Abstract
A core collection of Japanese wheat varieties (JWC) consisting of 96 accessions was established based on their passport data and breeding pedigrees. To clarify the molecular basis of the JWC collection, genome-wide single-nucleotide polymorphism (SNP) genotyping was performed using the genotyping-by-sequencing (GBS) approach. Phylogenetic tree and population structure analyses using these SNP data revealed the genetic diversity and relationships among the JWC accessions, classifying them into four groups; “varieties in the Hokkaido area”, “modern varieties in the northeast part of Japan”, “modern varieties in the southwest part of Japan” and “classical varieties including landraces”. This clustering closely reflected the history of wheat breeding in Japan. Furthermore, to demonstrate the utility of the JWC collection, we performed a genome-wide association study (GWAS) for three traits, namely, “days to heading in autumn sowing”, “days to heading in spring sowing” and “culm length”. We found significantly associated SNP markers with each trait, and some of these were closely linked to known major genes for heading date or culm length on the genetic map. Our study indicates that this JWC collection is a useful set of germplasm for basic and applied research aimed at understanding and utilizing the genetic diversity among Japanese wheat varieties.
Keywords: wheat, core collection, genotyping-by-sequencing (GBS), genetic diversity, genome-wide association study (GWAS), Japan
Introduction
Common wheat (Triticum aestivum L.) is a major staple food crop that is widely cultivated in many parts of the world. To meet the demands of a growing population and the challenges of global climate change, there is currently a strong need for the genetic improvement of wheat with the goals of achieving better quality, higher yield, adaptation to various environments, and tolerance to biotic stresses. The utilization of germplasms is the basis of wheat breeding and is therefore fundamental to sustaining global wheat production, and a core collection (Frankel 1984) or a mini core collection (Upadhyaya and Ortiz 2001) that represent the entire genetic diversity of wheat and its relatives are useful for mining accessions with desirable traits for breeding. However, the use of genetic resources in breeding is currently limited because the accessions preserved in gene banks have not been globally characterized, leading to a scarcity of genotypic and phenotypic information. In particular, there is a tremendous lack of genome-wide genotypic information due to the characteristics of the wheat genome. Wheat has a large genome, i.e., approximately 17 Gb, and is allohexaploid, with three homoeologous genomes (2n = 6x = 42, genome formula AABBDD) that originated from three ancestral parental species (Gill et al. 2004). This large size and polyploidy-related complexity have hampered genomic analyses for detecting the genome-wide molecular diversity of each wheat accession and elucidating the population structure of wheat collections.
The genotyping by sequencing (GBS) method using next-generation sequencing (NGS) technology is becoming increasingly popular for the detection of a large number of single-nucleotide polymorphisms (SNPs), which can be used to estimate chromosome-wide molecular diversity (Poland and Rife 2012). Indeed, GBS is designed for the efficient genotyping of large numbers of samples using NGS platforms and has several advantages: the genome complexity is reduced by methylation-sensitive restriction enzyme digestion; no preliminary sequence information is required; and all newly discovered markers originate from the population being genotyped. These attributes make this approach suitable for wheat because its genome is large and complex and a reference genome sequence is not currently available.
The NIAS (National Institute of Agrobiological Sciences) Genebank Plant Section is one of the largest collections of crop germplasms in the world (http://www.gene.affrc.go.jp/about-plant_en.php) and currently contains and maintains over 220,000 accessions including cereals, legumes, vegetables, and even tree crops. Wheat and its relatives constitute one of the important collections in the NIAS Genebank, and more than 15,000 accessions, including cultivars, breeding lines, landraces, and genetic stocks, are maintained. In 2012, a mini core collection of Japanese wheat accessions (NIAS Japanese Wheat Core Collection, JWC) was released. The collection consists of 96 accessions selected based on passport data from Japanese landraces and modern cultivars maintained at the NIAS Genebank and the National Institute of Crop Sciences. However, because the accessions were chosen based only on phenotypic characteristics and breeding pedigrees stated in the passport data, this mini core collection lacks important molecular information.
The objectives of this research were to apply recently developed genomics tools to wheat using NGS technology and to provide a molecular basis for the NIAS Japanese Wheat Core Collection. We applied the GBS method to develop large numbers of SNP datasets and then used these SNPs to elucidate the genome-wide molecular diversity and precise population structure among the accessions. In addition, a genome-wide association study (GWAS) was performed to validate the utility of the collection for identifying loci determining quantitative and qualitative traits.
Materials and Methods
Plant materials and phenotypic observation
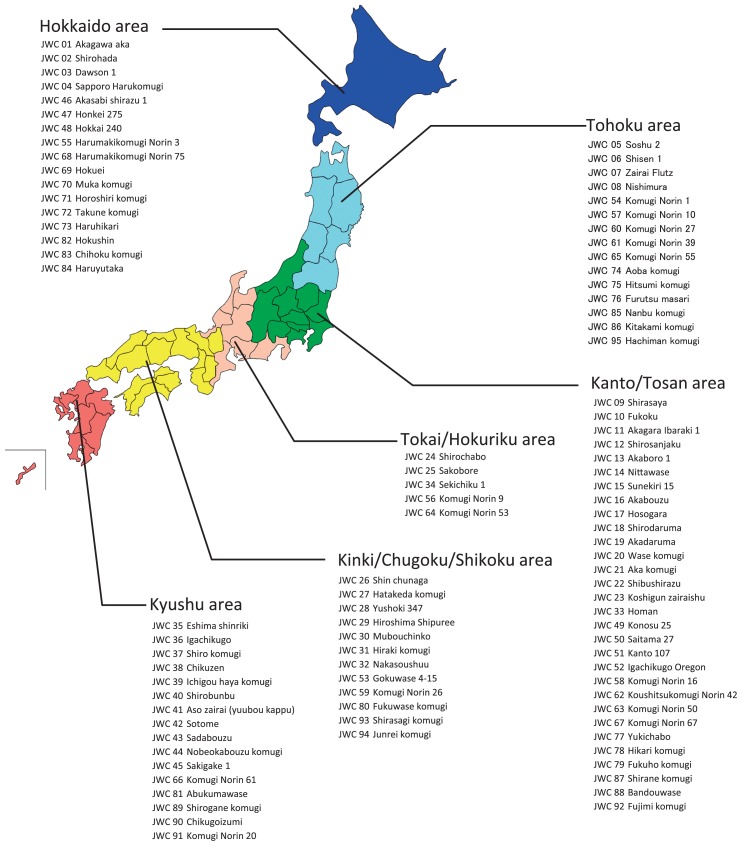
NIAS Japanese Wheat Core Collection (JWC) maintained at the Genebank Plant Section was used (http://www.gene.affrc.go.jp/databases-core_collections_jw_en.php). JWC consists of 96 accessions, including 75 breeders lines, 20 landraces and unknown lines, and the cv. Chinese Spring (CS) (Supplemental Table 1 and Fig. 1). For this study, the JWC accessions, with the exception of JWC96 (CS), were categorized into two groups: JWC01-45 were classified as classical varieties, including landraces, pure selected lines and unknown lines; whereas JWC46-95 were classified as modern varieties.
Fig. 1.
Collection or breeding area of the accessions in JWC used in this study.
To evaluate the phenotypic diversity of the collection, variations in days to heading in autumn sowing (DHA, in 2013), days to heading in spring sowing (DHS, in 2014), and culm length (CL, in 2013) were evaluated at the experimental field of NIAS (Tsukuba, Japan) under natural field conditions. The five plants of each accession were grown for the phenotypic evaluation. DHA and DHS were recorded as days after sowing date (October 25, 2012 for DHA, and May 2, 2014 for DHS). CL was measured as the average of three tallest individuals of each accession.
A mapping population of 210 recombinant inbred lines (RILs) derived from a cross between two common wheat cultivars, namely, CS and Mironovskaya 808 (M808) (Kobayashi et al. 2010), was also utilized to construct a genetic map with SNPs using GBS.
SNP discovery by GBS
Sequence libraries for GBS were constructed according to a previously described procedure (Poland et al. 2012), with some modifications. Total DNA was isolated from leaf tissues of individual plants using DNeasy Plant Maxi Kit (Qiagen, Hilden, Germany). The DNA (200 ng) was digested with two restriction enzymes, PstI and MspI, and then ligated with a barcoded PstI adapter and a common MspI adapter. A single pooled sample was produced from 48 ligated DNA samples and purified using AMPure XP (Beckman Coulter, Brea, CA, USA) to remove any unincorporated adapters. In the case of the RIL population, each pooled sample consisting of 48 DNA samples always included the two parental lines, resulting in nine replications for the sequencing of the parental lines. The purified samples were subjected to size selection using agarose gel electrophoresis and a MinElute Gel Extraction Kit (Qiagen, Hilden, Germany) to obtain DNA fragments of the appropriate size (200–500 bp) for sequencing. The size-selected samples were amplified by PCR with 16 cycles consisting of 95°C for 30 s, 62°C for 20 s, and 68°C for 30. The libraries were purified using AMPure XP beads (Beckman Coulter, Brea, CA, USA) and independently sequenced in a single lane of a HiSeq2000 system (Illumina, San Diego, CA, USA) at Hokkaido System Science Co., Ltd. (Sapporo, Hokkaido, Japan).
Data processing was performed using the TASSEL 3.0 UNEAK pipeline because of the non-reference GBS SNP calling (Bradbury et al. 2007). However, in the case of the core collection, data from five replicates of CS in five lanes of the RIL population were additionally used to complement any missing data for CS to obtain as much SNP data as possible.
For linkage analysis of the RIL population, SNP loci were selected through the following steps: a) high-confidence SNPs, i.e., no contradiction among data from five replicates of the parental lines; b) polymorphism between M808 and CS; and c) SNPs with less than 5% missing data. For data analysis of the core collection, data from nine replicates of CS from other experiments were additionally used to complement the missing data in CS to obtain as much SNP data as possible. The selection of SNP loci was conducted using the same protocol for the RIL population.
Homology search of the GBS tag sequences to repetitive sequences
To estimate how many GBS tag sequences were derived from repetitive sequences in wheat genome, a MEGABLAST search (Altschul et al. 1997) was performed using the database for the repetitive sequences in Poaceae, mipsREdat_9.0p_Poaceae (ftp://ftpmips.helmholtz-muenchen.de/plants/REdat/), with E-values <10−10. The tag sequences with SNPs, 300,818 and 211,131 from the RIL population and JWC, respectively, were used as queries.
Genetic map construction using the RIL population
A genetic map of M808 and CS was constructed using 210 RILs. In addition to the genotype data for the SNP loci generated by GBS, the data for 425 SSR loci was integrated for chromosomal assignment of linkage groups (Kobayashi et al. 2010). Loci were ordered using MapDisto version 1.8.1 (Lorieux 2012) and the genetic distances were calculated with the Kosambi function (Kosambi 1943).
Genetic diversity and population structure analysis
Evolutionary analyses were conducted using MEGA6 (Tamura et al. 2013), and the evolutionary history was inferred using the Neighbor-Joining (NJ) method (Saitou and Nei 1987). The tree is drawn to scale, and the branch lengths are presented in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the number of differences method (Nei and Kumar 2000), and they are presented in units of the number of base differences per sequence. All positions containing hetero sites, gaps and missing data were eliminated. The phylogenetic tree was modified using NJplot version 2.3 (Perrière and Gouy 1996) and TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Population structure analysis using a model-based clustering method was performed using STRUCTURE version 2.3.4 software (Pritchard et al. 2000). Because the accessions in the core collection are highly homozygous, we used a haploid setting. Genotypes A, T, G and C at each locus were coded as 1, 2, 3 and 4, respectively; any missing genotype scores were coded as -9. The algorithm was run with a burn-in length of 50,000 followed by 20,000 Markov Chain Monte Carlo simulations for estimating parameters. At least 20 runs were performed to estimate the mean likelihood for a number of populations (K) between 1 and 12 using the admixture model and correlated allele frequencies. The Structure Harvester version 0.6.94 software (Earl and von Holdt 2012) was used to calculate Evanno’s ΔK values, and CLUMPP version 1.1.2 (Jakobsson and Rosenberg 2007) was then applied to combine the outputs from STRUCTURE using the FullSearch algorithm.
Mapping with GWAS
To determine the chromosomal location of the SNPs, we only selected those from the GBS data of the core collection that successfully mapped to the genetic map of the RIL population resulting from the cross of CS and M808 (Kobayashi et al. 2010). Association analysis was performed mainly in accordance with a previous study (Uchiyama et al. 2013). To account for both population structure and marker-based kinship, we used the GWA function in the rrBLUP package, with some modifications (Endelman 2011); this function relies on a mixed model approach that incorporates both the population structure and marker-based kinship estimates (Q + K model). A marker-trait association (MTA) was considered when the P-value was significant at 0.01 [−log10(0.01) = 2]. Manhattan plots were generated based on association mapping results with SNPEVG (Wang et al. 2012).
Results
SNP identification and characterization by GBS
Sequences from the RIL mapping population and core collection were collected in seven 48-plex flow cells. On average, 184,312,348 sequence reads corresponding to 18,615 Mb were obtained per lane, and 97% of these were selected as high-quality reads. After trimming and filtering of the raw reads, an average of 3,609,645 reads were generated per line using the TASSEL program (Bradbury et al. 2007). Using the GBS method, 300,818 and 211,131 SNP loci were ultimately identified for the RIL population and core collection, respectively.
To verify the chromosomal locations of the detected SNP loci, GBS tag sequences with an SNP were mapped to the survey sequence data of CS established by the International Wheat Genome Sequencing Consortium (IWGSC 2014). We specified chromosomal assignments for 82,808 GBS tags in the SNP dataset of RILs, and 32,230 (39%), 41,233 (50%) and 9,345 (11%) of these tags were found to belong to chromosomes of the A, B and D genome, respectively (Table 1). This SNP distribution was consistent with that obtained in previous SNP-typing studies (Iehisa et al. 2014, Li et al. 2015, Poland et al. 2012). Chromosomal locations for the other 218,010 sequences could not be specified due to multiple locus hits or no hits with the survey sequences.
Table 1.
Number of SNPs on each chromosome arm identified by GBS analysis
| Chromosome arm | Number of SNPs in the RIL population | Number of SNPs in JWC |
|---|---|---|
| 1AS | 2,128 | 1,487 |
| 1AL | 2,436 | 2,149 |
| 1BS | 3,203 | 1,769 |
| 1BL | 2,464 | 1,883 |
| 1DS | 759 | 728 |
| 1DL | 464 | 474 |
| 2AS | 3,487 | 2,916 |
| 2AL | 3,792 | 2,957 |
| 2BS | 2,552 | 3,202 |
| 2BL | 3,848 | 3,898 |
| 2DS | 754 | 653 |
| 2DL | 798 | 815 |
| 3AS | 1,373 | 923 |
| 3AL | 1,661 | 1,365 |
| 3B | 10,653 | 8,575 |
| 3DS | 197 | 240 |
| 3DL | 367 | 393 |
| 4AS | 2,931 | 2,342 |
| 4AL | 3,667 | 3,084 |
| 4BS | 1,913 | 1,316 |
| 4BL | 1,443 | 1,142 |
| 4DS | 611 | 561 |
| 4DL | 855 | 786 |
| 5AS | 1,442 | 626 |
| 5AL | 1,710 | 1,322 |
| 5BS | 2,254 | 1,869 |
| 5BL | 5,198 | 4,522 |
| 5DS | 638 | 646 |
| 5DL | 779 | 854 |
| 6AS | 2,059 | 1,698 |
| 6AL | 2,197 | 1,624 |
| 6BS | 1,445 | 1,603 |
| 6BL | 2,243 | 2,178 |
| 6DS | 554 | 571 |
| 6DL | 855 | 827 |
| 7AS | 1,621 | 1,362 |
| 7AL | 1,726 | 1,586 |
| 7BS | 1,844 | 1,188 |
| 7BL | 2,173 | 1,664 |
| 7DS | 752 | 722 |
| 7DL | 962 | 857 |
|
| ||
| Total | 82,808 | 69,377 |
|
| ||
| A genome | 32,230 | 25,441 |
|
| ||
| B genome | 41,233 | 34,809 |
|
| ||
| D genome | 9,345 | 9,127 |
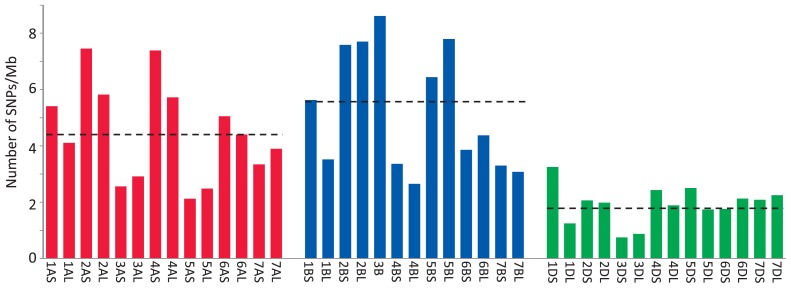
Concerning the core collection, the chromosomal locations of 69,377 tag sequences were determined. The number of SNPs on D-genome chromosomes (9,127; 13%) was lower than those on chromosomes of the A (25,441; 37%) or B genome (34,809; 50%) as well as the results obtained for the RIL population (Table 1). Based on the number of SNPs and physical size of the chromosome arms, the average SNP density was calculated as 4.1 SNPs/Mb, and this number was averaged from the values of 4.4, 5.5 and 1.8 SNPs/Mb obtained for the A, B and D genomes, respectively (Fig. 2). Within each sub-genome, some chromosomes (3A, 5A, 4B, 7B and 3D) and chromosome arms (1BL and 1DL) showed considerably lower SNP densities compared with the average SNP density (Fig. 2). This result suggests that these chromosomal regions are genetically conserved among Japanese wheat varieties.
Fig. 2.
Average SNP density in wheat chromosome arms. The number of SNPs per chromosome arm was calculated based on the estimated chromosome size described by Šafář et al. (2010). Chromosome 3B was not separated into two chromosome arms because the survey sequence data for chromosome 3B were generated as sequence data for the entire chromosome. The three dashed horizontal lines indicate the average SNP density in the three wheat sub-genomes.
About 11% and 12% of the SNP loci found in the RIL population and JWC, respectively, originated from repetitive sequences (Supplemental Table 2), which are apparently lower than the estimated proportion of the repetitive sequences in wheat genome (80–90%, Flavell et al. 1977). The LTR/Gypsy was most frequent in the tag sequences, followed by the CACTA and the LTR/Copia families. This distribution pattern is consistent between the RIL population and JWC (Supplemental Table 2). These results demonstrate that the SNPs were preferentially located in unique genomic regions, probably due to subtraction of the majority of repetitive sequences by the GBS method using a combination of two restriction enzymes.
Development of a genetic map using SNP information obtained by GBS
A genetic map using the RIL population between M808 and CS, which contains 425 SSR markers, was previously constructed (Kobayashi et al. 2010). Of the 300,818 SNPs obtained by GBS, 2,975 that were confident, polymorphic and had less than 5% missing data were used for linkage analysis; 425 SSR markers from a previous study were added to the analysis for chromosomal assignments of linkage groups. We established a genetic map with 24 linkage groups consisting of 1,682 loci (Supplemental Fig. 1), and each of three chromosomes, 3D, 5D and 7D, was consisted of two linkage groups (Table 2). The total length of the genetic map is 3,726.7 cM, indicating an average locus interval of 2.2 cM. The map resolution of linkage groups for D-genome chromosomes (4.5 cM/locus) was lower than those obtained for A- and B-genome chromosomes (1.7 and 1.8 cM/locus, respectively) (Table 2). This difference was due to the number of SNPs assigned to the sub-genomes: the number of SNP markers obtained for the D genome (324) was lower than those of the A (1,240) and B genomes (1,411) (Table 2).
Table 2.
Number of markers mapped to each chromosome of common wheat
| Chromosome | Map length (cM) | Number of SNP markers | Number of SSR markers | Number of loci | Map resolution (cM/locus) |
|---|---|---|---|---|---|
| 1A | 147.4 | 176 | 22 | 109 | 1.4 |
| 1B | 189.4 | 192 | 34 | 104 | 1.8 |
| 1D | 158.9 | 58 | 19 | 43 | 3.7 |
| 2A | 180.3 | 222 | 41 | 112 | 1.6 |
| 2B | 175.9 | 207 | 24 | 126 | 1.4 |
| 2D | 191.8 | 68 | 20 | 45 | 4.3 |
| 3A | 172.2 | 153 | 23 | 93 | 1.9 |
| 3B | 230.9 | 265 | 12 | 124 | 1.9 |
| 3D | 185.7 | 44 | 13 | 33 | 5.6 |
| 4A | 176.8 | 160 | 15 | 75 | 2.4 |
| 4B | 125.2 | 90 | 12 | 56 | 2.2 |
| 4D | 146.1 | 23 | 14 | 21 | 7.0 |
| 5A | 205.9 | 173 | 18 | 116 | 1.8 |
| 5B | 277.0 | 233 | 19 | 126 | 2.2 |
| 5D | 232.4 | 33 | 22 | 45 | 5.2 |
| 6A | 128.0 | 163 | 19 | 74 | 1.7 |
| 6B | 113.4 | 231 | 15 | 82 | 1.4 |
| 6D | 187.3 | 34 | 28 | 46 | 4.1 |
| 7A | 141.2 | 195 | 18 | 94 | 1.5 |
| 7B | 148.5 | 193 | 19 | 99 | 1.5 |
| 7D | 212.5 | 64 | 18 | 59 | 3.6 |
|
| |||||
| Total | 3726.7 | 2975 | 425 | 1682 | 2.2 |
|
| |||||
| A genome | 1151.7 | 1240 | 156 | 673 | 1.7 |
|
| |||||
| B genome | 1260.3 | 1411 | 135 | 717 | 1.8 |
|
| |||||
| D genome | 1314.7 | 324 | 134 | 292 | 4.5 |
Genetic diversity and population structure of the NIAS Japanese Wheat Core Collection
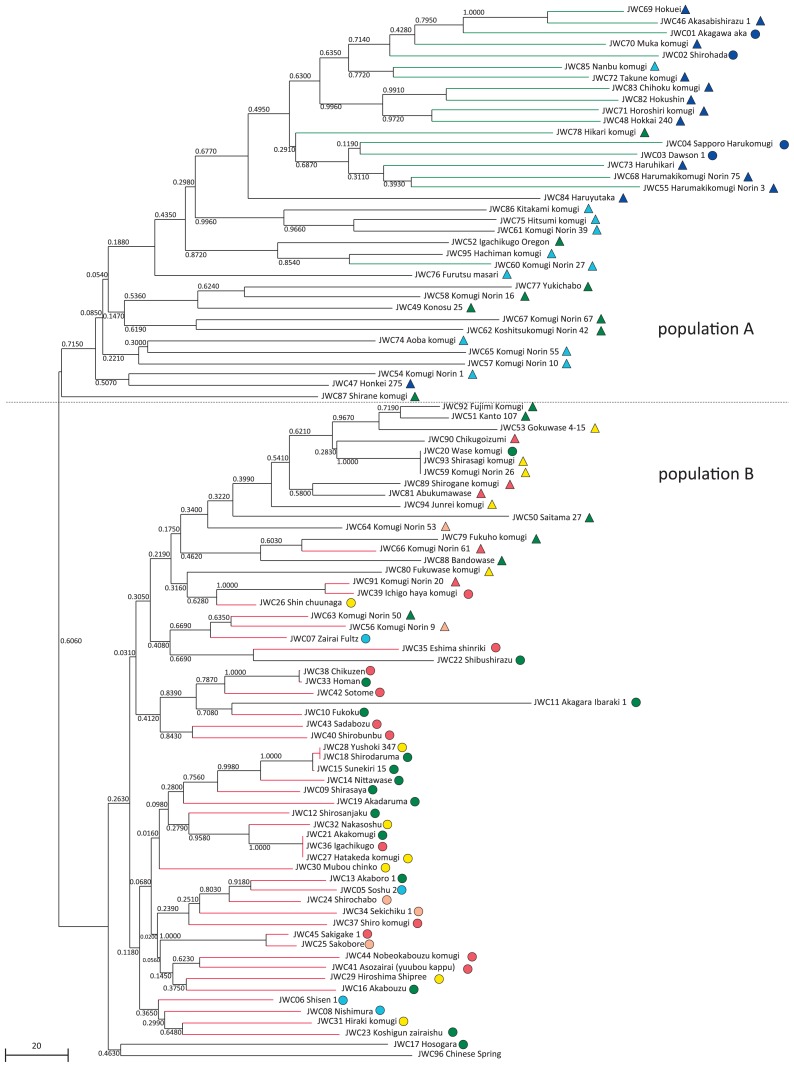
Genetic distances were calculated among the JWC accessions in the core collection, and a phylogenetic tree was constructed by the NJ method using 682 SNP loci with no missing data. The 96 accessions were divided into two large groups: populations A and B (Fig. 3). Population A consisted of 36 accessions including classical and modern varieties from Hokkaido and modern breeders lines from the Tohoku to Kanto/Tosan areas; population B was composed of CS and 59 JWC accessions including modern varieties from the Kanto to Kyushu areas and classical varieties except for Hokkaido accessions. This result revealed that the Hokkaido accessions are clearly distinct from classical Japanese varieties and that modern varieties are genetically divergent but classified into two major groups with the Kanto area as the boundary.
Fig. 3.
Phylogenetic relationship of the 96 JWC accessions based on the analysis of SNPs generated by GBS. The rooted genetic distance tree was constructed using the neighbor-joining method. Numbers above branches indicate bootstrap values. A circle or triangle on the right side of the variety name indicates its status, i.e., whether it is a classical or modern variety. The colors in the circles and triangles correspond to the colors for the collection/breeding area of the JWC accessions shown in Fig. 1. Green and red colors of branches for 18 and 41 accessions represent their classification groups obtained in the subsequent population structure analysis, populations I and III, respectively (Fig. 5).
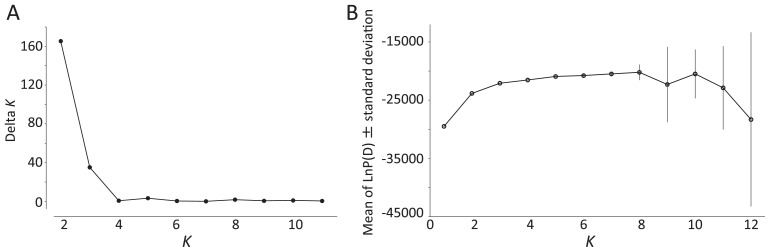
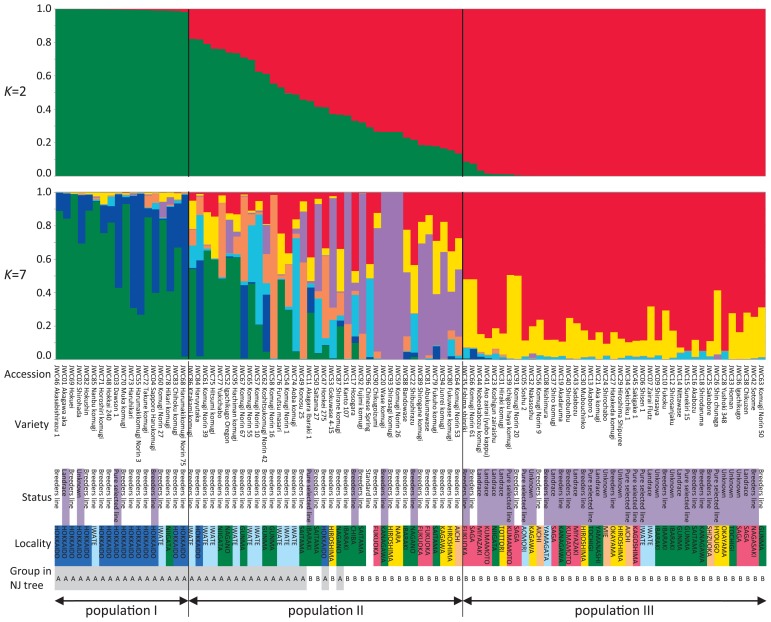
To study in greater detail the intraspecific differentiation and population structure of the Japanese wheat varieties, we analyzed the SNP dataset used for the construction of the phylogenetic tree via the model-based clustering method implemented in STRUCTURE (Pritchard et al. 2000). The optimal number of populations (K) was inferred by ΔK values (Evanno et al. 2005), and the calculation of ΔK based on the STRUCTURE output indicated an optimal K value of 2 (Fig. 4A). At K = 2, the Japanese wheat varieties were classified into three groups, populations I, II (admixtures) and III, using a threshold of 0.90 for Q statistics (Fig. 5). Population I (18 accessions) was mainly composed of accessions from Hokkaido, including two accessions from Iwate (Tohoku area) and one accession from Niigata (Kanto/Tosan area). Population II (37 accessions) consisted of 31 modern varieties, 4 classical varieties and CS; population III (41 accessions) included 38 classical varieties and 4 modern varieties. The accessions in populations II and III are distributed over wide range of Japan. Taken together with the results of the phylogenetic tree, populations I and III correspond to populations A and B, respectively, whereas population II includes accessions from both populations A and B (Figs. 3, 5). The bar plot with K = 7, which was inferred as the optimal K value based on the highest log-likelihood value (Fig. 4B), shows the population structure of the core collection in more detail, revealing that populations I and III have relatively simple genetic structures yet that of population II is complex and diverse (Fig. 5). This was in agreement with the result from the phylogenetic tree, in which the modern varieties are divided into two groups (populations A and B) without the creation of a single group (Fig. 3). For population II (Fig. 5), we found that 14 modern accessions carry a membership coefficient represented by a purple segment in common, a unique genetic structure that distinguishes these accessions from the others in the population II. In addition, all of the accessions with the purple-colored segment are classified into population B (Figs. 3, 5). This genetic structure represented as a purple segment for membership coefficient appeared to be one of the key factors responsible for the distinct genetic distance between the modern varieties of populations A and B observed in the phylogenetic tree (Fig. 3). Consequently, the Japanese wheat varieties could be divided into four groups: Hokkaido’s varieties (population I), modern varieties from Tohoku to Kanto/Tosan areas (hereafter designated as population IIa), modern varieties from Kanto to Kyushu areas (hereafter population IIb), and classical varieties (population III) (Fig. 5).
Fig. 4.
Inference of the optimal K value. Plots of ΔK (A) and mean likelihood (B) at each K obtained from STRUCTURE analysis of a dataset containing 96 JWC accessions.
Fig. 5.
Genetic relationship among 96 JWC accessions surveyed using STRUCTURE. The models with K = 2 and K = 7 showed the ΔK value and the highest log-likelihood value, respectively. A vertical axis indicates a membership coefficient in the each optimum number of K. For the individual bar plots, the corresponding accession number, variety name, status, locality and group, as classified by the phylogenetic tree, are indicated at the bottom of the plot. The colored status indicates the classical accessions JWC01-45. The colors of the localities correspond to the colors for the collection/breeding area of the JWC accessions shown in Fig. 1.
Genome-wide association studies (GWASs)
We performed a GWAS to test whether the natural diversity present in JWC could be exploited to identify related genes via association genetics. After removal of the markers with no frequency and imputation of the missing data, we used 2158 SNPs to identify alleles that affect two traits: “days to heading” and “culm length”. The phenotypic data used for the GWAS analyses were obtained from a field test conducted at our institute, NIAS (Tsukuba, Japan).
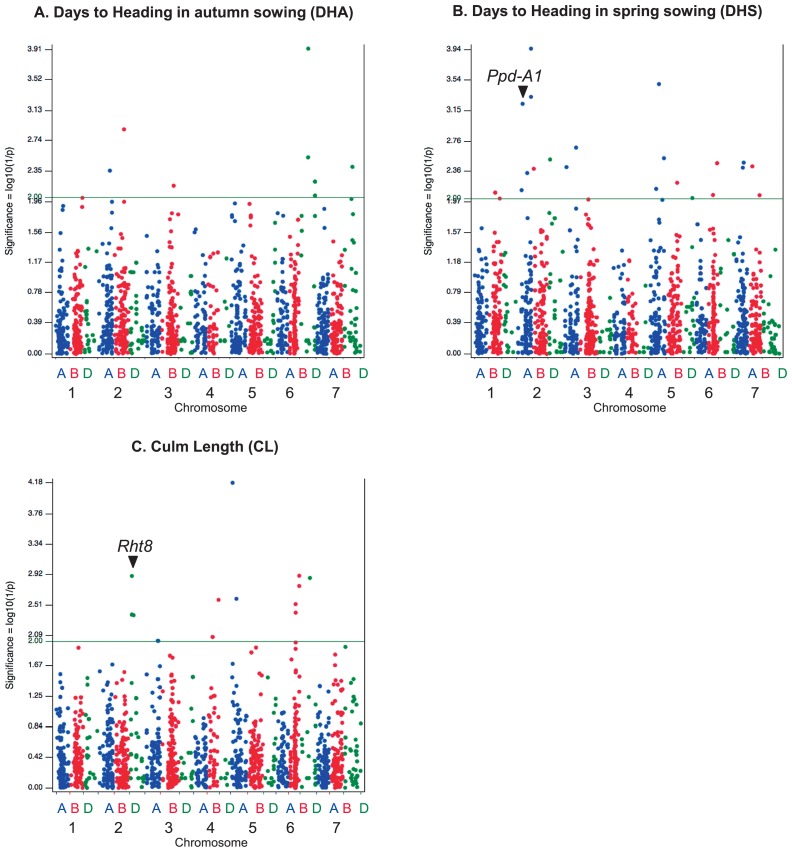
Days to heading is a key trait for wheat cultivation and is quantitatively controlled by the vernalization requirement, photoperiod sensitivity, and earliness per se of the accession (Worland and Snape 2001). We performed the following two independent experiments to measure days to heading: autumn-sowing and spring-sowing. A GWAS of “days to heading in autumn sowing” (DHA) revealed eleven significantly associated SNP markers (Supplemental Table 3). These were grouped into 6 QTLs located on 5 chromosomes 2A, 2B, 3B, 6D, and 7D, with chromosome 6D harboring the maximum number of markers associated with this trait (Fig. 6). Twenty-four markers yielding 19 QTLs were significantly associated with “days to heading in spring sowing” (DHS) (Supplemental Table 3), involving 11 chromosomes, and half of the significant markers are located on homoeologous group-2 (2A, 2B and 2D) and group-5 (5A, 5B and 5D) chromosomes (Fig. 6).
Fig. 6.
GWASs for three phenotypic traits from 96 JWC accessions. (A) Days to heading in autumn sowing (DHA). (B) Days to heading in spring sowing (DHS). (C) Culm length (CL). The vertical axis represents the −log10(P) values of the P-value obtained for the marker-trait association. The horizontal line in the plot indicates the significance level at 0.01 [−log10(0.01) = 2]. The SNPs in the vicinity of the Ppd-A1 and Rht8 genes are marked with arrowheads.
Culm length (CL) is an important trait in wheat and it is not only the factor that indicates the plant’s growth status from the perspective of basic biology but also an important target for applied aspects, including breeding. The results of the GWAS for CL displayed significant MTAs at 16 SNP sites grouped into 9 QTLs located on 6 chromosomes, 2D, 3A, 4B, 5A, 6B, and 6D (Fig. 6, Supplemental Table 3). Chromosome 2D was found to have the maximum number of markers associated with the CL trait.
Discussion
The Japanese wheat varieties in the core collection studies were classified into four groups via phylogenetic tree and population structure analyses using genome-wide SNP information generated by GBS (Figs. 3, 5). The clustering of the JWC accessions in this study well reflected the history of wheat breeding in Japan described by Hoshino and Seko (1996). Japan consists of a long series of islands stretching for 3,000 kilometers from north to south; therefore, the climate varies strongly among different regions. The northeast region has warm summers and very cold winters, with heavy snowfall next to the Sea of Japan and in mountainous areas. By contrast, summer in the southwest region is very hot and humid, with a relatively mild winter. Although many genetic resources including landraces and varieties introduced from foreign countries (mainly North America and Europe) have been used as breeding materials during the approximately 120-year history of Japanese wheat breeding, Japanese wheat breeders in northern areas have often used foreign varieties with superior traits due to the climate in that region; by contrast, landraces adapted to a climate with moderate winters and humid springs/summers have largely been used in the southern part of Japan. The phylogenetic tree of the JWC accessions, which resulted in two large groups, populations A and B, clearly supports this history from a genetic perspective (Fig. 3). Most accessions in the northeast region (Hokkaido, Tohoku and Kanto/Tosan areas) are included in population A. In addition, the genetic distances among them in the NJ tree are slightly longer than those for accessions in population B, which suggests that foreign varieties have been introduced into each breeding pedigree. Population B consists of modern varieties in southwest Japan (Kanto, Tokai/Hokuriku, Kinki/Chugoku/Shikoku and Kyushu areas) and classical cultivars including landraces. The modern varieties in this group are genetically close to a limited number of classical varieties, which indicates that they were generated from crosses with specific classical varieties. To date, differences between northeast and southwest varieties in Japan have been verified by analyses of various genes, such as necrosis genes (Ne1 and Ne2), a reduced height gene (Rht-1), a gametocidal inhibitor gene (Igc1), a vernalization requirement gene (Vrn-1), a photoperiod-responsive gene (Ppd-1) and a seed dormancy gene (MFT-3A), as well by isozyme analyses, such as a glutenin subunit (Glu-D1), gliadins, β-amylases, esterases and peroxidases (Chono et al. 2015, Ghimire et al. 2005, 2006, Gotoh 1979, Iwaki et al. 2000, Nakamura 2001, Seki et al. 2011, 2013, Tanaka et al. 2003, Tsujimoto et al. 1998, Tsunewaki and Nakai 1967, Yamada 1989). Here, we provide the first report of the genetic diversity among Japanese wheat varieties using genome-wide genotyping data, providing more reliable information for understanding the genetic diversity of Japanese wheat varieties because genome-wide analyses are less biased than analyses using the gene-based approaches.
The population structure analysis at K = 2 showed a clear difference in the genetic structure between accessions from the Hokkaido area (population I) and classical varieties (population III) (Fig. 5). As mentioned above, this result is in good agreement with the degree of contribution by foreign varieties to the breeding process. In northern Japan, the main breeding targets are cold tolerance and snow mold resistance (Hoshino and Seko 1996). To develop varieties with improved phenotypes, foreign varieties from Europe and North America, such as ‘Martin’s Amber’ and ‘Turkey Red II’, were introduced into the breeding program (Fukunaga and Inagaki 1985). The population I accessions have at least one of these two varieties as a crossing parent in their lineages, which well corresponds to the structure analysis results. The population structure at K = 7 showed a more detailed genetic structure of the JWC population (Fig. 5). The five accessions from Hokkaido, JWC03 (Dawson 1), JWC04 (Sapporo Harukomugi), JWC55 (Harumakikomugi Norin 3), JWC68 (Harumakikomugi Norin 75) and JWC73 (Haruhikari), showed a similar genetic structure carrying a large blue segment for membership coefficient, and formed a small cluster in the phylogenetic tree (Figs. 3, 5). All of these accessions are spring-type varieties, but each was independently developed in its specific lineage from an ancestral winter-type variety (Fukunaga and Inagaki 1985). In Hokkaido, spring-type varieties with late maturity were expected to increase the yield and extend the length of the harvest season in efforts to improve labor and management efficiency (Kato 2006), and the genetic diversity detected among these accessions appears to reflect the differences between winter- and spring-type varieties.
Population II showed an admixture pattern of genetic structure, which is well consistent with the breeding history demonstrating that most modern varieties were derived from hybridizations between Japanese landraces and varieties introduced from abroad (Fig. 5). This group was further separated into two subgroups, populations IIa and IIb, based on the phylogenetic tree and the population structure at K = 7 (Figs. 3, 5). Population IIa includes modern varieties in the northeast region of Japan and shows complex genetic structures, indicating the extensive mixture of various alleles introduced from Japanese landraces and foreign varieties, possibly leading to improvements in cold adaptation or disease resistance. Population IIb consists of modern varieties in the southwest part of Japan and exhibits a unique genetic structure, i.e., carrying a membership coefficient represented by a purple segment in common (Fig. 5). Among the 15 modern accessions in population IIb, nine accessions have ‘Norin 26’ (JWC59) as an ancestral crossing parent in their lineages. In addition, structure analysis showed that a large purple segment for membership coefficient (membership probability = 0.996) corresponds to ‘Norin 26’ (JWC59). These results suggest that the genetic structure composition of the nine accessions was substantially affected by the alleles introduced from ‘Norin 26’, which is known as an early-maturity variety. We found two early-maturity landraces, ‘Hayakomugi’ and ‘Sojukuakage’, in the lineage of ‘Norin 26’ (Fukunaga and Inagaki 1985) although it is not known whether these two landraces belonged to population IIb. Moreover, this group includes JWC80 (Fukuwase komugi) and JWC81 (Abukuma wase), which are also known as early-maturity varieties. In addition, other varieties in population IIb, such as JWC20 (Wase komugi), JWC53 (Gokuwase 4–15) and JWC88 (Bando wase), are thought to be early-maturity varieties because the names of these varieties include “wase”, which means “early-maturity” in Japanese. The main target of wheat breeding in the southern part of Japan was early-maturity, which would allow implementation of rice and wheat double cropping in a single year (Hoshino and Seko 1996). Therefore, population IIb can be characterized as the group of early-maturity varieties carrying a unique genetic structure, as represented by the common purple segment for membership coefficient.
CS (JWC96), which was used as a standard variety, is located in population B on the phylogenetic tree (Fig. 3) and in population IIb on the bar plot for K = 7 (Fig. 5). Sears and Miller (1985) described a landrace in Sichuan region of south China as the origin of CS, and previous studies have suggested genetic relationships between landraces in south China and southwest Japan (Ghimire et al. 2005, 2006, Iwaki et al. 2000). Our result for the position of CS obtained from the population structure analysis reveals the genetic similarities between CS and accessions in southwest Japan and supports these previous results.
Among the JWC accessions, we found that the SNP densities of several chromosomes and chromosome arms, namely, 3A, 5A, 4B, 7B, 3D, 1BL and 1DL, were lower than those of other chromosomes (Fig. 2). These results suggest that these chromosomes possess regions with important functions required for wheat varieties in Japan, such as adaptation to the Japanese climate and flour quality suitable for the end products. A lack of pre-harvest sprouting is one of the traits required for adaptation to the humid harvest season in Japan, and a major seed dormancy gene, MFT-3A, is located on chromosome 3A (Nakamura et al. 2011). Control of heading is also important for cropping systems in Japan, and vernalization is one of the factors for heading control. The vernalization requirement genes Vrn-A1 and Vrn-A2 are located on the long arm of chromosome 5A. Regarding flour quality, the high molecular weight glutenin subunit gene Glu-1 is located on the long arms of homoeologous group 1 chromosomes (McIntosh et al. 2013), which well correspond to the lower SNP densities of 1BL and 1DL. Further analyses are needed to evaluate relationships between specific chromosome regions and genes, which will reveal more detailed genomic features of these Japanese wheat varieties.
To demonstrate the suitability of JWC, we analyzed three traits, “days to heading in autumn sowing” (DHA), “days to heading in spring sowing” (DHS) and “culm length” (CL) (Fig. 6). “Days to heading” (DH) is one of the indicators for adaptation to the environment and is a complex trait affected by numerous QTLs. Many SNP markers were found to be associated with the traits, DHA and DHS (Fig. 6); indeed, we found 11 significant SNPs defining 6 QTLs and 24 MTAs yielding 19 QTLs for DHA and DHS, respectively. Some of these QTLs are located on chromosomes that were previously reported to harbor major genes associated with heading. In particular, the chromosomes of homoeologous groups 2 and 5 carry Ppd1 (a major gene for photoperiod response), and VRN1, VRN2 (genes for the vernalization response) and PhyC (a gene for flowering promotion), respectively (Chen et al. 2014, Cockram et al. 2007, Nishida et al. 2013). For the genetic map of CS/M808, one known SSR marker, wmc177, is located between two MTAs, TP355182 and TP64598 (Fig. 6, Supplemental Table 1), on chromosome 2A, which were determined from the GWAS for DHA with intervals of 5.1 cM and 8.2 cM, respectively. wmc177 is known to be closely linked to Ppd-A1 with a 2.2 cM interval (Wilhelm et al. 2009), and thus, the QTL defined by these two MTAs is suggested to be Ppd-A1.
For culm length (CL), we found 9 putative QTL regions located on chromosomes 2D, 3A, 4B, 5A, 6B and 6D, comprising 16 MTAs. Several “reduced-height genes” (Rht genes) have been introduced to modern wheat varieties. The most famous Rht gene is Rht-B1, which is located on the chromosome 4B, and this gene was used for the so-called “Green Revolution” (Hedden 2003). Rht8, also known as the semi-dwarf gene, is located on the short arm of chromosome 2D (2DS), and it was a target in the breeding of most dwarf and semi-dwarf European varieties (Borojevic and Borojevic 2005). Both of these Rht genes originate from Japanese varieties: Rht-B1 from Norin 10 and Rht8 from Akakomugi. We found one QTL comprising 5 MTAs on 2DS, 4 of which mapped to the same locus (24.2 cM) on 2DS in the CS/M808 genetic map, a site that is closely linked with the known SSR marker gwm261 (23.4 cM) (Fig. 6, Supplemental Fig. 1). gwm261 is a diagnostic marker for Rht8 located 0.6 cM proximal to the position of gwm261 (Korzun et al. 1998). This result suggests that the QTL found in this region corresponds to Rht8. We also identified 2 QTLs on the chromosome 4B, which may reflect the homoeologous copy of Rht1 on 4B (Rht-B1).
In this study, we used high-throughput genotyping data obtained by GBS to assess population structure patterns of JWC. The results revealed the genetic diversity contained within JWC, which is well coincident with the breeding history in Japan. We combined SNP data with a high-density genetic map to examine the linkage disequilibrium that we applied to GWAS, and the GWAS identified significant MTAs for model traits. Our results indicate that the JWC collection is a useful set of germplasm that can be used to understand and utilize the genetic diversity within Japanese wheat varieties in both basic and applied research. International efforts are currently addressing the reference genome sequencing of wheat (IWGSC, http://www.wheatgenome.org/). The availability of reference genome data for wheat would make the approach described herein easier to perform and would provide more precise information, which would hasten a better understanding of the genetic diversity of wheat and further enhance the use of wheat germplasm collections.
Supplementary Material
Acknowledgments
The authors wish to thank Hitoshi Matsunaka (NARO Kyushu Okinawa Agricultural Research Center) for valuable discussions and critical editing of the manuscript. We also grateful to Jun-ichi Yonemaru (National Institute of Agrobiological Sciences) and Shotaro Takenaka (Ryukoku University) for their technical assistances in the data analyses. This work was supported in part by a grant-in-aid from the National Institute of Agrobiological Sciences (NIAS Strategic Research Fund).
Literature Cited
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borojevic, K. and Borojevic, K. (2005) The transfer and history of “reduced height genes” (Rht) in wheat from Japan to Europe. J. Hered. 96: 455–459. [DOI] [PubMed] [Google Scholar]
- Bradbury, P.J., Zhang, Z., Kroon, D.E., Casstevens, T.M., Ramdoss, Y. and Buckler, E.S. (2007) TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- Chen, A., Li, C., Hu, W., Lau, M.Y., Lin, H., Rockwell, N.C., Martin, S.S., Jernstedt, J.A., Lagarias, J.C. and Dubcovsky, J. (2014) PHYTOCHROME C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc. Natl. Acad. Sci. USA 111: 10037–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chono, M., Matsunaka, H., Seki, M., Fujita, M., Kiribuchi-Otobe, C., Oda, S., Kojima, H. and Nakamura, S. (2015) Molecular and genealogical analysis of grain dormancy in Japanese wheat varieties, with specific focus on MOTHER OF FT AND TFL1 on chromosome 3A. Breed. Sci. 65: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram, J., Jones, H., Leigh, F.J., O’Sullivan, D., Powell, W., Laurie, D.A. and Greenland, A.J. (2007) Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J. Exp. Bot. 58: 1231–1244. [DOI] [PubMed] [Google Scholar]
- Earl, D.A. and vonHoldt, B.M. (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4: 359–361. [Google Scholar]
- Endelman, J.B. (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4: 250–255. [Google Scholar]
- Evanno, G., Regnaut, S. and Goudet, J. (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Flavell, R.B., Rimpau, J. and Smith, D.B. (1977) Repeated sequence DNA relationships in four cereal genomes. Chromosoma 63: 205–222. [Google Scholar]
- Frankel, O.H. (1984) Genetic perspectives of germplasm conservation. In: Arber, W.K., Limensee K., Peacock W.J. and Stralinger P. (eds.) Genetic manipulation: Impact on man and society, Cambridge University Press, Campridge, pp. 161–170. [Google Scholar]
- Fukunaga, K. and Inagaki, M. (1985) Genealogical pedigrees of Japanese wheat cultivars. Japan. J. Breed. 35: 89–92. [Google Scholar]
- Ghimire, S.K., Akashi, Y., Maitani, C., Nakanishi, M. and Kato, K. (2005) Genetic diversity and geographical differentiation in Asian common wheat (Triticum aestivum L.), revealed by the analysis of peroxidase and esterase isozymes. Breed. Sci. 55: 175–185. [Google Scholar]
- Ghimire, S.K., Akashi, Y., Masuda, A., Washio, T., Nishida, H., Zhou, Y.H., Yen, C., Qi, X., Li, Z., Yoshino, H.et al. (2006) Genetic diversity and phylogenetic relationships among East Asian common wheat (Triticum aestivum L.) populations, revealed by the analysis of five isozymes. Breed. Sci. 56: 379–387. [Google Scholar]
- Gill, B.S., Appels, R., Botha-Oberholster, A.M., Buell, C.R., Bennetzen, J.L., Chalhoub, B., Chumley, F., Dvořák, J., Iwanaga, M., Keller, B.et al. (2004) A work shop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics 168: 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh, T. (1979) Genetic studies on growth habit of some important spring wheat cultivars in Japan, with special reference to the identification of the spring gene involved. Japan. J. Breed. 29: 133–145. [Google Scholar]
- Hedden, P. (2003) The genes of the Green Revolution. Trends Genet. 19: 5–9. [DOI] [PubMed] [Google Scholar]
- Hoshino, T. and Seko, H. (1996) History of wheat breeding for a half century in Japan. Euphytica 89: 215–221. [Google Scholar]
- Iehisa, J.C.M., Ohno, R., Kimura, T., Enoki, H., Nishimura, S., Okamoto, Y., Nasuda, S. and Takumi, S. (2014) A high-density genetic map with array-based markers facilitates structural and quantitative trait locus analyses of the common wheat genome. DNA Res. 21: 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki, K., Nakagawa, K., Kuno, H. and Kato, K. (2000) Ecogeographical differentiation in east Asian wheat, revealed from the geographical variation of growth habit and Vrn genotype. Euphytica 111: 137–143. [Google Scholar]
- Jakobsson, M. and Rosenberg, N.A. (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. [DOI] [PubMed] [Google Scholar]
- Kato, K. (2006) Allelic variation of heading-trait-related genes, essential for wide adaptation of wheat, and its application to wheat breeding. Gamma Field Symposia 45: 23–34. [Google Scholar]
- Kobayashi, F., Takumi, S. and Handa, H. (2010) Identification of quantitative trait loci for ABA responsiveness at the seedling stage associated with ABA-regulated gene expression in common wheat. Theor. Appl. Genet. 121: 629–641. [DOI] [PubMed] [Google Scholar]
- Korzun, V., Röder, M.S., Ganal, M.W., Worland, A.J. and Law, C.N. (1998) Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 96: 1104–1109. [Google Scholar]
- Kosambi, D.D. (1943) The estimation of map distance from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Li, H., Vikram, P., Singh, R.P., Kilian, A., Carling, J., Song, J., Burgueno-Ferreira, J.A., Bhavani, S., Huerta-Espino, J., Payne, T.et al. (2015) A high density GBS map of bread wheat and its application for dissecting complex disease resistance traits. BMC Genomics 16: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux, M. (2012) MapDisto: fast and efficient computation of genetic linkage maps. Mol. Breed. 30: 1231–1235. [Google Scholar]
- McIntosh, R.A., Yamazaki, Y., Dubcovsky, J., Rogers, J., Morris, C., Apples, R. and Xia, X.C. (2013) Catalogue of gene symbols for wheat. [http://www.shigen.nig.ac.jp/wheat/komugi/genes/download.jsp].
- Nakamura, H. (2001) Genetic diversity of high-molecular-weight glutenin subunit compositions in landraces of hexaploid wheat from Japan. Euphytica 120: 227–234. [Google Scholar]
- Nakamura, S., Abe, F., Kawahigashi, H., Nakazono, K., Tagiri, A., Matsumoto, T., Utsugi, S., Ogawa, T., Handa, H., Ishida, H.et al. (2011) A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23: 3215–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M. and Kumar, S. (2000) Molecular Evolution and Phylogenetics. Oxford University Press, New York. [Google Scholar]
- Nishida, H., Ishihara, D., Ishii, M., Kaneko, T., Kawahigashi, H., Akashi, Y., Saisho, D., Tanaka, K., Handa, H., Takeda, K.et al. (2013) Phytochrome C is a key factor controlling long-day flowering in barley. Plant Physiol. 163: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrière, G. and Gouy, M. (1996) WWW-Query: An on-line retrieval system for biological sequence banks. Biochimie 78: 364–369. [DOI] [PubMed] [Google Scholar]
- Poland, J.A., Brown, P.J., Sorrells, M.E. and Jannink, J.L. (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 7: e32253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland, J.A. and Rife, T.W. (2012) Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 5: 92–102. [Google Scholar]
- Pritchard, J.K., Stephens, M. and Donnelly, P. (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šafář, J., Šimková, H., Kubaláková, M., Číhalíková, J., Suchánková, P., Bartoš, J. and Doležel, J. (2010) Development of chromosome-specific BAC resources for genomics of bread wheat. Cytogenet. Genome Res. 129: 211–223. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sears, E.R. and Miller, T.E. (1985) The history of Chinese Spring wheat. Cereal Res. Commun. 13: 261–263. [Google Scholar]
- Seki, M., Chono, M., Matsunaka, H., Fujita, M., Oda, S., Kubo, K., Kiribuchi-Otobe, C., Kojima, H., Nishida, H. and Kato, K. (2011) Distribution of photoperiod-insensitive alleles Ppd-B1a and Ppd-D1a and their effect on heading time in Japanese wheat cultivars. Breed. Sci. 61: 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Chono, M., Nishimura, T., Sato, M., Yoshimura, Y., Matsunaka, H., Fujita, M., Oda, S., Kubo, K., Kiribuchi-Otobe, C.et al. (2013) Distribution of photoperiod-insensitive allele Ppd-A1a and its effect on heading time in Japanese wheat cultivars. Breed. Sci. 63: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, H., Tomita, M., Tsujimoto, H. and Yasumuro, Y. (2003) Limited but specific variations of seed storage proteins in Japanese common wheat (Triticum aestivum L.). Euphytica 132: 167–174. [Google Scholar]
- The International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- Tsujimoto, H., Yamada, T. and Sasakuma, T. (1998) Pedigree of common wheat in East Asia deduced from distribution of the gametocidal inhibitor gene (Igc1) and β-amylsse isozymes. Breed. Sci. 48: 287–291. [Google Scholar]
- Tsunewaki, K. and Nakai, Y. (1967) Distribution of necrosis genes in wheat. II. Japanese local varieties of common wheat. Can. J. Genet. Cytol. 9: 75–78. [Google Scholar]
- Uchiyama, K., Iwata, H., Moriguchi, Y., Ujino-Ihara, T., Ueno, S., Taguchi, Y., Tsubomura, M., Mishima, K., Iki, T., Watanabe, A.et al. (2013) Demonstration of genome-wide association studies for identifying markers for wood property and male strobili traits in Cryptomeria japonica. PLoS ONE 8: e79866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya, H.D. and Ortiz, R. (2001) A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theor. Appl. Genet. 102: 1292–1298. [Google Scholar]
- Wang, S., Dvorkin, D. and Da, Y. (2012) SNPEVG: a graphical tool for GWAS graphing with mouse clicks. BMC Bioinformatics 13: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, E.P., Turner, A.S. and Laurie, D.A. (2009) Photoperid insensitive Ppd-A1 a mutations in tetraploid wheat (Triticum durum Desf.). Theor. Appl. Genet. 118: 285–294. [DOI] [PubMed] [Google Scholar]
- Worland, T. and Snape, J.W. (2001) Genetic basis of worldwide wheat varietal improvement. In: Bonjean, A.P. and Angus W.J. (eds.) The World Wheat Book, A History of Wheat Breeding, Lavoisier Publishing, Paris, pp. 59–100. [Google Scholar]
- Yamada, T. (1989) Identification of GA-insensitive Rht genes in Japanese modern varieties and landraces of wheat. Euphytica 43: 53–57. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.