Abstract
Research from the Multiethnic Cohort (MEC) demonstrated that, for the same quantity of cigarette smoking, African Americans and Native Hawaiians have a higher lung cancer risk than Whites, while Latinos and Japanese Americans are less susceptible. We collected urine samples from 2,239 cigarette smokers from five different ethnic groups in the MEC and analyzed each sample for S-phenylmercapturic acid (SPMA), a specific biomarker of benzene uptake. African Americans had significantly higher (geometric mean [SE] 3.69 [0.2], p<0.005) SPMA/ml urine than Whites (2.67 [0.13]) while Japanese Americans had significantly lower levels than Whites (1.65 [0.07], p<0.005). SPMA levels in Native Hawaiians and Latinos were not significantly different from those of Whites. We also conducted a genome-wide association study in search of genetic risk factors related to benzene exposure. The glutathione S-transferase T1 (GSTT1) deletion explained between 14.2–31.6% (p = 5.4x10-157) and the GSTM1 deletion explained between 0.2%-2.4% of the variance (p = 1.1x10-9) of SPMA levels in these populations. Ethnic differences in levels of SPMA remained strong even after controlling for the effects of these two deletions. These results demonstrate the powerful effect of GSTT1 status on SPMA levels in urine and show that uptake of benzene in African American, White, and Japanese American cigarette smokers is consistent with their lung cancer risk in the MEC. While benzene is not generally considered a cause of lung cancer, its metabolite SPMA could be a biomarker for other volatile lung carcinogens in cigarette smoke.
Introduction
Results from the Multiethnic Cohort (MEC) demonstrate that, for the same quantity of cigarettes smoked, particularly at lower levels of smoking, African Americans and Native Hawaiians had a higher risk for lung cancer than Whites while Latinos and Japanese Americans were less susceptible [1]. These variations were evident for all histologic types of lung cancer and in both women and men, but were not seen in non-smokers. Similar results from studies of different designs have been observed previously and the SEER data also demonstrate racial/ethnic differences in lung cancer incidence [2–9]. Multiple studies have examined genetic variation in carcinogen metabolizing genes and DNA repair pathways with respect to lung cancer incidence, with genome-wide association studies highlighting the region of the CHRNA5-CHRNA3-CHRNB4 gene cluster on chromosome 15q25 in association with quantity smoked and lung cancer risk [10–15]. These studies, which include ethnic-specific analyses, provide some important mechanistic leads but the degree to which these genetic factors contribute to ethnic differences in smoking-related lung cancer risk has not been established. Our approach uses a combination of carcinogen-specific phenotyping and a genome-wide association study (GWAS) in a subgroup of MEC participants who were cancer-free current smokers. In studies published to date, we have examined urinary biomarkers of uptake of nicotine, a tobacco-specific nitrosamine, acrolein, and crotonaldehyde in these smokers [16–19]. The focus of the study presented here is benzene, a representative volatile carcinogen in cigarette smoke.
Benzene is a human carcinogen and a recognized cause of acute myeloid leukemia and acute non-lymphocytic leukemia [20, 21]. The highest consistent non-occupational exposure to benzene occurs in cigarette smokers; the mainstream smoke of a single cigarette typically contains 15–59 micrograms of benzene [22]. It has been estimated that nearly 90% of benzene exposure in smokers is due to benzene in cigarette smoke [23]. When smokers stopped smoking cigarettes, benzene exposure as measured by urinary S-phenylmercapturic acid (SPMA) rapidly decreased by about 80% [24]. There also can be non-occupational exposures to benzene in high traffic areas and near gasoline filling stations, and from contaminated water and food. Occupational exposures to benzene can occur in a variety of settings including the petrochemical industry, around gasoline service stations, and in the rubber and paint industries, but these are relatively rare in the U.S. [20, 21, 23, 25]. Although benzene is not generally considered to play a major role in lung cancer etiology in smokers, compared for example to tobacco-specific nitrosamines and polycyclic aromatic hydrocarbons [26], it could be a biomarker for other volatile constituents of cigarette smoke involved in lung cancer etiology.
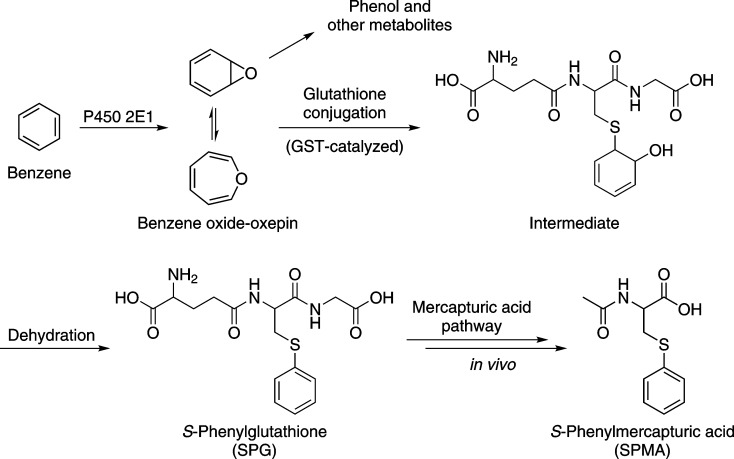
Benzene requires metabolic activation to exert its carcinogenic effects (Fig 1). The critical and requisite intermediate benzene oxide can be detoxified by reaction with glutathione, catalyzed by glutathione S-transferases (GSTs), such as GSTT1 and GSTM1. The glutathione conjugate of benzene oxide is processed by a series of enzymes resulting in the excretion of SPMA in urine. SPMA is an accepted and specific biomarker of benzene exposure [20, 27, 28].
Fig 1. Overview of benzene metabolism to SPMA.
Polymorphisms exist in GSTs, including deletion of the GSTT1 and GSTM1 genes, leading to the logical hypothesis that there could be corresponding effects on levels of SPMA in urine (reviewed in [29, 30]). With respect to GSTT1 polymorphisms and SPMA levels in urine, ten previous studies ranging in size from 37 to 386 subjects have reported an effect of GSTT1 null status on urinary SPMA levels. Consistently, higher levels of SPMA were found in the urine of subjects exposed occupationally or environmentally to benzene and who were GSTT1 positive compared to those with the null genotype [31–40]. Smaller effects on SPMA levels were observed in studies of individuals with the GSTM1 deletion [29].
In the study reported here of 2,239 cigarette smokers from the five ethnic groups of the MEC, we assessed benzene uptake by quantifying SPMA in urine. In tandem with these SPMA analyses, we carried out a GWAS exploring the effects of common genetic variants on urinary SPMA levels. This study is by far the largest yet reported to investigate SPMA levels in genotyped subjects, allowing us to confidently analyze ethnic differences in benzene uptake, as indicated by urinary SPMA, and determine the effects of genotype on SPMA levels. The results demonstrate both the power and the limitations of the SPMA biomarker while also providing new data on benzene uptake in smokers from populations with differing risks for lung cancer.
Materials and Methods
The Institutional Review Boards at the University of Southern California, the University of Hawaii, and the University of Minnesota approved of the study protocol. The participants provided written consent to participate in the study. The Institutional Review Boards at the University of Southern California and at the University of Hawaii approved of the consent procedure.
Study Population
The study subjects are MEC participants who were current smokers at the time of biospecimen collection. The MEC is a prospective cohort study established to investigate the association of lifestyle and genetic factors with chronic diseases [41] and is comprised of 215,251 men and women between the ages of 45 to 75 at baseline, primarily belonging to five ethnic/racial groups: African Americans, Native Hawaiians, Whites, Latinos, and Japanese Americans. Between 1993 and 1996, potential participants were identified in Hawaii and California (primarily Los Angeles County) through drivers’ license files, voter registration lists, and Health Care Financing Administration files. Each participant completed a mailed, self-administered questionnaire regarding demographic, dietary, lifestyle, and other exposure factors.
This specific study comprises a subgroup of the MEC participants who were cancer-free current smokers at the time of urine collection. Approximately 10 years after cohort entry, 2,393 current smokers with no cancer diagnosis participated in the MEC bio-specimen sub-cohort by providing a blood sample and overnight (subjects recruited in Hawaii—mostly Whites, Native Hawaiians and Japanese Americans) or first morning urine (subjects recruited in California–mostly African Americans and Latinos) and completing an epidemiologic questionnaire that included a history of daily cigarette smoking during the past two weeks, smoking duration, and a record of current medications. The overnight urine collection started between 5–9 pm (depending on the subject) and included all urine passed during the night as well as the first morning urine. All urine was kept on ice until processing. Aliquots were subsequently stored in a -80°C freezer until analysis.
Phenotype Measurements
Analysis of SPMA in urine was performed by liquid chromatography-tandem mass spectrometry essentially as described [42], with the following modifications: 1. [D5]SPMA (12.5 ng, Toronto Research Chemicals) was added to the urine samples as internal standard; 2. Following washing of the 96-well Oasis MAX plates with 0.7 ml of 30% methanol in 2% aqueous formic acid to elute 3-hydroxypropylmercapturic acid (3-HPMA) and 3-hydroxy-1-methylpropylmercapturic acid (HMPMA), the plates were washed with 0.7 ml 50% methanol in 2% aqueous formic acid and this wash was discarded. The plates were then washed with 0.7 ml of 90% methanol in 2% formic acid to collect the fraction containing SPMA and the internal standard; 3. The MS transitions monitored were m/z 238.05 → m/z 109.05 for SPMA and m/z 243.05 → m/z 114.05 for [D5]SPMA. The limit of quantitation was 0.1 pmol/ml.
Methods of analysis for total nicotine equivalents (TNE, the sum of nicotine, cotinine, 3′-hydroxycotinine and their glucuronides, and nicotine N-oxide), 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (total NNAL, a biomarker of uptake of the tobacco-specific carcinogen NNK), 3-HPMA, a biomarker of uptake of the volatile toxicant acrolein, and HMPMA, a biomarker of uptake of the volatile carcinogen and toxicant crotonaldehyde, in the urine of MEC subjects have been described [16, 42–44].
Individuals who smoked only to a limited degree (determined by TNE less than 1.4 nmol/ml, n = 80) were excluded from the study. Those with SPMA below the limit of detection (0.1 pmol/ml) were also excluded (n = 139).
Genotyping and Quality Control
A total of 2,418 current smokers were genotyped using the Illumina Human1M-Duo BeadChip (1,199,187 SNPs), as previously described [18]. The genotyping quality control consisted of 1) removing individual samples with ≥2% of genotypes not called (n = 8), 2) removing SNPs ≤98% call rate (n = 67,761), 3) removing known duplicate samples (n = 25), 4) excluding samples with close relatives (as determined by estimated IBD status in pair wise comparisons of samples; n = 59), and samples with conflicting or indeterminate sex (n = 7). Genotyping of the GSTT1 and GSTM1 deletions was performed by TaqMan and run on the 7900HT Fast Real-Time System (Life Technologies, Foster City, CA). Copy number counts were calculated using Life Technologies CopyCaller v2.0 software. Approximately 5% of blind duplicates were included for quality control. Genotyping of the GSTT1 and GSTM1 deletion polymorphisms was successful in 2,111, and 2,225 individuals, respectively. Test for Hardy Weinberg Equilibrium was met for all five populations for GSTM1 (p>0.05), Latinos did not meet these criteria for GSTT1 (p = 0.01).
Imputation to Estimate Unmeasured Genotypes
Imputation was performed using SHAPEIT [45] and IMPUTE2 [46] to a cosmopolitan reference panel from the 1000 Genomes Project (1KGP; March, 2012). We included SNPs with an IMPUTE2 info score of ≥0.30 and minor allele frequency (MAF) >1% in any MEC ethnic group. A total of 11,892,802 SNPs/indels with a frequency >1% in any single ethnic population (1,131,426 genotyped and 10,761,376 imputed) were included in the analysis.
Statistical Analysis
Least-square means (or geometric means) were estimated and compared between populations for the smoking variables. Principal components were estimated using 19,059 randomly selected autosomal SNPs with frequency ≥ 2% in the combined multiethnic sample [47]. The 10 leading eigenvectors from this matrix were included in the analysis to adjust for population stratification. The per allele association of each SNP/indel with geometric mean SPMA levels was evaluated using linear regression models, with adjustment for age at the time of urine collection, sex, reported ethnicity, TNE, BMI, and the first 10 principal components described above. A p-value cut-off of 5x10-8 was used to establish genome-wide significance. Conditional models were used for regions with multiple associated variants (at p<5x10-8). Ethnic-specific analyses were performed to search for loci that may be important in individual populations and tests of heterogeneity by ethnic group were performed by including an interaction term between ethnicity and variant in regression models. We also conducted analyses among subjects homozygous for the GSTT1 and GSTM1 non-null alleles to examine associations with variants located in the deleted region. We also report on the associations with variants in candidate gene regions known to be involved in benzene metabolism (e.g., CYP2E1). R2 value was used to assess the percentage of variation of SPMA accounted for by the variants examined. To examine correlations between SPMA and other biomarkers, Pearson’s partial correlations (r) were reported and adjusted for age, gender, BMI and race. Genomic control [48] (estimation of over-dispersion parameter λ) was used to assess adequacy of control for population stratification and other aspects of the behavior of tests SNP effects in the GWAS data.
Results
A total of 2,239 smokers (364 African Americans, 311 Native Hawaiians, 437 Whites, 453 Latinos, and 674 Japanese Americans) were included in the main analysis (Table 1). Significant differences in smoking, as expressed in cigarettes per day, among the ethnic groups in this sample have been reported previously [16–18]. Among both men and women, Whites reported smoking the highest number of cigarettes per day followed by Native Hawaiians, Japanese Americans, African Americans and Latinos (Table 1).
Table 1. Descriptive characteristics of the multiethnic sample of smokers.
| n = 2,239 | African Americans | Native Hawaiians | Whites | Latinos | Japanese Americans | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean [SE] | n | Mean [SE] | n | Mean [SE] | n | Mean [SE] | n | Mean [SE] | ||
| Age (yrs) | All | 364 | 64.86 [0.38] * | 311 | 61.36 [0.41] *** | 437 | 63.69 [0.35] | 453 | 65.53 [0.34] *** | 674 | 63.60 [0.29] |
| Male | 111 | 63.49 [0.65] | 114 | 63.08 [0.64] | 190 | 63.36 [0.50] | 237 | 66.43 [0.45] | 388 | 63.62 [0.36] | |
| Female | 253 | 65.39 [0.47] | 197 | 60.28 [0.54] | 247 | 63.92 [0.48] | 216 | 64.58 [0.51] | 286 | 63.69 [0.47] | |
| CPDa | All | 364 | 11.72 [0.48] *** | 311 | 15.52 [0.52] *** | 437 | 18.02 [0.43] | 453 | 9.31 [0.43] *** | 674 | 13.76 [0.37] *** |
| Male | 111 | 11.98 [0.91] *** | 114 | 17.00 [0.91] ** | 190 | 21.00 [0.70] | 237 | 10.87 [0.64] *** | 388 | 15.58 [0.51] *** | |
| Female | 253 | 10.79 [0.53] *** | 197 | 14.27 [0.61] | 247 | 15.47 [0.53] | 216 | 8.14 [0.57] *** | 286 | 12.05 [0.53] *** | |
| TNEb | All | 364 | 44.94 [2.00] *** | 311 | 30.61 [1.48] | 437 | 34.44 [1.38] | 453 | 31.28 [1.25] | 674 | 25.08 [0.86] *** |
| Male | 111 | 47.97 [3.85] | 114 | 33.04 [2.65] * | 190 | 42.55 [2.64] | 237 | 33.76 [1.91] * | 388 | 27.99 [1.25] *** | |
| Female | 253 | 41.20 [2.16] *** | 197 | 28.86 [1.71] | 247 | 28.90 [1.51] | 216 | 29.65 [1.67] | 286 | 22.76 [1.20] ** | |
| Creatininec | All | 364 | 90.18 [2.87]*** | 311 | 59.08 [2.03] | 437 | 57.66 [1.66] | 453 | 70.63 [2.01]*** | 674 | 52.66 [1.24]* |
| Male | 111 | 110.4 [6.44]*** | 114 | 72.88 [4.22] | 190 | 71.85 [3.26] | 237 | 84.24 [3.43]** | 388 | 65.66 [2.07] | |
| Female | 253 | 75.46 [2.81]*** | 197 | 49.37 [2.08] | 247 | 47.70 [1.78] | 216 | 61.03 [2.44]*** | 286 | 43.18 [1.53] | |
| SPMA/mld | All | 354 | 3.69 [0.2] *** | 296 | 2.42 [0.14] | 418 | 2.67 [0.13] | 431 | 2.86 [0.14] | 601 | 1.65 [0.07] *** |
| Male | 109 | 4.52 [0.44]** | 110 | 2.42 [0.24] | 182 | 3.09 [0.24] | 225 | 2.78 [0.19] | 355 | 1.73 [0.09] *** | |
| Female | 245 | 3.34 [0.22]** | 186 | 2.33 [0.17] | 236 | 2.42 [0.16] | 206 | 2.93 [0.20] * | 246 | 1.60 [0.10] *** | |
| SPMA/mge | All | 354 | 4.06 [0.19] | 296 | 4.06 [0.21] | 418 | 4.56 [0.19] | 431 | 3.96 [0.17] * | 601 | 3.05 [0.11] *** |
| Male | 109 | 4.10 [0.34] | 110 | 3.32 [0.28] * | 182 | 4.16 [0.28] | 225 | 3.20 [0.19] ** | 355 | 2.56 [0.12] *** | |
| Female | 245 | 4.38 [0.24] | 186 | 4.68 [0.29] | 236 | 5.05 [0.28] | 206 | 4.75 [0.28] | 246 | 3.61 [0.20] *** | |
a CPD = cigarettes/day; least square means for CPD have been adjusted for BMI, age (and gender where appropriate).
b TNE (total nicotine equivalents) is the sum of total nicotine, total cotinine, total 3′-hydroxycotinine, and nicotine N-oxide expressed as nmol/ml; geometric least square means for TNE have been adjusted for BMI, age (and gender where appropriate).
c Creatinine expressed as mg/dl; geometric least square means of creatinine have been adjusted for BMI, age, TNE (and gender where appropriate).
d SPMA (S-Phenyl mercapturic Acid) expressed as pmol/ml; geometric least square means of SPMA have been adjusted for BMI, age, TNE (and gender where appropriate).
e SPMA expressed as pmol/mg creatinine; geometric least square means of SPMA have been adjusted for BMI, age, TNE (and gender where appropriate).
* P-values across ethnic groups (with Whites as the reference) were indicated where significant as *p < 0.05
**p<0.005 and
***p<0.0005.
As also reported previously, levels of TNE were highest among the African Americans and lowest in the Japanese Americans compared to the Whites (Table 1) [16, 18]. We also noted significantly higher levels of creatinine in African Americans (p = 1x10-24) and Latinos (p = 8x10-7) compared to Whites with significantly lower levels observed in Japanese (p = 0.015) (Table 1). For simplicity, because of this large variability in creatinine levels across populations, SPMA was expressed per ml of urine rather than per mg of creatinine.
SPMA was significantly correlated with several other urinary biomarkers in the MEC subjects: TNE, total NNAL, 3-HPMA, and HMPMA (Table 2). The strongest correlations were with TNE and total NNAL, but all were highly significant.
Table 2. Partial Correlations Among SPMA and Other Biomarkers.
| N | SPMA & TNEa | N | SPMA & Total NNALa | N | SPMA & 3-HPMAa | N | SPMA & HMPMAa | |
|---|---|---|---|---|---|---|---|---|
| Overall | 2100 | 0.57 | 2048 | 0.47 | 2070 | 0.26 | 2070 | 0.28 |
| African Americans | 354 | 0.61 | 343 | 0.47 | 346 | 0.34 | 346 | 0.34 |
| Native Hawaiians | 296 | 0.52 | 290 | 0.39 | 294 | 0.26 | 294 | 0.31 |
| European Americans | 419 | 0.63 | 408 | 0.54 | 410 | 0.21 | 410 | 0.26 |
| Latinos | 431 | 0.66 | 418 | 0.57 | 422 | 0.30 | 422 | 0.29 |
| Japanese Americans | 601 | 0.42 | 589 | 0.36 | 598 | 0.19 | 598 | 0.23 |
| GSTT1 = 0 | 524 | 0.53 | 515 | 0.49 | 523 | 0.22 | 523 | 0.24 |
| GSTT1 = 1 | 937 | 0.71 | 912 | 0.6 | 919 | 0.31 | 919 | 0.36 |
| GSTT1 = 2 | 514 | 0.74 | 498 | 0.62 | 504 | 0.31 | 504 | 0.35 |
| GSTM1 = 0 | 1005 | 0.54 | 973 | 0.48 | 992 | 0.18 | 992 | 0.25 |
| GSTM1 = 1 | 861 | 0.63 | 847 | 0.54 | 850 | 0.28 | 850 | 0.29 |
| GSTM1 = 2 | 221 | 0.63 | 215 | 0.49 | 215 | 0.24 | 215 | 0.27 |
a All partial correlations are significant at p < 0.0001 and have been adjusted for age, gender, BMI (and race where appropriate).
We observed large differences in mean SPMA levels per ml urine across populations, even after adjusting for TNE, with African Americans having 38% higher levels (p = 1.4x10-5) and Japanese Americans having 38% lower levels (p = 2x10-13) than Whites. Similar results were obtained when the data were expressed as median SPMA levels. Given the variability in creatinine, lesser differences in SPMA were observed when adjusted for creatinine levels (Table 1). Still however, levels in Japanese remained significantly lower than levels in Whites.
In the GWAS analysis of SPMA we observed little evidence of inflation in the test statistic in the overall multiethnic sample (λ = 1.0; S1 Fig) or in any single ethnic group (0.97 ≤ λ’s ≤ 1.0). We detected associations at p<5x10-8 with 403 variants located between 24.2–24.4 Mb near the GSTT1 gene on chromosome 22q11 (S2 Fig) and 1 variant near the GSTM1 gene at 1p13 (S3 Fig; S1 Table). The highly significant association observed at 22q11 was explained by the GSTT1 deletion (n = 1,975, beta per allele = 2.06 pmol/ml, p = 6.0x10-107; Table 3). The r2 between the deletion and the associated variants (P<5x10-8) ranged from 0.02 to 0.43 in the multiethnic sample and no secondary signals (at p<1x10-3) were detected after conditioning on the deletion in the multiethnic sample or in any ethnic group (S4 Fig). The deletion allele, which is associated with lower SPMA levels, varies in frequency across populations from 0.40 in Latinos to 0.66 in Japanese (Table 3). We also did not detect any highly significant associations (p<1.7x10-7) with SNPs or indels among those without the GSTT1 deletion (n = 514 homozygotes) which suggests that alternate forms of functional variation in the region are likely to have only a minor impact on the regulation or activity of GSTT1. We also performed the analysis of the genetic data with SPMA levels expressed as pmol/mg creatinine; the results vary only marginally (S2 Table).
Table 3. SPMA levels by GST deletion genotype.
| SPMA (pmol/mL) | |||||||||||||
| GSTT1 deletion | African Americans | Native Hawaiians | Whites | Latinos | Japanese Americans | Total | P-hetb | ||||||
| n | Meana | n | Meana | n | Meana | n | Meana | n | Meana | n | Meana | ||
| All | 329 | 3.71 | 265 | 2.43 | 404 | 2.69 | 415 | 2.88 | 562 | 1.66 | 1975 | ||
| 0 | 68 | 1.56 | 69 | 1.03 | 68 | 1.09 | 73 | 1.04 | 246 | 0.9 | 524 | 1.1 | |
| 1 | 174 | 4.10 | 130 | 3.07 | 198 | 2.47 | 182 | 2.65 | 253 | 2.33 | 937 | 2.87 | |
| 2 | 87 | 5.89 | 66 | 4.01 | 138 | 4.59 | 160 | 4.79 | 63 | 3.53 | 514 | 4.56 | |
| Null Frequency | 0.47 | 0.51 | 0.41 | 0.40 | 0.66 | ||||||||
| Beta per allele | 1.84 | 1.97 | 2.07 | 1.98 | 2.18 | 2.06 | |||||||
| P-trendb | 8.0x10-14 | 4.9x10-18 | 8.8x10-25 | 2.8x10-23 | 1.2x10-36 | 6.0x10-107 | 0.328 | ||||||
| Variation Explained | 14.24% | 31.56% | 23.87% | 20.45% | 27.18% | 20.88% | |||||||
| SPMA (pmol/mL) | |||||||||||||
| GSTM1 deletion | African Americans | Native Hawaiians | Whites | Latinos | Japanese Americans | Total | P-hetb | ||||||
| n | Meana | n | Meana | n | Meana | n | Meana | n | Meana | n | Meana | ||
| All | 353 | 3.73 | 290 | 2.46 | 417 | 2.69 | 430 | 2.88 | 597 | 1.66 | 2087 | ||
| 0 | 103 | 3.2 | 185 | 2.15 | 220 | 2.33 | 196 | 2.58 | 301 | 1.41 | 1005 | 2.23 | |
| 1 | 168 | 3.84 | 91 | 3.2 | 158 | 3.10 | 192 | 3.06 | 252 | 1.97 | 861 | 2.95 | |
| 2 | 82 | 4.2 | 14 | 2.66 | 39 | 3.35 | 42 | 3.77 | 44 | 1.86 | 221 | 3.14 | |
| Null Frequency | 0.53 | 0.79 | 0.72 | 0.68 | 0.72 | ||||||||
| Beta per allele | 1.13 | 1.29 | 1.28 | 1.2 | 1.24 | 1.2 | |||||||
| P-trendb | 0.13 | 9.2x10-3 | 1.4x10-3 | 0.02 | 1.6x10-3 | 3.3x10-9 | 0.896 | ||||||
| Variation Explained | 0.20% | 1.44% | 1.34% | 1.46% | 2.40% | 1.26% | |||||||
a All Means are expressed as geometric least squares means of SPMA adjusted for age, gender, total nicotine equivalents, BMI and race (where appropriate).
b P-values for trend and heterogeneity have been adjusted for age, gender, total nicotine equivalents, BMI, PCs and race (where appropriate).
The second region of association was at 1p13 where only a single imputed variant (indel at position 110223001 bp; info score = 0.8) was found to be associated with SPMA at p<5x10-8 in the multiethnic sample (frequency range 0.27–0.47 across populations, beta = 0.81 pmol/mL per allele, p = 1.48x10-10; S3 Fig). This variant was correlated with the GSTM1 deletion polymorphism (r2 of 0.58 in the multiethnic sample), which was similarly associated with lower SPMA levels (n = 2,087, beta per allele = 1.20 pmol/mL, p = 3.3x10-9; Table 3). The indel was no longer significantly associated with SPMA after conditioning on the large deletion polymorphism (S5 Fig). The deletion allele, which is associated with lower SPMA levels, varies in frequency across populations from 0.53 in African Americans to 0.79 in Native Hawaiians. As with 22q11 and the GSTT1 deletion, we did not detect any significant associations (p<3.4x10-7) with SNPs or indels among those without the GSTM1 deletion (n = 221 homozygotes).
In ethnic-specific analyses, a cluster of ten highly correlated variants (r2 ≥0.8) were significant at p<5x10-8 within POU4F1-AS1 at chromosome 13q31 in Latinos. All variants were imputed (imputation quality, info scores ≥ 0.94) and are common in all five populations (freq>0.7); however, these SNPs were only associated with SPMA levels in Latinos (beta>0.62, p ≥ 1.85x10-8; beta>1.01 and p-value>0.23 in all other populations).
Given the importance of CYP2E1 in benzene metabolism and the previously reported associations with polymorphisms in CYP2E1 and benzene metabolite levels we also examined variation at this locus. We observed little evidence of an association with common alleles in this region (within 200 kb of CYP2E1). Through a literature review, we created a composite list of 13 SNPs reported to be associated with benzene metabolism; none of these SNPs were found to be associated with SPMA (p< 0.05; S3 Table). The results were similar among those with or without the GSTT1 deletion polymorphism (data not shown).
Combined, the baseline covariates age, sex, BMI, TNE, cigarettes per day, ethnicity and the first 10 principal components explained 37% of variability in SPMA. Ethnicity and principal components accounted for ~6% of the variability (Table 4). When adjusted for these baseline covariates, cigarettes per day and TNE were both highly associated with SPMA (p = 2.0x10-14 and p = 2.1x10-176, respectively), though cigarettes per day only explains ~2.5% of the variability in SPMA, whereas TNE explains 29.4%. In the multivariate model, the GSTT1 deletion accounted for an additional 20.9% of the variability in levels of SPMA in smokers, with the proportion explained ranging from 14.2% in African Americans to 31.6% in Native Hawaiians (phet = 0.33; Table 3). Although genome-wide significant, the contribution of the GSTM1 deletion was more modest, and could explain only 1.3% of the variation in the multiethnic sample (range across populations: 0.2–2.4%). Together, the GSTT1 and GSTM1 deletion polymorphisms explain ~22% of the variation in SPMA levels in this multiethnic sample of smokers, which ranges across ethnic groups from 14.4% in African Americans to 33.0% in Native Hawaiians (Table 3).
Table 4. Geometric least square means of SPMA by population and percent variation explained by smoking and GST genotypes.
| SPMA (pmol/mL) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | Overall Percent Variation Explained | African Americans | Native Hawaiians | Whites | Latinos | Japanese Americans | ||||||
| n | Meana | n | Meana | n | Meana | n | Meana | n | Meana | |||
| Sex + Age + BMI + TNEb + CPDb | 37.21% | 329 | 3.9 | 264 | 2.42 | 403 | 2.6 | 414 | 2.86 | 562 | 1.6 | |
| P-values | 4.2x10-7 | 4.2x10-7 | 0.38 | 0.38 | - | - | 0.26 | 0.26 | 6.3x10-13 | 6.3x10-13 | ||
| + GSTT1 deletion | 20.87% | 329 | 3.62 | 264 | 2.42 | 403 | 2.28 | 414 | 2.47 | 562 | 2.02 | |
| P-values | 5.7x10-12 | 5.7x10-12 | 0.39 | 0.39 | - | - | 0.22 | 0.22 | 0.05 | 0.05 | ||
| + GSTM1 deletion | 1.40% | 329 | 3.38 | 264 | 2.52 | 403 | 2.31 | 414 | 2.46 | 562 | 2.05 | |
| P-values | 1.1x10-8 | 1.1x10-8 | 0.21 | 0.21 | - | - | 0.32 | 0.32 | 0.04 | 0.04 | ||
a All Means are expressed as geometric least squares means of SPMA adjusted for age, gender, BMI, TNE & CPD.
b TNE,- total nicotine equivalents; CPD, cigarettes per day.
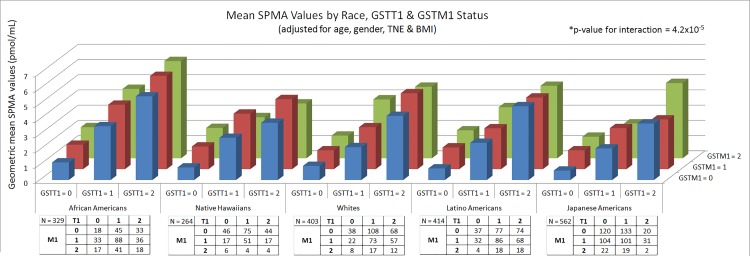
In examining the combined effects of the GSTT1 and GSTM1 deletions, SPMA values were lowest among Japanese Americans with null genotypes for both deletions (0.62 pmol/mL) and highest amongst African Americans who were wild-type (6.4 pmol/mL), a 10-fold difference (Fig 2). We observed modest evidence of a statistical interaction between the GSTT1 and GSTM1 deletions with SPMA levels (p = 1.2x10-5), though the interaction only explained 0.86% of the variability in SPMA. Overall, ~ 60% of the variability in SPMA could be accounted for by the covariates and both deletion polymorphisms.
Fig 2. GSTT1 and GSTM1 genotype and SPMA levels in each population.
Despite the highly significant association between the GSTT1 deletion polymorphism (and to a lesser degree the GSTM1 polymorphism) and SPMA levels, and the variability in the prevalence of these polymorphisms across populations, they could not account for the large ethnic differences in SPMA levels (Table 4, Fig 2). Compared to Whites, SPMA levels in African Americans remained higher (p<0.0001) while levels in Japanese remained lower (p = 0.04), with the magnitude of these differences not being substantially altered when adjusting for the GST deletions. SPMA levels in Latinos and Native Hawaiians were similar to those of Whites.
Discussion
As in our studies demonstrating statistically higher levels of TNE and total NNAL in African Americans and lower levels in Japanese Americans than in Whites [16, 17], we report here that levels of SPMA, an established and specific biomarker of uptake of the volatile human carcinogen benzene, are significantly higher in African American smokers than in White smokers and significantly lower in Japanese American smokers than in White smokers, even after correction for the effects of variants in the GSTT1 and GSTM1 genes. While benzene is well established as a cause of leukemia, it is not generally considered a cause of lung cancer in humans. However, it does cause lung tumors (as well as tumors at other sites) in mice, and some studies indicate its possible involvement in human lung cancer etiology [20]. Perhaps more importantly, benzene uptake as indicated by urinary SPMA, could be a biomarker for other volatile carcinogens in cigarette smoke, such as 1,3-butadiene, which causes lung tumors in mice and has been evaluated as an important carcinogen in cigarette smoke [49–51]. Thus, a single analysis of SPMA could potentially replace multiple analyses of other volatile carcinogen metabolite biomarkers.
The U.S. National Toxicology Program conducted two year carcinogenesis studies of benzene in F-344 rats and B6C3F1 mice. The doses used were 0, 25, 50, or 100 mg/kg body weight of benzene, administered by gavage in corn oil 5 days per week for 103 weeks. Significant incidences of tumors compared to vehicle controls were observed at multiple sites including the hematopoietic system in both rats and mice. Among these, lung tumors were observed only in mice. Significantly increased incidences of alveolar/bronchiolar carcinomas and adenomas were reported, mainly in the mice treated with the highest dose [52]. While statistically significant, the carcinogenic effect of benzene to the rodent lung is far weaker than that of NNK or NNAL [53].
A major finding of the GWAS presented here was the highly significant association of the GSTT1 deletion on chromosome 22q11 and SPMA levels, which explained up to 31.6% of the variation in SPMA levels, depending on the ethnic group. SPMA is a specific biomarker of benzene uptake, formed by glutathione detoxification of the requisite intermediate benzene oxide, followed by normal metabolic processing of the glutathione conjugate (Fig 1). While this effect of genotype has been noted before, our study is the largest and most definitive [31–40]. The stronger effect of GSTT1 than GSTM1 deletion observed here is consistent with our metabolic studies which demonstrate that GSTT1 is a better catalyst of benzene oxide conjugation than GSTM1 [54]. The size of our study allowed us to analyze ethnic differences in SPMA levels correcting for each genotype. As summarized in Table 4 and Fig 2, even after this correction, SPMA levels were significantly higher in African-Americans than in Whites and significantly lower in Japanese Americans than in Whites.
Ethnic differences in GSTT1 have been observed previously [55]. The prevalence of the null genotype was 64.4% in Chinese, 60.2% in Koreans, 21.8% in African Americans, 20.4% in Caucasians, and 9.7% in Mexican Americans. These results are generally consistent with ours (Table 3) in which the highest null frequency was observed in Japanese Americans (66%) and Native Hawaiians (51%).
The strong effect of GSTT1 genotype on SPMA levels presents a potential problem in smaller studies interpreting this biomarker as related to benzene uptake. GSTT1 catalysis of the reaction between benzene oxide and glutathione is a detoxification mechanism for benzene, as benzene oxide is widely recognized as a significant and critical intermediate in benzene carcinogenesis [56, 57]. SPMA levels are affected both by benzene exposure and GSTT1 genotype. Higher benzene exposure leads to higher levels of urinary SPMA, but GSTT1 null status, which should increase risk for benzene induced toxicity and carcinogenicity (because more benzene oxide will be available to express its deleterious cellular effects) will decrease levels of urinary SPMA, as clearly seen in this study. This conundrum could be a problem in smaller studies or those in which genotyping information is not available. An alternate measure of benzene exposure is urinary benzene, which compares well to SPMA in specificity to benzene exposure, but is more difficult to quantify because of its volatility [30].
In this multiethnic sample, which is modest in size for a GWAS, we had limited statistical power to detect a genetic factor that accounts for a small fraction of the variation (R2) in SPMA levels. For example, in the entire sample of 2,239 smokers, we had 80% power to detect an R2 of 1.8%, at p<5x10-8 (allowing for multiple comparisons). The ethnic group specific sample sizes ranged from 311–674 participants so that detectable R2 values in any one ethnic group ranged from 6% to 11%. Revealing additional common variants that convey modest effects or less common alleles <5% that may be ethnic specific and which may contribute to population differences in SPMA levels will require substantially larger studies in these racial/ethnic populations.
There were some other limitations to our study. Slightly different urine collection methods–overnight for most of the Native Hawaiians, Whites and Japanese Americans versus first morning for the other two groups–were used. It is possible that these differences might have affected the levels of SPMA in these two groups relative to the others. However, SPMA values did not differ according to collection method when comparing 96 Japanese collected in Los Angeles using first morning urines to the Japanese samples (N = 578) from Hawaii measured from overnight urine collection. In addition, levels of SPMA strongly correlated with those of 3-HPMA and HMPMA (Table 2), yet 3-HPMA and HMPMA were as high in Native Hawaiians as in African Americans [19], while SPMA was significantly lower in Native Hawaiians than in African Americans. Another limitation relates to expressing the results per ml urine rather than per mg creatinine. This mode of expression, which can introduce unwanted variability related to extent of hydration, was necessary because of the wide differences in creatinine seen among some of the ethnic groups collected in the same location (e.g., between African Americans and Latinos collected in Los Angeles, or between Whites and Japanese Americans, collected primarily in Hawaii) which could not be explained by differences in collection method. Twenty-four hour urine samples would have been the preferable method of comparing SPMA levels, but these were not available.
In summary, the results of this study demonstrate that uptake in smokers of the volatile cigarette smoke constituent benzene, as measured by the specific biomarker SPMA, is highest in African Americans, intermediate in Whites, and lowest in Japanese Americans, consistent with their previously determined levels of TNE and total NNAL. Our GWAS convincingly demonstrated the strong effect of GSTT1 genotype on urinary levels of SPMA, but this did not affect our conclusion regarding ethnic differences in benzene uptake among the MEC smokers.
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(CSV)
(XLSX)
(CSV)
Acknowledgments
We thank Bob Carlson for editorial assistance and Dr. Maarit Tiirikainen for the GSTT1 and GSTM1 genotype analysis.
Data Availability
Data will be available from the dbGAP repository. Once data has been processed, the authors will post a comment to the published article with the accession numbers necessary to access the data.
Funding Statement
This work was supported by the National Institutes of Health [5P01CA138338-05 to S.S.H., 5U01CA164973-02 to L.L.M.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006;354(4):333–42. Epub 2006/01/27. doi: 354/4/333 [pii] 10.1056/NEJMoa033250 . [DOI] [PubMed] [Google Scholar]
- 2.Gadgeel SM, Severson RK, Kau Y, Graff J, Weiss LK, Kalemkerian GP. Impact of race in lung cancer: analysis of temporal trends from a surveillance, epidemiology, and end results database. Chest. 2001;120(1):55–63. [DOI] [PubMed] [Google Scholar]
- 3.Harris RE, Zang EA, Anderson JI, Wynder EL. Race and sex differences in lung cancer risk associated with cigarette smoking. Int J Epidemiol. 1993;22:592–9. [DOI] [PubMed] [Google Scholar]
- 4.Hinds MW, Stemmermann GN, Yang HY, Kolonel LN, Lee J, Wegner E. Differences in lung cancer risk from smoking among Japanese, Chinese and Hawaiian women in Hawaii. Int J Cancer. 1981;27(3):297–302. [DOI] [PubMed] [Google Scholar]
- 5.Le Marchand L, Wilkens LR, Kolonel LN. Ethnic differences in the lung cancer risk associated with smoking. Cancer Epidemiol Biomarkers Prev. 1992;1(2):103–7. Epub 1992/01/01. . [PubMed] [Google Scholar]
- 6.Schwartz AG, Swanson GM. Lung carcinoma in African Americans and whites. A population-based study in metropolitan Detroit, Michigan. Cancer. 1997;79(1):45–52. [DOI] [PubMed] [Google Scholar]
- 7.Sobue T, Yamamoto S, Hara M, Sasazuki S, Sasaki S, Tsugane S. Cigarette smoking and subsequent risk of lung cancer by histologic type in middle-aged Japanese men and women: the JPHC study. Int J Cancer. 2002;99(2):245–51. [DOI] [PubMed] [Google Scholar]
- 8.Stellman SD, Takezaki T, Wang L, Chen Y, Citron ML, Djordjevic MV, et al. Smoking and lung cancer risk in American and Japanese men: An international case-control study. Cancer Epidemiol Biomarkers Prev. 2001;10:1193–9. [PubMed] [Google Scholar]
- 9.Blot WJ, Cohen SS, Aldrich M, McLaughlin JK, Hargreaves MK, Signorello LB. Lung cancer risk among smokers of menthol cigarettes. J Natl Cancer Inst. 2011;103(10):810–6. 10.1093/jnci/djr102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LS, Saccone NL, Culverhouse RC, Bracci PM, Chen CH, Dueker N, et al. Smoking and genetic risk variation across populations of European, Asian, and African American ancestry—a meta-analysis of chromosome 15q25. Genet Epidemiol. 2012;36(4):340–51. 10.1002/gepi.21627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhan P, Wang Q, Qian Q, Wei SZ, Yu LK. CYP1A1 MspI and exon7 gene polymorphisms and lung cancer risk: an updated meta-analysis and review. J Exp Clin Cancer Res. 2011;30:99 10.1186/1756-9966-30-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B, Qiu LX, Li Y, Xu W, Wang XL, Zhao WH, et al. The CYP1B1 Leu432Val polymorphism contributes to lung cancer risk: evidence from 6501 subjects. Lung Cancer. 2010;70(3):247–52. 10.1016/j.lungcan.2010.03.011 [DOI] [PubMed] [Google Scholar]
- 13.Carpenter CL, Yu MC, London SJ. Dietary isothiocyanates, glutathione S-transferase M1 (GSTM1), and lung cancer risk in African Americans and Caucasians from Los Angeles County, California. Nutr Cancer. 2009;61(4):492–9. 10.1080/01635580902752270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiyohara C, Takayama K, Nakanishi Y. Lung cancer susceptibility and hOGG1 ser326Cys polymorphism: a meta-analysis. Cancers (Basel). 2010;2(4):1813–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truong T, Sauter W, McKay JD, Hosgood HD III, Gallagher C, Amos CI, et al. International Lung Cancer Consortium: coordinated association study of 10 potential lung cancer susceptibility variants. Carcinogenesis. 2010;31(4):625–33. 10.1093/carcin/bgq001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy SE, Park SS, Thompson EF, Wilkens LR, Patel Y, Stram DO, et al. Nicotine N-glucuronidation relative to N-oxidation and C-oxidation and UGT2B10 genotype in five ethnic/racial groups. Carcinogenesis. 2014;35(11):2526–33. Epub 2014/09/23. 10.1093/carcin/bgu191 bgu191 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SL, Carmella SG, Ming X, Vielguth E, Stram DO, Le Marchand L, et al. Variation in levels of the lung carcinogen NNAL and its glucuronides in the urine of cigarette smokers from five ethnic groups with differing risks for lung cancer. Cancer Epidemiol Biomarkers Prev. 2015;24(3):561–9. Epub 2014/12/30. 10.1158/1055-9965.EPI-14-1054 1055-9965.EPI-14-1054 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel YM, Stram DO, Wilkens LR, Park SS, Henderson BE, Le Marchand L, et al. The contribution of common genetic variation to nicotine and cotinine glucuronidation in multiple ethnic/racial populations. Cancer Epidemiol Biomarkers Prev. 2014. Epub 2014/10/09. doi: 1055-9965.EPI-14-0815 [pii] 10.1158/1055-9965.EPI-14-0815 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SL, Carmella SG, Chen M, Patel Y, Stram DO, Haiman CA, et al. Mercapturic acids derived from the toxicants acrolein and crotonaldehyde in the urine of cigarette smokers from five ethnic groups with differing risks for lung cancer. PLoS One. 2015;10(6):e0124841 10.1371/journal.pone.0124841 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.International Agency for Research on Cancer. A Review of Human Carcinogens: Chemical Agents and Related Occupations IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol 100F Lyon, FR: IARC; 2012. p. 249–94. [PMC free article] [PubMed] [Google Scholar]
- 21.National Toxicology Program. 12th Report on Carcinogens. Washington DC: US Department of Health and Human Services; 2011. 2011. [Google Scholar]
- 22.Roemer E, Stabbert R, Rustemeier K, Veltel DJ, Meisgen TJ, Reininghaus W, et al. Chemical composition, cytotoxicity and mutagenicity of smoke from US commercial and reference cigarettes smoked under two sets of machine smoking conditions. Toxicology. 2004;195(1):31–52. [DOI] [PubMed] [Google Scholar]
- 23.Wallace L. Environmental exposure to benzene: an update. Environ Health Perspect. 1996;104 Suppl 6:1129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carmella SG, Chen M, Han S, Briggs A, Jensen J, Hatsukami DK, et al. Effects of smoking cessation on eight urinary tobacco carcinogen and toxicant biomarkers. Chem Res Toxicol. 2009;22(4):734–41. 10.1021/tx800479s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams PR. An analysis of violations of Osha's (1987) occupational exposure to benzene standard. J Toxicol Environ Health B Crit Rev. 2014;17(5):259–83. 10.1080/10937404.2014.924452 . [DOI] [PubMed] [Google Scholar]
- 26.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91(14):1194–210. Epub 1999/07/21. . [DOI] [PubMed] [Google Scholar]
- 27.Hecht SS. Human urinary carcinogen metabolites: biomarkers for investigating tobacco and cancer. Carcinogenesis. 2002;23:907–22. [DOI] [PubMed] [Google Scholar]
- 28.Hecht SS, Yuan J- M, Hatsukami DK. Applying tobacco carcinogen and toxicant biomarkers in product regulation and cancer prevention. Chem Res Toxicol. 2010;23:1001–8. 10.1021/tx100056m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dougherty D, Garte S, Barchowsky A, Zmuda J, Taioli E. NQO1, MPO, CYP2E1, GSTT1 and GSTM1 polymorphisms and biological effects of benzene exposure—a literature review. Toxicol Lett. 2008;182(1–3):7–17. 10.1016/j.toxlet.2008.09.008 [DOI] [PubMed] [Google Scholar]
- 30.Hoet P, De Smedt E, Ferrari M, Imbriani M, Maestri L, Negri S, et al. Evaluation of urinary biomarkers of exposure to benzene: correlation with blood benzene and influence of confounding factors. Int Arch Occup Environ Health. 2009;82(8):985–95. 10.1007/s00420-008-0381-6 . [DOI] [PubMed] [Google Scholar]
- 31.Scheepers PT, Coggon D, Knudsen LE, Anzion R, Autrup H, Bogovski S, et al. BIOMarkers for occupational diesel exhaust exposure monitoring (BIOMODEM)—a study in underground mining. Toxicol Lett. 2002;134(1–3):305–17. [DOI] [PubMed] [Google Scholar]
- 32.Sorensen M, Poole J, Autrup H, Muzyka V, Jensen A, Loft S, et al. Benzene exposure assessed by metabolite excretion in Estonian oil shale mineworkers: influence of glutathione S-transferase polymorphisms. Cancer Epidemiol Biomarkers Prev. 2004;13(11 Pt 1):1729–35. [PubMed] [Google Scholar]
- 33.Qu Q, Shore R, Li G, Su L, Jin X, Melikian AA, et al. Biomarkers of benzene: urinary metabolites in relation to individual genotype and personal exposure. Chem BiolInteract. 2005;153–154:85–95. [DOI] [PubMed] [Google Scholar]
- 34.Avogbe PH, Ayi-Fanou L, Autrup H, Loft S, Fayomi B, Sanni A, et al. Ultrafine particulate matter and high-level benzene urban air pollution in relation to oxidative DNA damage. Carcinogenesis. 2005;26(3):613–20. [DOI] [PubMed] [Google Scholar]
- 35.Kim S, Lan Q, Waidyanatha S, Chanock S, Johnson BA, Vermeulen R, et al. Genetic polymorphisms and benzene metabolism in humans exposed to a wide range of air concentrations. Pharmacogenet Genomics. 2007;17(10):789–801. [DOI] [PubMed] [Google Scholar]
- 36.Lin LC, Chen WJ, Chiung YM, Shih TS, Liao PC. Association between GST genetic polymorphism and dose-related production of urinary benzene metabolite markers, trans, trans-muconic acid and S-phenylmercapturic acid. Cancer Epidemiol Biomarkers Prev. 2008;17(6):1460–9. 10.1158/1055-9965.EPI-08-0160 [DOI] [PubMed] [Google Scholar]
- 37.Manini P, De Palma G, Andreoli R, Mozzoni P, Poli D, Goldoni M, et al. Occupational exposure to low levels of benzene: Biomarkers of exposure and nucleic acid oxidation and their modulation by polymorphic xenobiotic metabolizing enzymes. Toxicol Lett. 2010;193(3):229–35. 10.1016/j.toxlet.2010.01.013 [DOI] [PubMed] [Google Scholar]
- 38.Mansi A, Bruni R, Capone P, Paci E, Pigini D, Simeoni C, et al. Low occupational exposure to benzene in a petrochemical plant: modulating effect of genetic polymorphisms and smoking habit on the urinary t,t-MA/SPMA ratio. Toxicol Lett. 2012;213(1):57–62. 10.1016/j.toxlet.2011.02.001 [DOI] [PubMed] [Google Scholar]
- 39.Carrieri M, Bartolucci GB, Scapellato ML, Spatari G, Sapienza D, Soleo L, et al. Influence of glutathione S-transferases polymorphisms on biological monitoring of exposure to low doses of benzene. Toxicol Lett. 2012;213(1):63–8. 10.1016/j.toxlet.2011.11.031 [DOI] [PubMed] [Google Scholar]
- 40.Egner PA, Chen JG, Zarth AT, Ng D, Wang J, Kensler KH, et al. Rapid and sustainable detoxication of airborne pollutants by broccoli sprout beverage: results of a randomized clinical trial in china. Cancer Prev Res (Phila). 2014. Epub 2014/06/11. doi: canprevres.0103.2014 [pii] 1940-6207.CAPR-14-0103 [pii] 10.1158/1940-6207.CAPR-14-0103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, Pike MC, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151(4):346–57. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carmella SG, Chen M, Zarth A, Hecht SS. High throughput liquid chromatography-tandem mass spectrometry assay for mercapturic acids of acrolein and crotonaldehyde in cigarette smokers' urine. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;935:36–40. Epub 2013/08/13. 10.1016/j.jchromb.2013.07.004 S1570-0232(13)00371-1 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Liang Q, Mendes P, Sarkar M. Is 24h nicotine equivalents a surrogate for smoke exposure based on its relationship with other biomarkers of exposure? Biomarkers. 2011;16(2):144–54. Epub 2011/02/18. 10.3109/1354750X.2010.536257 . [DOI] [PubMed] [Google Scholar]
- 44.Carmella SG, Ming X, Olvera N, Brookmeyer C, Yoder A, Hecht SS. High throughput liquid and gas chromatography-tandem mass spectrometry assays for tobacco-specific nitrosamine and polycyclic aromatic hydrocarbon metabolites associated with lung cancer in smokers. Chem Res Toxicol. 2013;26(8):1209–17. Epub 2013/07/11. 10.1021/tx400121n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–81. 10.1038/nmeth.1785 [DOI] [PubMed] [Google Scholar]
- 46.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529 Epub 2009/06/23. 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. Epub 2010/12/21. 10.1016/j.ajhg.2010.11.011 S0002-9297(10)00598-7 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. . [DOI] [PubMed] [Google Scholar]
- 49.Fowles J, Dybing E. Application of toxicological risk assessment principles to the chemical constituents of cigarette smoke. Tob Control. 2003;12(4):424–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burns DM, Dybing E, Gray N, Hecht S, Anderson C, Sanner T, et al. Mandated lowering of toxicants in cigarette smoke: a description of the World Health Organization TobReg proposal. Tob Control. 2008;17(2):132–41. 10.1136/tc.2007.024158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.International Agency for Research on Cancer. A review of human carcinogens: chemical agents and related occupations IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol 100F Lyon, FR: IARC; 2012. p. 309–38. [PMC free article] [PubMed] [Google Scholar]
- 52.National Toxicology Program. NTP toxicology and carcinogenesis studies of benzene (CAS No. 71-43-2) in F344/N rats and B6C3F1 mice (Gavage Studies). Natl Toxicol Program Tech Rep Ser. 1986;289:1–277. . [PubMed] [Google Scholar]
- 53.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem ResToxicol. 1998;11:559–603. [DOI] [PubMed] [Google Scholar]
- 54.Zarth AT, Murphy SE, Hecht SS. Benzene oxide is a substrate for glutathione S-transferases. Chem Biol Interact. 2015;242:390–5. 10.1016/j.cbi.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson HH, Wiencke JK, Christiani DC, Cheng TJ, Zuo ZF, Schwartz BS, et al. Ethnic differences in the prevalence of the homozygous deleted genotype of glutathione S-transferase theta. Carcinogenesis. 1995;16(5):1243–5. . [DOI] [PubMed] [Google Scholar]
- 56.Monks TJ, Butterworth M, Lau SS. The fate of benzene-oxide. Chem Biol Interact. 2010;184(1–2):201–6. 10.1016/j.cbi.2009.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McHale CM, Zhang L, Smith MT. Current understanding of the mechanism of benzene-induced leukemia in humans: implications for risk assessment. Carcinogenesis. 2012;33(2):240–52. 10.1093/carcin/bgr297 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(CSV)
(XLSX)
(CSV)
Data Availability Statement
Data will be available from the dbGAP repository. Once data has been processed, the authors will post a comment to the published article with the accession numbers necessary to access the data.