Abstract
Previous studies showed that the yellow seed color gene of a yellow mustard was located on the A09 chromosome. In this study, the sequences of the molecular markers linked to the yellow seed color gene were analyzed, the gene was primarily mapped to an interval of 23.304 to 29.402M. Twenty genes and eight markers’ sequences in this region were selected to design the IP and SCAR primers. These primers were used to screen a BC8S1 population consisting of 1256 individuals. As a result, five IP and five SCAR markers were successfully developed. IP4 and Y1 were located on either side of the yellow seed color gene at a distance of 0.1 and 0.3 cM, respectively. IP1, IP2 and IP3 derived from Bra036827, Bra036828, Bra036829 separately, co-segregated with the target gene. BLAST analysis indicated that the sequences of newly developed markers showed good collinearity with those of the A09 chromosome, and that the target gene might exist between 27.079 and 27.616M. In light of annotations of the genes in this region, only Bra036828 is associated with flavonoid biosynthesis. This gene has high similarity with the TRANSPARENT TESTA6 gene, Bra036828 was hence identified as being the gene possibly responsible for yellow seed color, in our research.
Keywords: Brassica juncea, yellow seed color gene, fine mapping, IP and SCAR
Introduction
Rapeseed is one of the most important oil crops in China. The total growing area of rapeseed exceeds 100 million acres in China every year, but it still cannot satisfy the demands of the Chinese market. Every year the Chinese government imports millions of tons of vegetable oil from other countries (Wang 2006). So the main goal of rapeseed breeding in China is to improve oil production per unit area. Oil content is one component of oil production, many studies have shown that the oil content of yellow seeded rapeseed is higher than that of black seeded rapeseed with the same genetic background (Abraham and Bhatia 1986). Therefore, yellow seed breeding is considered as one effective way to improve oil content. However, few studies of pure yellow seeded materials in B. napus have been reported, due to limited sources of yellow seeded rapeseed germplasm in B. napus worldwide. B. juncea, an allotetraploid species, possesses some pure yellow seed types. A yellow mustard is the main rapeseed variety in the northwest of China. The yellow seed color of this mustard was controlled by a single recessive gene, which was mapped to the A09 chromosome in B. rapa (Huang et al. 2012).
Some studies have been conducted on the inheritance and mapping of the yellow seed color genes in Brassica (Rahman et al. 2007, Xiao et al. 2007). Xiao et al. (2012) identified 17 AFLP and SSR markers linked to a yellow seed color gene from “Dahuang”, which is a B. rapa landrace in Qinghai-Tibetan plateau, China. Liu et al. (2005) constructed a genetic map around the yellow seed color gene in a resynthesized B. napus, lines No. 2127-17, the yellow seed color gene was mapped in a region of 7.0 cM. However, all of the yellow seed color genes above were derived from the A genome in Brassica. Research about yellow seed color genes in the B genome can be rarely found. Padmaja et al. (2005) studied a B. juncea yellow seeded line, the yellow seed color was controlled by two independent loci (BjSc1 and BjSc2), the two yellow seed color genes were mapped to two linkage groups, LG A9 and B3, respectively.
Before the publication of the B. rapa genome sequence, gene mapping and cloning in Brassica were mainly based on the published genome of Arabidopsis, which is a member of the Brassica family and has been serving as a model species. High levels of synteny and remarkably conserved genome structure have been found between Arabidopsis and Brassica genomes (Mun et al. 2009). In Arabidopsis, there are some TT (TRANSPARENT TESTA) or TTG (TRANSPARENT TESTA GLABRA) genes that are involved in seed coat pigmentation, a process involving flavonoid biosynthesis (Debeaujon et al. 2001, Routaboul et al. 2006). It is suggested that the information of these TT or TTG genes is helpful for studying yellow seed in Brassica. TTG1 and TT8 in B. rapa, TT10 in the eight Brassica lines that have been cloned successfully (Li et al. 2012, Zhang et al. 2009, 2013).
Thanks to the power of next generation sequencing technology, the B. rapa genome has been completely sequenced and published in the public domain (Wang et al. 2011). The sequence information can be employed freely in developing markers linked to genes of interest, and in fine mapping or cloning target genes in Brassica. Developing Intron Polymorphism (IP) markers is a very effective method for gene mapping, which has already exhibited its powerful application specifically in narrowing the chromosome region of a target gene. Lei et al. (2007) developed a dominant amplified consensus genetic marker (ACGM, equivalent to IP marker), which was more closely linked to BnMs2, and this marker reduced the candidate chromosome interval. Ban et al. (2013) developed three IP markers based on the sequences of Arabidopsis and B. rapa to narrow down the mapping region of the yellow seed color gene.
In this paper, we studied a yellow seeded B. juncea line popular in northern Shaanxi, China. A previous study (Huang et al. 2012) reported that the yellow seed color gene was located on the A09 chromosome in B. rapa. In order to fine map the yellow seed color gene, we made use of the genome sequences of B. rapa to develop the IP and SCAR markers. We then fine mapped the yellow seed color gene and predicted the possible genes responsible for yellow seed color. This study will provide a useful clue for cloning the yellow seed color gene.
Materials and Methods
Plant materials and population construction
A BC8S1 population consisting of 1256 individuals derived from a yellow seeded landrace ‘Wuqi mustard’ and a brown seeded landrace ‘Wugong mustard’ was developed for gene mapping. The ‘Wuqi mustard” was used as the recurrent parent to cross with the brown seeded individuals of the BC population in every generation. Finally, a brown seeded plant in the BC8 population was selected for selfing to produce a BC8S1 population. Every individual in BC8S1 was selfed to produce BC8S2 seeds and each BC8S2 line was sown in field to produce BC8S3 seeds. The genotypes underlying seed coat color of BC8S2 individuals were determined according to the phenotypes of the BC8S3 seeds. Similarly, the genotypes of every BC8S1 individual were inferred. Three genotypes in the BC8S1 population (homozygous brown seeded, heterozygous brown seeded and yellow seeded plants) were found and distinguished. All of the field experiments were carried out in the field of Northwest A&F University, Yangling, Shaanxi, China.
DNA extraction and bulked segregant analysis
Genomic DNA was extracted from young leaves of BC8S1 individuals at the seedling stage by the CTAB method (Doyle and Doyle 1990). Equal quantities of DNA from twelve yellow seeded BC8S1 plants and twelve brown seeded BC8S1 plants were pooled to form the yellow bulk (BY) and brown bulk (BB), respectively. The final DNA concentration was adjusted to 50 ng/μl.
Gene location analysis on the A genome
Huang et al. (2012) identified 23 SSR and AFLP markers linked to the yellow seed color gene, which was mapped to the A09 chromosome in B. rapa. In our research, we first used the markers identified by Huang et al. (2012) to screen a partial mapping population of 96 BC8S1 individuals. The AFLPs and SSRs amplifications were performed as described by Vos et al. (1995) and Lowe et al. (2002), respectively. The PCR products were separated on a 6% denaturing polyacrylamide gel. Silver staining was carried out using the method reported by Yi et al. (2006). Mapmaker 3.0 software (Lander et al. 1987, Lincoln et al. 1992) was used to analyze the linkage between markers and the gene. Secondly we sequenced AFLP and SSR markers used in this study and BLAST searched the B. rapa genome (http://brassicadb.org/brad, version 1.5).
Development of IP and SCAR markers linked to the yellow seed color gene
Through analyzing previous markers’ sequences, we found that the gene was located between 23.304 and 29.402M on the A09 chromosome in B. rapa (Fig. 1 and Table 1). We randomly selected genes in this region and designed primers according to the sequences of these genes. At the same time, we also used the markers’ sequences of this region published on the website: http://brassicadb.org/brad to develop new SCAR markers. Finally, we selected 20 genes and 8 markers’ sequences to design 28 pairs of primers. These primers were used to screen a small segregated population comprised of 12 yellow seeded individuals (yy) and 12 homozygous brown seeded plants (YY) in BC8S1 population. The PCR products were separated on a 6% denaturing polyacrylamide gel to test whether or not there was polymorphism between the phenotypes. The specific fragments that showed reproducible polymorphism between brown seeded and yellow seeded individuals were regarded as markers linked to the yellow seed color gene.
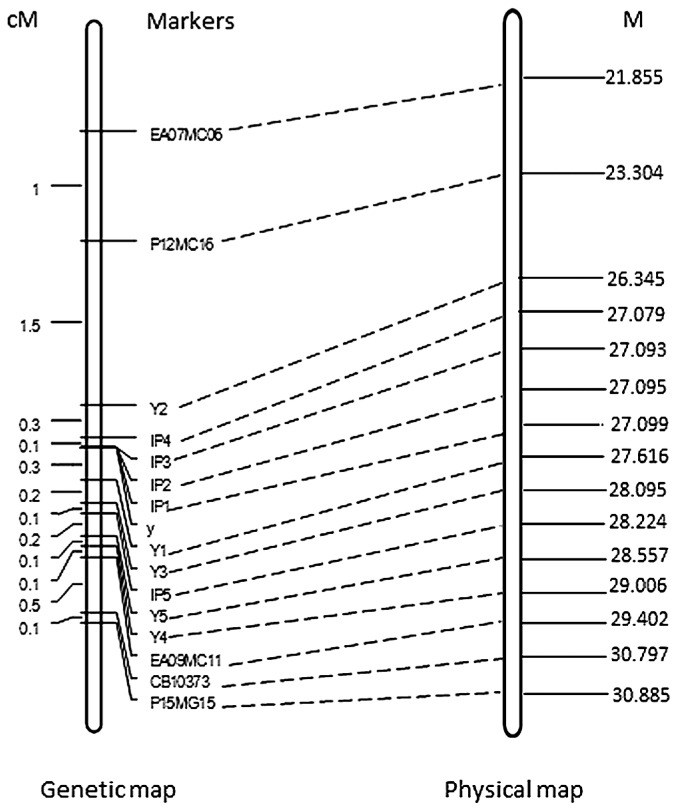
Fig. 1.
The left map is a genetic map surrounding the yellow seed color gene (y) from the BC8S1 population; the right map is a partial physical map of the A09 chromosome of B. rapa showing the homologues of mapped markers sequences. Dotted lines indicate the relationship of the two maps.
Table 1.
Characterization of molecular markers linked to yellow seed color gene
| Markers | Sequences (5′-3′) | Homologous B. rapa gene and E value | Locations on A09 |
|---|---|---|---|
| Y1 | TAAAGGGGTGGACAATAACA/GGTCAGAAGTGTTACGGGT | Bra006909, 2e-33 | 27616367..27616462 |
| Y2 | TCTACCCCATAACTGCATTC/CAGCTAATGAAGCCAAAACT | Bra036036, 1e-72 | 26345808..26345945 |
| Y3 | GGTGGCGTATCCGTAAAGGTAGA/CGCCGTCGCTGCTCCACT | Bra006970, 1e-67 | 28095119..28095357 |
| Y4 | TCCCGTATCAATGGCGTAACAG/CGATGGTGACATTATTGTGGCG | Bra007133, 3e-08 | 29006078..29006215 |
| Y5 | TCAAGCTACTACCTTTCAAGC/TTGACTCACTTCGAGTCTGA | Bra007049, 2e-45 | 28557476..28557567 |
| IP1 | GGCTTAAGAGGGGAAACGAG/ACCGAACCAGTGATGAGTCC | Bra036827,7e-47 | 27099538..27099895 |
| IP2 | CTTACCTCCTGAGGAGAAACTCA/CCGTGAGTAGTCTCTGTTTC | Bra036828, e-38 | 27095848..27096349 |
| IP3 | GAGGCTGTCACTCTCTTCGG/CGAGGAGCTGAGACAAAACC | Bra036829, 1e-63 | 27093092..27093399 |
| IP4 | ATCAAATGGAAACGACCTGC/CCAACGGATTTTGCTTGTTT | Bra036832, 4e-74 | 27079608..27079959 |
| IP5 | GAATCAAGTGCTAAGAGAGATGGTC/CATCTGAACCATCATATGCGAAC | Bra006991, 5e-25 | 28224201-28224487 |
| CB10373 | CGGTCAGATTCCAACAGA/GCCATCTCAGAGACGACA | Bra007453, 2e-65 | 30797156..30797514 |
| EA09MC11 | GACTGCGTACCAATTCACA/GATGAGTCCTGAGTAACCC | Bra007194, 2e-32 | 29402040..29402207 |
| EA07MC06 | GACTGCGTACCAATTCATC/GATGAGTCCTGAGTAACTT | Bra032392, 7e-56 | 21855926..21856106 |
| P15MG15 | GACTGCGTACATGCAGACA/GATGAGTCCTGAGTAAGGC | Bra007469, 7e-71 | 30885797..30885935 |
| P12MC16 | GACTGCGTACATGCAGTGA/GATGAGTCCTGAGTAACGG | Bra032885, 2e-55 | 23304264..23304372 |
Linkage analysis
The newly developed markers in this experiment and the previously identified markers (Huang et al. 2012) were used to amplify BC8S1 populations (1256 individuals), the results were analyzed using MAPMAKER/EXP 3.0 program (Lander et al. 1987, Lincoln et al. 1992). A minimum LOD score of 3.0 was used for map construction. Map distances were calculated using Kosambi’s (1943) mapping function.
Construction of the physical map around the yellow seed color gene
Mapdraw 2.9 (Liu et al. 2003) was used to construct a high resolution genetic map around the target gene. All of the markers’ sequences were BLAST analyzed against the B. rapa genome (http://brassicadb.org/brad, version 1.5). The collinearity between the sequences of the genetic markers and those of the A09 chromosome was compared using the BLAST tool found on the website: http://brassicadb.org/brad. All of the genes’ annotations within the mapping region were retrieved from the publicly accessible Brassica genetic database (http://brassicadb.org/brad). The genes related to flavonoid biosynthesis were selected for assumption as the possible genes responsible for yellow seed color in B. juncea.
Results
Genetics of yellow seed color in B. juncea
There were 1256 individuals in the BC8S1 population, which included 305 homozygous brown seeded individuals, 661 heterozygous brown seeded and 290 yellow seeded plants. This segregation of 3 genotypes was consistent with the expected ratio of 1:2:1 (χ2 = 3.83, and p > 0.05), indicating that yellow seed color is controlled by a single Mendelian recessive gene.
Preliminary analysis of the gene’s location on the A genome
In order to confirm the primary location of the yellow seed color gene on the A genome, five molecular markers (P15MG15, CB10373, EA09MC11, EA07MC06 and P12MC16) located on the either side of gene (Huang et al. 2012) were sequenced and used to screen 96 plants from the BC8S1 population. The results showed that P12MC16 and EA09MC11 were the two closest markers linked to the yellow seed color gene. Through comparing the markers’ sequences with the B. rapa genome, these five markers’ sequences showed good collinearity with the A09 chromosome of B. rapa (Fig. 1). The homologues of P12MC16 and EA09MC11 were located between 23.304 and 29.402M on the A09 chromosome. Therefore, we primarily justified that the gene responsible for yellow seed color might also exist between 23.304 and 29.402M on the A09 chromosome.
Development of IP and SCAR markers linked to the yellow seed color gene
28 pairs of primers were designed according to the sequences of the region from 23.304 to 29.402M on the A09 chromosome. Five pairs of primers IP1, IP2, IP3, IP4 and IP5 derived from Bra036827, Bra036828, Bra036829, Bra036832 and Bra006991, respectively, could show polymorphisms between 12 yellow seeded individuals (yy) and 12 homozygous brown seeded plants (YY). All of them were co-dominant markers that could distinguish the heterozygous brown seeded (Yy) and homozygous brown seeded plants (YY) (Fig. 2). Five pairs of primers Y1, Y2, Y3, Y4 and Y5 derived from BrID10539, BrID10955, 8C0462, BRMS016 and BrID10541 respectively (http://brassicadb.org/brad) could also amplify the polymorphic bands in the small population comprised of 12 yellow seeded individuals (yy) and 12 homozygous brown seeded plants (YY). These five pairs of primers were verified as the SCAR markers linked to the yellow seed color gene. All of the newly developed markers’ information is in Table 1.
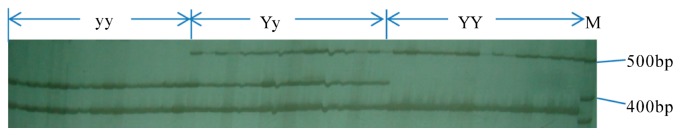
Fig. 2.
Analysis of the PCR products obtained using IP2 on BC8S1 plants. The BC8S1 individuals are represented as yellow seeds (yy), brown seeds (Yy, heterozygous and YY, homozygous). M: 100 bp DNA ladder.
Linkage analysis
The two farthest markers, P15MG15 and EA07MC06, were used to screen 1256 individuals in the BC8S1 population, resulting in identification of 18 recombinants (Fig. 3). Next, all of the markers, including the newly developed markers and the other three AFLP and SSR markers previously used in our study, were used to screen these recombinants. As a result, a high resolution genetic map around the yellow seed color gene was constructed. There were 15 molecular markers on this map, spanning a region of 4.5 cM, with an average genetic distance of 0.3 cM between the two adjacent markers. Three IP markers derived from Bra036827, Bra036828 and Bra036829 were co-segregated with the target gene. IP4 and Y1 were located on either side of the gene, and the genetic distance was only 0.1 and 0.3 cM, respectively (Fig. 1).
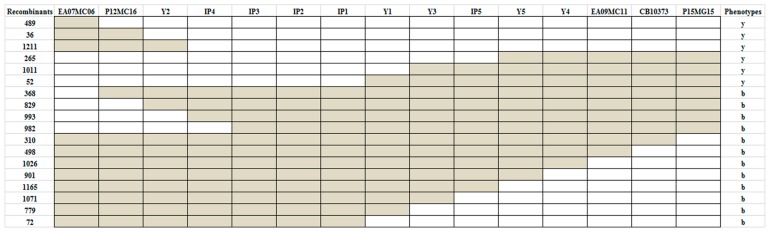
Fig. 3.
Genotypes and phenotypes of recombinants selected from the BC8S1 population. Recombinants and phenotypes (y for yellow seed, b for brown seed) are denoted on the left and right, respectively, with marker names at the top. The yellow seeded alleles are denoted in white and brown seeded alleles in gray. IP1, IP2, and IP3 have no recombinant.
Construction of the physical map around the yellow seed color gene
All of the molecular markers’ sequences on the genetic map around the yellow seed color gene showed good collinearity with those of the A09 chromosome, and the gene was located between 27.079 and 27.616M on the A09 chromosome. This region was approximately 0.54M with only 95 genes residing. The locations of IP1, IP2 and IP3 were in 27.099M (Bra036827), 27.095M (Bra036828) and 27.093M (Bra036829), respectively, these three markers co-segregated with the target gene (Fig. 1). The functions of 95 genes in the gene region were analyzed according to the annotations published on the website: http://brassicadb.org/brad. Because Bra036828 is associated with pigment formation such as flavones, and it is highly homologous with F3H (TRANSPARENT TESTA 6, tt6, E value, 0.0), it is possible that Bra036828 is one of the genes responsible for yellow seed color in B. juncea in our research.
Discussion
IP markers in gene fine mapping
Mapping the yellow seed color gene to the A09 chromosome was the key step to fine mapping the yellow seed color gene (Huang et al. 2012). Through analyzing the sequences of previously published molecular markers linked to the target gene, the yellow seed color gene was mapped in a region of 6M on the A09 chromosome. However, it was extremely challenging to narrow down the mapping region due to the unavailability of the B. rapa genomic sequences before publication of the mapping results. Utilization of the Arabidopsis genome as a reference didn’t improve the situation on account of the differences in genome size and genome sequences between the two species. Fortunately, the whole genome sequence of B. rapa (Chiifu-401) was published by the multinational Brassica genome project (BrGSP) (Wang et al. 2011), which provided vital information for gene mapping on the A genome. Some studies showed that the A genome had retained a high degree of collinearity between Brassica species (Parking et al. 2005, Suwabe et al. 2008). Therefore, marker development or candidate gene prediction in B. juncea could likely be carried out according to the whole genome sequence of B. rapa. IP marker discovery was a very effective way to develop the linked markers. Sometimes, the sequence of an exon in a gene is conserved, but its intron sequence varies. If a pair of primers whose sequences were located in two different exons, were used to amplify the introns, the products may display differences in band size between the yellow seeded and the brown seeded individuals. This approach has been successfully employed by some researchers. Xia et al. (2012) developed nine IP markers linked to a sterile gene in B. napus using the sequences of Arabidopsis and B. rapa. In previous research, the yellow seed color gene was mapped to a region of 6M on the A09 chromosome through analyzing the AFLP and SSR markers’ sequences. Now that the sequence of the gene region has been completely published, it becomes practically possible to develop dense markers to narrow down the genomic sequence region where the yellow seed color gene is localized. Eventually, five IP markers and five SCAR markers were developed successfully. The gene region was reduced to approximate 0.54M and three co-segregated IP markers in this region were developed. Through analyzing a bigger population, it is believed that we can confirm linkage between these three co-segregated IP markers and the yellow seed color gene, therefore our next work is to use different genetic populations or a bigger population to determine the gene region.
Co-dominant markers in MAS
Because of the rarity and instability of yellow seeds in B. napus, it is essential to transfer yellow seeded germplasms from B. juncea or B. rapa to B. napus. Because conventional breeding in field is complicated and time-consuming, it takes a long time and a large space to improve the yellow seed in B. napus. The co-dominant markers linked to the yellow seed color gene are helpful for the selection of the yellow seed color gene in a segregated population. IP1, IP2, IP3, IP4 and IP5 identified in this study are co-dominant markers linked to the yellow seed color gene and three of them are co-segregated with the yellow seed color gene. These markers can distinguish three genotypes in yellow seed locus (yy, Yy and YY), which can help breeders accurately select yellow seed individuals in separated populations. The utilization of these molecular markers combined with comprehensive trait observation in field will not only save time and space for breeders, but also the accuracy of selection will be increased substantially.
Prediction of possible genes responsible for yellow seed color in B. juncea
In Arabidopsis, mutants of the genes for flavonoid biosynthesis produce transparent testa (tt) and yellow seed. Some genes regulating flavonoid biosynthesis have been found in Brassica species and close allies of Arabidopsis. Zhang et al. (2009) reported on the TTG1 gene responsible for the yellow seeded trait in Chinese cabbage; there was a 94-base deletion for this gene in the yellow seeded line. Li et al. (2012) found that a large insertion of transposable elements in the BrTT8 gene in yellow sarson caused the yellow seed. It is possible that some TT genes are also responsible for yellow seed synthesis in this study. Through analyzing the annotations of genes in the mapping region, a gene, Bra036828, is related to the flavones pathway and there is high homologue with the F3H gene (TT6). Therefore it was tempting to predict that Bra036828 was one gene responsible for yellow seed color formation. However, this prediction should be confirmed by a functional complementation experiment, which is currently an on-going project in our lab.
Analysis of yellow seed color gene in Brassica genomes
B. juncea is an allotetraploid species containing genome A and B. It is reasonable to predict that multiple genes from both genomes are responsible for yellow seed color in B. juncea. Padmaja et al. (2005) mapped two yellow seed color genes to A9 and B3 through QTL mapping. However, our meticulous study on the inheritance of yellow seed color in B. juncea through classical Mendel statistical analysis revealed that yellow seed color was controlled by a single gene, which was consistent with a previous publication (Xu et al. 2010). In order to investigate the location of the underlying gene in Brassica genomes, the sequences of the newly developed markers for yellow seed color gene were submitted to the BRAD website for BLAST searches in a hope to estimate the location of the gene. All the BLAST hits with high similarity turned out to fall in a region of 0.54M on chromosome A09. Due to the unavailability of B genome sequences in the public domain, it is unlikely to perform BLAST analysis of these marker sequences against the B genome, hence the possibility of the presence of orthologous genes in the B genome cannot be ruled out at this moment. However, numerous orthologous genes were identified on different chromosomes of B. oleracea through BLAST search. For instance, Bra036828 in B. rapa exhibits high similarity with a sequence (34.01M) on the C08 chromosome (E value = 109, 4e−22), indicative of a homologous gene present in the C genome. It is tempting to assume that multiple genes located in the A and B genomes contribute to the formation of yellow color, and identification and characterization of the genes and subsequent investigation on their interaction becomes key to shedding light on the mechanism behind yellow color formation. A tremendous effort is still directed to addressing this in our lab right now.
Acknowledgement
The research was supported by the High-Tech “863” Program (2011AA10A104), Chinese Doctoral Education Fund Ministry (20110204120015), Natural Science Foundation of Yangling (2014NY-19), the Tang Zhong Ying Breeding Fund and the Science and Technology Funding of Shaanxi Province (2011KTZB02-01-03).
Literature Cited
- Abraham, V. and Bhatia, C.R. (1986) Development of strains with yellow seedcoat in Indian mustard (Brassica juncea Czern. & Coss.). Plant Breed. 97: 86–88. [Google Scholar]
- Ban, Y.Y., Huang, Z., Xu, A.X., Zhang, X.X., Ding, J., Liu, L. and Gao, L.S. (2013) Development of intron polymorphism markers linked to the yellow-seeded gene in Brassica juncea of Northern Shannxi. Acta Agriculturae Boreali-Occidentalis Sinica 22: 37–41. [Google Scholar]
- Debeaujon, I., Peeters, A.J.M., Léon-Kloosterziel, K.M. and Koornneef, M. (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. The Plant Cell 13: 853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J.J. and Doyle, J.L. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Huang, Z., Ban, Y.Y., Yang, L., Zhang, Y., Li, H.Q., Xiao, E.S., Xu, A.X. and Zhang, D.H. (2012) Fine mapping of the yellow seed locus in Brassica juncea L. Genome 55: 8–14. [DOI] [PubMed] [Google Scholar]
- Kosambi, D.D. (1943) The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S. and Newburg, L. (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lei, S.L., Yao, X.Q., Yi, B., Chen, W., Ma, C.Z., Tu, J.X. and Fu, T.D. (2007) Towards map-based cloning: Fine mapping of a recessive genic male-sterile gene (BnMs2) in Brassica napus L. and syntenic region identification based on the Arabidopsis thaliana genome sequences. Theor. Appl. Genet. 115: 643–651. [DOI] [PubMed] [Google Scholar]
- Li, X., Chen, L., Hong, M., Zhang, Y., Zu, F., Wen, J., Yi, B., Ma, C.Z., Shen, J.X., Tu, J.X.et al. (2012) A large insertion in bHLH transcription factor BrTT8 resulting in yellow seed coat in Brassica rapa. PLoS ONE 7: e44145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, S., Daly, M. and Lander, E. (1992) Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead institute technical report, 3rd edn Whitehead Technical Institute, Cambridge, MA. [Google Scholar]
- Liu, R.H. and Meng, J.L. (2003) Mapdraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25: 317–321. [PubMed] [Google Scholar]
- Liu, Z.W., Fu, T.D., Tu, J.X. and Chen, B.Y. (2005) Inheritance of seed colour and identification of RAPD and AFLP markers linked to the seed colour gene in rapeseed (Brassica napus L.). Theor. Appl. Genet. 110: 303–310. [DOI] [PubMed] [Google Scholar]
- Lowe, A.J., Jones, A.E., Raybould, A.F., Trick, M., Moule, C.J. and Edwards, K.J. (2002) Transferability and genome specificity of a new set of microsatellite primers among Brassica species of the U triangle. Mol. Ecol. Notes 2: 7–11. [Google Scholar]
- Mun, J.H., Kwon, S.J., Yang, T.J., Seol, Y.J., Jin, M., Kim, J.A., Lim, M.H., Kim, J.S., Baek, S., Choi, B.S.et al. (2009) Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 10: R111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmaja, K.L., Arumugam, N., Gupta, V., Mukhopadhyay, A., Sodhi, Y.S., Pental, D. and Pradhan, A.K. (2005) Mapping and tagging of seed coat colour and the identification of microsatellite markers for marker assisted manipulation of the trait in Brassica juncea. Theor. Appl. Genet. 111: 8–14. [DOI] [PubMed] [Google Scholar]
- Parkin, I.A.P., Gulden, S.M., Sharpe, A.G., Lukens, L., Trick, M., Osborn, T.C. and Lydiate, D.J. (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, M., McVetty, P.B.E. and Li, G.Y. (2007) Development of SRAP, SNP and multiplexed SCAR molecular markers for the major seed coat color gene in Brassica rapa L. Theor. Appl. Genet. 115: 1101–1107. [DOI] [PubMed] [Google Scholar]
- Routaboul, J.M., Kerhoas, L., Debeaujon, I., Pourcel, L., Caboche, M., Einhorn, J. and Lepiniec, L. (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224: 96–107. [DOI] [PubMed] [Google Scholar]
- Suwabe, K., Morgan, C. and Bancroft, L. (2008) Integration of Brassica A genome genetic linkage map between Brassica napus and B. rapa. Genome 51: 169–176. [DOI] [PubMed] [Google Scholar]
- Vos, P., Hogers, R., Bleeker, M., Reijians, M., VandeLee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M.et al. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.W., Wang, H.Z., Wang, J., Sun, R.F., Wu, J., Liu, S.Y., Bai, Y.Q., Mun, J.H., Bancroft, I., Cheng, F.et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Wang, Y.G. (2006) Edible vegetable oil consumption growth in China and its influencing factors [J]. Agricultural Technology and Economy 6: 54–59. [Google Scholar]
- Xia, S.Q., Cheng, L., Zu, F., Dun, X.L., Zhou, Z.F., Yi, B., Wen, J., Ma, C.Z., Shen, J.X., Tu, J.X.et al. (2012) Mapping of BnMs4 and BnRf to a common microsyntenic region of Arabidopsis thaliana chromosome 3 using intron polymorphism markers. Theor. Appl. Genet. 124: 1193–1200. [DOI] [PubMed] [Google Scholar]
- Xiao, L., Zhao, Z., Du, D.Z., Yao, Y.M., Xu, L. and Tang, G.Y. (2012) Genetic characterization and fine mapping of a yellow-seeded gene in Dahuang (a Brassica rapa landrace). Theor. Appl. Genet. 124: 903–909. [DOI] [PubMed] [Google Scholar]
- Xiao, S.S., Xu, J.S., Li, Y., Zhang, L., Shi, S.J., Shi, S.W., Wu, J.S. and Liu, K.D. (2007) Generation and mapping of SCAR and CAPS markers linked to the seed coat color gene in Brassica napus using a genome-walking technique. Genome 50: 611–618. [DOI] [PubMed] [Google Scholar]
- Yi, B., Chen, Y.N., Lei, S.L., Tu, J.X. and Fu, T.D. (2006) Fine mapping of the recessive genic male-sterile gene (Bnms1) in Brassica napus L. Theor. Appl. Genet. 113: 643–650. [DOI] [PubMed] [Google Scholar]
- Zhang, J.F., Lu, Y., Yuan, Y.X., Zhang, X.W., Geng, J.F., Chen, Y., Cloutier, S., McVetty, P.B.E. and Li, G.Y. (2009) Map-based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa. Plant Mol. Biol. 69: 553–563. [DOI] [PubMed] [Google Scholar]
- Zhang, K., Lu, K., Qu, C., Liang, Y., Wang, R., Chai, Y.R. and Li, J.N. (2013) Gene silencing of BnTT10 family genes causes retarded pigmentation and lignin reduction in the seed coat of Brassica napus. PLoS ONE 8: e61247. [DOI] [PMC free article] [PubMed] [Google Scholar]