Abstract
Black spot disease, which is caused by the Japanese pear pathotype of the filamentous fungus Alternaria alternata (Fries) Keissler, is one of the most harmful diseases in Japanese pear cultivation. We mapped a gene for susceptibility to black spot disease in the Japanese pear (Pyrus pyrifolia Nakai) cultivar ‘Kinchaku’ (Aki gene) at the top of linkage group 11, similar to the positions of the susceptibility genes Ani in ‘Osa Nijisseiki’ and Ana in ‘Nansui’. Using synteny-based marker enrichment, we developed novel apple SSR markers in the target region. We constructed a fine map of linkage group 11 of ‘Kinchaku’ and localized the Aki locus within a 1.5-cM genome region between SSR markers Mdo.chr11.28 and Mdo.chr11.34. Marker Mdo.chr11.30 co-segregated with Aki in all 621 F1 plantlets of a ‘Housui’ × ‘Kinchaku’ cross. The physical size of the Aki region, which includes three markers (Mdo.chr11.28, Mdo.chr11.30, and Mdo.chr11.34), was estimated to be 250 Kb in the ‘Golden Delicious’ apple genome and 107 Kb in the ‘Dangshansuli’ Chinese pear genome. Our results will help to identify the candidate gene for susceptibility to black spot disease in Japanese pear.
Keywords: Alternaria alternata, black spot disease, Pyrus pyrifolia, fine mapping, comparative genomics
Introduction
Pears (Pyrus spp.) have been cultivated in East Asia, Europe, and North America for more than 3000 years and are among the most important fruits in over 50 countries in temperate zones (Bell 1990, Bell et al. 1996). The Japanese pear (Pyrus pyrifolia Nakai), European pear (P. communis L.), and Chinese pears (P. bretschneideri Rehd. and P. ussuriensis Maxim.) are the major edible species commercially grown for fruit production (Bell et al. 1996). The Japanese and Chinese pears are cultivated in East Asia, and the European pear is grown in Europe, North America, and temperate regions of the southern hemisphere. All Pyrus species are inter-crossable and there are no major incompatibility barriers to interspecific hybridization in this genus (Westwood and Bjornsta 1971).
Black spot disease, caused by the Japanese pear pathotype of the filamentous fungus Alternaria alternata (Fries) Keissler (previously A. kikuchiana Tanaka), is one of the most serious diseases of Japanese pear cultivated in Asia. Previously, black spot disease was not observed in North America or Europe (Sanada et al. 1988). However, this disease had already occurred in ‘Nijisseiki’ pear orchard in Europe (Baudry et al. 1993, Kohmoto et al. 1992). Bagging and fungicide spraying to prevent infection by A. alternata are costly and labor-intensive (Kozaki 1973). Alternaria fungi produce host-selective toxins (HSTs), the structures of which have been determined in Japanese pear, strawberry, tangerine, apple, tomato, and rough lemon pathotype (Tsuge et al. 2013, Walton 1996). The AK-toxin, which is specifically produced by the Japanese pear pathotype, causes necrosis, early leaf fall, and low yield in Japanese pears, especially in commercial cultivars ‘Nijisseiki’, ‘Shinsui’, and ‘Nansui’ (Nakashima et al. 1985). The range of pear cultivars sensitive to AK-toxin is the same as the host range of the pathogen (Otani et al. 1985). Susceptibility to black spot disease is controlled by the single dominant gene A (Kozaki 1973). Susceptible cultivars are heterozygous (A/a), and homozygous (A/A) cultivars have not been identified. The inactivation of this gene has been attempted to induce disease-resistant mutant. A resistant mutant, γ-1-1 (current cultivar name ‘Gold Nijisseiki’), was selected from ‘Nijisseiki’ plants after chronic irradiation of gamma-rays (Sanada et al. 1988).
Banno et al. (1999) identified random amplified polymorphic DNA (RAPD) markers associated with susceptibility to black spot in the Japanese pear ‘Osa Nijisseiki’; one of these markers, CMNB41, was linked to the susceptibility gene at a genetic distance of 3.1 cM. ‘Kinchaku’, an old Japanese pear cultivar of unknown parentage, is also susceptible to black spot. A genetic linkage map of ‘Kinchaku’ was constructed with RAPD markers (120 loci in 18 linkage groups), and the susceptibility gene was mapped (Iketani et al. 2001). However, these studies did not anchor these linkage groups to the reference linkage maps, and little information was available on molecular markers linked to the gene. In a previous study, we obtained simple sequence repeat (SSR) markers linked to the susceptibility genes in the Japanese pear cultivars ‘Osa Nijisseiki’ (gene Ani) and ‘Nansui’ (gene Ana) (Terakami et al. 2007). We used two mapping populations derived from crosses between ‘Osa Nijisseiki’ and ‘Okusankichi’ (110 F1 plantlets) and between ‘Oushuu’ and ‘Nansui’ (40 F1 plantlets) for genetic analysis of susceptibility to black spot. The SSR markers CH04h02 and CH03d02 were tightly linked to both Ani and Ana. Although Ani and Ana are derived from different cultivars, both genes are located at the top of linkage group 11 (Terakami et al. 2007).
Pears and apples (Malus × domestica Borkh.) belong to the family Rosaceae, subfamily Amygdaloideae, tribe Pyreae, and the two species have the same basic chromosome number (x = 17). The family Rosaceae includes many well-known and beloved species of economic importance, in particular fruits (Janick 2005) and ornamentals, but also some timber crops and many medicinal and nutraceutical plants (Hummer and Janick 2009). It includes from 95 to >100 genera and 2830–3100 species (Judd et al. 1999, Mabberley 1987). Velasco et al. (2010) reported a high-quality draft genome sequence of the domesticated apple. A hybrid assembly of Sanger and 454 reads produced 122,146 contigs, 103,076 of which were assembled into 1,629 metacontigs. The total length of available contigs (603.9 Mb) covers about 81.3% of the estimated apple genome size (742.3 Mb). Whole genome sequences have also been reported for Chinese pear (Wu et al. 2013) and European pear (Chagné et al. 2014). The draft genome sequence of the Chinese pear was determined using a combination of BAC-by-BAC and next-generation sequencing, and the assembled genome consists of 2103 scaffolds with an N50 of 540.8 Kb, totaling 512.0 Mb with 194× coverage, close to the estimated size of 527.0 Mb (Wu et al. 2013). Transferability of SSR information has been reported within the Pyreae (previously Maloideae sensu lato) (Liebhard et al. 2002, Silfverberg-Dilworth et al. 2006, Yamamoto et al. 2001), indicating the collinear synteny between pear and apple in all 17 linkage groups. These published genome sequences of the Pyreae could support and accelerate the genome analysis, including marker development and gene prediction, in Japanese pear.
Alternaria blotch, caused by the apple pathotype of A. alternata (previously A. mali Roberts), is an economically important disease in apple production in Japan and other Asian countries. Saito and Takeda (1984) suggested that susceptibility to Alternaria blotch in apple might be controlled by a single dominant gene, Alt, and that the most susceptible cultivars are heterozygous. SSR markers linked to the susceptibility genes of ‘Starking Delicious’ and ‘Golden Delicious’ have been reported (Li et al. 2011, Moriya et al. 2013).
In this study, we constructed a genetic map of linkage group 11 of ‘Kinchaku’ and identified the exact position of the gene for susceptibility to black spot disease using a large mapping population derived from a ‘Housui’ (syn. ‘Hosui’) × ‘Kinchaku’ cross. ‘Housui’, a leading Japanese pear cultivar, is resistant to black spot disease and has a homozygous recessive genotype (a/a), whereas ‘Kinchaku’ is susceptible and has a heterozygous genotype (A/a). We refer to the susceptibility gene of ‘Kinchaku’ as Aki; therefore, the genotype of ‘Kinchaku’ was considered as Aki/a. Using a comparative genomic approach, we developed a number of apple SSR markers in the region containing the susceptibility gene Aki and used them to fine-map the gene. Novel markers tightly linked to Aki were obtained, and one of them co-segregated with Aki in 621 F1 plantlets.
Materials and Methods
Plant materials and DNA extraction
A mapping population was derived from a cross between Japanese pear cultivars ‘Housui’ and ‘Kinchaku’ and included 621 F1 plantlets. Ungrafted progeny was grown in plastic pot (18 cm diameter × 16 cm height), and maintained in the greenhouse at the NARO Institute of Fruit Tree Science (Tsukuba).
Frozen young leaves (50 mg) were homogenized for 20 s in a Shake Master Auto (Bio Medical Science). Genomic DNA was extracted with a NucleoMag 96 Plant kit (Macherey-Nagel) according to the manufacturer’s instructions, except that lysis buffer MC1 contained 2% 2-mercaptoethanol.
Evaluation of black spot susceptibility or resistance
Responses to black spot disease were evaluated by using the spore inoculation test (Hayashi et al. 1990). Both parental cultivars and all F1 progeny were inoculated with spores of the virulent isolate No. 15A of A. alternata, which was kindly provided by Dr. T. Tsuge (Nagoya University). The isolate was cultured in potato dextrose broth without shaking for 10 days at 25°C. Mycelial mats were washed with tap water to remove culture medium and maintained at 25°C in the dark. The spores formed were collected, suspended in distilled water, and diluted to a concentration of approximately 5 × 105 spores/mL. The spore suspension was sprayed with a glass atomizer onto two young leaves detached from each plant. The inoculated leaves were incubated in a moist chamber at 25°C for 48 h. Leaves were classified into two groups: resistant (no disease symptoms) and susceptible (necrotic symptoms). All inoculation tests were duplicated for all plantlets.
Positional identification of the susceptibility gene Aki
Because the black spot susceptibility genes Ani and Ana were both located in the same region of linkage groups 11, we suspected that Aki might also be in that region. To confirm this, all F1 plantlets of the ‘Housui’ × ‘Kinchaku’ cross were analyzed using SSR markers CH04h02 and CH03d02 (mapped on linkage group 11), which show significant linkage to both Ani and Ana (Terakami et al. 2007).
A total of 18 SSR markers previously mapped on linkage group 11 of pear or apple (Liebhard et al. 2002, Terakami et al. 2014) were tested on the mapping population. These SSRs included 7 apple SSRs (CH02d08, CH02d12, CH03d02, CH04a12, CH04g07, CH04d10, and CH04d07; Liebhard et al. 2002) and 11 pear SSRs (NB111a, RLG1, NH024b, NB118a, NB105a, NH030a, NB135a, IPPN02, IPPN14, TsuENH044, and TsuENH102; Inoue et al. 2007, Nishitani et al. 2009, Yamamoto et al. 2002a, 200b, 2002c).
PCR analysis was performed in a total volume of 10 μL containing 5 μL of 2× GoTaq Colorless Master Mix (Promega), 0.4 μM each of forward and reverse primers, and 2 ng of genomic DNA. DNA was amplified in a GeneAmp PCR system 9700 (Life Technologies) programmed as follows: an initial denaturation step at 94°C for 5 min; 35 cycles of 94°C for 1 min (denaturation), 55°C for 1 min (annealing), and 72°C for 1 min (extension); and a final extension at 72°C for 7 min. Amplified DNA fragments were separated using an Applied Biosystems 3130xl Genetic Analyzer (Life Technologies) with an internal size standard (GeneScan HD 400 ROX; Life Technologies). Data were collected and analyzed using the GeneMapper software version 5.0 (Life Technologies).
Development of novel SSR markers from apple genome sequences
SSR markers for fine mapping of the Aki gene were developed on the basis of synteny between the pear and apple genomes. The data sets for Malus × domestica Whole Genome v1.0 were downloaded from the Genome Database for Rosaceae (https://www.rosaceae.org/species/malus/malus_x_domestica/genome_v1.0). Apple contigs containing CH04h02 and CH03d02 regions were identified by BLASTN. The contigs between CH04h02 and CH03d02 were filtered out according to the GFF file. The identified contigs were searched for potential SSR sequences using the Tandem Repeats Finder software version 4.07b (Benson 1999) with the following alignment parameters: match = 2, mismatch = 11, and indel = 11. We detected di-nucleotide (at least 10 repeats) and tri-nucleotide (at least 6 repeats) SSR motifs. Based on these SSRs, 45 primer pairs (Table 1) were designed with the Primer3 software version 2.3.6 (Koressaar and Remm 2007, Untergasser et al. 2012) using the following parameters: length of 18–23 bp (optimum 20 bp), primer Tm of 56–63°C (optimum 60°C), a maximum Tm difference of 1°C, a primer GC content of 45–65% (optimum 55%), and a product size range of 90–360 bp. Novel apple SSR markers developed from the draft sequence of ‘Golden Delicious’ are indicated as Mdo.chr11. SSR analyses were performed as described above.
Table 1.
Characteristics of novel SSR markers developed from apple contigs
| Marker name | Primer sequence (5′ to 3′) | Origin of apple contig | Contig start position (bp) | SSR start position in contig (bp) | Motif typea | Copy numbera |
|---|---|---|---|---|---|---|
| Mdo.chr11.1 | Forward: CGAACTTCAGGTGAGTGGGT Reverse: AAGCATACATGTGCATCCCA |
MDC002629.387 | 2,513,421 | 3,294 | GT | 14.5 |
| Mdo.chr11.2 | Forward: ACACCGAGAGGACTCGAAGA Reverse: CTGTCTGCTTAGAGACGGGG |
MDC002629.387 | 2,513,421 | 17,035 | AT | 16 |
| Mdo.chr11.3 | Forward: TTCATGACCAACCCAGTTGA Reverse: AACAGACTGACGAACCGACC |
MDC018137.214 | 3,057,173 | 7,237 | GA | 11 |
| Mdo.chr11.4 | Forward: GTACCGGGTGCTTTTCGTTA Reverse: GTCTTGTGCTTGAGAGCGTG |
MDC011303.296 | 3,506,644 | 10,412 | TC | 10 |
| Mdo.chr11.5 | Forward: CCTTCATGAGGAACTCCATCC Reverse: TTGAGTTTGCAGCCATTGAG |
MDC011303.296 | 3,506,644 | 20,684 | TC | 18 |
| Mdo.chr11.6 | Forward: ATTGGGAGGGAAAAACAAGG Reverse: ACATTACTACGCCCTGCCAC |
MDC010086.240 | 4,014,253 | 12,847 | AG | 11 |
| Mdo.chr11.7 | Forward: TTGTTTGTCTGGGAAAAGGG Reverse: GAATGTACGCACGGCTTTCT |
MDC019643.199 | 4,485,593 | 939 | CAC | 6.7 |
| Mdo.chr11.8 | Forward: CACTCGCTATCCTCCTCCTG Reverse: GCAGAGTGGTGGGGTTTAGA |
MDC019643.199 | 4,485,593 | 11,432 | CT | 14 |
| Mdo.chr11.9 | Forward: TCTGTTTGAACCTCCATCCC Reverse: TTGTTGGCAACCTCACAAAC |
MDC020062.119 | 5,061,058 | 13,509 | GA | 13 |
| Mdo.chr11.10 | Forward: CAACCAACTGCTCAAGTCAA Reverse: TGCTGCTACTGTGAAGTTCG |
MDC020062.119 | 5,061,058 | 15,569 | AC | 22.5 |
| Mdo.chr11.11 | Forward: GGAAACCCTAACCATGCAAA Reverse: AGAGATACCAGAGGGGCGAT |
MDC020062.119 | 5,061,058 | 27,436 | AG | 22.5 |
| Mdo.chr11.12 | Forward: AGCCATGGCCACAGTTTAAT Reverse: GGCCTGTTCGATATCTTTGC |
MDC012329.239 | 5,445,700 | 23,490 | ATG | 7.7 |
| Mdo.chr11.13 | Forward: GCACAATTTCCCAAACAAGG Reverse: CGATATGCGTGCTAGGGAGT |
MDC019380.167 | 1,999,875 | 3,661 | AT | 23.5 |
| Mdo.chr11.14 | Forward: CCATGCATGACCTATTTCCC Reverse: TTCGATAAGTTGGCACGTCA |
MDC040598.17 | 2,138,383 | 10,758 | AT | 22 |
| Mdo.chr11.15 | Forward: CTGCGTTCGGTTAGATCACA Reverse: TTGCCTTATCGCCCATAATC |
MDC005041.466 | 2,154,005 | 3,099 | TA | 11 |
| Mdo.chr11.16 | Forward: CAAGAGGACTCGAAGATGCC Reverse: TCCGTTCCTCTCCGATACAC |
MDC015929.398 | 2,282,115 | 4,552 | AT | 12 |
| Mdo.chr11.17 | Forward: TACAAGGGTTTTGGGTCTCG Reverse: ATTGGTGGTGATTGGTTGGT |
MDC004271.236 | 2,444,754 | 3,766 | AG | 14 |
| Mdo.chr11.18 | Forward: TACCTAAAACCTGGAGGGGC Reverse: TACCACAAGGATTCCGGCTA |
MDC020758.380 | 2,460,173 | 5,968 | AT | 10.5 |
| Mdo.chr11.19 | Forward: CGCTAGAGGATGGGTTTTGC Reverse: GGCGTTGGAAGACCTATTGC |
MDC010399.168 | 2,470,302 | 1,066 | TA | 19 |
| Mdo.chr11.20 | Forward: GTGGAAACGGAAGTTGTGGT Reverse: TATCGGGTTCCAGACCACTC |
MDC009710.295 | 2,554,186 | 3,571 | TA | 15.5 |
| Mdo.chr11.21 | Forward: CACCAACGGTGTACGATGAG Reverse: CCACTAGATACGCACAAAACCA |
MDC024524.27 | 2,613,603 | 20,758 | AT | 26 |
| Mdo.chr11.22 | Forward: ATGATTTATGAAGGCAGCCG Reverse: GGTCCTCATCCGAAACTGAA |
MDC012704.279 | 2,659,409 | 14,275 | TA | 14.5 |
| Mdo.chr11.23 | Forward: GAGATATTGCCCTCCATTCG Reverse: CAAGACACTGTTGGATTCAGTC |
MDC022706.249 | 2,698,057 | 22,759 | AT | 11 |
| Mdo.chr11.24 | Forward: GTTCATGTTACGGATTGCAG Reverse: GAAAGCCTAGCCTCCTAGTCC |
MDC002601.127 | 2,773,258 | 11,709 | AT | 12 |
| Mdo.chr11.25 | Forward: CATGTTGTAAGCCCTCGGAT Reverse: ACTCCCACCAAGGAAGAGGT |
MDC016602.78 | 2,811,403 | 13,177 | AG | 21.5 |
| Mdo.chr11.26 | Forward: AGGGGATCCAATTCCTAACG Reverse: AAATCGTCACTGGAACTCCG |
MDC018345.161 | 2,881,408 | 6,536 | TC | 10.5 |
| Mdo.chr11.27 | Forward: TGACTTCAGCCTGCTAAACCT Reverse: TCACTCTCCCTTTTCATGCAC |
MDC018345.160 | 2,897,025 | 6,262 | AT | 16 |
| Mdo.chr11.28 | Forward: CAGATTCACCGAGTTGCAGA Reverse: AGGGTTTTGGTTGACAGTCG |
MDC001444.132 | 2,910,449 | 4,945 | AG | 15 |
| Mdo.chr11.29 | Forward: ACGCTCTTCCCAGCAAAATA Reverse: CCGTGGCGAAAATACAACTT |
MDC003953.308 | 2,947,393 | 3,151 | AG | 15.5 |
| Mdo.chr11.30 | Forward: TAACCGTTGCAAACCCTCTC Reverse: GATGGATGGAAACAAATGGG |
MDC017308.389 | 3,030,417 | 10,676 | TC | 11 |
| Mdo.chr11.31 | Forward: GCAAGGAACCAGCTGACAGT Reverse: CAGCACCGTCGTCTGATCTA |
MDC019519.287 | 3,049,141 | 607 | TA | 13.5 |
| Mdo.chr11.32 | Forward: TCCTAAAGCAGCCACTCCTC Reverse: GAGATGCCAACCAGGAAAAG |
MDC027417.11 | 3,109,934 | 9,953 | TAA | 8 |
| Mdo.chr11.33 | Forward: GAGATCTCCTGCGTTTCTGG Reverse: TTCCCGCCCCTATCTCTATT |
MDC021160.220 | 3,150,224 | 6,672 | GA | 15.5 |
| Mdo.chr11.34 | Forward: AACAACCGAACCGATCTTTG Reverse: CGACGCGTACAATTCGTTAT |
MDC021160.236 | 3,155,605 | 10,288 | GA | 15 |
| Mdo.chr11.35 | Forward: TGATTTTTGGTGGAAGGCTC Reverse: GATCTCAATCAGGGACCCAA |
MDC001844.336 | 3,186,360 | 14,311 | TC | 23 |
| Mdo.chr11.36 | Forward: TGGACTCCAAACACCGAATAG Reverse: CCAAGAGGGACAAATTGGAA |
MDC016570.325 | 3,342,482 | 8,236 | TA | 19.5 |
| Mdo.chr11.37 | Forward: CTTTAGCGGTATGGCTTTGG Reverse: GAGGGCAGACAATTCACGAG |
MDC005623.477 | 3,352,403 | 7,157 | CT | 14.5 |
| Mdo.chr11.38 | Forward: TCCGAAGTGAGGCTACAAAA Reverse: CTTCTTCCAACTTGTTCGCC |
MDC003776.129 | 3,542,189 | 22,300 | TA | 11.5 |
| Mdo.chr11.39 | Forward: ATGTGGTGTCCTTTTGAGCC Reverse: GAATGTCTTGCTAGCCTCGG |
MDC013005.433 | 3,579,304 | 30,338 | AG | 23.5 |
| Mdo.chr11.40 | Forward: GGGTTTTCATGGGTGATGTT Reverse: TAAACCCGACCCGTTTACAG |
MDC005218.335 | 3,632,162 | 13,823 | AT | 16 |
| Mdo.chr11.41 | Forward: CCAACAAAGCACTCACATGG Reverse: TGTGCTCAAAAAGTGGATGC |
MDC013149.562 | 3,661,752 | 8,716 | AT | 11.5 |
| Mdo.chr11.42 | Forward: CGGTCCACTACTAGCCCTCA Reverse: GACCACATTGGTTTGAGAGTGA |
MDC022698.364 | 3,695,978 | 5,597 | TA | 17 |
| Mdo.chr11.43 | Forward: ATCGGTTACGTTTGCTTGGT Reverse: ATGAAGGAGTGGCTGCTTGT |
MDC010666.357 | 3,929,341 | 4,280 | TC | 16 |
| Mdo.chr11.44 | Forward: CCGGAAGGGTATTGTGAAAT Reverse: TGGCCAAGTATCAATGTGGA |
MDC003759.133 | 3,948,061 | 4,779 | AT | 24.5 |
| Mdo.chr11.45 | Forward: CCCTCGACAAGAATTGGGTA Reverse: CCTAACCGCCAGAAAAATCA |
MDC012710.225 | 3,984,326 | 12,533 | GA | 10.5 |
Motif type and copy number are estimated on apple genome sequences.
Linkage analysis and construction of genetic linkage group 11
Linkage group 11 of ‘Kinchaku’ was constructed using JoinMap 4.1 software (Van Ooijen 2006), and a pseudo-testcross strategy was applied to create genetic linkage maps (Grattapaglia and Sederoff 1994). An independence LOD score of 10.0 was used to define linkage groups. The regression mapping algorithm was selected for building linkage maps, and map distances were calculated according to Kosambi’s mapping function (Kosambi 1943). The linkage map was drawn using MapChart 2.2 software (Voorrips 2002).
Homologous sequences around the Aki gene in the Chinese pear genome
We conducted BLASTN searches against a high-quality draft genome sequence of the diploid Chinese pear ‘Dangshansuli’ (Wu et al. 2013) (Pbr_v1.0; http://peargenome.njau.edu.cn/) to identify scaffolds with homology to the 1.5-cM region that includes the Aki locus of ‘Kinchaku’. Apple genome sequences amplified by three primer pairs (Mdo.chr11.28, Mdo.chr11.30, and Mdo.chr11.34) were used as queries; an e-value cutoff of 1e-30 was used.
Results
Evaluation of black spot susceptibility or resistance
The parental cultivars and all F1 progeny were evaluated for susceptibility or resistance to black spot. Identical results were obtained in all duplicate tests. The susceptible parent ‘Kinchaku’ showed severe symptoms, whereas the resistant parent ‘Housui’ showed no symptoms (Fig. 1).
Fig. 1.
Black spot disease symptoms of ‘Housui’ and ‘Kinchaku’ at 48 hours after inoculation with spore suspension of A. alternata. The resistant cultivar ‘Housui’ shows no visible disease symptoms (A), while necrotic lesions on leaves were observed in the susceptible cultivar ‘Kinchaku’ (B).
Of 621 F1 progeny of the ‘Housui’ × ‘Kinchaku’ cross, 309 showed no symptoms and 312 showed necrotic symptoms. Resistant and susceptible progeny could be clearly distinguished because no plantlets showed an intermediate response. The segregation ratio of resistant to susceptible plants fitted the expected ratio of 1:1 in the Chi-square test (χ2 = 0.014, P value = 0.904), in good accordance with previous reports, which used different combinations of susceptible and resistant parents (Kozaki 1973).
Linkage of Aki to SSR markers on linkage group 11
The SSR markers CH04h02 and CH03d02, developed from the apple genome sequence (Liebhard et al. 2002), show significant linkage to susceptibility genes Ani (in ‘Osa Nijisseiki’) and Ana (in ‘Nansui’) (Terakami et al. 2007). In the present study, CH03d02 showed scorable polymorphism, i.e., a heterozygous genotype in ‘Kinchaku’ and polymorphic band patterns between ‘Housui’ and ‘Kinchaku’, whereas CH04h02 was not polymorphic and therefore could not be scored. CH03d02 was significantly linked to Aki with a genetic distance of 17.6 cM (LOD score: 64.5). This result indicated that Aki was located at the top of linkage group 11, i.e., its position was very similar to those of Ani and Ana.
Map construction and fine mapping of Aki
Our results indicated that the susceptibility gene Aki was located between CH04h02 and CH03d02. To fine-map the Aki locus, we developed apple SSRs located between the two markers. In BLASTN searches, CH04h02 and CH03d02 showed high sequence similarity to apple genome contigs MDC010914.246 (e-value: 1e-116) and MDC005828.284 (1e-100), respectively. MDC010914.246 and MDC005828.284 are located on apple chromosome 11 (231,017–250,688 bp and 8,976,681–8,984,436 bp, respectively). We searched an approximately 8.7 Mb region on apple chromosome 11 (231,017–8,984,436 bp; 1,996 contigs) for di- and tri-nucleotide SSRs and designed 45 primer sets from those contigs (Table 1).
We also tested 18 SSR markers mapped previously on linkage groups 11 of pear or apple (Liebhard et al. 2002, Terakami et al. 2014) to establish linkage group 11 in ‘Kinchaku’. Eight markers including CH03d02 were polymorphic in the mapping population and were mapped on linkage group 11 of ‘Kinchaku’. Out of the 45 new apple SSR markers, 16 (36%) were polymorphic and were mapped on linkage group 11 of ‘Kinchaku’; 14 markers (31%) showed no polymorphisms, 10 markers (22%) did not amplify bands of the expected size, and 5 markers (11%) were polymorphic but were not mapped on linkage group 11 of ‘Kinchaku’.
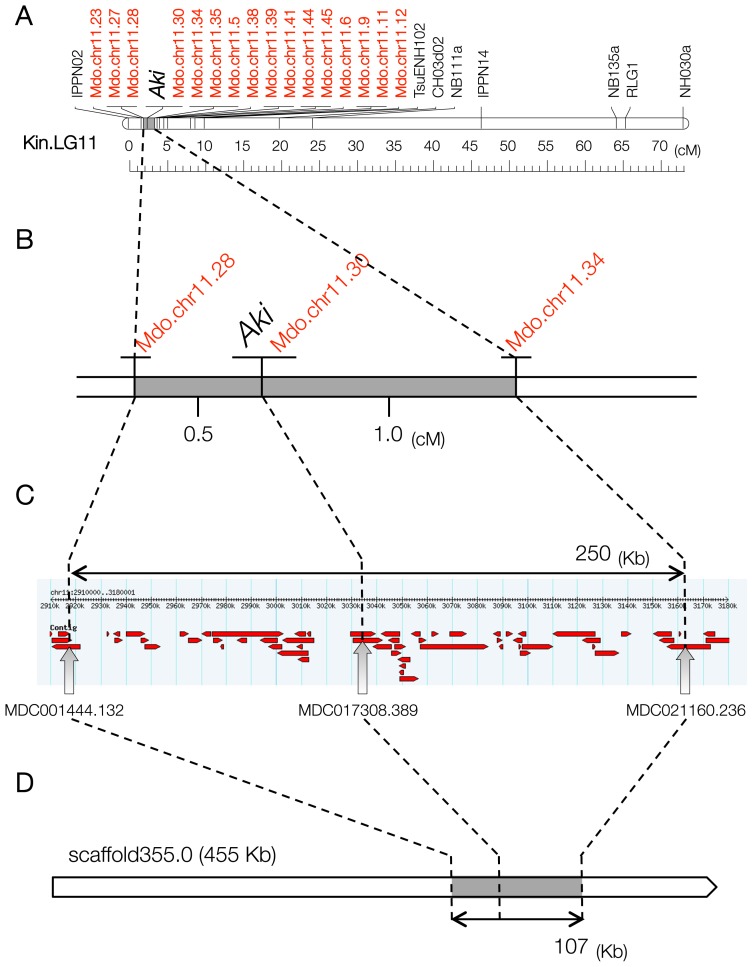
Thus, the map of linkage group 11 of ‘Kinchaku’ consisted of 24 SSR markers and the black spot susceptibility gene (Fig. 2A). It spanned 73.1 cM and had an average marker density of 2.9 cM per marker (Supplemental Fig. 1). Aki was located within a 1.5-cM genome region between the Mdo.chr11.28 and Mdo.chr11.34 markers (Fig. 2B). In the mapping population, three progeny were identified with recombination between Mdo.chr11.28 and Aki and six between Mdo.chr11.34 and Aki. No double recombination events were detected between Mdo.chr11.28 and Mdo.chr11.34. Mdo.chr11.30 co-segregated with Aki in all 621 F1 plantlets. The segregation ratio was not distorted at any locus. The positions of novel apple SSR markers were well conserved in the Japanese pear (Table 1, Fig. 2A).
Fig. 2.
Comparative map of Japanese pear, apple, and Chinese pear. A. Linkage group 11 in the Japanese pear ‘Kinchaku’. Aki, the black spot susceptibility gene. Mdo.chr11, novel apple SSR markers developed from the ‘Golden Delicious’ draft genome sequence. The downside scale marks genetic distance (cM). B. Fine map of Aki and flanking regions. The numbers between markers indicate genetic distance (cM). C. Physical map of an apple genome segment (250 Kb) between Mdo.chr11.28 and Mdo.chr11.34 on chromosome 11 of ‘Golden Delicious’. A screen snapshot of the Malus × domestica Whole Genome v1.0 view in GBrowse (https://www.rosaceae.org/gb/gbrowse/malus_x_domestica/). Red arrows indicate contigs. D. A scaffold of Chinese pear (‘Dangshansuli’) anchored to the region containing the Aki locus by using closely linked SSR markers (Mdo.chr11.28, Mdo.chr11.30, and Mdo.chr11.34). Block arrow indicates strand polarity.
To verify that Mdo.chr11.30 was tightly linked to the genes for susceptibility to black spot disease, we carried out genotyping with Mdo.chr11.30 in 110 F1 plantlets derived from a cross between ‘Osa Nijisseiki’ and ‘Okusankichi’, and found co-segregation of Mdo.chr11.30 with Ani (data not shown).
Aki was located between Mdo.chr11.28 and Mdo.chr11.34, which were developed from contigs MDC001444.132 and MDC021160.236, respectively (Table 2). The physical size of the region between these markers was approximately 250 Kb (2,915–3,165 Kb), including several gaps in the apple genome (Fig. 2C).
Table 2.
DNA sequences in the Chinese pear genome (Pbr_v1.0) homologous to novel apple SSR markers
| SSR locus in the apple genome | Homologous scaffold in the Chinese pear genome | E-value | Alignment start position in Chinese pear scaffold (bp) | Alignment end position in Chinese pear scaffold (bp) |
|---|---|---|---|---|
| Mdo.chr11.28 | scaffold289.0 | 1e-80 | 347,627 | 347,400 |
| scaffold355.0 | 2e-73 | 272,825 | 272,626 | |
| Mdo.chr11.30 | scaffold355.0 | 4e-64 | 311,449 | 311,613 |
| Mdo.chr11.34 | scaffold355.0 | 1e-71 | 379,953 | 379,705 |
| scaffold153.0 | 1e-71 | 484,759 | 484,511 |
Scaffold containing Aki in the Chinese pear genome
Although a high-quality draft genome sequence of the diploid Chinese pear ‘Dangshansuli’ has been released and its 2103 scaffolds are available, their chromosomal positions are unknown. We surveyed Chinese pear scaffolds corresponding to apple genome sequences that include markers Mdo.chr11.28, Mdo.chr11.30, and Mdo.chr11.34.
Apple genome sequences amplified by the primer pairs corresponding to these three markers (Table 1) were used as queries in a BLASTN search against the Chinese pear genome (Table 2). Mdo.chr11.28 and Mdo.chr11.34 showed sequence similarity to multiple genome regions. The best hit for Mdo.chr11.28 was a 228-bp region in scaffold289.0 and the second best hit was in scaffold355.0 (272,626–272,825 bp; e-value: 2e-73). The latter scaffold also contained hits for Mdo.chr11.30 (a 165-bp region; 311,449–311,613 bp) and for Mdo.chr11.34 (a 249-bp region; 379,705–379,953 bp) (Table 2). These results indicated that the Aki gene is located in a 107-Kb region of scaffold355.0 (272,626–379,953 bp) of the Chinese pear genome (Fig. 2D).
Discussion
Using several anchored SSRs previously mapped in apple and pear (Silfverberg-Dilworth et al. 2006, Terakami et al. 2014), we constructed a genetic linkage map of linkage group 11 of the Japanese pear ‘Kinchaku’ that included the susceptibility gene Aki; the map consisted of 25 loci and spanned 73.1 cM (Fig. 2A, Supplemental Fig. 1). The position of Aki was identified at the top of linkage group 11. The marker CH03d02, which is linked to the susceptibility genes Ani of ‘Osa Nijisseiki’ and Ana of ‘Nansui’ (Terakami et al. 2007), was also significantly linked to Aki of ‘Kinchaku’. The reference linkage maps of pear and apple were considered to be saturated (Silfverberg-Dilworth et al. 2006, Terakami et al. 2014). Comparison of the positions of anchored SSRs shows that the linkage map of ‘Kinchaku’ covers most of linkage group 11. Several RAPD markers linked to the susceptibility gene of ‘Kinchaku’ have been previously obtained and a genetic linkage map of ‘Kinchaku’ was constructed (120 loci, 18 linkage groups, 768 cM; Iketani et al. 2001). However, the linkage groups were not anchored to the reference linkage maps of pear or apple. The current study is the first to identify the position of Aki.
Several virulent and non-virulent isolates of A. alternata were reported, and strain No. 15A, used in this study, is an AK-toxin producer (Hayashi et al. 1990). All isolates of the pathogen that produce HSTs are pathogenic to the specific host; all isolates that fail to produce HSTs lack pathogenicity to the host plants (Tsuge et al. 2013). The range of pear cultivars sensitive to AK-toxin and spore suspension is the same as the host range of the pathogen (Kozaki 1974, Otani et al. 1985). The segregation ratio of resistant and susceptible progeny fitted the expected ratio of 1:1 in the Chi-square test. This result is in good agreement with a previous report that susceptibility of ‘Kinchaku’ to black spot is controlled by a single dominant gene (Kozaki 1973). In our previous study, we proposed different names (Ani and Ana) for the susceptibility genes in ‘Osa Nijisseiki’ and ‘Nansui’, respectively, because these genes are derived from different cultivars (Terakami et al. 2007). ‘Osa Nijisseiki’ is a self-compatible mutant of the native Japanese cultivar ‘Nijisseiki’ and is susceptible to black spot. ‘Nansui’ and ‘Doitsu’ are considered to carry the same susceptibility gene, Ana, because ‘Nansui’ is derived from the native Japanese cultivar ‘Doitsu’ (Terakami et al. 2007). In this study, we named the susceptibility gene of ‘Kinchaku’ as Aki; the parentage of this native Japanese pear cultivar has not been identified. All three susceptibility genes, Aki, Ani, and Ana, have been mapped at the top of linkage group 11, indicating that they are located very close to each other or have the same origin. Fine mapping of the Aki gene and determining nucleotide sequences flanking this gene were the starting point for a positional cloning of genes for susceptibility to black spot disease. The cloning of the susceptibility gene, Aki, represents the basis for further investigation of the susceptibility mechanism. It also takes a step toward identifying the homology and the functional relationships among susceptibility genes, Aki, Ana, and Ani.
In a genome-wide association study in the Japanese pear, Iwata et al. (2013) showed that the SSR marker CH04h02 was significantly associated with resistance to black spot. Although 76 Japanese pear varieties (31 modern elite cultivars, 19 old cultivars, 17 indigenous cultivars, and 9 breeding lines) were used in this study, ‘Kinchaku’ was not included. The 172-bp allele of CH04h02, which is linked in coupling phase with the susceptibility allele Ani, had the second-largest negative effect on resistance (Iwata et al. 2013). The Aki was located at the top of linkage group 11 and ‘Kinchaku’ has the 172-bp allele of CH04h02 support the results of Iwata et al. (2013).
Several apple SSR markers have been successfully used in pears such as P. pyrifolia, P. bretschneideri, P. ussuriensis, P. communis, and P. calleryana (Yamamoto et al. 2001). Transferability of apple SSR information has been reported in some other species such as Amelanchier canadensis, Cotoneaster dammeri, Cydonia oblonga, Sorbus domestica, Prunus armeniaca, P. persica, and P. salicina (Liebhard et al. 2002). Similarly, SSR markers derived from pear and Sorbus torminalis were mapped on apple reference maps (Silfverberg-Dilworth et al. 2006). Pear reference linkage maps were constructed with apple SSR markers, and several apple SSRs were mapped on the same linkage groups in pear and apple (Terakami et al. 2009, Yamamoto et al. 2007). In addition, SSR loci were identified in similar regions of linkage groups, indicating collinear synteny between pear and apple in all 17 linkage groups.
Using a comparative genomic approach, we developed several apple SSR markers in the target region (the top of linkage group 11 in pear). Novel apple SSR markers developed from the draft sequence of ‘Golden Delicious’ are indicated as Mdo.chr11 (Fig. 2A). Of 45 novel apple SSR markers, 3 markers (Mdo.chr11.28, Mdo.chr11.30, and Mdo.chr11.34) were closely linked to the gene for susceptibility to black spot disease. The susceptibility gene, Aki, was located within a 1.5-cM genome region between Mdo.chr11.28 and Mdo.chr11.34, whereas Mdo.chr11.30 co-segregated with Aki in all 621 F1 plantlets of the ‘Housui’ × ‘Kinchaku’ cross (Fig. 2B).
Alternaria blotch of apple, black spot of Japanese pear, and five other diseases are now known to be caused by A. alternata (Tsuge et al. 2013). Alternaria blotch, caused by the apple pathotype of A. alternata, is a destructive disease that can greatly reduce apple quality and yield. The degree of resistance or susceptibility of many cultivars has been previously reported (Abe et al. 2010, Saito and Takeda 1984). Saito and Takeda (1984) proposed that a single dominant gene, Alt, controls susceptibility to this disease and that the genotype of resistant cultivars is alt/alt. Using two populations of 57 each F1 individuals derived from a cross between ‘Starking Delicious’ (susceptible, Alt/alt) and ‘Jonathan’ (resistant, alt/alt), Moriya et al. (2013) constructed linkage group 11 of ‘Starking Delicious’, which contained 12 markers spanning 22.1 cM, with an average of 1.8 cM between markers; Alt was located 6.7 cM from the top of linkage group 11 (Moriya et al. 2013). Alternaria blotch and black spot disease may have a common genetic basis in the two host species: host susceptibility is controlled by single dominant genes (Alt or A) located in similar positions at the top of linkage group 11 in both Japanese pear and apple.
A total of 54 contigs and several gaps were reported between MDC001444.132 and MDC021160.236, and 35 putative genes were predicted in the data sets for Malus × domestica Whole Genome v1.0. The susceptibility gene derived from ‘Golden Delicious’ is linked to the SSR marker CH05g07 at 5.6 cM (Li et al. 2011), but this marker was mapped on two linkage groups, 12 and 14 (Silfverberg-Dilworth et al. 2006). These results contradict the results of Moriya et al. (2013). This inconsistency may be caused by the use of different susceptible cultivars and inoculation methods (Moriya et al. 2013): Li et al. (2011) defined ‘Golden Delicious’ as susceptible, whereas other studies have found it to be resistant to Alternaria blotch (Abe et al. 2010, Saito and Takeda 1984). For these reasons, it is difficult to identify and characterize the gene for susceptibility to black spot disease using a comparative genomic approach on the basis of the published apple genome.
The Chinese pear genome size (527.0 Mb) is smaller than that of apple (742.3 Mb; Velasco et al. 2010), and the current version of the Chinese pear genome (Pbr_v1.0) is more informative for comparative genomics in the Japanese pear. The draft genome of the Chinese pear ‘Dangshansuli’, obtained using a combination of BAC-by-BAC and next-generation sequencing technologies, consists of 2103 scaffolds with an N50 of 540.8 Kb, and the total 512.0-Mb sequence corresponds to 97.1% of the estimated genome size (Wu et al. 2013). The draft genome of the European pear (Bartlett v1.0) has also been reported (Chagné et al. 2014), but this sequence is fragmented and consists of 142,083 scaffolds. Because the scaffolds of the Chinese pear genome are not anchored to chromosomes, we used the apple genome for synteny-based marker enrichment. Our results show that the susceptibility gene (Aki) is located within a 1.5-cM region between two apple SSR markers, Mdo.chr11.28 and Mdo.chr11.34 (Fig. 2B). We also surveyed the Chinese pear scaffold containing the Aki locus. A BLAST search against the Chinese pear genome revealed that all three markers showed high similarity to scaffold355.0 (Table 2). The physical size of the Aki locus defined by the three markers is 107 Kb (272,626–379,953 bp of scaffold355.0) in the Chinese pear genome (Fig. 2D).
Linkage maps were constructed to use for anchoring and orienting the scaffolds. A large number of single-nucleotide polymorphism (SNP) and SSR markers have been developed from the genome sequence of the Chinese pear, and several scaffolds have been anchored to 17 linkage groups (Chen et al. 2015, Wu et al. 2014). Wu et al. (2014) constructed a high-density linkage map of the Chinese pear using restriction-associated DNA sequencing technology, and a SNP marker developed from scaffold355.0 was mapped on linkage group 11. Their report supports our results. Five putative genes were predicted in the 107-Kb region, but those genes are not annotated in the Chinese pear genome. As information about the response of ‘Dangshansuli’ to black spot disease is no information, it is difficult to predict the candidate gene for the Aki locus in the Chinese pear genome.
Pear scab (caused by Venturia nashicola) is one of the most harmful diseases of pears, especially Japanese and Chinese pear species. V. nashicola is pathogenic only for Asian pears, and is not pathogenic for European pears (Bell et al. 1996, Ishii et al. 2002). None of the major commercial Japanese pear cultivars are resistant to scab disease caused by V. nashicola (Bell et al. 1996, Ishii et al. 1992), but no scab symptoms were observed on the indigenous Japanese pear ‘Kinchaku’ (Abe and Kotobuki 1998, Ishii et al. 1992). Inheritance analysis indicated that this resistance is controlled by a single dominant gene (Abe and Kotobuki 1998). Six DNA markers, showing close linkages to a resistance gene, were identified, and the resistance gene was mapped in the central region of linkage group 1 (Terakami et al. 2006). Marker-assisted selection approach with regard to the pear scab resistance of ‘Kinchaku’ has been successfully tested (Gonai et al. 2009). However, ‘Kinchaku’ is susceptibility to black spot disease, while is resistance to pear scab. When ‘Kinchaku’ is used as the parent cultivar, about half of the progeny showed susceptibility to black spot disease. Molecular markers closely linked to the gene for susceptibility to black spot disease and the scab resistance gene will be useful for improving pear breeding by marker-assisted selection.
Supplementary Material
Acknowledgments
We are grateful to Mss. F. Hosaka, N. Shigeta, H. Oshino, and N. Yagihashi for their technical assistance. The virulent isolate No. 15A of A. alternata was provided by Dr. Takashi Tuge, Nagoya University, Japan. This work was partially supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics-based Technology for Agricultural Innovation, HOR-2001). Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
Literature Cited
- Abe, K. and Kotobuki, K. (1998) Inheritance of high resistance to Venturia nashicola Tanaka et Yamamoto in Japanese pear (Pyrus pyrifolia Nakai) and Chinese pear (P. ussuriensis Maxim.). J. Japan. Soc. Hort. Sci. 67: 677–680. [Google Scholar]
- Abe, K., Iwanami, H., Kotoda, N., Moriya, S. and Takahashi, S. (2010) Evaluation of apple genotypes and Malus species for resistance to Alternaria blotch caused by Alternaria alternata apple pathotype using detached-leaf method. Plant Breed. 129: 208–218. [Google Scholar]
- Banno, K., Ishikawa, H., Hamauzu, Y. and Tabira, H. (1999) Identification of a RAPD marker linked to the susceptible gene of black spot disease in Japanese pear. J. Japan. Soc. Hort. Sci. 68: 476–481. [Google Scholar]
- Baudry, A., Morzières, J.P. and Larue, P. (1993) First report of Japanese pear black spot caused by Alternaria kikutiana in France. Plant Disease 77: 428. [Google Scholar]
- Bell, R.L. (1990) Pears (Pyrus). In: Moore, J.N. and Ballington J.R. Jr. (eds.) Genetic resources of temperate fruit and nut crops I, International Society for Horticultural Science, Wageningen, The Netherlands, pp. 655–697. [Google Scholar]
- Bell, R.L., Quamme, H.A., Layne, R.E.C. and Skirvin, R.M. (1996) Pears. In: Janick, J. and Moore J.N. (eds.) Fruit breeding, vol. I: Tree and tropical fruits, John Wiley & Sons, London, pp. 441–514. [Google Scholar]
- Benson, G. (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagné, D., Crowhurst, R.N., Pindo, M., Thrimawithana, A., Deng, C., Ireland, H., Fiers, M., Dzierzon, H., Cestaro, A., Fontana, P.et al. (2014) The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’). PLoS ONE 9: e92644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Song, Y., Li, L.T., Khan, M.A., Li, X.G., Korban, S.S., Wu, J. and Zhang, S.L. (2015) Construction of a high-density simple sequence repeat consensus genetic map for pear (Pyrus spp.). Plant Mol. Biol. Rep. 33: 316–325. [Google Scholar]
- Gonai, T., Terakami, S., Nishitani, C., Yamamoto, T. and Kasumi, M. (2009) The validity of marker-assisted selection using DNA markers linked to a pear scab resistance gene (Vnk) in two populations. J. Japan. Soc. Hort. Sci. 78: 49–54. [Google Scholar]
- Grattapaglia, D. and Sederoff, R. (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137: 1121–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, N., Tanabe, K., Tsuge, T., Nishimura, S., Kohmoto, K. and Otani, H. (1990) Determination of host-selective toxin production during spore germination of Alternaria alternata by high-performance liquid chromatography. Phytopathology 80: 1088–1091. [Google Scholar]
- Hummer, K.E. and Janick, J. (2009) Rosaceae: Taxonomy, economic importance, genomics. In: Gardiner, S.E. and Folta K.M. (eds.) Plant Genetics/Genomics vol 6: Genetics and genomics of Rosaceae, Springer, New York, pp. 1–17. [Google Scholar]
- Iketani, H., Abe, K., Yamamoto, T., Kotobuki, K., Sato, Y., Saito, T., Terai, O., Matsuta, N. and Hayashi, T. (2001) Mapping of disease-related genes in Japanese pear using a molecular linkage map with RAPD markers. Breed. Sci. 51: 179–184. [Google Scholar]
- Inoue, E., Matsuki, Y., Anzai, H. and Evans, K. (2007) Isolation and characterization of microsatellite markers in Japanese pear (Pyrus pyrifolia Nakai). Mol. Ecol. Notes 7: 445–447. [Google Scholar]
- Ishii, H., Udagawa, H., Nishimoto, S., Tsuda, T. and Nakashima, H. (1992) Scab resistance in pear species and cultivars. Acta Phytopathol. Entomol. Hung. 27: 293–298. [Google Scholar]
- Ishii, H., Watanabe, H. and Tanabe, K. (2002) Venturia nashicola: Pathological specialization on pears and control trial with resistance inducers. Acta Hortic. 587: 613–621. [Google Scholar]
- Iwata, H., Hayashi, T., Terakami, S., Takada, N., Sawamura, Y. and Yamamoto, T. (2013) Potential assessment of genome-wide association study and genomic selection in Japanese pear Pyrus pyrifolia. Breed. Sci. 63: 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janick, J. (2005) The origins of fruits, fruit growing, and fruit breeding. Plant Breed. Rev. 25: 255–320. [Google Scholar]
- Judd, W.S., Campbell, C.S., Kellogg, E.A. and Stevens, P.F. (1999) Phylogenetic relationships of angiosperms. Plant Systematics: A phylogenetic approach, Sinauer Associates, Sunderland, pp. 290–306. [Google Scholar]
- Kohmoto, K., Otani, H., Cavanni, P. and Bugiani, R. (1992) Occurrence of the Japanese pear pathotype of Alternaria alternata in Japanese pear orchards in Italy. Phytopathol. Mediterr. 31: 141–147. [Google Scholar]
- Koressaar, T. and Remm, M. (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23: 1289–1291. [DOI] [PubMed] [Google Scholar]
- Kosambi, D.D. (1943) The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Kozaki, I. (1973) Black spot disease resistance in Japanese pear. I. Heredity of the disease resistance. Bull. Hortic. Res. Stn. (Minist. Agric. For.) Ser. A 12: 17–27. [Google Scholar]
- Kozaki, I. (1974) Black spot disease resistance in Japanese pear. II. Early evaluation of the disease resistance. Bull. Fruit Tree Res. Stn. Ser. A 1: 13–24. [Google Scholar]
- Li, Y., Zhang, L.Y., Zhang, Z., Cong, P.H. and Cheng, Z.M. (2011) A simple sequence repeat marker linked to the susceptibility of apple to alternaria blotch caused by Alternaria alternata apple pathotype. J. Amer. Soc. Hort. Sci. 136: 109–115. [Google Scholar]
- Liebhard, R., Gianfranceschi, L., Koller, B., Ryder, C.D., Tarchini, R., Van de Weg, E. and Gessler, C. (2002) Development and characterisation of 140 new microsatellites in apple (Malus × domestica Borkh.). Mol. Breed. 10: 217–241. [Google Scholar]
- Mabberley, D.J. (1987) The Plant Book, Cambridge University Press, Cambridge, U.K., pp. 506–507. [Google Scholar]
- Moriya, S., Terakami, S., Iwanami, H., Haji, T., Okada, K., Yamamoto, T. and Abe, K. (2013) Genetic mapping and marker-assisted selection of the gene conferring susceptibility to Alternaria blotch caused by Alternaria alternata apple pathotype in apple. Acta Hortic. 976: 555–560. [Google Scholar]
- Nakashima, T., Ueno, T., Fukami, H., Taga, T., Masuda, H., Osaki, K., Otani, H., Kohmoto, K. and Nishimura, S. (1985) Isolation and structures of AK-toxin I and II host-specific phytotoxic metabolites produced by Alternaria alternata Japanese pear pathotype. Agr. Biol. Chem. 49: 807–815. [Google Scholar]
- Nishitani, C., Terakami, S., Sawamura, Y., Takada, N. and Yamamoto, T. (2009) Development of novel EST-SSR markers derived from Japanese pear (Pyrus pyrifolia). Breed. Sci. 59: 391–400. [Google Scholar]
- Otani, H., Kohmoto, K., Nishimura, S., Nakashima, T., Ueno, T. and Fukami, H. (1985) Biological activities of AK-toxins I and II, host-specific toxins from Alternaria alternata Japanese pear pathotype. Ann. Phytopathol. Soc. Jpn. 51: 285–293. [Google Scholar]
- Saito, K. and Takeda, K. (1984) Genetic analysis of resistance to Alternaria blotch (Alternaria mali Roberts) in apple. Japan. J. Breed. 34: 197–209. [Google Scholar]
- Sanada, T., Nishida, T. and Ikeda, F. (1988) Resistant mutant to black spot disease of Japanese pear ‘Nijisseiki’ induced by gamma-rays. J. Japan. Soc. Hort. Sci. 57: 159–166. [Google Scholar]
- Silfverberg-Dilworth, E., Matasci, C.L., Van de Weg, W.E., Van Kaauwen, M.P.W., Walser, M., Kodde, L.P., Soglio, V., Gianfranceschi, L., Durel, C.E., Costa, F.et al. (2006) Microsatellite markers spanning the apple (Malus × domestica Borkh.) genome. Tree Genet. Genomes 2: 202–224. [Google Scholar]
- Terakami, S., Shoda, M., Adachi, Y., Gonai, T., Kasumi, M., Sawamura, Y., Iketani, H., Kotobuki, K., Patocchi, A., Gessler, C.et al. (2006) Genetic mapping of the pear scab resistance gene Vnk of Japanese pear cultivar Kinchaku. Theor. Appl. Genet. 113: 743–752. [DOI] [PubMed] [Google Scholar]
- Terakami, S., Adachi, Y., Iketani, H., Sato, Y., Sawamura, Y., Takada, N., Nishitani, C. and Yamamoto, T. (2007) Genetic mapping of genes for susceptibility to black spot disease in Japanese pears. Genome 50: 735–741. [DOI] [PubMed] [Google Scholar]
- Terakami, S., Kimura, T., Nishitani, C., Sawamura, Y., Saito, T., Hirabayashi, T. and Yamamoto, T. (2009) Genetic linkage map of the Japanese pear ‘Housui’ identifying three homozygous genomic regions. J. Japan. Soc. Hort. Sci. 78: 417–424. [Google Scholar]
- Terakami, S., Nishitani, C., Kunihisa, M., Shirasawa, K., Sato, S., Tabata, S., Kurita, K., Kanamori, H., Katayose, Y., Takada, N.et al. (2014) Transcriptome-based single nucleotide polymorphism markers for genome mapping in Japanese pear (Pyrus pyrifolia Nakai). Tree Genet. Genomes 10: 853–863. [Google Scholar]
- Tsuge, T., Harimoto, Y., Akimitsu, K., Ohtani, K., Kodama, M., Akagi, Y., Egusa, M., Yamamoto, M. and Otani, H. (2013) Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 37: 44–66. [DOI] [PubMed] [Google Scholar]
- Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B.C., Remm, M. and Rozen, S.G. (2012) Primer3-new capabilities and interfaces. Nucleic Acids Res. 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen, J.W. (2006) JoinMap 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen, Netherlands. [Google Scholar]
- Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., Fontana, P., Bhatnagar, S.K., Troggio, M., Pruss, D.et al. (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42: 833–839. [DOI] [PubMed] [Google Scholar]
- Voorrips, R.E. (2002) MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Walton, J.D. (1996) Host-selective toxins: Agents of compatibility. Plant Cell 8: 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood, M.N. and Bjornstad, H.O. (1971) Some fruit characteristics of interspecific hybrids and extent of self-sterility in Pyrus. Bull. Torrey Bot. cClub 98: 22–24. [Google Scholar]
- Wu, J., Wang, Z.W., Shi, Z.B., Zhang, S., Ming, R., Zhu, S.L., Khan, M.A., Tao, S.T., Korban, S.S., Wang, H.et al. (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 23: 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Li, L.T., Li, M., Khan, M.A., Li, X.G., Chen, H., Yin, H. and Zhang, S.L. (2014) High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. J. Exp. Bot. 65: 5771–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T., Kimura, T., Sawamura, Y., Kotobuki, K., Ban, Y., Hayashi, T. and Matsuta, N. (2001) SSRs isolated from apple can identify polymorphism and genetic diversity in pear. Theor. Appl. Genet. 102: 865–870. [Google Scholar]
- Yamamoto, T., Kimura, T., Sawamura, Y., Manabe, T., Kotobuki, K., Hayashi, T., Ban, Y. and Matsuta, N. (2002a) Simple sequence repeats for genetic analysis in pear. Euphytica 124: 129–137. [Google Scholar]
- Yamamoto, T., Kimura, T., Shoda, M., Ban, Y., Hayashi, T. and Matsuta, N. (2002b) Development of microsatellite markers in the Japanese pear (Pyrus pyrifolia Nakai). Mol. Ecol. Notes 2: 14–16. [Google Scholar]
- Yamamoto, T., Kimura, T., Shoda, M., Imai, T., Saito, T., Sawamura, Y., Kotobuki, K., Hayashi, T. and Matsuta, N. (2002c) Genetic linkage maps constructed by using an interspecific cross between Japanese and European pears. Theor. Appl. Genet. 106: 9–18. [DOI] [PubMed] [Google Scholar]
- Yamamoto, T., Kimura, T., Terakami, S., Nishitani, C., Sawamura, Y., Saito, T., Kotobuki, K. and Hayashi, T. (2007) Integrated reference genetic linkage maps of pear based on SSR and AFLP markers. Breed. Sci. 57: 321–329. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.