the treatment (Dillin et al. 2002; Lee et al. 2003; Rea et al. 2007). Even from these early studies, it was apparent that the relationship between reduced ETC function and increased longevity was not always a simple one of reduced ROS resulting in increased longevity (Dillin et al. 2002; Lee et al. 2003; Rea et al. 2007).
Similarly, lifespan increases due to RNAi knock down of other mitochondrial ETC subunits show either a cryptic relationship with oxidative stress (Lee et al. 2003) or are largely independent of ROS damage, quantified via carbonylated protein content (Rea et al. 2007; Yang et al. 2007). A much more decisive factor in whether RNAi knock down of ETC subunits extended lifespan was identified to be the timing of the knock down, with down regulation during a specific developmental window being necessary and sufficient to increase adult lifespan (Dillin et al. 2002). Reducing ETC subunit expression only during adult stages, after completion of the critical L3/L4 larval stages, caused similar reductions in oxidative phosphorylation (OXPHOS) (measured by ATP levels) as life-long knock downs, but did not result in longer lifespans (Dillin et al. 2002). The fact that the timing of ETC activity decrease is a much stronger determinant of longevity compared to the degree of OXPHOS reduction strongly indicates that the FRTA is insufficient to explain these findings.
More recent studies in C. elegans have yielded further evidence contrary to the FRTA. Worms with reduced ETC activity, either by RNAi knock down or hypomorphic mutation of ETC complex subunits, have extended longevities that are dependent on increased ROS levels (Lee et al. 2010; Yang and Hekimi 2010a; Yee et al. 2014). An RNAi knock down screen in C. elegans for increased activity of hypoxia-inducible factor 1 (HIF-1) prominently featured ETC genes, including nuo-1, another complex I subunit (Lee et al. 2010). Although nuo-1 was not specifically tested, reduced expression of other proteins involved in ETC function, including a complex III subunit (isp-1), were shown to have increased HIF-1 activity and extended longevity largely as a result of mitochondrial hormesis or “mitohormesis” (Yun and Finkel 2014) induced by increased ROS levels (Lee et al. 2010). A different study carefully examined the downstream pro-longevity mechanisms of reduced complex I activity and showed that mutating and knocking down expression of a complex I subunit, nuo-6, extend lifespan through independent mechanisms (Yang and Hekimi 2010b). Recent work from the same lab has revealed that unlike RNAi knock down of ETC complex subunits, mutation of ETC complex subunits cause increased ROS production which results in activation of a different stress response pathway, the intrinsic apoptosis pathway (Yee et al. 2014). Lifespan extensions in both cases are, however, attributable to mitohormetic increases in ROS production, such that both manipulations can be phenocopied in wild-type animals by supplementation with low doses of pro-oxidants such as paraquat (Lee et al. 2010; Yang and Hekimi 2010a).
Studies in Drosophila largely involved knocking down different complex I subunits than those in the C. elegans studies, in part due to the developmental lethality of reduced expression of CG12079 and CG5548, the D. melanogaster homologs of C. elegans nuo-2 and D2030.4, respectively (Copeland et al. 2009), and the lack of a Drosophila homolog of nuo-6. Instead, work in our lab and others focused on two other complex I subunits that extended lifespan in flies, CG9762 (mammalian NADH dehydrogenase 1 beta subcomplex 5, ndufb5) and CG9172 (mammalian NADH dehydrogenase iron-sulfur protein 7, ndufs7), whereas the Tricoire lab examined the effects of reduced CG9140 (mammalian NADH dehydrogenase flavoprotein 1, ndufv1) (Rera et al. 2010) and the Perrimon lab focused on the muscle specific knock down of CG2286 (mammalian NADH dehydrogenase iron-sulfur protein 1, ndufs1) (Owusu-Ansah et al. 2013).
In line with the results of reduced expression of complex I subunits in C. elegans, reduced expression of two different complex I subunits in flies (CG9172 and CG9762) caused increased lifespan and decreased fertility (Copeland et al. 2009). Knocking down complex I subunits in flies, however, also showed significant differences. First, unlike knock down of nuo-2 or D2030.4 in C. elegans, where reduced ETC subunit expression consistently resulted in decreased ATP levels, conflicting results were obtained with respect to ATP levels in knock downs of complex I subunits in flies, showing a full range of effects, from decreases (CG9140) (Rera et al. 2010), to no change (CG9172), to increases (CG9762) (Copeland et al. 2009). Other measures related to organismal physiology, such as developmental timing, were also inconsistent, with CG9762 reduction resulting in no difference in development time (Copeland et al. 2009) but CG9140 showing significant delay (Rera et al. 2010). Second, while knock down of the nuo-2 subunit at a specific developmental time window was shown to be required to extend lifespan in C. elegans, knock down of CG9762 or CG9172 only during adulthood was sufficient to extend lifespan in flies (Copeland et al. 2009). Moreover, reduced expression of a complex I subunit which had been associated with lifespan extension in C. elegans mutant studies (CG9140, homolog of C. elegans nuo-1) (Tsang et al. 2001) throughout developmental stages did not extend lifespan over a broad range of knock down levels in flies (Rera et al. 2010), showing that reduced expression of the same subunit can have different species-specific effects on lifespan. Finally, it was shown that CG2286, CG9763, or CG9162 knock down-induced longevity is tissue dependent. By taking advantage of the availability of different tissue specific manipulations in flies, knocking down CG9763 or CG9162 only in neurons was shown to be sufficient for lifespan extension (Copeland et al. 2009). Similarly, RNAi knock down of CG2286 only in muscles during development was also sufficient to extend lifespan (Owusu-Ansah et al. 2013). Thus, reducing expression of different subunits of complex I have complex effects, possibly in a species- and tissue-specific manner.
Therefore, reducing complex I subunit expression, can prolong lifespan in both worms and flies, despite the distinct requirements for different subunits in different model systems. It is clear, however, that the relationship between complex I function and aging is more complicated than a simple one of reduced function resulting in reduced OXPHOS, reduced ROS levels, and increased lifespans. For some subunits in C. elegans, reducing ETC function at a specific stage of development was vital to lifespan extension, with no increase in lifespans with similar reductions in adulthood (Dillin et al. 2002). Other subunits, tested in flies, showed lifespan extension regardless of time of knock down (Copeland et al. 2009). Direct measurements of oxidative damage over a range of knock downs that produce lifespan extensions in C. elegans did not yield a clear relationship between ROS damage and longevity (Rea et al. 2007) and reduced complex I activity either by subunit mutation or RNAi knock down were shown to increase lifespan by increasing ROS production (Lee et al. 2010; Yang and Hekimi 2010b). Moreover, reducing expression of specific subunits was also seen to have surprisingly diverse effects in overall OXPHOS activity, in at least one case resulting in increased overall ATP levels (Copeland et al. 2009). Even reduced expression of an orthologous subunit in different model organisms was shown to have different effects in terms of longevity (Rera et al. 2010; Owusu-Ansah et al. 2013). These results speak to the complex interactions that govern the relationships among the complex I subunits, ETC activity, and longevity.
Increasing complex I and lifespan
An intriguing result of studies of dietary restriction (DR), the most well recognized and studied intervention for extending lifespans in mammals (Mair and Dillin 2008), has been the revelation that DR may increase mitochondrial activity relative to age-matched controls (Guarente 2008; Anderson and Weindruch 2010). Moreover, a gradual decline in mitochondrial homeostasis has been linked with aging (Wallace 2005), prompting a search for ways to directly test the effects of increased mitochondrial activity or improved mitochondrial homeostasis on aging. Simply overexpressing complex I to increase activity is currently technically impossible, as complex I is a holoenzyme comprised of more than 40 subunits, some of which are encoded in the mitochondrial genome, making it difficult if not impossible to overexpress the holo-enzyme in a coordinated manner (Carroll et al. 2006). Nevertheless, our group and others have examined the impact of stimulating mitochondrial activity in aging flies by two independent approaches—by up-regulating the Drosophila PGC-1 homolog, and by transgenic expression of an alternative single-subunit complex I enzyme.
In mammals, the PGC-1 family of transcriptional coactivators plays a central role in the regulation of mitochondrial biogenesis and activity (Lin et al. 2005). Recently, our group reported that overexpression of the fly PGC-1 homolog (dPGC-1) results in significant increases in multiple markers of complex I biogenesis and function (Rera et al. 2011). Interestingly, lifespan was only extended when dPGC-1 was overexpressed in the fly intestine (Rera et al. 2011), a tissue that has recently come into focus as an important mediator of systemic aging (Rera et al. 2013; Wang et al. 2014). In addition to increased metabolic output and increased lifespans, dPGC-1 overexpression reduced ROS levels and improved tissue homeostasis in the aging intestine. However, as PGC-1 can impact diverse metabolic processes, it is important to note that effects of overexpression cannot be attributed solely to any single downstream effect of dPGC-1 expression, such as tissue-specific up-regulation of complex I activity.
S. cerevisiae utilize single-subunit enzymes to perform complex I-like functions in its ETC (Bakker et al. 2001). One of these, NADH dehydrogenase internal 1 (ndi1) is functional in higher metazoans, including mammals, acting as a non-proton translocating NADH dehydrogenase/ubiquinone oxidoreductase in the mitochondrial matrix (Yagi et al. 2006). Transgenic expression of ndi1 in D. melanogaster rescues loss of endogenous complex I (Cho et al. 2012) and expression in flies without complex I defects supplements and boosts endogenous NADH dehydrogenase/ubiquinone oxidoreductase activity in a rotenone-insensitive manner (Sanz et al. 2010; Bahadorani et al. 2010; Hur et al. 2013). Critically, unlike dPGC-1, this increase in complex I-like activity was achieved without significant changes to other ETC complexes (Sanz et al. 2010; Bahadorani et al. 2010). Like dPGC-1, expression of ndi1 in flies resulted in increased markers of complex I-like activity, extended longevity, and reduced ROS levels (Sanz et al. 2010; Bahadorani et al. 2010; Hur et al. 2013). Moreover, tissue specific expression of ndi1, in the nervous system or intestine, resulted in increased lifespan but ubiquitous expression did not (Bahadorani et al. 2010; Hur et al. 2013).
Due to the more complex nature of overexpressing a complex multi-subunit holoenzyme, current results regarding longevity effects of increased complex I activity are limited. Still, up-regulation of a metabolic master regulator (PGC-1) and transgenic expression of a protein with complex I-like functions (NDI1) have suggested positive longevity effects of increased complex I-like activity. Unfortunately, comprehensive studies that examine the relationship among complex I activity levels, ROS, and lifespan are not available. The dPGC-1 and ndi1 results outlined above suggest that increasing mitochondrial activity may increase lifespan by simply increasing the pool of functional mitochondria over time, as well as by decreasing ROS that is generated as a result of age-related mitochondrial dysfunction (Wallace 2005). The tissue specific requirements of the pro-longevity effects of increased metabolism and complex I-like activities, however, argue against a simple relationship between complex I and aging.
Mitochondrial homeostasis—untangling complex-I-ty?
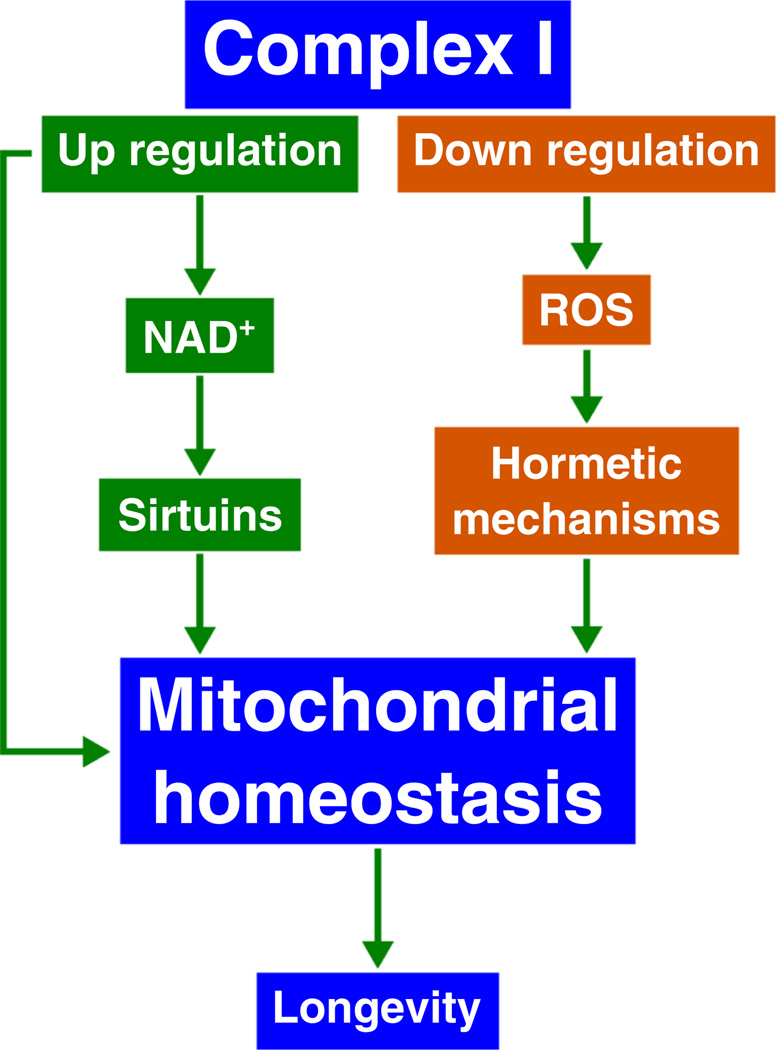
A unifying theme that has emerged relatively recently in extension of lifespan via alterations of mitochondrial activity, including complex I activity, is the central role of mitochondrial homeostasis in aging (Fig. 1). Proteomic analysis of protein aggregates in C. elegans revealed that the protein products of genes involved in mitochondrial respiration, including those involved in complex I, are significantly over-represented in populations of proteins prone to misfolding and forming inclusion bodies (David et al. 2010). Supporting the consensus that loss of mitochondrial homeostasis contributes to aging, overexpression of parkin, a gene involved in mitochondrial quality control, increased lifespan and mitochondrial activity, including complex I activity (Rana et al. 2013).We hypothesize that the previous examples of lifespan extensions resulting from both decreased and increased complex I function are due, at least in part, to increased mitochondrial homeostatic activities that may result from the diverse manipulations. In addition to simply increasing mitochondrial function, increased complex I activity could improve mitochondrial homeostasis through increases in NAD+ levels, which have been shown to increase mitochondrial stress responses through activation of sirtuins (Mouchiroud et al. 2013). Increasing mitochondrial homeostasis by ETC knock down may occur in at least three different ways—by depleting the pool of proteins that are prone to aggregation and forming inclusion bodies in mitochondria, hormeticially increasing mitochondrial turn over, and by altering the ratio of mitochondrial and nuclear protein ratios which has been shown to hormetically induce stress response mechanisms (Houtkooper et al. 2013).
Fig. 1.
Complex I, mitochondrial homeostasis, and aging. An increase in complex I activity increases mitochondrial homeostasis directly and also results in increased NAD+ levels, which activates sirtuins and stimulates mitochondrial homeostasis. A decrease in complex I activity, by mutation or reduced expression, increases ROS levels, resulting in induction of mitochondrial stress response mechanisms that help maintain mitochondrial homeostasis over time
Complex I subunits appear to be particularly prone to forming protein aggregates in C. elegans, as evidenced by their dramatic over-representation in the set of insoluble proteins (David et al. 2010). Increased aggregation of insoluble proteins with aging, in addition to direct effects of the aggregated proteins, can deplete proteostatic machinery, affecting functionality of proteins that do not show age-dependence in aggregate formation (Ben-Zvi et al. 2009). Knocking down complex I subunits, by decreasing overall levels of aggregation prone proteins, may simply shift the equilibrium away from further accumulation of protein aggregates, alleviating stress on the proteostatic pathways that become overloaded with age and improving mitochondrial health (Rana et al. 2013; Ben-Zvi et al. 2009).
Both RNAi knock down (Lee et al. 2010) and mutation (Yang and Hekimi 2010a) of complex I subunits induces a mitohormetic response by increasing ROS that results in activation of stress response pathways. Reduced expression of subunits by RNAi knock down has been shown to induce a hypoxic response, increasing HIF-1 activity (Lee et al. 2010), which has been shown to increase tolerance to proteotoxic stress (Mehta et al. 2009) and activate autophagy (Bohensky et al. 2007), both of which may contribute to mitochondrial homeostasis. Mutation of complex I subunits also results in increased ROS levels which causes activation of a different mitochondrial stress response mechanism, the intrinsic apoptosis pathway (Yee et al. 2014). Therefore, reduced complex I function, by mutation or RNAi knock down, independently extend lifespan by activating different mitochondrial stress response pathways.
Given the intricate coordination that is required to assemble large holoenzymes like complex I (Mimaki et al. 2012), it is likely that reduced expression of single subunits will cause imbalances in the steady state levels of enzyme subunits, including those between mitochondrial and nuclear protein levels. An imbalance between nuclear and mitochondrial mRNA levels has also been shown to be sufficient to cause induction of mitochondrial stress response mechanisms (Gomes et al. 2013). These stress response mechanisms, such as the mitochondrial unfolded protein response (mitoUPR) have been shown to be important for the extended lifespans in studies of knock down of certain ETC genes (Owusu-Ansah et al. 2013; Houtkooper et al. 2013; Durieux et al. 2011). However, more recently, the role of mitoUPR in ETC-mediated life extension has been challenged (Bennett et al. 2014). Indeed, Bennett et al. show that induction of mitoUPR alone is insufficient to extend lifespan, dissociating mitoUPR and increased lifespan due to ETC complex subunit knock down. Regardless of the specific involvement of mitoUPR in lifespan extension however, it is clear from the various knock down and mutation experiments that maintenance of mitochondrial integrity by other mitochondrial stress response mechanisms (Lee et al. 2010; Yee et al. 2014) can play key roles in lifespan extensions due to complex I down regulation, and that imbalanced nuclear and mitochondrial mRNA levels may be sufficient to trigger some of these (Gomes et al. 2013). Fly studies also support a role for improved mitochondrial homeostasis in increased longevity, as reduced expression of complex I subunits (CG2286 and CG9762) causes improved mitochondrial homeostasis over time (Owusu-Ansah et al. 2013).
Increasing complex I activity also improves mitochondrial homeostasis similarly to that caused by reduced complex I expression or function. Interventions that increase complex I activity, either by boosting wholesale metabolic activity with overexpression of the fly PGC-1 homolog (Rera et al. 2011), or by transgenic expression of foreign NADH dehydrogenases like ndi-1 (Yagi et al. 2006; Sanz et al. 2010; Bahadorani et al. 2010; Hur et al. 2013) are reasonably expected to increase ETC activity and increase NAD+ concentrations as NADH dehydrogenase activity increases. As a potent modulator of sirtuin activity, NAD+ concentrations, altered both genetically and pharmacologically, have been shown to be critical regulators of mitochondrial gene expression (Mouchiroud et al. 2013; Gomes et al. 2013). Therefore, increasing complex I activity may extend lifespan by both increasing the amount of functional mitochondrial proteins over time, and by triggering improved mitochondrial homeostasis through increased NAD+ levels.
Conclusion
Increases in mitochondrial homeostasis mechanisms have been linked to enhanced lifespan in both worms (Yuan et al. 2012; Munkácsy and Rea 2014) and flies (Rana et al. 2013; Cho et al. 2011). Similarly, broad evidence exists for increased lifespans caused by both increasing and decreasing mitochondrial activity, including complex I activity. How manipulations that increase or decrease complex I activity both result in increased longevity has recently been clarified by the discovery of pathways that link both ETC down regulation and high NAD+ concentrations to improved mitochondrial homeostasis, though the exact role of at least one homeostatic mechanism, mitoUPR, remains to be further clarified (Owusu-Ansah et al. 2013; Houtkooper et al. 2013; Bennett et al. 2014). Evidence suggests that much of the increased longevity due to decreased mitochondrial subunit expression results from hormetic effects that result in mitochondrial stress responses, with FRTA, as originally outlined, playing a relatively minor role (Lapointe and Hekimi 2010; Yang et al. 2007; Yang and Hekimi 2010a, b; Yee et al. 2014; Durieux et al. 2011; Doonan et al. 2008; Ristow and Zarse 2010). Further details of what these mechanisms are and what contributions they may make to lifespan determination in other organisms, including mammals, remains to be determined. However, insights from the association of both increased and decreased complex I activity with improved mitochondrial homeostasis suggest that mitochondrial decline remains a primary cause of aging and valuable target for therapeutic intervention.
Acknowledgments
We apologize to our colleagues whose work we could not cite due to space limitations. DWW is supported by the National Institute on Aging (R01 AG037514, R01 AG040288).
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Contributor Information
Jae H. Hur, Email: jaehur@gmail.com, Department of Biology, Harvey Mudd College, Claremont, CA 91711, USA.
Devon A. Stork, Department of Biology, Harvey Mudd College, Claremont, CA 91711, USA
David W. Walker, Email: davidwalker@ucla.edu, Department of Physiological Science, University of California, Los Angeles, Los Angeles, CA 90095, USA; Molecular Biology Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
References
- Anderson RM, Weindruch R. Metabolic reprogramming, caloric restriction and aging. Trends Endocrinol Metab. 2010;21:134–141. doi: 10.1016/j.tem.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahadorani S, Cho J, Lo T, Contreras H, Lawal HO, Krantz DE, Bradley TJ, Walker DW. Neuronal expression of a single-subunit yeast NADH-ubiquinone oxidoreductase (Ndi1) extends Drosophila lifespan. Aging Cell. 2010;9:191–202. doi: 10.1111/j.1474-9726.2010.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker BM, Overkamp KM, van Maris AJ, Kötter P, Luttik MA, van Dijken JP, Pronk JT. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- Barja G. Mitochondrial oxygen radical generation and leak: sites of production in states 4 and 3, organ specificity, and relation to aging and longevity. J Bioenerg Biomembr. 1999;31:347–366. doi: 10.1023/a:1005427919188. [DOI] [PubMed] [Google Scholar]
- Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat Commun. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohensky J, Shapiro IM, Leshinsky S, Terkhorn SP, Adams CS, Srinivas V. HIF-1 regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy. 2007;3:207–214. doi: 10.4161/auto.3708. [DOI] [PubMed] [Google Scholar]
- Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine complex I is a complex of 45 different subunits. J Biol Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- Cho J, Hur JH, Walker DW. The role of mitochondria in Drosophila aging. Exp Gerontol. 2011;46:331–334. doi: 10.1016/j.exger.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Hur JH, Graniel J, Benzer S, Walker DW. Expression of yeast NDI1 rescues a Drosophila complex i assembly defect. PLoS One. 2012;7:e50644. doi: 10.1371/journal.pone.0050644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clason T, Ruiz T, Schägger H, Peng G, Zickermann V, Brandt U, Michel H, Radermacher M. The structure of eukaryotic and prokaryotic complex I. J Struct Biol. 2010;169:81–88. doi: 10.1016/j.jsb.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T, Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Kotarsky H, Fellman V, Moraes CT. Mitochondrial disorders caused by mutations in respiratory chain assembly factors. Semin Fetal Neonatal Med. 2011;16:197–204. doi: 10.1016/j.siny.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Doonan R, McElwee JJ, Matthijssens F, Walker GA, Houthoofd K, Back P, Matscheski A, Vanfleteren JR, Gems D. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on lifespan in Caenorhabditis elegans. Genes Dev. 2008;3:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- Gabaldón T, Rainey D, Huynen MA. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (complex I) J Mol Biol. 2005;348:857–870. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- Gems D, Doonan R. Antioxidant defense and aging in C. elegans: is the oxidative damage theory of aging wrong? Cell Cycle. 2009;8:1681–1687. doi: 10.4161/cc.8.11.8595. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Mitochondria–a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JH, Bahadorani S, Graniel J, Koehler CL, Ulgherait M, Rera M, Jones DL, Walker DW. Increased longevity mediated by yeast NDI1 expression in Drosophila intestinal stem and progenitor cells. Aging (Albany) 2013;5:662–681. doi: 10.18632/aging.100595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Hekimi S. When a theory of aging ages badly. Cell Mol Life Sci. 2010;67:1–8. doi: 10.1007/s00018-009-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- Mehta R, Steinkraus KA, Sutphin GL, Ramos FJ, Shamieh LS, Huh A, Davis C, Chandler-Brown D, Kaeberlein M. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 2009;324:1196–1198. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta. 2012;1817:851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Cantó C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, Guarente L, Auwerx J. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkácsy E, Rea SL. The paradox of mitochondrial dysfunction and extended longevity. Exp Gerontol. 2014 doi: 10.1016/j.exger.2014.03.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa S, Rasmo DD, Technikova-Dobrova Z, Panelli D, Signorile A, Scacco S, Petruzzella V, Papa F, Palmisano G, Gnoni A, Micelli L, Sardanelli AM. Respiratory chain complex I, a main regulatory target of the cAMP/PKA pathway is defective in different human diseases. FEBS Lett. 2012;586:568–577. doi: 10.1016/j.febslet.2011.09.019. [DOI] [PubMed] [Google Scholar]
- Pearl R. The rate of living, being an account of some experimental studies on the biology of life duration. Knopf, New York: 1928. [Google Scholar]
- Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci U S A. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Monnier V, Tricoire H. Mitochondrial electron transport chain dysfunction during development does not extend lifespan in Drosophila melanogaster. Mech Ageing Dev. 2010;131:156–164. doi: 10.1016/j.mad.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jr, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Azizi MJ, Walker DW. Organ-specific mediation of lifespan extension: more than a gut feeling? Ageing Res Rev. 2013;12:436–444. doi: 10.1016/j.arr.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: the concept of mitochondrial hormesis (mitohormesis) Exp Gerontol. 2010;2:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Ross WD. Works of Aristotle. Clarendon, Oxford: 1952. [Google Scholar]
- Rubner M. Das Problem det Lebensdaur und seiner beziehunger zum Wachstum und Ernarnhung. Munich, Germany: Oldenberg; 1908. [Google Scholar]
- Sanz A, Soikkeli M, Portero-Otín M, Wilson A, Kemppainen E, McIlroy G, Ellilä S, Kemppainen KK, Tuomela T, Lakanmaa M, Kiviranta E, Stefanatos R, Dufour E, Hutz B, Naudí A, Jové M, Zeb A, Vartiainen S, Matsuno-Yagi A, Yagi T, Rustin P, Pamplona R, Jacobs HT. Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proc Natl Acad Sci U S A. 2010;107:9105–9110. doi: 10.1073/pnas.0911539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Sayles LC, Grad LI, Pilgrim DB, Lemire BD. Mitochondrial respiratory chain deficiency in Caenorhabditis elegans results in developmental arrest and increased life span. J Biol Chem. 2001;276:32240–32246. doi: 10.1074/jbc.M103999200. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A Mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Karpac J, Jasper H. Promoting longevity by maintaining metabolic and proliferative homeostasis. J Exp Biol. 2014;217:109–118. doi: 10.1242/jeb.089920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann A. Essays upon heredity and kindred biological problems. Clarendon, Oxford: 1889. [Google Scholar]
- Yagi T, Seo BB, Nakamaru-Ogiso E, Marella M, Barber-Singh J, Yamashita T, Matsuno-Yagi A. Possibility of transkingdom gene therapy for complex I diseases. Biochim Biophys Acta. 2006;1757:708–714. doi: 10.1016/j.bbabio.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010a;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell. 2010b;9:433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- Yang W, Li J, Hekimi S. A Measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–2074. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee C, Yang W, Hekimi S. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell. 2014;157:897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Kadiyala CS, Ching TT, Hakimi P, Saha S, Xu H, Yuan C, Mullangi V, Wang L, Fivenson E, Hanson RW, Ewing R, Hsu AL, Miyagi M, Feng Z. Enhanced energy metabolism contributes to the extended life span of calorie-restricted Caenorhabditis elegans. J Biol Chem. 2012;287:31414–31426. doi: 10.1074/jbc.M112.377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Finkel T. Mitohormesis. Cell Metab. 2014;19:757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]