Abstract
The liver performs a large number of essential synthetic and regulatory functions that are acquired during fetal development and persist throughout life. Their disruption underlies a diverse group of heritable and acquired diseases that affect both pediatric and adult patients. Although experimental analyses used to study liver development and disease are typically performed in cell culture models or rodents, the zebrafish is increasingly used to complement discoveries made in these systems. Forward and reverse genetic analyses over the past two decades have shown that the molecular program for liver development is largely conserved between zebrafish and mammals, and that the zebrafish can be used to model heritable human liver disorders. Recent work has demonstrated that zebrafish can also be used to study the mechanistic basis of acquired liver diseases. Here, we provide a comprehensive summary of how the zebrafish has contributed to our understanding of human liver development and disease.
Introduction
The human liver is an essential organ that performs a large number of physiological roles. The liver is the primary site of synthesis for a number of serum proteins, such as clotting factors and transport proteins, as well as essential lipids, such as cholesterol and phospholipids. The liver also plays a central role in nutrient and intermediary metabolism, and is the major site of detoxification for xenobiotics, toxins, drugs, and other exogenous substances. In humans, the liver is targeted by developmental, infectious, immune-mediated, metabolic, and neoplastic diseases, all of which are associated with significant morbidity and mortality. The liver has a remarkable regenerative capacity compared to most other organs; however, this capacity is finite and often inadequate to compensate for severe acute or chronic injury, both of which can progress to organ failure. Since there is a limited supply of donor organs for orthotopic liver transplantation, and no artificial organ-replacement therapy has been devised to treat liver failure, understanding the mechanisms of liver disease is important for the development of novel treatments for liver failure.
Human liver diseases have been successfully modeled using in vitro cell culture systems; however, there are significant advantages to using model organisms for this purpose. Each of the commonly used model organisms has particular strengths and weaknesses. The fruit fly Drosophila melanogaster and the worm Caenorhabditis elegans are well-characterized developmental models that are inexpensive to maintain and amenable to both high throughput mutagenesis screens and gene targeting methodologies. However, both models lack key features of vertebrate anatomy and physiology that are critical to understanding liver disease. Indeed, neither organism has a tissue that can be considered a true liver homolog. Thus, while both models provide powerful methods to genetically dissect conserved molecular signaling pathways, it can be difficult to place the data derived from these studies in an appropriate physiological context. In contrast to flies and worms, vertebrate developmental models, such as the frog, Xenopus laevis, and the chicken have the advantage of highly conserved physiology and are well suited to embryological manipulations. However, neither model has well developed genetic methods. Mice and other mammalian models are most similar to humans in anatomy and physiology and are amenable to genetic manipulation. However, high throughput genetic analyses are hampered by the difficulty in generating large numbers of progeny, in utero embryonic development (precluding easy observation and manipulation), and expense of husbandry. In addition, inbred mice and rats can show strain-specific differences in physiology. Thus, while mammalian models are often considered the “gold standard” for human disease, such models rarely show identical clinical presentation and natural history to the corresponding human disease. Indeed, as no single animal model is likely to replicate all the clinically relevant features of a human disease, it is becoming increasingly evident that “simpler” model organisms can be of great benefit for disease-oriented research. A remarkable example of this is the use of genetic analyses in yeast to identify novel gene candidates for human neurodegenerative diseases (22). Similarly, the zebrafish is now recognized as being a more useful model for melanoma research than the mouse (144).
Zebrafish as a model for human liver development and disease
The ability to conduct detailed embryological and genetic analyses using the zebrafish makes it a particularly useful model system for studying human liver development and disease (Table 1). The zebrafish was originally developed as a model system to study vertebrate development because it shares many of the features of the powerful fruit fly and worm models. Like these organisms, zebrafish are relatively inexpensive to maintain and breed, such that hundreds to thousands of embryos can be obtained for analysis on a daily to weekly basis. Zebrafish embryos also develop externally and are optically transparent during the first weeks of life, thus allowing direct observation of liver development throughout organogenesis using light and fluorescence microscopy. Zebrafish embryos and larvae develop rapidly, with progression from one-cell embryo to free-swimming, feeding larva by the fifth day postfertilization (dpf). At this developmental stage, the liver is comprised of well-differentiated hepatocytes coupled to a network of intraheptic biliary channels that are contiguous with gallbladder and intestine via larger extrahepatic bile ducts. Hepatocyte synthetic and secretory functions are well developed at this stage. In addition, the most commonly used laboratory strains of zebrafish are out-bred, leading to less strain-specific effects than seen in inbred rodent strains.
Table 1.
Advantages and Disadvantages of Zebrafish as a Model for Human Liver Pathophysiology
| Advantages | Disadvantages |
|---|---|
| Vertebrate body plan | Partial genome duplication in teleosts |
| Ease of husbandry | Differences in microanatomy and liver architecture |
| Inexpensive to maintain | Less conserved physiology than mammalian models |
| Large numbers of embryos produced rapidly | Less conserved morphogenesis than mammals |
| External development | Less developed cell culture technology |
| Optical clarity during development | Poorly developed embryonic stem cell technology |
| Zebrafish liver not required for fetal hematopoiesis | |
| Amenable to forward and reverse genetics | |
| Molecular conservation of development | |
| Amenable to high-throughput screening | |
| • Phenotype assessment | |
| • Drug/chemical screening |
A distinguishing feature of the zebrafish system is that it is amenable to forward genetic analyses. Large-scale screens for mutations induced by ethylnitrosourea (ENU) or retroviral insertion, first published in the mid-1990s, have yielded hundreds of mutations affecting development of all organ systems, including the digestive and hepatobiliary system (2,11,63,141). Molecular characterization of these mutations has in most instances confirmed that the molecular mechanisms regulating hepatobiliary development are broadly conserved between zebrafish, rodents, and humans. In addition, new insights have emerged from these studies that would not have been discovered without the use of a nonbiased forward genetic approach. The mutagenesis screens have also uncovered mutant alleles of the zebrafish orthologs of human genes responsible for heritable hepatobiliary diseases. Phenotypic analyses of larvae carrying these mutations have confirmed the utility of the zebrafish for modeling these conditions.
In addition to unbiased forward genetic screens, the zebrafish can be used for targeted gene manipulations. Perhaps the simplest method involves use of morpholino oligonu-cleotides (MOs), which are synthetic nucleotide analogs designed to bind and inhibit pre-mRNA splicing or the translation of mature mRNA (7). When injected into early-stage (1–8 cell) embryos, MOs can generate global dose-dependent inhibition of gene expression during embryonic and larval development (up to ~6–7 dpf) depending on endogenous levels of the targeted mRNA. Morpholinos are relatively inexpensive to produce; however, their utility can be limited by off-target effects (40,155) which often precludes their ability to achieve full gene inactivation.
Stable manipulation of gene expression can be achieved through transgenesis, first described in zebrafish in 1990 (176). The most commonly employed strategy involves flanking a transgenic construct with Tol2 transposon repeats, and coinjection of the construct into newly fertilized embryos with mRNA encoding the Tol2 Transposase, which mediates mosaic insertion of the construct into the genome of the developing embryo (91). Using the Tol2 system, a significant percentage of embryos within an injected clutch show widespread mosaic gene expression. This enables experimental analyses to be performed in the F0 generation, thus saving both time and expense needed to generate germline transgenics. A number of well-characterized promoters have been described for studying hepatobiliary gene function, including promoters and regulatory elements driving gene expression within the endoderm of the developing gut tube (48), in differentiating hepatocytes (97, 187), and in developing biliary epithelial cells (71, 108, 111, 130, 142, 171, 192).
Up until recently, stable disruption of gene function was not feasible in zebrafish owing to the lack of a reliable embryonic stem cell methodology. Culture of zebrafish embryonic stem cells has been achieved, along with successful homologous recombination-mediated gene targeting (42, 43); however, the technique has not gained broad use within the zebrafish community. More recently, targeted stable gene disruption in zebrafish has been achieved through the use of two techniques (105). The first approach, Targeting Induced Local Lesions in Genomes, or TILLING (93, 175) involves sequencing mutagenized sperm to detect nonsense mutations or critical splice junction mutations that disrupt a targeted gene’s coding sequence. A portion of the mutagenized sperm is frozen ahead of time, prior to sequencing, thus allowing recovery of the mutagenized allele. Large scale screens to identify null mutations in all zebrafish protein coding genes using the TILLING methodology have been initiated (123, 196). TILLING can also be used to identify novel missense mutations that fail to fully complement null mutations. Discovery of such hypomorphic alleles are a great advantage of ENU mutagenesis in the zebrafish compared with gene targeting strategies in the mouse, which most often generate null alleles.
The second approach that has been successfully used to target genes in zebrafish is through the use of zinc-finger nucleases (ZFNs) (41). ZFNs are engineered restriction nucleases that are comprised of customized arrays of zinc fingers fused to the FokI endonuclease. Each finger in the array binds a three-nucleotide recognition motif. Typically three fingers are joined to generate DNA sequence target specificity of nine nucleotides. Since Fok1 must dimerize to cleave DNA, the ZFN methodology employs pairs of fingers that recognize nonpalindromic DNA loci. The nucleotide specificity of the DNA binding domain of the fingers allows Fok1 to create double-strand breaks within a specific site in the genome. Mutagenesis at the targeted locus is achieved through the inherent infidelity of the nonhomologous end joining mechanism used to repair the DNA break. This leads to small random insertions and/or deletions at the DNA break site that typically generate frameshift and/or nonsense mutations. The high cost and difficulty of ZFN design has limited its widespread use, although significant effort has been made to simplify the ZFN design process (49, 164). Reports of off-target effects arising from the use of ZFNs has also been reported (61). Most recently, the related Transcription Activator-Like Effector Nuclease (TALEN) gene targeting methodology has been reported to be a highly efficient alternate means of generating targeted gene knockouts in the zebrafish (8, 18, 83, 124, 163).
Zebrafish are now recognized as an important model system for drug discovery and validation. Because of their small size, zebrafish embryos and larvae are amenable to high throughput assays that can be used to screen large numbers of chemical compounds for medically relevant effects on a broad range of physiological processes (145, 179). To date, compounds affecting hematopoietic stem cells, pancreatic beta cells, cancer cells (melanoma), liver injury, bone morphogenetic proteins (BMP) signaling, behavior and intestinal lipid absorption have been identified (134, 135, 154,157,188,194).
The ability to conduct in vivo analyses is an essential feature of the zebrafish system; however, it is often necessary to combine these studies with detailed mechanistic analyses that are most efficiently undertaken using in vitro models. While many investigators elect to pursue these studies using mammalian cell lines, zebrafish fibroblast and hepatocyte cell lines are available for these and other studies (21, 54).
Conserved and divergent aspects of human-zebrafish liver anatomy
The basic structural unit of the human liver is the lobule. In two-dimensional tissue sections the lobule is typically depicted as being bounded by portal tracts positioned at hexagonal vertices. Each portal tract is comprised of a branch of the portal vein and the hepatic artery, and one or two interlobular bile ducts. At the center of the lobule is the central vein, the most proximal branch of the hepatic vein. Bicellular cords of hepatocytes are radially arrayed between the portal tracts and the central vein. Afferent blood enters the lobule from portal vein and hepatic artery branches in the portal tracts and flows centripetally to hepatocytes via sinusoids, which are capillary-like channels lined by fenestrated endothelium. Efferent blood flows out of the lobule through the central veins, and then into the systemic circulation via the hepatic venous system. Hepatocytes interface with sinusoidal blood flow at their basal surface, and excrete bile into microscopic channels formed by the apical plasma membranes of apposed hepatocytes that are referred to as canaliculi. Canaliculi are typically described as being bicellular, as they arise from the apical cell membranes of paired hepatocytes apposed to one another in the hepatic cords. While lobular blood flow is centripetal, bile flow is centrifugal, traveling within the canalicular system to interlobular bile ducts within portal tracts via canals of Hering, small bicellular biliary channels that are lined by hepatocytes and cholangiocytes, and then to small ductules comprised exclusively of cholangiocytes (9). In addition to hepatocytes, biliary epithelial cells, and vascular and sinusoidal endothelium, other hepatic cell types include Kupffer cells (sinusoidal macrophage-like cells), stellate cells, portal macrophages, and portal fibroblasts. Another cell type seen in injured livers is the oval cell, a transit amplifying cell derived from hepatocytes, cholangiocytes and rarely bone marrow stem cells that are thought to account for at least part of the liver’s regenerative capacity (34, 46, 190).
Although the anatomical organization of the zebrafish liver anatomy has not been fully determined, detailed studies have been performed in other teleosts (67). These studies have suggested that fish have a tubular as opposed to lobular liver architecture. In contrast to mammals, hepatocytes in fish are arranged in muralia, or tube-like cords with a central biliary channel. Canaliculi arise as invaginations of the apical hepatocyte membrane that are contiguous with the central biliary channels within the hepatocyte tubules. These small intraheptic bile channels, which have also been referred to as preductules (67) and are very difficult to recognize in histological sections, join to larger ducts that can be identified in the hepatic parenchyma by their prominent cuboidal epithelium. As in the mammalian liver, bile is excreted from hepatocytes into the biliary ductal network via transporters embedded in apical canaliculi. The basal border of the hepatic tubules are lined by sinusoidal endothelial cells that form channels (sinusoids) which receive blood directly from portal vein radicles with which they are contiguous (191), along with hepatic artery radicles. Sinusoidal blood is drained from the liver via central veins that join the hepatic venous system. Recognizable portal tracts are not found in the teleost liver since the portal vein and hepatic artery radicles and the interlobular bile ducts are independently distributed in the liver parenchyma. A recent study reports the presence of stellate cells in the zebrafish liver (192), but the presence of Kupffer cells has not been reported. Our own studies of larval zebrafish are consistent with this anatomical organization (112). A recent study of adult liver anatomy was supportive of this model, although the presence of intercellular canaliculi was suggested (191).
Liver Development in Zebrafish
The liver arises from the anterior foregut endoderm, and therefore, proper maintenance and specification of the endoderm is required for normal liver development. Studies in zebrafish have confirmed that many of the signaling pathways (Nodal, BMPs) and transcription factors (Gata4–6, Foxa factors, Sox32/17) shown to play important roles in mammalian endoderm development are also vital for endoderm formation in zebrafish (152, 183). A discussion of early endoderm development is beyond the scope of this review. However, extensive reviews of this topic have previously been published (136).
Liver development in zebrafish (and mammals) can be divided into three phases: specification, differentiation, and growth. During the earliest phase, specification, the region of anterior endoderm that has capacity to give rise to progenitor cells committed to form liver (hepatoblasts), rather than other endoderm-derived organs [intestine, pancreas, hepatopancreatic ductal (HPD) system, or pneumatic duct and swim bladder (a lung homolog)] is determined. Liver progenitors in the zebafish embryos can be recognized as early as 22–24 hpf, when they begin to express the markers hhex and prox-1 (185, 186). Hepatoblasts arise in a lateral region of anterior endoderm that also has potential to form pancreas (16), and these lateral endoderm columns maintain the ability to respond to liver-inducing signals up to 34 hpf (171).
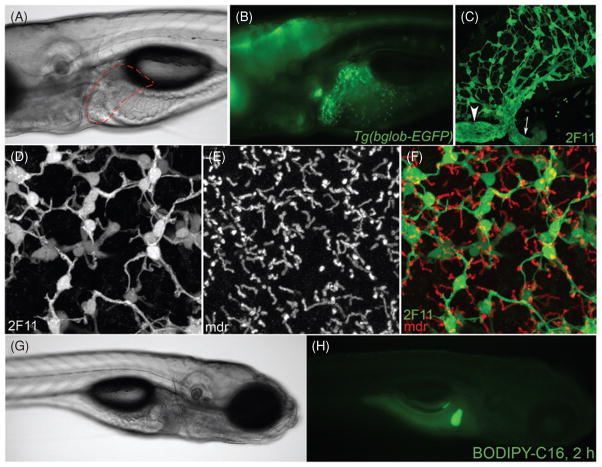
Differentiation of liver progenitor cells (hepatoblasts) within the hepatic anlage occurs between 24 and 50 hpf (48), when expression of molecular markers of mature hepatocytes and biliary epithelial cells is first detected. Albumin and alpha-fetoprotein are two of the most characteristic markers of hepatocytes in mammalian embryos; however, genes encoding clear orthologs of these proteins are not present in the zebrafish genome (133). Ceruloplasmin is a hepatocyte marker used in many zebrafish studies that is expressed in the liver primordium by 32 hpf, although its expression can be detected at earlier stages in nonhepatic endoderm (96). Other hepatocyte markers used to identify hepatocytes in zebrafish are Transferrin and Liver fatty acid binding protein (L-FABP) (Fig. 1), which are expressed beginning at 48 hpf (73, 127). Biliary epithelial progenitors can be detected by the monoclonal antibody 2F11 at 36 hpf (111), and biliary epithelial cells are also distinguished from hepatocytes by expression of cytokeratins, particularly Keratin-18 (112). Biliary growth and maturation largely occur after hepatocyte differentiation, between 3 and 5 dpf (Fig. 2A–F). During this period, biliary epithelial cells differentiate, proliferate with the growing liver, then remodel into a functional network capable of bile excretion by 5 dpf. Bile excretion can be assessed in vivo by monitoring the processing of ingested fluorescently tagged lipid reporters (PED-6, BODIPY-FL C16, and others). When added at low concentration to the aqueous media in which the larvae are reared, fluorescent metabolites of the lipid reporters are initially visualized in bile stored within the gallbladder, which is subsequently excreted into the intestinal lumen (Fig. 2G–H) (44). Failure to accumulate fluorescent material in the gallbladder at 5 dpf often indicates defective biliary development, and has been used to identify mutants in a forward genetic screen (39).
Figure 1.
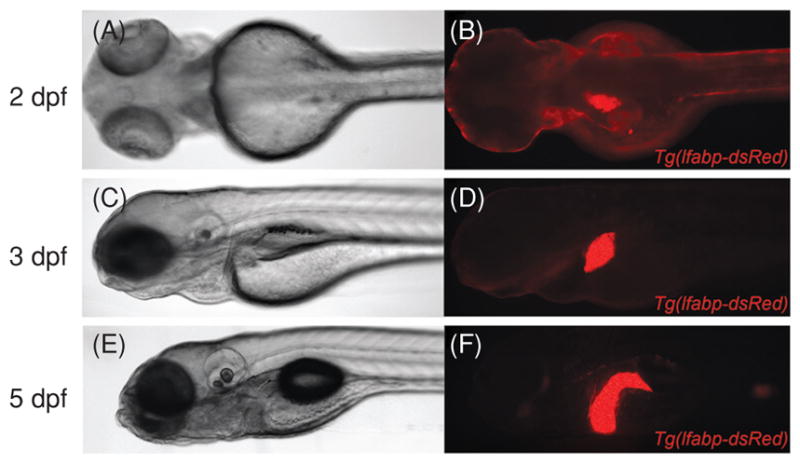
Brightfield images (A, C, and E) and corresponding whole-mount fluorescent images (B, D, and F) of live Tg(L-FABP-dsRed) embryos and larvae, demonstrating liver growth between 2 dpf and 5 dpf.
Figure 2.
Overview of zebrafish biliary development and function. (A) Brightfield high-magnification image of a live 5 dpfTg(bglob-EGFP)larva, raised in 1-phenyl-2-thiourea (PTU) to inhibit melanophore development. Liver (red dashed line) is anterior to intestinal bulb and swim bladder. (B) The same larva imaged by whole-mount fluorescence microscopy. Developing biliary cells in the liver express the Notch-responsive reporter gene encoding EGFP. (C) Confocal projection of fixed wild-type liver, stained with monoclonal antibody 2F11 to visualize biliary tree, including gallbladder (arrowhead) and extrahepatic bile duct (arrow). (D–F) High-magnification confocal projection through the liver of a 5 dpf wild-type larvae immunstained with the 2F11 antibody showing the relationship between bile ducts (labeled by 2F11) and canaliculi (labeled by anti-Mdr antibody). (G) Brightfield image of live 5 dpf wild-type larva in right lateral view, showing right liver lobe, gallbladder, and pancreas. (H) Whole-mount fluorescent image of the same larva, two hours following application of BODIPY-FL C16 to the aqueous media. Fluorescent lipid is secreted into bile and accumulated in the gallbladder.
Growth of the hepatic anlage is most pronounced from 50 hpf onward, and continues in juvenile fish until they are fully developed (48). During these stages, hepatocytes proliferate and become polarized, and there is a pronounced expansion of the biliary system as well as growth of the hepatic vasculature. During early stages of differentiation, the zebrafish liver is essentially avascular, but growth from 55 to 72 hpf requires the presence of endothelial cells, which play an important role in cell signaling, while at later time points a functional vasculature is required for circulatory support of tissue growth (97). One particularly important role of vascular endothelial cells during liver differentiation is to provide signals required to polarize adjacent hepatocytes, as well as to promote biliary development (162). Recent studies of liver growth in zebrafish have shown that it is regulated by signaling pathways known to play a role in mammalian liver development, such as hepatocyte growth factor (HGF)-c-Met pathway (102). Novel factors affecting liver growth have also been reported (10,12). Table 2 lists genes that have been shown to play a role in zebrafish liver development, many of which are discussed in further detail below.
Table 2.
Signals Involved in Zebrafish Liver Development
| Stage | Hours postfertilization (hpf) | Genes/signals involved |
|---|---|---|
| Endoderm specification | <18 | nodal |
| gata5 | ||
| sox32/17 | ||
| BMP | ||
| Hepatic specification | 18–24 | wnt2bb |
| hhex | ||
| prox1 | ||
| BMP -> alk8 | ||
| FGF | ||
| vhnf1 | ||
| gata4 or gata6 | ||
| hdac1/3 | ||
| miR-122 | ||
| rbp4 (from YSL) | ||
| fgf10a—inhibitory | ||
| Hepatic/biliary differentiation | 24–48 | wnt2 -> fzd5 |
| Notch/jagged | ||
| oc3 -> vhnf1 | ||
| pk1a -> vhnf1 | ||
| vps33b/vipar | ||
| vps18 | ||
| miR-30a | ||
| DNA methylation | ||
| Hepatobiliary outgrowth and maturation | 48–120 | gata4/6 |
| uhrf1 -> top2a | ||
| copeb | ||
| grnA -> met | ||
| npo | ||
| leg-1 | ||
| mmp23b | ||
| α2-mg | ||
| snx7 | ||
| zfblp-1 | ||
| zfmcl-1a | ||
| val | ||
| heg | ||
| Notch | ||
| sox9b | ||
| oc3 -> hnf6 -> vhnf1 | ||
| snapc4/c2 | ||
| cldn15lb |
Signaling pathways in zebrafish liver development
Bone morphogenetic proteins
BMPs, members of the transforming growth factor (TGF) family of secreted ligands, signal through Type I and II BMP receptors and Smad transcription factors to regulate a large number of developmental processes (37,95). During zebrafish liver development, BMPs are first crucial for endoderm patterning (183). bmp2b functions as a ventralizing signal, and inhibits expression of her5, a regulator of antero-posterior endoderm patterning; chordin (a BMP inhibitor) has the opposing function. Mutant alleles of these genes (swirl and chordino, respectively) disrupt endodermal patterning, but not endodermal induction. Each mutation disrupts formation of anterior endoderm precursors, reflected in subsequent abnormalities in liver/pancreas situs and pancreas size (183).
Following specification of the anterior endoderm, BMP signaling plays a role in liver specification. Inducible expression of a dominant-negative BMP receptor (dnBMPR) just prior to liver specification (~18 hpf), and mutation of the type I BMP receptor alk8, both result in decreased expression of hepatic progenitor cells expressing hhex and prox1 and decreased expression of ceruloplasmin, a marker of differentiated hepatocytes (171). Interestingly, while the BMP signaling cascades that drive liver specification normally occur between 18 and 26 hpf, the prehepatic anterior endoderm maintains the capacity to respond to BMP-Alk8 induction beyond this stage (171).
In mammalian embryos, BMP signals for hepatogenesis originate from the septum transversum (156), a mesenchymal tissue that has no clear homolog in the zebrafish. Instead, these signals that direct hepatic specification of anterior endoderm in zebrafish arise from BMP-expressing cells in the adjacent lateral plate mesoderm (LPM). Lineage-tracing experiments show that lateral endoderm adjacent to the LPM contributes to liver and pancreas, while more medial columns of endoderm are fated to become pancreas and intestine. Bmp2b signaling from the LPM (through the Alk8 receptor) is a positive signal for liver specification, and transgenic overexpression of bmp2b results in liver expansion at the expense of pancreas (16). Inhibition of this signaling pathway inhibits liver formation, with expansion of pdx-1-expressing pancreatic progenitors. The importance of BMP signaling from the LPM in zebrafish liver development was highlighted by analysis of mypt1 mutants, which have a liverless phenotype (82). mypt1, encoding a myosin phosphatase targeting subunit, is essential for proper actin bundling and mesendodermal cell movement. Mutant embryos fail to correctly position bmp2a-expressing LPM cells adjacent to anterior endoderm, with subsequent failure of hepatoblast proliferation and maintenance.
Fibroblast growth factors
Fibroblast growth factors (FGFs) are a large family of secreted ligands that signal through receptor tyrosine kinases (FGFRs) and a number of downstream intracellular signaling cascades, including Ras-MAP kinase and PI3K-Akt (87). Studies in mouse embryos have shown that FGF signaling plays many of the same roles as BMP signaling in liver development, namely, that FGFs are required for early embryogenesis, and then are used later as an inductive signal for liver specification and outgrowth (88, 195). In mammals, hepatogenic FGF signaling arises from cardiac mesoderm, which is adjacent to hepatic endoderm during embryogenesis (88). In the zebrafish embryo, the cardiac mesoderm and anterior endoderm are widely dispersed from each other during hepatic induction. The precise origin of the FGF signal that is required for hepatic induction has not been reported.
Loss of function studies performed using an inducible dominant negative FGF receptor (dnFgfr) have confirmed that FGF signaling promotes liver specification in zebrafish. Expression of the dnFgfr cDNA at 18 hpf reduced expression of hhex and prox1 at 30 hpf, and ceruloplasmin expression at 40 hpf (171). Inhibition of FGF signaling did not induce loss of previously specified hepatoblasts, thus indicating that it is not required to sustain their development. Liver specification defects caused by dnFgfr were partially compenstated by inducible overexpression of bmp2b, suggesting that FGF signaling does not function downstream of BMP signaling in this process (171). However, the epistatic relationship between BMP and FGF signaling during zebrafish liver specification has not been definitively determined. Interestingly, similar findings concerning the relationship of BMP and FGF signaling during mammalian liver development were observed in mouse tissue explants (88, 156).
In addition to its role in promoting liver specification in zebrafish, a negative regulatory role for FGF signaling during liver development has also been identified. fgf10a is expressed in mesenchyme surrounding the HPD system and pancreatic anlage from 30 to 80 hpf (33,170). Mutation of fgf10a causes hypoplasia of the HPDs and anterior intestine at the level of duct insertion into the intestine (the ampulla). Ectopic hepatocyte and pancreatic endocrine cell differentiation within the presumptive HPD domain and intestine were also observed in the fgf10a mutants (33). Based on these findings, it was hypothesized that Fgf10a “protects” a midline domain of endoderm from liver and pancreas specification, allowing it to form the ductal system. Signaling via Wnt8a can also induce ectopic liver specification within endoderm posterior to the liver-forming domain and this is inhibited by a constitutively active Fgfr transgene and potentiated by MO-induced knockdown of fgf10a or a dnRas transgene (170). These studies highlight both developmental plasticity in foregut development, as well as the spatiotemporal complexity of signaling in that process.
Wnt/β-catenin
Wnts comprise a diverse family of secreted ligands that signal through Frizzled (Fzd) cell surface receptors, and effect a variety of cellular processes including transcription and cytoskeletal dynamics (101). Canonical Wnt signals increase β-catenin-mediated transcription by repressing the β-catenin destruction complex (APC, Axin, and GSK-3β).
As with BMP and FGF signaling, Wnt signaling plays a complex role in embryo patterning and liver development. In zebrafish, the first indication of this role was the molecular characterization of the wnt2bbprometheus mutant (prt), which exhibits delay and/or failure of hepatic specification and outgrowth. Like bmp2b, wnt2bb is expressed in LPM, and MO-induced knockdown of wnt2bb or transgenic overexpression of dnTcf3 (a transcriptional partner of β-catenin) copies the prt phenotype (137). Further experiments have outlined a biphasic role for Wnt/β-catenin signaling in liver specification and outgrowth; Wnt/β-catenin signaling inhibits anterior (foregut) endoderm specification during somitogenesis, but promotes hepatoblast specification and proliferation from 18 hpf onward (58, 147). One target of wnt2bb in this process is nav3a, a zebrafish homolog of C. elegans unc-53, important in axon guidance. nav3 is expressed in gut endoderm and subsequently in the liver bud, and is induced by Wnt signaling independent of BMP/FGF signaling. Inhibition of nav3a leads to hypoplasia of endodermal derivatives (liver, pancreas, and swim bladder), while overexpression causes formation of ectopic endodermal anlagen (94).
In addition to wnt2bb, wnt2 also plays a crucial role in early zebrafish liver development. wnt2 is normally expressed in LPM following hepatoblast specification. MO knockdown of wnt2 on its own leads to decreased liver size and hepatoblast proliferation, thus indicating that the Wnt2 signal is required for normal hepatoblast proliferation. Knockdown of wnt2 in wnt2bb-mutant embryos leads to uniform failure of liver specification, as well as frequent failure of extrahepatic duct morphogenesis (147). These results indicate that wnt2 can compensate for loss of wnt2bb during hepatoblast specification. Normal function of sox32 in the endoderm is required for this compensation. Epistasis experiments indicate that wnt2bb/wnt2 both signal through the Fzd receptor fzd5. Interestingly, liver specification defects in wnt2bb mutants that lack wnt2 are partially rescued by inhibition of fgf10a (147). These latter findings indicate that Wnt and FGF signaling combine to control the regulative capacity of endoderm to form liver (172).
Notch/Jagged
In contrast to BMP, FGF, and Wnt, all of which are secreted ligands, Notch receptors and their ligands (Jagged and Delta) are transmembrane proteins, and signal through cell-cell interactions. Upon ligand engagement with the extracellular domain of Notch, the intracellular domain is cleaved and translocates to the nucleus, where it interacts with a complex of other factors to affect transcription (5). During development, Notch signaling has been shown to play both inductive and inhibitory roles in cell fate determination (20, 69). In the mammalian liver, JAGGED and NOTCH genes are expressed in portal mesenchyme and endothelial cells adjacent to hepatoblasts, and play an inductive role in biliary differentiation (198). Haploinsufficiency of JAGGED1 is responsible for most cases of Alagille syndrome, a multiorgan disorder associated with cholestasis that is caused by intrahepatic bile duct paucity (106, 138).
Zebrafish express multiple jagged and notch genes in the developing liver, and MO-mediated knockdown of several of these genes, either individually or in pairs, causes bile duct paucity in developing larvae (112). Hepatocytes in these morphants also expressed biliary cell markers. This suggests that the biliary lineage normally develops from a bipotential liver progenitor cell that also gives rise to hepatocytes. Similar findings have been observed in the mammalian liver (198). Interestingly, overexpression of the activated Notch1 intracellular domain at 3 dpf leads to ectopic bile duct formation (112), similar to what has been reported in mice (198). Thus, Notch signaling in hepatic progenitor cells is required and sufficient for biliary specification. Modulation of Notch signaling in zebrafish does not affect development of the extrahepatic ductal system, a finding that is also observed in the Alagille syndrome (1).
Following biliary specification, Notch signaling is again used to promote remodeling of immature biliary epithelial cells into an extended and functional biliary network (111). Utilizing the pharmacologic Notch inhibitor N-[N-(3,5-Difluorophenacetyl)-1-alanyl]-S-phenylglycine t-butyl ester (DAPT), and visualizing the biliary network via monoclonal antibody 2F11 or a Notch-responsive GFP reporter line, biliary epithelial cells were shown to be responsive to Notch signaling throughout development (2–5 dpf). Disruption of Notch signaling not only affected biliary growth and remodeling, but also canalicular development within hepatocytes (assessed by immunostaining for Mdr/P-glycoprotein, a canalicular protein) (111). A recent publication has shown that Sox9b interacts with Notch signaling during biliary morphogenesis (31). sox9b is expressed in the developing HPD system in zebrafish. Mutants homozygous for a loss-of-function allele (sox9bfh313) are viable to adulthood, but show bile-stained organs with biliary and pancreatic duct malformations by 5 months of age (31). sox9bfh313 mutants have normal biliary specification, proliferation, and survival, but biliary morphogenesis is altered. This causes a phenotype resembling the phenotype of larvae treated with DAPT following biliary specification (111). Indeed, sox9b has positive feedback with Notch: sox9b expression is directly correlated with Notch signaling in developing larvae, and sox9bfh313 larvae have decreased Notch signaling during biliary morphogenesis (31).
Live imaging of GFP-expressing biliary epithelial cells in Notch reporter fish also provided insights into the process of biliary remodeling in zebrafish. This showed that nascent biliary cells extend filopodial processes toward each other and that fusion of the filopodia leads to formation of a network of contiguous cells. Lumenogenesis within the developing ductal network appeared to involve the fusion of intracytoplasmic vacuoles (111). In contrast, morphogenesis of the interlobular bile ducts in the mammalian liver arises from the sequential radial differentiation of biliary cells within the ductal plate (198). The morphogenetic processes that drive formation of the canals of Hering and biliary ductules in the mammalian liver have not been characterized.
Other signaling mechanisms active during liver development
Several other signaling pathways have been shown to play a role in zebrafish liver development. Pharmacological inhibition of Hedgehog signaling prior to liver specification blocks formation of the liver primordium (185). A similar defect was reported when Hedgehog signaling was inhibited by mutation of the smoothened transmembrane receptor (158). Another study examined the role of Retinol binding protein 4 (rbp4), which is expressed in adult liver, but during development is expressed in the yolk syncytial layer (YSL) prior to liver specification. Morpholino-mediated knockdown of rbp4 leads to abnormal yolk extension and formation of two liver buds, indicating a possible role in liver progenitor cell migration or specification (109).
Inhibition of a number of factors in zebrafish results in normal hepatocyte specification but impaired liver growth. These include met, the tyrosine kinase receptor for HGF (102), and progranulin A (grnA) which likely functions through a met-dependent pathway (107). Matrix metalloproteinase activity promotes hepatocyte proliferation upstream of tumor necrosis factor (TNF) signaling, as demonstrated by MO-mediated knockdown of mmp23b (149). Liver growth is also impaired in α2-macroglobulin-like deficient embryos (78), and is arrested at 72 hpf by mutation of nil per os (npo), an RNA binding protein (121). Endothelial cells and blood circulation are required for liver growth, as previously mentioned (97).
Apoptosis plays an essential role in many facets of animal development (52). However, an exact spatiotemporal role for apoptosis in liver development has not been demonstrated. It has been inferred that liver development requires physiologic levels of apoptosis, based on zebrafish experiments that modulate levels of apoptosis-related factors. Hepatoblasts undergo massive apoptosis in response to MO-mediated inhibition of the antiapoptotic factor snx7 (189), and liver hyperplasia results from transgenic overexpression of the antiapoptotic Bcl-2-related genes zfblp1 and zfmcl-1a (72).
In addition to their role in promoting liver growth, en-dothelial signals are required for proper hepatocyte polarization and proper biliary localization (162). Genes involved in establishing planar cell polarity, including prickle-1a (pk1a), are also required for proper biliary development via their effects on cytoskeletal elements (24). pk1a-deficient larvae have decreased biliary proliferation, altered biliary morphology and function, and are phenocopied by chemical inhibitors of Rho GTPase, c-Jun N-terminal kinase, and cytoskeletal function. Interestingly, planar cell polarity signaling appears to function upstream of hepatocyte nuclear factor (HNF) signaling; vhnf1 expression is increased in pk1a deficient larvae, which in turn are rescued by overexpression of vhnf1 mRNA (24).
Transcriptional Control of Zebrafish Liver Development
Gata family members
As is the case for signaling pathways, a significant number of transcription factors essential for zebrafish endoderm development also play a later role in liver development. One example is that of the GATA family of transcription factors. gata5 (faust) is required for normal endoderm development (152), and the related factors gata4 and gata6 are required for proper liver development. MO-mediated knockdowns show that gata4 or gata6 are required for liver specification and that both are required for subsequent liver growth (77).
Hepatocyte nuclear factor
HNFs are a group of transcription factor families, whose expression is enriched in liver compared to other organs. Five classes of HNFs are recognized in mammals, whereas in zebrafish there are four (128). The HNF1 family is comprised of two homeodomain containing members in zebrafish, HNF1α and HNF1β (also known as variant HNF1 or vHNF1), each of which can bind DNA as either a homodimer or heterodimer. The HNF3 family is comprised of three factors that bind DNA as a monomer through a winged-helix domain that has homology to Drosophila forkhead; HNF3α (also known as Foxa1), HNF3β (Foxa2), and HNF3γ (Foxa3) play important roles in liver development and physiology (51). HNF4 is a family of three steroid hormone nuclear receptors (HNF4α-γ) with unknown ligands, each of which binds DNA as a dimer. The HNF6 family has three members that each contain a variant homeodomain and a single cut domain; HNF6 is also known as Onecut1 (Oc1), with the remaining members known as Onecut2 (Oc2) and Onecut3 (Oc3). CCAAT/enhancer binding protein (C/EBP) is a family of two leucine-zipper DNA-binding proteins (C/EBPα and C/EBPβ). All of these factors are thought to function as a hierarchical and regulatory network in a number of processes related to liver development, including foregut endoderm differentiation, liver specification, liver outgrowth, and biliary differentiation (128).
In zebrafish, most studies have focused on the role of HNF1 and HNF6 in liver and biliary development. hnf1α and hnf1β (or vhnf1) are both expressed in the liver, as well as in the pancreas, gut, and kidney (60). vhnf1 is also expressed prior to liver specification in a variety of tissues, including anterior endoderm (177). A vhnf1 allele recovered in an insertional mutagenesis screen causes defects in endodermal patterning and liver specification, differentiation, and growth (177). Mutant embryos also have severe defects in biliary differentiation and remodeling (119).
Biliary development is also controlled by zebrafish hnf6 (oc1) and oc3 (teleosts do not appear to have a homolog of mammalian Oc2). Morpholino knockdown of hnf6 results in normal-appearing biliary epithelial cells at 3 dpf, but failure of billiary extension and remodeling at 4–5 dpf leads to dilated ducts and a paucity of interconnecting and terminal ducts (119). In contrast, knockdown of oc3 induces a more severe biliary phenotype, with earlier onset (by early 4 dpf) than seen in hnf6 morphants (118).
Several lines of evidence indicate that the model of HNF signaling in a hierarchical and self-reinforcing regulatory network is also valid in zebrafish. During biliary development, both overexpression and inhibition of hnf6 or vhnf1 results in biliary defects (119). Inhibition of oc3 causes downregulation of both hnf6 and vhnf1, while oc3 expression is downregulated in hnf6 morphants (118). The oc3 morphant phenotype can be partially rescued by coexpression of hnf6 mRNA, and the hnf6 morphant phenotype can be partially rescued by co-expression of vhnf1 mRNA (118, 119). These observations support a model where biliary development is directed by oc3 upstream of hnf6, which is upstream of vhnf1. In addition, biliary defects caused by inhibition of planar cell polarity are associated with downregulation of vhnf1, and can be partially rescued by coexpression of vhnf1 mRNA (24).
Gene expression data and promoter analysis also support the HNF network model of liver development in zebrafish. Analyses of 51 zebrafish liver-enriched gene promoters showed that the majority contain binding sites for two or more families of HNF factors (12). For example, a 435 bp distal enhancer in the L-FABP promoter contains both vhnf1-and hnf3β-binding sites (75). Liver-specific expression of L-FABP is decreased by loss of one of the binding sites, but completely eliminated by loss of both sites. Studies in mice and zebrafish have also shown that hnf6 directly regulates the microRNA miR-122, and that Hnf6-miR-122 form a positive feedback loop to promote expression of hepatocyte-specific genes (103). In fact, hnf6 directly binds the promoters of all hepatocyte-specific genes that are stimulated by miR-122 (103).
Additional transcription factors involved in liver development
Two of the earliest markers of liver specification in zebrafish are the transcription factors hhex and prox1, both of which are expressed in the liver primordium by 22 hpf (185). hhex is a divergent homeobox gene, expressed in the extra-embryonic YSL prior to 22 hpf. Deletion of hhex in mouse embryos leads to failure of liver differentiation and growth, despite normal patterning of the foregut endoderm (92,114). Similarly, MO-mediated hhex inhibition in zebrafish embryos leads to dose-dependent reduction or absence of the liver at 50 hpf, as well as reductions in exocrine pancreas size and randomization of gut looping (186). Interestingly, liver organogenesis requires hhex activity within embryonic tissues, while determination of gut chirality is dependent on YSL hhex expression.
Impaired liver outgrowth is also seen in embryos with MO-induced knockdown of copeb, the zebrafish homolog of the mammalian zinc-finger transcription factor Klf6 (197). Morphant embryos have proper endoderm and liver specification, differentiation, and vascularization, but show decreased hepatocyte proliferation and defective outgrowth of several endoderm-derived organs. One role of copeb in normal liver growth is as a transcriptional inhibitor of cdkn1a, a cell-cycle inhibitor, as morphants show increased cdkn1a levels. Notably, liver defects in mouse Klf6 mutants are confounded by defective hematopoiesis and angiogenesis (115). This does not occur in the zebrafish copeb morphants because hematopoietic development does not occur in the embryonic liver.
Additional regulators of liver development
There is an increasing appreciation for the role of epigenetic and posttranscriptional regulation in the control of gene expression. A major mechanism of epigenetic regulation is via the acetylation and methylation of histones and nucleic acids. Two separate studies have outlined a role for histone deacetylases (HDACs) in liver development. hdac1 mutant embryos have expanded foregut endoderm, with impaired liver (and exocrine pancreas) specification and differentiation, ectopic endocrine pancreas, and defective extrahepatopancreatic duct morphogenesis (132). Similar effects were seen in hdac1 morphants, while hdac3 morphants had more specific defects in liver formation (45). One role of hdac3 in promoting liver development is inhibition of gdf11, a TGFβ family member.
The role of DNA methylation in liver development has also been examined in zebrafish. Morpholino knockdown of dnmt2, a DNA methyltransferase, interferes with liver differentiation. However, in zebrafish liver Dnmt2 appears to function as a tRNA methylase, and nuclear activity (DNA methylation) is dispensable for liver development (150). MO-mediated knockdown of dnmt1 or suv39h1 (a histone H3K9 methyltransferase) had no effect on liver differentiation but did disrupt differentiation of intestine and exocrine pancreas (151). In contrast, a separate study showed defects in biliary development and function in ducttrip mutants, dnmt1 morphants, and embryos treated with azacytidine, all of which have defects in DNA methylation (116, 117). The ducttrip mutation disrupts an important enzyme of the methionine metabolism pathway, S-adenosylhomocysteine hydrolase (Ahcy). This elevates levels of S-adenosylhomocysteine, a potent inhibitor of a wide range of methyltransferases, including DNA methyltransferases.
A forward genetic screen for liver growth and development also uncovered a role for uhrf1 in liver development (161). uhrf1 functions in the maintenance of DNA methylation; zebrafish mutants show lower levels of cytosine methylation than wild-type fish (47, 184), and have impaired liver outgrowth and embryonic survival (161). Studies in human tumor cells have implicated UHRF1 as a transcriptional activator of topoisomerase 2α (TOP2A) (79), and in a separate screen, mutation of top2a was shown to be embryonic lethal, in part from decreased cell proliferation (35). However, a separate study in cultured human cells with knockdown of UHRF1 showed no inhibition of TOP2A (182).
MicroRNAs (miRNA) are short nontranscribed RNA molecules that bind and regulate posttranscriptional stability and translation of target mRNAs. Numerous studies have linked miRNAs to regulation of development and pathophysiology in virtually all organ systems, including the liver (98). Two studies in zebrafish have detected a role for miRNA in liver development. As described above, miR-122 participates in a feedback loop with hnf6 to regulate a variety of hepatocyte-specific genes during development (103). A separate microarray study in late fetal and neonatal mice identified miR-30a specifically expressed in the ductal plate. MO-mediated knockdown of miR-30a in zebrafish resulted in defective biliary development (66).
Another mechanism of posttranscriptional regulation is at the level of pre-mRNA splicing, which is regulated by a large ribonucleoprotein complex (the spliceosome) comprised of 5 small nuclear RNAs (U1, U2, U4, U5, and U6 snRNA). Transcription of the snRNA genes is mediated by the snRNA-activating protein complex (SNAPc). A recessive lethal mutation in one member of the complex, snapc4, disrupted survival of intrahepatic biliary epithelial cells in zebrafish larvae, and was associated with decreased production of selected snR-NAs (U4 and U5) (165). The mutant could be phenocopied by MO-mediated knockdown of snapc2, another member of the complex.
The tight junction protein Claudin 15-like b (Cldn15lb) has also been associated with defective biliary development (13). cldn15lb is expressed in hepatocytes and biliary epithelial cells at 3 dpf, then is restricted to biliary cells by 4 dpf. Gene expression studies in vhnf1 mutant embryos showed severe downregulation of cldn15lb as compared to wild type, and a cldn15lb TILLING mutant showed defects in hepatocyte polarity and biliary remodeling between 80 and 100 hpf (13).
Modeling human liver disease mechanisms in zebrafish
Both congenital and acquired liver diseases can be successfully modeled in zebrafish. Here, we discuss examples from several classes of conditions (Table 3).
Table 3.
Human Liver Diseases and Processes Successfully Modeled in Zebrafish
| Cholestatic diseases of infancy |
| Alagille syndrome |
| Arthrogryposis-renal dysfunction-cholestasis syndrome |
| Extrahepatic biliary atresia |
| Choledochal cyst |
| Alcoholic liver disease |
| Nonalcoholic fatty liver disease |
| Viral hepatitis |
| Hepatitis C |
| Malignancy |
| Hepatocellular carcinoma |
| Cholangiocarcinoma |
| Liver metastases |
| Metabolic derangement |
| Obesity |
| Starvation |
| Inborn errors of metabolism |
| Ferroportin disease (Type 4 hemochromatosis) |
| Erythropoietic protoporphyria |
| Disorders of methionine metabolism |
| Disorders of ketone body metabolism |
| Liver regeneration |
| Partial hepatectomy |
| Mitochondrial hepatopathy |
| APAP toxicity and other drug-induced acute liver failure |
Cholestatic liver diseases of infancy
A wide number of infantile liver disorders are first recognized by the presence of jaundice (6,159). The most common cause of pathologic jaundice is extrahepatic biliary atresia (EHBA), a fibroinflammatory disorder of unknown etiology that destroys the extrahepatic bile ducts and often the gallbladder. Over time, intrahepatic bile ducts are also affected in children with EHBA. Other causes of infantile cholestasis include mutations in genes encoding canalicular membrane channel proteins (progressive familial intrahepatic cholestases, cystic fibrosis), the protease inhibitor alpha-1-Antitrypsin, inborn errors of metabolism (bile acid synthetic disorders), or toxicity from total parenteral nutrition. Unfortunately, the etiology and pathophysiological mechanisms that underlie these rare disorders are either unknown or poorly understood.
Alagille syndrome is an autosomal dominant disorder, caused by haploinsufficiency of JAGGED1 or NOTCH2, that leads to defects in cardiac, craniofacial, and liver development, with variable effects on a number of other organs (1, 146). Unlike obstructive jaundice (such as EHBA) where there is intrahepatic bile duct proliferation, Alagille syndrome (AGS) is marked by absence or loss of interlobular bile ducts. AGS can be modeled in zebrafish by morpholino knockdown of JAGGED and NOTCH gene orthologs (112). Jagged-mediated Notch signaling in zebrafish is required for biliary epithelial development, as well as remodeling that shapes nascent biliary epithelial cells into a functional network (111, 112). It is also required for proper cardiac, craniofacial, and pancreatic development, which is consistent with clinical spectrum of AGS (112). Together, these studies indicate that the findings in AGS patients may be due to both a lack of sufficient fetal biliary progenitors (bile duct paucity), as well as insufficient postnatal growth or remodeling of the biliary tree. Studies performed in mouse models have also supported these findings (198).
Arthrogryposis-renal dysfunction-cholestasis syndrome (ARC) is a severe autosomal recessive disorder that manifests with paucity of bile ducts, cholestasis, musculoskeletal contractures, and abnormal renal function (38). The majority of cases are caused by mutations in the class C vacuolar sorting protein Vps33b, responsible for regulation of intracellular vesicle trafficking and protein localization (57). Zebrafish vps33b is expressed in developing bile ducts, and MO-mediated knockdown causes defects in biliary morphogenesis and function at 5 dpf (120). Ultrastructural studies suggested defective vesicle trafficking in biliary cells as the cause of this disorder (120). vps33b is a direct target of vhnf1, and the vps33b morphant biliary phenotype is similar to that of vhnf1 mutants and hnf6 morphants. Vps33b forms a functional complex with VIPAR (VPS33-interacting protein involved in polarity and apical protein restriction), and vipar knockdown by MO in zebrafish also shows defects in membrane polarity, biliary morphology, and biliary function (26). At least 7 ARC patients who lack mutations in VPS33B have mutations in VIPAR (26, 39, 76, 160, 167).
As described above, EHBA has no single identified etiology (62). However, gene expression studies in EHBA patients have shown increased expression of genes regulated by interferon-γ (a proinflammatory cytokine) and DNA methylation. ducttrip larvae, which carry a mutation in S-adenosylhomocysteine hydrolase (ahcy), have hepatic steatosis and liver degeneration, as well as impaired biliary development, relative DNA hypomethylation, and increased INFγ signaling (116, 117). The biliary defects in these mutants are phenocopied by treatment with the DNA demethylating agent azacytadine, and are corrected by treatment with corticosteroids (an anti-inflammatory). A recent study reported reduced DNA methylation in biliary epithelial cells of children with EHBA compared with other cholestatic liver disorders (116). This suggests that corticosteroid treatment could be used to block intrahepatic biliary fibrosis that occurs in most EHBA patients following Kasai portoenterostomy. A National Institutes of Health-sponsored clinical trial to test this hypothesis is ongoing (131).
Choledochal cysts are malformations of the extrahepatic biliary tree and gallbladder. They can be clinically silent, but can also lead to obstructive cholestasis in childhood or beyond, and they are a risk factor for cholangiocarcinoma (174). Unlike EHBA, surgical correction of the choledochal cyst is usually sufficient to cure cholestasis, without ongoing intra-hepatic biliary disease. A mutation in the tumor suppressor gene nf2 has been reported to cause a choledochal cyst-like phenotype with epithelial hyperplasia in zebrafish larvae with hepatomegaly (160).
Mowat-Wilson syndrome (MWS) is caused by mutations in the zinc-finger transcription factor ZFHX1B, and is characterized by facial dysmorphism, microcephaly, mental retardation, Hirschsprung disease, and cardiac anomalies (199). While the syndrome is not typically associated with liver disease, one patient with MWS also had intrahepatic biliary defects, and MO-mediated knockdown of zebrafish zfhx1b led to defects in biliary development and function (25). Similarly, while studies in zebrafish have elucidated a role for vhnf1 in biliary development (119), liver disease is not typically associated with human mutations in VHNF1, which cause mature onset diabetes of the young (MODY5) and familial glomerulocystic kidney disease. However, zebrafish vhnf1 mutants do have pronephric defects, consistent with the human disease (177).
Additional congenital liver diseases
Autosomal dominant polycystic liver disease (ADPLD) is caused by mutations in one of two endoplasmic reticulum-associated proteins, Prkcsh or Sec63. Large liver cysts can also be seen in patients with autosomal dominant polycystic kidney disease (ADPKD), caused by mutations in TRPP2 (encodes Polycystin-2). prkcsh-deficient zebrafish larvae have pronephric cysts and situs inversus, but lack obvious liver pathology (53). The same study indicated a common mechanism for cyst formation in ADPLD and ADPKD, namely, that Prkcsh acts as a chaperone for Trpp2, preventing its ubiquitination and endoplasmic reticulum (ER)-associated degradation.
Hemochromatosis is a group of diseases that are characterized by mutations in genes responsible for iron metabolism, leading to iron overload in a variety of tissues (173). Type 4 hemochromatosis is caused by mutations in the cell membrane iron export protein Ferroportin, and a mutation in ferroportin1 is causative of the zebrafish weissherbst anemia mutant (50). Mutant larvae rescued with iron injections eventually develop intestinal and hepatic iron overload with iron-deficiency anemia as adults (50). A variety of point mutations have been isolated in patients with ferroportin disease, leading to clinical heterogeneity; the function of at least two mutant proteins has been tested by expression in zebrafish (56). One caveat of these models is that the regulatory mechanisms of iron metabolism are slightly different in zebrafish and mammals, in particular the transcriptional regulation of hepcidin, a key iron regulatory hormone (55).
Porphyrias are a group of diseases caused by defects in heme synthesis and metabolism, at least some of which develop liver disease due to hepatic accumulation of iron, heme metabolites, and degenerating erythrocytes (148). The zebrafish mutant dracula is caused by a recessive mutation in ferratochelatase, which catalyzes the iron-binding step in heme synthesis, and is the causative mutation in erythropoietic protoporphyria (14). Dracula mutants show light-dependent liver disease, characterized by hepatic inclusions comprised of degraded erythrocytes and protoporphyrin IX crystals (heme precursor metabolite).
Alcoholic liver disease
Alcohol abuse is a major public health problem. In the liver, acute and/or chronic exposure to alcohol can lead to steatohepatitis and cirrhosis, and it is a significant risk factor for the development of hepatocellular carcinoma (HCC). Two recent studies have shown that zebrafish larvae are a viable model system for alcoholic liver disease (80). Acute exposure of 4 dpf larvae to 2% ethanol leads to hepatomegaly, hepatic steatosis, and gene expression profiles consistent with increased hepatic lipid metabolism (143). Acute alcoholic steatosis in this model is dependent on Srepb activity, as is chronic alcoholic steatosis. An ultrastructural and gene expression study showed that the same level of alcohol exposure leads to ER dilation and a gene profile consistent with the unfolded protein response, which has been implicated as a mechanism of alcohol-induced liver damage (81). Zebrafish exposed to ethanol have not been reported to develop steatohepatitis, liver fibrosis, or cirrhosis. However, in a recent publication describing zebrafish HSC, exposure of 4 dpf larvae to 2% ethanol caused an increase in HSC proliferation rate and cell number, as well as increased laminin deposition (192). Similar mechanisms, including HSC activation, proliferation, and extracellular matrix deposition have been described in early-stage mammalian hepatic fibrosis (126).
Nonalcoholic fatty liver disease (NAFLD)
A variety of metabolic perturbations can lead to hepatic steatosis aside from ethanol consumption, including dietary intake, toxin exposure, mitochondropathies, and defects in intermediary and lipid metabolism. Clinically, NAFLD leads to the same liver pathology as alcoholic liver disease—steatosis, steatohepatitis, liver fibrosis leading to cirrhosis, and risk of HCC (104). Studies in zebrafish have isolated several models of hepatic steatosis, with or without inflammation and liver degeneration.
One model involves treatment of larvae with thioacetamide (TAA), which induces hepatic steatosis, apoptosis, and induction of lipid peroxidation (3). Continuous treatment with TAA for 12 weeks leads to development of hepatic fibrosis and induction of HCC (153). TAA-induced steatosis has been successfully treated by administration of taurine, which functions at least in part, by decreasing oxidative stress (65).
As previously noted, ducttrip mutants carrying defects in ahcy have impaired methionine metabolism and DNA/protein methylation, leading to hepatic steatosis and degeneration (117). Disruption of methionine metabolism also causes hepatic steatosis in mice, and has long been linked to steatosis associated with alcohol induced liver disease (64). The steatosis in ducttrip is mediated through proinflammatory TNF-α signaling, as steatosis and hepatic degeneration are inhibited in this model by MO-mediated tnfa inhibition. Adult heterozygote ducttrip mutants have a normal lifespan but develop hepatic steatosis (117). This suggests that changes in Ahcy activity could be a risk factor for steatosis in humans. Although ducttrip mutant larvae do not develop overt steatohepatitis, they do have severe liver injury, in contrast to larvae that develop steatosis associated with ethanol exposure.
Two additional zebrafish mutants, foie gras (foigr) and cdipt, also develop hepatic steatosis with signs of ER stress (160, 181). Cdipt mutants have defects in de novo phosphatidylinositol synthesis, while foigr encodes a novel but conserved protein of unknown function. The role of chronic ER stress in hepatic steatosis was confirmed by treatment of zebrafish with tunicamycin, which blocks ER protein export and phenocopies foigr (17). Tunicamycin-mediated steatosis is inhibited in atf6 morphants, which are deficient in one mediator of the ER-associated unfolded protein response (17).
Gankyrin is an ankyrin-repeat protein overexpressed in HCC, and gankyrin transgenic zebrafish show increased larval liver cell proliferation, juvenile/adult hepatic steatosis, and increased hepatic apoptosis leading to death at 10 months of age (74). While the exact mechanism for these findings is unknown, transgenic fish show altered expression levels of multiple lipid metabolism genes and four miRNAs, including miR-122.
Viral hepatitis
At least six hepatotropic viruses cause acute or chronic hepatitis in humans, in addition to viral hepatitis caused by nonhepatotropic viruses (e.g., cytomegalovirus and Epstein-Barr virus). Of these, most morbidity and mortality is due to chronic hepatitis B (HBV) or hepatitis C (HCV) infection, leading to fibrosis, liver failure, and risk of HCC. While these viruses are not known to infect zebrafish, transgenic expression of viral genes can be used to model certain features of diseases caused by chronic infection of humans. Chronic HCV hepatitis is often marked by steatosis and a high risk of HCC. While TAA-treated, wild-type zebrafish develop hepatic steatosis and HCC in 12 weeks of treatment, zebrafish transgenic for HCV core protein progress twice as fast, developing HCC in 6 weeks of drug treatment (153). A bipartite subreplicon model of HCV showed amplification of core RNA and protein, with concomitant gene expression changes similar to those seen in human HCV-infected hepatocytes (32). Subreplicon amplification could be inhibited by treatment with ribavirin and oxymatrine, known antiviral agents with clinical activity against HCV. Transgenic overexpression of HBV X protein caused hepatic steatosis or steatohepatitis in a majority of animals by 4 months of age, eventually progressing to liver degeneration (169). Consistent with the histologic changes, gene expression studies showed induction of genes involved in lipogenesis, cellular stress, and apoptosis. Chronic viral hepatitis has also been associated with certain subtypes of intrahepatic cholangiocarcinoma (168). A recent publication showed that bitransgenic overexpression of both HBV X protein and HCV core protein in zebrafish led to the TGFβ-dependent development of intrahepatic cholangiocarcinoma and tumor-associated fibrosis by 3 months of age (110).
Hepatocellular carcinoma and other hepatic malignancies
Chronic hepatitis of many etiologies leads to hepatocellular damage and proliferation, which increases the risk for development of HCC. Not surprisingly, these tumors are associated with dysregulation of signaling pathways that control cellular proliferation or apoptosis. Numerous studies in zebrafish have shown that constitutive or inducible transgenic expression of oncogenic forms of the Ras/mitogen-activated protein kinase signaling cascade, often activated in human HCC, can induce zebrafish liver tumors. Expression of k-ras (V12), human ΔRAF1, and constitutively active xmrk (zebrafish homolog of epidermal growth factor receptor) lead to the development of HCC that is treatable with MEK/ERK and PI3K/AKT inhibitors (71, 108, 129, 130). Comparative gene expression studies have shown that human and zebrafish HCCs have generally conserved expression profiles, including similar markers of tumor progression (71, 99, 100, 129).
Zebrafish have several technical advantages for the study of malignancy. The optical clarity of embryos and larvae allows for in vivo monitoring of proliferation, metastasis, neovascularization, and treatment response of fluorescent-labeled zebrafish and human tumor implants (85, 113). Although the lack of inbred zebrafish lines can impair serial tumor transplantation studies; a recent method for developing gynogenetic clonal diploid lines has been described. This enabled serial transgenerational passage of multiple chemically induced tumor types (including liver tumors) within a syngeneic line (122). In another in vivo cancer study, high-resolution microscopic ultrasound was used to follow progression of adult liver tumors, and to guide fine-needle aspiration of tumors in live animals (59). Finally, forward genetic screens have uncovered genomic instability (gin) mutants that develop spontaneous tumors (including liver tumors) at up to ten times the rate of wild-type animals (125). When molecularly characterized, these mutants will likely further our understanding of tumor initiation mechanisms.
Hepatic response to obesity and starvation
Conditions of nutrient excess and deprivation (overfeeding and starvation) lead to profound metabolic changes that impact virtually all organ systems. Obesity has become a major health problem in the developed world, and there is currently a great deal of research investigating the genetic and environmental factors that impact the development of obesity. The liver is the primary organ in intermediary metabolism, and plays a central role in adapting to changing nutrient availability. Recently, several studies have shown that zebrafish may be a good system for modeling metabolic processes, including fatty acid metabolism and hepatic steatosis (166).
One study of diet-induced obesity in zebrafish showed increased body mass index, hypertriglyceridemia, and hepatic steatosis over an 8-week feeding regimen, with changes in gene expression that mimic those seen in mammalian obesity (139). These changes are reversible by a 2 week period of calorie restriction, or treatment with extracts of selected foods (178). A separate overfeeding model used a proteomic approach to show decreased levels of oxygen binding proteins (including iron ion-binding proteins) in overfed fish, similar to a response seen in obese humans (89). A 3-week starvation model showed hepatic gene expression changes consistent with decreased metabolism and increased gluconeogenesis (36), while a separate fasting/refeeding model showed modulation of energy-responsive signaling intermediates and transcription factors similar to that seen in mammals (23). In addition, one “proof of principle” study showed that zebrafish are amenable to metabolomic profiling using similar laboratory techniques (NMR, GC/MS, and LC/MS) as employed for human metabolic studies (140).
Combining these attributes with the power of zebrafish genetics will lead to further understanding of the role of the liver in metabolism. One example is the recent report of red moon (rmn), a mutant that develops steatosis only following yolk depletion or during fasting (86). rmn carries a loss-of-function mutation in slc16a6, encoding a transmembrane protein that functions as a hepatic exporter of the major ketone body β-hydroxybutyrate. Instead of being exported as fuel for other organs during fasting, these ketone bodies are trapped in hepatocytes and lead to gene expression changes that result in de novo lipogenesis and resulting hepatic steatosis (86).
Liver regeneration
Compared to most organs, the liver has a remarkable ability to regenerate in response to cell injury from a variety of etiologies. As the only available organ-replacement therapy at this time is liver transplantation, understanding the regulation of this process may improve our ability to treat liver failure. Several studies have shown that zebrafish models of liver injury and regeneration are amenable to study, and have been reviewed in detail elsewhere (15, 29, 70). Generally, these studies have indicated that the programs that regulate liver development are also important for regeneration.
One adult model for liver regeneration is partial hepatectomy. At least three studies have shown that partial hepatectomy is feasible in adult zebrafish. As described previously, top2a and its regulator uhrf1 are recessive lethal mutants and are both required for embryonic hepatic outgrowth. Adult heterozygotes for either mutation are viable, but show impaired regeneration in response to liver resection (35, 161). Interestingly, in response to partial hepatectomy, uhrf1+/− fish fail to increase expression of top2a, as predicted by previous studies in both zebrafish and humans (161). A separate study performed with transgenic dnBmp or dnFgf animals showed that these two pathways involved in liver development are also necessary for remnant proliferation and regeneration following partial hepatectomy (90).
Acute liver failure is often the result of overwhelming toxic insult, leading to massive hepatic cell death. In some cases, sufficient regenerative capacity remains following insult to allow for functional recovery. At least three models have been reported that may mimic this process. Transgenic overexpression of the enzyme nitroreductase in the liver causes hepatic cell death following administration of the antimicrobial metronidazole (MTZ), which is converted to a cytotoxic DNA cross-linking agent only in cells expressing the transgene (27,30). Withdrawal of MTZ leads to reversal of the process, allowing for proliferation and regeneration of surviving cells. A second reversible model was indicated by the recessive mutant oliver, which shows hepatocyte cell death following liver differentiation. The mutation is caused by a missense mutation in tomm22, encoding an outer mitochondrial membrane translocase (28). tomm22 morphants also show hepatocyte apoptosis that peaks at 5 dpf, but as the morpholino effect is lost, proliferation of remaining viable cells leads to liver repopulation by 8 dpf. Inhibitor studies show that this regeneration is dependent on Wnt and FGF signaling (28). A third model is acetaminophen (APAP) toxicity, the most common cause of medication-induced liver failure in the United States. Treatment of zebrafish embryos with APAP caused dose-dependent hepatic necrosis and exhaustion of reduced glutathione stores, treatable with N-acetylcysteine (NAC), the clinically used medication for APAP toxicity (134). A chemical screen discovered that prostaglandin E2 (PGE2) induced Wnt signaling to also inhibit APAP-mediated hepatic necrosis; in adult fish PGE2 and NAC had similar effects when used individually, but in combination were synergistic (134).
One outstanding question that may be addressed by future studies of this kind is the identity of the cell(s) that repopulate the liver following insult. Differentiated hepatocytes have the capacity to proliferate in response to certain types of liver injury, but mammalian studies have also shown that oval cells (in the canals of Hering) may function as a facultative hepatic stem cell in other types of injury (180,193). In zebrafish, bile preductular cells are postulated to have a similar identity as oval cells based on ultrastructural studies (191), but this has not been experimentally confirmed.
Conclusions and future directions
The zebrafish is increasingly recognized as an important model system for studying liver development and human liver disease. The versatility of the model system will be enhanced by several developments. In the future, the ability to combine high throughput forward genetics with efficient gene targeting in this hardy, economical, and rapidly developing organism offers a tremendous opportunity for discovery research. As previously described, improved methods for insertional mutagenesis (4), gene targeting (18, 164), and the molecular analysis of chemically induced mutations via deep sequencing will allow for the identification of an expanding group of physiologically and medically relevant liver phenotypes. The development of enhanced mutant screening tools (such as through the use of fluorescent transgenic reporters or organ-specific chemical assays) will allow for identification of alleles that are functionally important, but not detectable by traditional light microscopic screening.
One area of concern in modeling human liver development and disease in zebrafish is the degree to which these models faithfully recapitulate the human condition. While the molecular determinants of development and disease are generally well conserved, differences in morphogenesis and anatomy (particularly microanatomy) can be associated with differences in physiology. As more human diseases are modeled in fish, it will be of interest to note the degree to which they model the progression of disease—not only acute or developmental injury to hepatocytes and bile ducts, but progression to inflammation, fibrosis, organ failure, and malignancy. It is also important to consider the possibility that for some conditions, the zebrafish models may prove to be more useful than existing mammalian models, as is the case with malignant melanoma (144).
Despite these concerns, the major advantage of zebrafish as a model system for hepatic processes is the ability to perform screening (genetic or chemical) in a vertebrate organism. This type of unbiased approach allows for discovery or “hypothesis generating research” in a way that mammalian models often cannot. For example, the inductive role of Wnt signaling in liver specification (137), and the ability of PGE2 to synergize with NAC in the treatment of APAP-induced liver injury (134), were both found through this type of unbiased approach. Our lab discovered a selective effect of the DNA methyltransferase inhibitor azacytidine on biliary development through analyses of ducttrip mutants (116). This led to the discovery of DNA methylation defects in patients with biliary atresia (116). Our lab has also used the zebrafish to biochemically characterize a novel plant toxin that is responsible for a naturally occurring epidemic form of biliary atresia in livestock (68) (K. Lorent and M. Pack, submitted). In these roles, the zebrafish complements studies using cell culture and rodent models, and/or human tissue. The data from zebrafish studies can also generate new hypotheses that can be tested in mammalian animal or cell culture systems. In addition to its use in “discovery research” the zebrafish is also well suited for in vivo confirmatory studies, such as the functional annotation of novel disease gene candidates identified in human genome-wide association studies.
Finally, zebrafish have been validated as models for in vivo drug discovery (19,84,134), and methods for automated high-throughput chemical library screening continue to evolve. Here, the zebrafish can be used to test both the therapeutic effects and toxicity of lead compounds in a more physiologically relevant context than is possible using cultured cells or cell-free systems.
References
- 1.Alagille D, Estrada A, Hadchouel M, Gautier M, Odievre M, Dommergues JP. Syndromic paucity of interlobular bile ducts (Alagille syndrome or arteriohepatic dysplasia): Review of 80 cases. J Pediatr. 1987;110:195–200. doi: 10.1016/s0022-3476(87)80153-1. [DOI] [PubMed] [Google Scholar]
- 2.Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141–3155. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- 3.Amali AA, Rekha RD, Lin CJ, Wang WL, Gong HY, Her GM, Wu JL. Thioacetamide induced liver damage in zebrafish embryo as a disease model for steatohepatitis. J Biomed Sci. 2006;13:225–232. doi: 10.1007/s11373-005-9055-5. [DOI] [PubMed] [Google Scholar]
- 4.Amsterdam A, Varshney GK, Burgess SM. Retroviral-mediated insertional mutagenesis in zebrafish. Methods Cell Biol. 2011;104:59–82. doi: 10.1016/B978-0-12-374814-0.00004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson ER, Sandberg R, Lendahl U. Notch signaling: Simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 6.Bezerra JA, Balistreri WF. Cholestatic syndromes of infancy and childhood. Semin Gastrointest Dis. 2001;12:54–65. [PubMed] [Google Scholar]
- 7.Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpentier R, Suner RE, van Hul N, Kopp JL, Beaudry JB, Cordi S, Antoniou A, Raynaud P, Lepreux S, Jacquemin P, Leclercq IA, Sander M, Lemaigre FP. Embryonic ductal plate cells give rise to cholangiocytes, periportal hepatocytes, and adult liver progenitor cells. Gastroenterology. 2011;141:1432–1438. e1431–1434. doi: 10.1053/j.gastro.2011.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C, Hu M, Zhu Z, Lo LJ, Chen J, Peng J. liver-enriched gene 1a and 1b encode novel secretory proteins essential for normal liver development in zebrafish. PLoS One. 2011;6:e22910. doi: 10.1371/journal.pone.0022910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Nusslein-Volhard C. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- 12.Cheng W, Guo L, Zhang Z, Soo HM, Wen C, Wu W, Peng J. HNF factors form a network to regulate liver-enriched genes in zebrafish. Dev Biol. 2006;294:482–496. doi: 10.1016/j.ydbio.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Cheung ID, Bagnat M, Ma TP, Datta A, Evason K, Moore JC, Lawson ND, Mostov KE, Moens CB, Stainier DY. Regulation of intrahepatic biliary duct morphogenesis by Claudin 15-like b. Dev Biol. 2012;361:68–78. doi: 10.1016/j.ydbio.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Childs S, Weinstein BM, Mohideen MA, Donohue S, Bonkovsky H, Fishman MC. Zebrafish dracula encodes ferrochelatase and its mutation provides a model for erythropoietic protoporphyria. Curr Biol. 2000;10:1001–1004. doi: 10.1016/s0960-9822(00)00653-9. [DOI] [PubMed] [Google Scholar]
- 15.Chu J, Sadler KC. New school in liver development: Lessons from zebrafish. Hepatology. 2009;50:1656–1663. doi: 10.1002/hep.23157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung WS, Shin CH, Stainier DY. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell. 2008;15:738–748. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cinaroglu A, Gao C, Imrie D, Sadler KC. Activating transcription factor 6 plays protective and pathological roles in steatosis due to endoplasmic reticulum stress in zebrafish. Hepatology. 2011;54:495–508. doi: 10.1002/hep.24396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark KJ, Voytas DF, Ekker SC. A TALE of two nucleases: Gene targeting for the masses? Zebrafish. 2011;8:147–149. doi: 10.1089/zeb.2011.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clifton JD, Lucumi E, Myers MC, Napper A, Hama K, Farber SA, Smith AB, 3rd, Huryn DM, Diamond SL, Pack M. Identification of novel inhibitors of dietary lipid absorption using zebrafish. PLoS One. 2010;5:e12386. doi: 10.1371/journal.pone.0012386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen M, Georgiou M, Stevenson NL, Miodownik M, Baum B. Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition. Dev Cell. 2010;19:78–89. doi: 10.1016/j.devcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Collodi P, Kamei Y, Ernst T, Miranda C, Buhler DR, Barnes DW. Culture of cells from zebrafish (Brachydanio rerio) embryo and adult tissues. Cell Biol Toxicol. 1992;8:43–61. doi: 10.1007/BF00119294. [DOI] [PubMed] [Google Scholar]
- 22.Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, Epstein J, Chiang A, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Solski JA, Williams KL, Mojsilovic-Petrovic J, Ingre C, Boylan K, Graff-Radford NR, Dickson DW, Clay-Falcone D, Elman L, McCluskey L, Greene R, Kalb RG, Lee VM, Trojanowski JQ, Ludolph A, Robberecht W, Andersen PM, Nicholson GA, Blair IP, King OD, Bonini NM, Van Deerlin V, Rademakers R, Mourelatos Z, Gitler AD. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig PM, Moon TW. Fasted zebrafish mimic genetic and physiological responses in mammals: a model for obesity and diabetes? Zebrafish. 2011;8:109–117. doi: 10.1089/zeb.2011.0702. [DOI] [PubMed] [Google Scholar]
- 24.Cui S, Capecci LM, Matthews RP. Disruption of planar cell polarity activity leads to developmental biliary defects. Dev Biol. 2011;351:229–241. doi: 10.1016/j.ydbio.2010.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui S, Erlichman J, Russo P, Haber BA, Matthews RP. Intrahepatic biliary anomalies in a patient with Mowat-Wilson syndrome uncover a role for the zinc finger homeobox gene zfhx1b in vertebrate biliary development. J Pediatr Gastroenterol Nutr. 2011;52:339–344. doi: 10.1097/MPG.0b013e3181ff2e5b. [DOI] [PubMed] [Google Scholar]
- 26.Cullinane AR, Straatman-Iwanowska A, Zaucker A, Wakabayashi Y, Bruce CK, Luo G, Rahman F, Gurakan F, Utine E, Ozkan TB, Denecke J, Vukovic J, Di Rocco M, Mandel H, Cangul H, Matthews RP, Thomas SG, Rappoport JZ, Arias IM, Wolburg H, Knisely AS, Kelly DA, Muller F, Maher ER, Gissen P. Mutations in VIPAR cause an arthrogryposis, renal dysfunction and cholestasis syndrome phenotype with defects in epithelial polarization. Nat Genet. 2010;42:303–312. doi: 10.1038/ng.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: A new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 28.Curado S, Ober EA, Walsh S, Cortes-Hernandez P, Verkade H, Koehler CM, Stainier DY. The mitochondrial import gene tomm22 is specifically required for hepatocyte survival and provides a liver regeneration model. Dis Model Mech. 2010;3:486–495. doi: 10.1242/dmm.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curado S, Stainier DY. deLiver’in regeneration: Injury response and development. Semin Liver Dis. 2010;30:288–295. doi: 10.1055/s-0030-1255357. [DOI] [PubMed] [Google Scholar]
- 30.Curado S, Stainier DY, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: A spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3:948–954. doi: 10.1038/nprot.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L, Ma TP, Farber SA, Moens CB, Stainier DY. Sox9b is a key regulator of pan-creaticobiliary ductal system development. PLoS Genet. 2012;8:e1002754. doi: 10.1371/journal.pgen.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding CB, Zhang JP, Zhao Y, Peng ZG, Song DQ, Jiang JD. Zebrafish as a potential model organism for drug test against hepatitis C virus. PLoS One. 2011;6:e22921. doi: 10.1371/journal.pone.0022921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DY. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- 34.Dor Y, Stanger BZ. Regeneration in liver and pancreas: Time to cut the umbilical cord? Sci STKE. 2007:pe66. doi: 10.1126/stke.4142007pe66. [DOI] [PubMed] [Google Scholar]
- 35.Dovey M, Patton EE, Bowman T, North T, Goessling W, Zhou Y, Zon LI. Topoisomerase II alpha is required for embryonic development and liver regeneration in zebrafish. Mol Cell Biol. 2009;29:3746–3753. doi: 10.1128/MCB.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drew RE, Rodnick KJ, Settles M, Wacyk J, Churchill E, Powell MS, Hardy RW, Murdoch GK, Hill RA, Robison BD. Effect of starvation on transcriptomes of brain and liver in adult female zebrafish (Danio rerio) Physiol Genomics. 2008;35:283–295. doi: 10.1152/physiolgenomics.90213.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutko JA, Mullins MC. SnapShot: BMP signaling in development. Cell. 2011;145:636, e631–e632. doi: 10.1016/j.cell.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Eastham KM, McKiernan PJ, Milford DV, Ramani P, Wyllie J, van’t Hoff W, Lynch SA, Morris AA. ARC syndrome: An expanding range of phenotypes. Arch Dis Child. 2001;85:415–420. doi: 10.1136/adc.85.5.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.EauClaire SF, Cui S, Ma L, Matous J, Marlow FL, Gupta T, Burgess HA, Abrams EW, Kapp LD, Granato M, Mullins MC, Matthews RP. Mutations in vacuolar H+-ATPase subunits lead to biliary developmental defects in zebrafish. Dev Biol. 2012;365:434–444. doi: 10.1016/j.ydbio.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisen JS, Smith JC. Controlling morpholino experiments: Don’t stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 41.Ekker SC. Zinc finger-based knockout punches for zebrafish genes. Zebrafish. 2008;5:121–123. doi: 10.1089/zeb.2008.9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan L, Collodi P. Zebrafish embryonic stem cells. Methods Enzymol. 2006;418:64–77. doi: 10.1016/S0076-6879(06)18004-0. [DOI] [PubMed] [Google Scholar]
- 43.Fan L, Moon J, Crodian J, Collodi P. Homologous recombination in zebrafish ES cells. Transgenic Res. 2006;15:21–30. doi: 10.1007/s11248-005-3225-0. [DOI] [PubMed] [Google Scholar]
- 44.Farber SA, Pack M, Ho SY, Johnson ID, Wagner DS, Dosch R, Mullins MC, Hendrickson HS, Hendrickson EK, Halpern ME. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292:1385–1388. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- 45.Farooq M, Sulochana KN, Pan X, To J, Sheng D, Gong Z, Ge R. Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev Biol. 2008;317:336–353. doi: 10.1016/j.ydbio.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 46.Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech Dev. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 47.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- 49.Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS One. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fraenkel PG, Traver D, Donovan A, Zahrieh D, Zon LI. Ferroportin1 is required for normal iron cycling in zebrafish. J Clin Invest. 2005;115:1532–1541. doi: 10.1172/JCI23780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao H, Wang Y, Wegierski T, Skouloudaki K, Putz M, Fu X, Engel C, Boehlke C, Peng H, Kuehn EW, Kim E, Kramer-Zucker A, Walz G. PRKCSH/80K-H, the protein mutated in polycystic liver disease, protects polycystin-2/TRPP2 against HERP-mediated degradation. Hum Mol Genet. 2010;19:16–24. doi: 10.1093/hmg/ddp463. [DOI] [PubMed] [Google Scholar]
- 54.Ghosh C, Zhou YL, Collodi P. Derivation and characterization of a zebrafish liver cell line. Cell Biol Toxicol. 1994;10:167–176. doi: 10.1007/BF00757560. [DOI] [PubMed] [Google Scholar]
- 55.Gibert Y, Lattanzi VJ, Zhen AW, Vedder L, Brunet F, Faasse SA, Babitt JL, Lin HY, Hammerschmidt M, Fraenkel PG. BMP signaling modulates hepcidin expression in zebrafish embryos independent of hemojuvelin. PLoS One. 2011;6:e14553. doi: 10.1371/journal.pone.0014553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Girelli D, De Domenico I, Bozzini C, Campostrini N, Busti F, Castagna A, Soriani N, Cremonesi L, Ferrari M, Colombari R, McVey Ward D, Kaplan J, Corrocher R. Clinical, pathological, and molecular correlates in ferroportin disease: A study of two novel mutations. J Hepatol. 2008;49:664–671. doi: 10.1016/j.jhep.2008.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gissen P, Johnson CA, Morgan NV, Stapelbroek JM, Forshew T, Cooper WN, McKiernan PJ, Klomp LW, Morris AA, Wraith JE, Mc-Clean P, Lynch SA, Thompson RJ, Lo B, Quarrell OW, Di Rocco M, Trembath RC, Mandel H, Wali S, Karet FE, Knisely AS, Houwen RH, Kelly DA, Maher ER. Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome. Nat Genet. 2004;36:400–404. doi: 10.1038/ng1325. [DOI] [PubMed] [Google Scholar]
- 58.Goessling W, North TE, Lord AM, Ceol C, Lee S, Weidinger G, Bourque C, Strijbosch R, Haramis AP, Puder M, Clevers H, Moon RT, Zon LI. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 59.Goessling W, North TE, Zon LI. Ultrasound biomicroscopy permits in vivo characterization of zebrafish liver tumors. Nat Methods. 2007;4:551–553. doi: 10.1038/nmeth1059. [DOI] [PubMed] [Google Scholar]
- 60.Gong HY, Lin CJ, Chen MH, Hu MC, Lin GH, Zhou Y, Zon LI, Wu JL. Two distinct teleost hepatocyte nuclear factor 1 genes, hnf1alpha/tcf1 and hnf1beta/tcf2, abundantly expressed in liver, pancreas, gut and kidney of zebrafish. Gene. 2004;338:35–46. doi: 10.1016/j.gene.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 61.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haber BA, Russo P. Biliary atresia. Gastroenterol Clin North Am. 2003;32:891–911. doi: 10.1016/s0889-8553(03)00049-9. [DOI] [PubMed] [Google Scholar]
- 63.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 64.Halsted CH, Medici V. Aberrant hepatic methionine metabolism and gene methylation in the pathogenesis and treatment of alcoholic steatohepatitis. Int J Hepatol. 2012;2012:959746. doi: 10.1155/2012/959746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hammes TO, Pedroso GL, Hartmann CR, Escobar TD, Fracasso LB, da Rosa DP, Marroni NP, Porawski M, da Silveira TR. The effect of taurine on hepatic steatosis induced by thioacetamide in zebrafish (Danio rerio) Dig Dis Sci. 2012;57:675–682. doi: 10.1007/s10620-011-1931-4. [DOI] [PubMed] [Google Scholar]
- 66.Hand NJ, Master ZR, Eauclaire SF, Weinblatt DE, Matthews RP, Friedman JR. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136:1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hardman RC, Volz DC, Kullman SW, Hinton DE. An in vivo look at vertebrate liver architecture: Three-dimensional reconstructions from medaka (Oryzias latipes) Anat Rec (Hoboken) 2007;290:770–782. doi: 10.1002/ar.20524. [DOI] [PubMed] [Google Scholar]
- 68.Harper P, Plant JW, Unger DB. Congenital biliary atresia and jaundice in lambs and calves. Aust Vet J. 1990;67:18–22. doi: 10.1111/j.1751-0813.1990.tb07385.x. [DOI] [PubMed] [Google Scholar]
- 69.Hartman BH, Reh TA, Bermingham-McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc Natl Acad Sci U S A. 2010;107:15792–15797. doi: 10.1073/pnas.1002827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hata S, Namae M, Nishina H. Liver development and regeneration: From laboratory study to clinical therapy. Dev Growth Differ. 2007;49:163–170. doi: 10.1111/j.1440-169X.2007.00910.x. [DOI] [PubMed] [Google Scholar]
- 71.He S, Krens SG, Zhan H, Gong Z, Hogendoorn PC, Spaink HP, Snaar-Jagalska BE. A DeltaRaf1-ER-inducible oncogenic zebrafish liver cell model identifies hepatocellular carcinoma signatures. J Pathol. 2011;225:19–28. doi: 10.1002/path.2936. [DOI] [PubMed] [Google Scholar]
- 72.Her GM, Cheng CH, Hong JR, Sundaram GS, Wu JL. Imbalance in liver homeostasis leading to hyperplasia by overexpressing either one of the Bcl-2-related genes, zfBLP1 and zfMcl-1a. Dev Dyn. 2006;235:515–523. doi: 10.1002/dvdy.20624. [DOI] [PubMed] [Google Scholar]
- 73.Her GM, Chiang CC, Chen WY, Wu JL. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio) FEBS Lett. 2003;538:125–133. doi: 10.1016/s0014-5793(03)00157-1. [DOI] [PubMed] [Google Scholar]
- 74.Her GM, Hsu CC, Hong JR, Lai CY, Hsu MC, Pang HW, Chan SK, Pai WY. Overexpression of gankyrin induces liver steatosis in zebrafish (Danio rerio) Biochim Biophys Acta. 2011;1811:536–548. doi: 10.1016/j.bbalip.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 75.Her GM, Yeh YH, Wu JL. 435-bp liver regulatory sequence in the liver fatty acid binding protein (L-FABP) gene is sufficient to modulate liver regional expression in transgenic zebrafish. Dev Dyn. 2003;227:347–356. doi: 10.1002/dvdy.10324. [DOI] [PubMed] [Google Scholar]
- 76.Ho SY, Lorent K, Pack M, Farber SA. Zebrafish fat-free is required for intestinal lipid absorption and Golgi apparatus structure. Cell Metab. 2006;3:289–300. doi: 10.1016/j.cmet.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development. 2005;132:4005–4014. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- 78.Hong SK, Dawid IB. Alpha2 macroglobulin-like is essential for liver development in zebrafish. PLoS One. 2008;3:e3736. doi: 10.1371/journal.pone.0003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- 80.Howarth DL, Passeri M, Sadler KC. Drinks like a fish: Using zebrafish to understand alcoholic liver disease. Alcohol Clin Exp Res. 2011;35:826–829. doi: 10.1111/j.1530-0277.2010.01407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Howarth DL, Vacaru AM, Tsedensodnom O, Mormone E, Nieto N, Costantini LM, Snapp EL, Sadler KC. Alcohol disrupts endoplasmic reticulum function and protein secretion in hepatocytes. Alcohol Clin Exp Res. 2012;36:14–23. doi: 10.1111/j.1530-0277.2011.01602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang H, Ruan H, Aw MY, Hussain A, Guo L, Gao C, Qian F, Leung T, Song H, Kimelman D, Wen Z, Peng J. Mypt1-mediated spatial positioning of Bmp2-producing cells is essential for liver organogenesis. Development. 2008;135:3209–3218. doi: 10.1242/dev.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 84.Huang X, Nguyen AT, Li Z, Emelyanov A, Parinov S, Gong Z. One step forward: The use of transgenic zebrafish tumor model in drug screens. Birth Defects Res C Embryo Today. 2011;93:173–181. doi: 10.1002/bdrc.20208. [DOI] [PubMed] [Google Scholar]
- 85.Huang X, Zhou L, Gong Z. Liver tumor models in transgenic zebrafish: An alternative in vivo approach to study hepatocarcinogenes. Future Oncol. 2012;8:21–28. doi: 10.2217/fon.11.137. [DOI] [PubMed] [Google Scholar]
- 86.Hugo SE, Cruz-Garcia L, Karanth S, Anderson RM, Stainier DY, Schlegel A. A monocarboxylate transporter required for hepatocyte secretion of ketone bodies during fasting. Genes Dev. 2012;26:282–293. doi: 10.1101/gad.180968.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Itoh N. The Fgf families in humans, mice, and zebrafish: Their evolutional processes and roles in development, metabolism, and disease. Biol Pharm Bull. 2007;30:1819–1825. doi: 10.1248/bpb.30.1819. [DOI] [PubMed] [Google Scholar]
- 88.Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- 89.Jury DR, Kaveti S, Duan ZH, Willard B, Kinter M, Londraville R. Effects of calorie restriction on the zebrafish liver proteome. Comp Biochem Physiol Part D Genomics Proteomics. 2008;3:275–282. doi: 10.1016/j.cbd.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kan NG, Junghans D, Izpisua Belmonte JC. Compensatory growth mechanisms regulated by BMP and FGF signaling mediate liver regeneration in zebrafish after partial hepatectomy. Faseb J. 2009;23:3516–3525. doi: 10.1096/fj.09-131730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- 92.Keng VW, Yagi H, Ikawa M, Nagano T, Myint Z, Yamada K, Tanaka T, Sato A, Muramatsu I, Okabe M, Sato M, Noguchi T. Homeobox gene Hex is essential for onset of mouse embryonic liver development and differentiation of the monocyte lineage. Biochem Biophys Res Commun. 2000;276:1155–1161. doi: 10.1006/bbrc.2000.3548. [DOI] [PubMed] [Google Scholar]
- 93.Kettleborough RN, Bruijn E, Eeden F, Cuppen E, Stemple DL. High-throughput target-selected gene inactivation in zebrafish. Methods Cell Biol. 2011;104:121–127. doi: 10.1016/B978-0-12-374814-0.00006-9. [DOI] [PubMed] [Google Scholar]
- 94.Klein C, Mikutta J, Krueger J, Scholz K, Brinkmann J, Liu D, Veerkamp J, Siegel D, Abdelilah-Seyfried S, le Noble F. Neuron navigator 3a regulates liver organogenesis during zebrafish embryogenesis. Development. 2011;138:1935–1945. doi: 10.1242/dev.056861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kondo M. Bone morphogenetic proteins in the early development of zebrafish. Febs J. 2007;274:2960–2967. doi: 10.1111/j.1742-4658.2007.05838.x. [DOI] [PubMed] [Google Scholar]
- 96.Korzh S, Emelyanov A, Korzh V. Developmental analysis of ceruloplasmin gene and liver formation in zebrafish. Mech Dev. 2001;103:137–139. doi: 10.1016/s0925-4773(01)00330-6. [DOI] [PubMed] [Google Scholar]
- 97.Korzh S, Pan X, Garcia-Lecea M, Winata CL, Pan X, Wohland T, Korzh V, Gong Z. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev Biol. 2008;8:84. doi: 10.1186/1471-213X-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lakner AM, Bonkovsky HL, Schrum LW. microRNAs: Fad or future of liver disease. World J Gastroenterol. 2011;17:2536–2542. doi: 10.3748/wjg.v17.i20.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lam SH, Gong Z. Modeling liver cancer using zebrafish: A comparative oncogenomics approach. Cell Cycle. 2006;5:573–577. doi: 10.4161/cc.5.6.2550. [DOI] [PubMed] [Google Scholar]
- 100.Lam SH, Wu YL, Vega VB, Miller LD, Spitsbergen J, Tong Y, Zhan H, Govindarajan KR, Lee S, Mathavan S, Murthy KR, Buhler DR, Liu ET, Gong Z. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat Biotechnol. 2006;24:73–75. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- 101.Langdon YG, Mullins MC. Maternal and zygotic control of zebrafish dorsoventral axial patterning. Annu Rev Genet. 2011;45:357–377. doi: 10.1146/annurev-genet-110410-132517. [DOI] [PubMed] [Google Scholar]
- 102.Latimer AJ, Jessen JR. Hgf/c-met expression and functional analysis during zebrafish embryogenesis. Dev Dyn. 2008;237:3904–3915. doi: 10.1002/dvdy.21794. [DOI] [PubMed] [Google Scholar]
- 103.Laudadio I, Manfroid I, Achouri Y, Schmidt D, Wilson MD, Cordi S, Thorrez L, Knoops L, Jacquemin P, Schuit F, Pierreux CE, Odom DT, Peers B, Lemaigre FP. A feedback loop between the liver-enriched transcription factor network and mir-122 controls hepatocyte differentiation. Gastroenterology. 2012;142:119–129. doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 104.Law K, Brunt EM. Nonalcoholic fatty liver disease. Clin Liver Dis. 2010;14:591–604. doi: 10.1016/j.cld.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 105.Lawson ND, Wolfe SA. Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Dev Cell. 2011;21:48–64. doi: 10.1016/j.devcel.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 106.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont ME, Rand EB, Piccoli DA, Hood L, Spinner NB. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 107.Li YH, Chen MH, Gong HY, Hu SY, Li YW, Lin GH, Lin CC, Liu W, Wu JL. Progranulin A-mediated MET signaling is essential for liver morphogenesis in zebrafish. J Biol Chem. 2010;285:41001–41009. doi: 10.1074/jbc.M110.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Z, Huang X, Zhan H, Zeng Z, Li C, Spitsbergen JM, Meierjohann S, Schartl M, Gong Z. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. J Hepatol. 2012;56:419–425. doi: 10.1016/j.jhep.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 109.Li Z, Korzh V, Gong Z. Localized rbp4 expression in the yolk syncytial layer plays a role in yolk cell extension and early liver development. BMC Dev Biol. 2007;7:117. doi: 10.1186/1471-213X-7-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu W, Chen JR, Hsu CH, Li YH, Chen YM, Lin CY, Huang SJ, Chang ZK, Chen YC, Lin CH, Gong HY, Lin CC, Kawakami K, Wu JL. A zebrafish model of intrahepatic cholangiocarcinoma by dual expression of hepatitis B virus X and hepatitis C virus core protein in liver. Hepatology. 2012;56:2268–2276. doi: 10.1002/hep.25914. [DOI] [PubMed] [Google Scholar]
- 111.Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239:855–864. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development. 2004;131:5753–5766. doi: 10.1242/dev.01411. [DOI] [PubMed] [Google Scholar]
- 113.Marques IJ, Weiss FU, Vlecken DH, Nitsche C, Bakkers J, Lagendijk AK, Partecke LI, Heidecke CD, Lerch MM, Bagowski CP. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer. 2009;9:128. doi: 10.1186/1471-2407-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martinez Barbera JP, Clements M, Thomas P, Rodriguez T, Meloy D, Kioussis D, Beddington RS. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development. 2000;127:2433–2445. doi: 10.1242/dev.127.11.2433. [DOI] [PubMed] [Google Scholar]
- 115.Matsumoto N, Kubo A, Liu H, Akita K, Laub F, Ramirez F, Keller G, Friedman SL. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood. 2006;107:1357–1365. doi: 10.1182/blood-2005-05-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Matthews RP, Eauclaire SF, Mugnier M, Lorent K, Cui S, Ross MM, Zhang Z, Russo P, Pack M. DNA hypomethylation causes bile duct defects in zebrafish and is a distinguishing feature of infantile biliary atresia. Hepatology. 2011;53:905–914. doi: 10.1002/hep.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Matthews RP, Lorent K, Manoral-Mobias R, Huang Y, Gong W, Murray IV, Blair IA, Pack M. TNFalpha-dependent hepatic steatosis and liver degeneration caused by mutation of zebrafish S-adenosylhomocysteine hydrolase. Development. 2009;136:865–875. doi: 10.1242/dev.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Matthews RP, Lorent K, Pack M. Transcription factor onecut3 regulates intrahepatic biliary development in zebrafish. Dev Dyn. 2008;237:124–131. doi: 10.1002/dvdy.21407. [DOI] [PubMed] [Google Scholar]
- 119.Matthews RP, Lorent K, Russo P, Pack M. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev Biol. 2004;274:245–259. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 120.Matthews RP, Plumb-Rudewiez N, Lorent K, Gissen P, Johnson CA, Lemaigre F, Pack M. Zebrafish vps33b, an ortholog of the gene responsible for human arthrogryposis-renal dysfunction-cholestasis syndrome, regulates biliary development downstream of the onecut transcription factor hnf6. Development. 2005;132:5295–5306. doi: 10.1242/dev.02140. [DOI] [PubMed] [Google Scholar]
- 121.Mayer AN, Fishman MC. Nil per os encodes a conserved RNA recognition motif protein required for morphogenesis and cytodifferentiation of digestive organs in zebrafish. Development. 2003;130:3917–3928. doi: 10.1242/dev.00600. [DOI] [PubMed] [Google Scholar]
- 122.Mizgirev I, Revskoy S. Generation of clonal zebrafish lines and transplantable hepatic tumors. Nat Protoc. 2010;5:383–394. doi: 10.1038/nprot.2010.8. [DOI] [PubMed] [Google Scholar]
- 123.Moens CB, Donn TM, Wolf-Saxon ER, Ma TP. Reverse genetics in zebrafish by TILLING. Brief Funct Genomic Proteomic. 2008;7:454–459. doi: 10.1093/bfgp/eln046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moore JL, Rush LM, Breneman C, Mohideen MA, Cheng KC. Zebrafish genomic instability mutants and cancer susceptibility. Genetics. 2006;174:585–600. doi: 10.1534/genetics.106.059386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mormone E, George J, Nieto N. Molecular pathogenesis of hepatic fibrosis and current therapeutic approaches. Chem Biol Interact. 2011;193:225–231. doi: 10.1016/j.cbi.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mudumana SP, Wan H, Singh M, Korzh V, Gong Z. Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: Liver, intestine, and exocrine pancreas. Dev Dyn. 2004;230:165–173. doi: 10.1002/dvdy.20032. [DOI] [PubMed] [Google Scholar]
- 128.Nagaki M, Moriwaki H. Transcription factor HNF and hepatocyte differentiation. Hepatol Res. 2008;38:961–969. doi: 10.1111/j.1872-034X.2008.00367.x. [DOI] [PubMed] [Google Scholar]
- 129.Nguyen AT, Emelyanov A, Koh CH, Spitsbergen JM, Lam SH, Mathavan S, Parinov S, Gong Z. A high level of liver-specific expression of oncogenic Kras(V12) drives robust liver tumorigenesis in transgenic zebrafish. Dis Model Mech. 2011;4:801–813. doi: 10.1242/dmm.007831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nguyen AT, Emelyanov A, Koh CH, Spitsbergen JM, Parinov S, Gong Z. An inducible krasV12 transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis Model Mech. 2012;5:63–72. doi: 10.1242/dmm.008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) ClinicalTrials.gov [Internet] Bethesda, MD: National Library of Medicine (US); Feb 21, 2006. [18 June 2012]. A Randomized, Double-Blinded, Placebo-Controlled Trial of Corticosteroid Therapy Following Portoenterostomy in Infants With Biliary Atresia. Available at: http://clinicaltrials.gov/ct2/show/NCT00294684. [Google Scholar]
- 132.Noel ES, Casal-Sueiro A, Busch-Nentwich E, Verkade H, Dong PD, Stemple DL, Ober EA. Organ-specific requirements for Hdac1 in liver and pancreas formation. Dev Biol. 2008;322:237–250. doi: 10.1016/j.ydbio.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Noel ES, Reis MD, Arain Z, Ober EA. Analysis of the Albumin/alpha-Fetoprotein/Afamin/Group specific component gene family in the context of zebrafish liver differentiation. Gene Expr Patterns. 2010;10:237–243. doi: 10.1016/j.gep.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 134.North TE, Babu IR, Vedder LM, Lord AM, Wishnok JS, Tannenbaum SR, Zon LI, Goessling W. PGE2-regulated wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proc Natl Acad Sci U S A. 2010;107:17315–17320. doi: 10.1073/pnas.1008209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, Fitzgerald GA, Daley GQ, Orkin SH, Zon LI. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ober EA, Field HA, Stainier DY. From endoderm formation to liver and pancreas development in zebrafish. Mech Dev. 2003;120:5–18. doi: 10.1016/s0925-4773(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 137.Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 138.Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, Chandrasekharappa SC. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 139.Oka T, Nishimura Y, Zang L, Hirano M, Shimada Y, Wang Z, Umemoto N, Kuroyanagi J, Nishimura N, Tanaka T. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 2010;10:21. doi: 10.1186/1472-6793-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ong ES, Chor CF, Zou L, Ong CN. A multi-analytical approach for metabolomic profiling of zebrafish (Danio rerio) livers. Mol Biosyst. 2009;5:288–298. doi: 10.1039/b811850g. [DOI] [PubMed] [Google Scholar]
- 141.Pack M, Solnica-Krezel L, Malicki J, Neuhauss SC, Schier AF, Stemple DL, Driever W, Fishman MC. Mutations affecting development of zebrafish digestive organs. Development. 1996;123:321–328. doi: 10.1242/dev.123.1.321. [DOI] [PubMed] [Google Scholar]
- 142.Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, Leach SD. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Passeri MJ, Cinaroglu A, Gao C, Sadler KC. Hepatic steatosis in response to acute alcohol exposure in zebrafish requires sterol regulatory element binding protein activation. Hepatology. 2009;49:443–452. doi: 10.1002/hep.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Patton EE, Mathers ME, Schartl M. Generating and analyzing fish models of melanoma. Methods Cell Biol. 2011;105:339–366. doi: 10.1016/B978-0-12-381320-6.00014-X. [DOI] [PubMed] [Google Scholar]
- 145.Peterson RT, Fishman MC. Designing zebrafish chemical screens. Methods Cell Biol. 2011;105:525–541. doi: 10.1016/B978-0-12-381320-6.00023-0. [DOI] [PubMed] [Google Scholar]
- 146.Piccoli DA, Spinner NB. Alagille syndrome and the Jagged1 gene. Semin Liver Dis. 2001;21:525–534. doi: 10.1055/s-2001-19036. [DOI] [PubMed] [Google Scholar]
- 147.Poulain M, Ober EA. Interplay between Wnt2 and Wnt2bb controls multiple steps of early foregut-derived organ development. Development. 2011;138:3557–3568. doi: 10.1242/dev.055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Puy H, Gouya L, Deybach JC. Porphyrias. Lancet. 2010;375:924–937. doi: 10.1016/S0140-6736(09)61925-5. [DOI] [PubMed] [Google Scholar]
- 149.Qi F, Song J, Yang H, Gao W, Liu NA, Zhang B, Lin S. Mmp23b promotes liver development and hepatocyte proliferation through the tumor necrosis factor pathway in zebrafish. Hepatology. 2010;52:2158–2166. doi: 10.1002/hep.23945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21:261–266. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, Cairns BR, Jones DA. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol. 2006;26:7077–7085. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reiter JF, Kikuchi Y, Stainier DY. Multiple roles for Gata5 in zebrafish endoderm formation. Development. 2001;128:125–135. doi: 10.1242/dev.128.1.125. [DOI] [PubMed] [Google Scholar]
- 153.Rekha RD, Amali AA, Her GM, Yeh YH, Gong HY, Hu SY, Lin GH, Wu JL. Thioacetamide accelerates steatohepatitis, cirrhosis and HCC by expressing HCV core protein in transgenic zebrafish Danio rerio. Toxicology. 2008;243:11–22. doi: 10.1016/j.tox.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 154.Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, Schier AF. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rovira M, Huang W, Yusuff S, Shim JS, Ferrante AA, Liu JO, Parsons MJ. Chemical screen identifies FDA-approved drugs and target pathways that induce precocious pancreatic endocrine differentiation. Proc Natl Acad Sci U S A. 2011;108:19264–19269. doi: 10.1073/pnas.1113081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Roy S, Qiao T, Wolff C, Ingham PW. Hedgehog signaling pathway is essential for pancreas specification in the zebrafish embryo. Curr Biol. 2001;11:1358–1363. doi: 10.1016/s0960-9822(01)00402-x. [DOI] [PubMed] [Google Scholar]
- 159.Russo P, Magee JC, Boitnott J, Bove KE, Raghunathan T, Finegold M, Haas J, Jaffe R, Kim GE, Magid M, Melin-Aldana H, White F, Whitington PF, Sokol RJ. Design and validation of the biliary atresia research consortium histologic assessment system for cholestasis in infancy. Clin Gastroenterol Hepatol. 2011;9:357–362. e352. doi: 10.1016/j.cgh.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- 161.Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci U S A. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, Joung JK. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schaub M, Nussbaum J, Verkade H, Ober EA, Stainier DY, Sakaguchi TF. Mutation of zebrafish Snapc4 is associated with loss of the intra-hepatic biliary network. Dev Biol. 2012;363:128–137. doi: 10.1016/j.ydbio.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schlegel A, Stainier DY. Lessons from “lower” organisms: What worms, flies, and zebrafish can teach us about human energy metabolism. PLoS Genet. 2007;3:e199. doi: 10.1371/journal.pgen.0030199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schonthaler HB, Fleisch VC, Biehlmaier O, Makhankov Y, Rinner O, Bahadori R, Geisler R, Schwarz H, Neuhauss SC, Dahm R. The zebrafish mutant lbk/vam6 resembles human multisystemic disorders caused by aberrant trafficking of endosomal vesicles. Development. 2008;135:387–399. doi: 10.1242/dev.006098. [DOI] [PubMed] [Google Scholar]
- 168.Sempoux C, Jibara G, Ward SC, Fan C, Qin L, Roayaie S, Fiel MI, Schwartz M, Thung SN. Intrahepatic cholangiocarcinoma: New insights in pathology. Semin Liver Dis. 2011;31:49–60. doi: 10.1055/s-0031-1272839. [DOI] [PubMed] [Google Scholar]
- 169.Shieh YS, Chang YS, Hong JR, Chen LJ, Jou LK, Hsu CC, Her GM. Increase of hepatic fat accumulation by liver specific expression of Hepatitis B virus X protein in zebrafish. Biochim Biophys Acta. 2010;1801:721–730. doi: 10.1016/j.bbalip.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 170.Shin D, Lee Y, Poss KD, Stainier DY. Restriction of hepatic competence by Fgf signaling. Development. 2011;138:1339–1348. doi: 10.1242/dev.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DY. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- 172.Shin D, Weidinger G, Moon RT, Stainier DY. Intrinsic and extrinsic modifiers of the regulative capacity of the developing liver. Mech Dev. 2012;128:525–535. doi: 10.1016/j.mod.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Siddique A, Kowdley KV. Review article: The iron overload syndromes. Aliment Pharmacol Ther. 2012;35:876–893. doi: 10.1111/j.1365-2036.2012.05051.x. [DOI] [PubMed] [Google Scholar]
- 174.Singham J, Yoshida EM, Scudamore CH. Choledochal cysts: Part 1 of 3: Classification and pathogenesis. Can J Surg. 2009;52:434–440. [PMC free article] [PubMed] [Google Scholar]
- 175.Sood R, English MA, Jones M, Mullikin J, Wang DM, Anderson M, Wu D, Chandrasekharappa SC, Yu J, Zhang J, Paul Liu P. Methods for reverse genetic screening in zebrafish by resequencing and TILLING. Methods. 2006;39:220–227. doi: 10.1016/j.ymeth.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 176.Stuart GW, Vielkind JR, McMurray JV, Westerfield M. Stable lines of transgenic zebrafish exhibit reproducible patterns of transgene expression. Development. 1990;109:577–584. doi: 10.1242/dev.109.3.577. [DOI] [PubMed] [Google Scholar]
- 177.Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tainaka T, Shimada Y, Kuroyanagi J, Zang L, Oka T, Nishimura Y, Nishimura N, Tanaka T. Transcriptome analysis of anti-fatty liver action by Campari tomato using a zebrafish diet-induced obesity model. Nutr Metab (Lond) 2011;8:88. doi: 10.1186/1743-7075-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tan JL, Zon LI. Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol. 2011;105:493–516. doi: 10.1016/B978-0-12-381320-6.00021-7. [DOI] [PubMed] [Google Scholar]
- 180.Tanaka M, Itoh T, Tanimizu N, Miyajima A. Liver stem/progenitor cells: Their characteristics and regulatory mechanisms. J Biochem. 2011;149:231–239. doi: 10.1093/jb/mvr001. [DOI] [PubMed] [Google Scholar]
- 181.Thakur PC, Stuckenholz C, Rivera MR, Davison JM, Yao JK, Amsterdam A, Sadler KC, Bahary N. Lack of de novo phosphatidylinositol synthesis leads to endoplasmic reticulum stress and hepatic steatosis in cdipt-deficient zebrafish. Hepatology. 2011;54:452–462. doi: 10.1002/hep.24349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tien AL, Senbanerjee S, Kulkarni A, Mudbhary R, Goudreau B, Ganesan S, Sadler KC, Ukomadu C. UHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem J. 2011;435:175–185. doi: 10.1042/BJ20100840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tiso N, Filippi A, Pauls S, Bortolussi M, Argenton F. BMP signalling regulates anteroposterior endoderm patterning in zebrafish. Mech Dev. 2002;118:29–37. doi: 10.1016/s0925-4773(02)00252-6. [DOI] [PubMed] [Google Scholar]
- 184.Tittle RK, Sze R, Ng A, Nuckels RJ, Swartz ME, Anderson RM, Bosch J, Stainier DY, Eberhart JK, Gross JM. Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens. Dev Biol. 2011;350:50–63. doi: 10.1016/j.ydbio.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wallace KN, Pack M. Unique and conserved aspects of gut development in zebrafish. Dev Biol. 2003;255:12–29. doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- 186.Wallace KN, Yusuff S, Sonntag JM, Chin AJ, Pack M. Zebrafish hhex regulates liver development and digestive organ chirality. Genesis. 2001;30:141–143. doi: 10.1002/gene.1050. [DOI] [PubMed] [Google Scholar]
- 187.Wang R, Li Z, Wang Y, Gui JF. An Apo-14 promoter-driven transgenic zebrafish that marks liver organogenesis. PLoS One. 2011;6:e22555. doi: 10.1371/journal.pone.0022555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, Chen F, Long HK, Kramer M, Datta S, Neuberg D, Granter S, Young RA, Morrison S, Wheeler GN, Zon LI. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xu L, Yin W, Xia J, Peng M, Li S, Lin S, Pei A, Shu X. An anti-apoptotic role of SNX7 is required for liver development in zebrafish. Hepatology. 2012;55:1985–1993. doi: 10.1002/hep.25560. [DOI] [PubMed] [Google Scholar]
- 190.Yanger K, Stanger BZ. Facultative stem cells in liver and pancreas: Fact and fancy. Dev Dyn. 2011;240:521–529. doi: 10.1002/dvdy.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yao Y, Lin J, Yang P, Chen Q, Chu X, Gao C, Hu J. Fine structure, enzyme histochemistry, and immunohistochemistry of liver in zebrafish. Anat Rec (Hoboken) 2012;295:567–576. doi: 10.1002/ar.22416. [DOI] [PubMed] [Google Scholar]
- 192.Yin C, Evason KJ, Maher JJ, Stainier DY. The basic helix-loop-helix transcription factor, heart and neural crest derivatives expressed transcript 2, marks hepatic stellate cells in zebrafish: Analysis of stellate cell entry into the developing liver. Hepatology. 2012;56:1958–1970. doi: 10.1002/hep.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yovchev MI, Grozdanov PN, Zhou H, Racherla H, Guha C, Dabeva MD. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology. 2008;47:636–647. doi: 10.1002/hep.22047. [DOI] [PubMed] [Google Scholar]
- 194.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 196.Zebrafish Mutation Project. Cambridge: Wellcome Trust Sanger Institute; Cited on 18 June 2012. Available at: http://www.sanger.ac.uk/Projects/Drerio/zmp/ [Google Scholar]
- 197.Zhao X, Monson C, Gao C, Gouon-Evans V, Matsumoto N, Sadler KC, Friedman SL. Klf6/copeb is required for hepatic outgrowth in zebrafish and for hepatocyte specification in mouse ES cells. Dev Biol. 2010;344:79–93. doi: 10.1016/j.ydbio.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zweier C, Albrecht B, Mitulla B, Behrens R, Beese M, Gillessen-Kaesbach G, Rott HD, Rauch A. “Mowat-Wilson” syndrome with and without Hirschsprung disease is a distinct, recognizable multiple congenital anomalies-mental retardation syndrome caused by mutations in the zinc finger homeo box 1B gene. Am J Med Genet. 2002;108:177–181. [PubMed] [Google Scholar]