Abstract
A rise in invasive diseases due to Neisseria meningitidis C:2b:P1.5 with decreased penicillin susceptibility occurred in Italy during the last 2 years. Real-time PCR identified the Peni phenotype, and the penA sequence revealed the mosaicism of the gene. Molecular analyses assigned the isolates to a single emergent clone of the hypervirulent A4 cluster.
In Italy, the incidence of meningococcal disease has been consistently low (about 0.3 to 0.4 per 100,000 inhabitants per year). Throughout the 1990s, Neisseria meningitidis serogroup C strains represented fewer than 30% of all isolated meningococci. In 2002 to 2003, we observed an increase in serogroup C meningococcal disease, which became responsible for 42.5% of culture-confirmed cases. Interestingly, most of these isolates have the antigenic phenotype C:2b:P1.5 and show intermediate susceptibility to penicillin (Peni) (0.06 μg/ml > MIC < 1 μg/ml), due to sequence changes in the penicillin binding protein 2 penA gene. Peni meningococci have been reported and are being monitored in several countries (3, 5, 8). The attention is driven by the concept that strains with decreased susceptibility may still be evolving and in the process of acquiring further alterations in the penA gene, anticipating the appearance of resistant strains, as observed for Streptococcus pneumoniae and Neisseria gonorrhoeae (9, 12). In Italy, before 2002 Peni meningococci accounted for 7.5% of the isolates, but since then the percentage has risen significantly (to 27.4%). All Peni strains showed a mosaic structure in the transpeptidase region of the penA gene (1), and a recently validated real-time PCR assay (10) has been used for rapid confirmation of this phenotype. In this study, all C:2b:P1.5 Peni meningococci isolated in Italy were analyzed by penA sequencing, multilocus sequence typing, and pulsed-field gel electrophoresis (PFGE) to determine whether the recent increase in the number of cases due to this phenotype correlates with the emergence of a single clone and whether it circulates in other European countries.
A total of 214 invasive N. meningitidis strains were received at the reference laboratory of the National Surveillance of Meningococcal Meningitis between January 2002 and December 2003. All isolates were typed (7), and of 214 N. meningitidis isolates received, 91 were serogroup C and 38 of these (42.2%) belonged to phenotype C:2b:P1.5. Among the remaining serogroup C strains, 21% had the phenotype C:2a:P1.5, which up to then was the most frequent phenotype among C strains, and the others showed a variety of different sero- and subtypes.
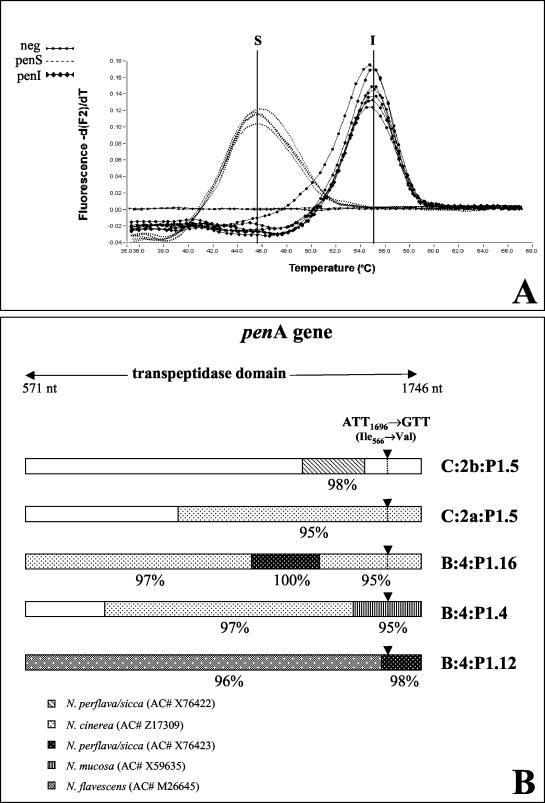
The recently described real-time PCR (10) was used to discriminate between penicillin-susceptible (Pens) and Peni strains. Two hybridization probes were used to distinguish the wild-type penA gene in the Pens meningococci from the mutated gene at codon 566 in Peni strains. Thermal analysis of probe hybridization revealed melting temperatures of 45.5 and 55°C for the Pens and Peni strains, respectively, as shown in Fig. 1A. MICs of penicillin were also assessed by use of the E-test (AB Biodisk) on Mueller-Hinton agar (Oxoid) supplemented with 5% sheep blood, and the majority of C:2b:P1.5 strains (78.4%) showed MICs of ≥0.094 μg/ml, whereas a few serogroup C strains (24%) with other sero- or subtypes were Peni.
FIG. 1.
(A) Examples of Tm curves for Pens and Peni N. meningitidis phenotypes obtained by real-time PCR assay with the mutated 566 codon in the penA gene. (B) Mosaic penA genes of Peni N. meningitidis strains. Each line indicates the proposed origin of the exogenous DNA blocks in the transpeptidase domain of the gene. The arrowheads show the position of the mutated 566 codon used as a marker of penA translocation in the real-time PCR assay. nt, nucleotides; AC#, accession number.
The Peni phenotype was finally confirmed by sequencing the penA PCR products in the transpeptidase domain of the encoding gene. Analysis of the sequences was performed with the Accelrys Wisconsin Genetics Computer Group package.
The penA sequence analysis showed a short DNA region, between nucleotides 1364 and 1545, with 98% identity to the sequence derived from Neisseria perflava/sicca (accession number X76422) and 100% homology with all penA sequences from Spanish C:2b:P1.5,2 Peni strains deposited in the National Center for Biotechnology Information databank (http://www.ncbi.nlm.nih.gov). Conversely, the penA genes of Peni meningococci with other serotypes or serosubtypes appear to have acquired larger blocks of DNA, often from more than one commensal Neisseria species along all of the transpeptidase domain (Fig. 1B). The replacement of the short region between nucleotides 1364 and 1545 in the C:2b:P1.5 strains seems to be sufficient to produce a form of the protein with a lower affinity to penicillin.
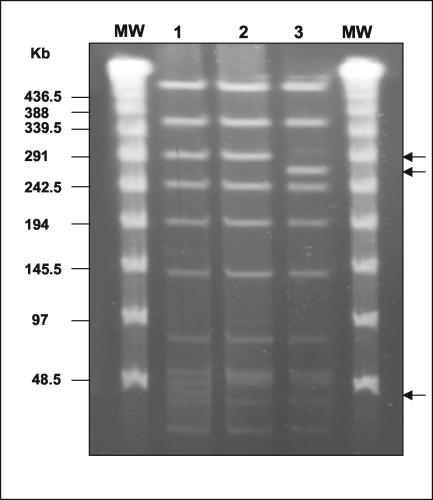
The spread of Peni C:2b:P1.5 meningococci over the country was more remarkable in the first 6 months of 2003, when twice as many were isolated as in the previous year. This sharp increase led us to carry out a molecular analysis of these strains to confirm the circulation of a clone. According to multilocus sequence typing results, obtained by the methodology described by Maiden et al. (6) (http://neisseria.org/nm/typing/mlst/), all of the C:2b:P1.5 Peni meningococci were assigned to the ST8/A4 cluster (Table 1), which is one of the two hypervirulent lineages responsible for most of serogroup C disease worldwide (the other is ST11). DNA macrorestriction fragments generated with the NheI restriction enzyme and analyzed by PFGE, as previously described (4), showed one main pulse type (PTA) and two subclones with two or three minor differences attributable to point mutations, named PTA1 and PTA2 (Fig. 2). When BglII was used, all of the strains showed the same pulse type (data not shown). One Peni strain with phenotype C:2b:nst, isolated in the first months of 2003, belonged to the same clone. The lack of identification of a serosubtype in this strain was due to the presence of IS1301 in the porA gene and its subsequent inactivation, as demonstrated by sequencing (data not shown).
TABLE 1.
Characteristics of N. meningitidis C:2b:P1.5 strains belonging to the ST8/A4 cluster isolated between January 2002 and December 2003
| No. of strains | Yr of isolation | Patient age(s) (yr) | MIC of penicillin (μg/ml) | Pulse type |
|---|---|---|---|---|
| 1 | 2002 | 1 | 0.094 | PTA |
| 4 | 2002 | 3, 4, 17, 19 | 0.125 | PTA |
| 1 | 2002 | 18 | 0.125 | PTA1 |
| 2 | 2002 | 4, 7 | 0.19 | PTA |
| 6 | 2003 | <1, 4, 15, 24, 49, NKa | 0.094 | PTA |
| 1 | 2003 | <1 | 0.094 | PTA2 |
| 5 | 2003 | 4, 11, 12, 17, 35 | 0.125 | PTA |
| 1 | 2003 | <1 | 0.125 | PTA2 |
| 2 | 2003 | 23, 46 | 0.125 | PTA1 |
| 4 | 2003 | 8, 30, 60, 62 | 0.19 | PTA |
| 2 | 2003 | 21, NK | 0.25 | PTA |
| 1 | 2003 | 18 | 0.38 | PTA |
NK, not known.
FIG. 2.
PFGE profiles of genomic DNAs from N. meningitidis C:2b:P1.5 strains after digestion with endonuclease NheI. The lambda ladder DNA marker (New England Biolabs) (lane MW) was used as a molecular size standard (48.5 kb). Lane 1, pulse type PTA; lane 2, PTA1; lane 3, PTA2. The arrows identify the band differences.
Interestingly, the pulse type PTA found in most of the examined C:2b:P1.5 strains is identical to fingerprint pattern 2 (PT2) described by Arreaza et al. (2) for C:2b:P1.5,2 epidemic Spanish strains, just as our PTA1 resembles PT1 described in the same paper. Unlike the Spanish PT1 strains, the PTA1 strains are able to cause disease in all age groups and not predominantly in children under 2 years of age. In contrast, the subclone PTA2 is a unique profile found only in two strains, both of which were responsible for meningitis in infants.
All of these findings seem to confirm that the recent increase of meningococcal disease caused by N. meningitidis C:2b:P1.5 in Italy is due to the spread of a single emergent clone with decreased penicillin susceptibility and belonging to the hypervirulent cluster A4. Although the first strain C:2b:P1.5, ST8/A4 cluster appeared in Italy in 1998, in the following 3 years all meningococci with phenotype C:2b:P1.5 belonged to ST1860, a new ST of the ET37 complex detected in Italy (11). None of these strains were Peni, in contrast to most of the serogroup C ET37 strains isolated in other countries. In 2002, Peni meningococci of phenotype C:2b:P1.5, ST8/A4 cluster suddenly reappeared and rapidly spread all over the country.
We speculate that the Spanish clone might have been imported in the second half of the 1990s. In fact, one Peni strain with phenotype C:2b:P1.5,2 ST8/A4 and pulse type PTA1 was isolated in Italy in 1996, and it showed the same molecular characteristics as the Spanish strains. After this episode, for reasons difficult to understand and linked to the evolution and dynamics of the meningococcal population, it took some years to settle in, with a minor modification in the outer membrane proteins encoded by the porA gene. However, once established, the phenotype C:2b:P1.5 has become so fit as to spread very rapidly and to successfully compete with the other phenotypes. The acquisition of a very small exogenous DNA fragment might have provided an advantage in terms of fitness compared to the other Peni phenotypes. It is important to underline that there is a direct relationship between the increase in serogroup C strains causing meningococcal disease in Italy and the eightfold increase of meningococci with decreased susceptibility to penicillin as a result of the spread of this virulent clone. It will be of extreme importance to closely monitor the endemic circulation of this phenotype in order to plan specific vaccination programs in a timely fashion.
Acknowledgments
We thank the microbiologists of the hospital laboratories participating in the Italian National Surveillance of Bacterial Meningitis for isolating the strains and sending them to the Reference Laboratory of the Istituto Superiore di Sanita' (Rome). This study made use of the Neisseria MultiLocus Sequence Typing website (http://neisseria.mlst.net) developed by Man-Suen Chan and sited at the University of Oxford.
The development of the Neisseria MultiLocus Sequence Typing website is funded by the Wellcome Trust. This work was partially funded by Ministero della Salute (Italy), Programma per la Ricerca Finalizzata 2002.
REFERENCES
- 1.Antignac, A., P. Kriz, G. Tzanakaki, J. M. Alonso, and M. K. Taha. 2001. Polymorphism of Neisseria meningitidis penA gene associated with reduced susceptibility to penicillin. J. Antimicrob. Chemother. 47:285-296. [DOI] [PubMed] [Google Scholar]
- 2.Arreaza, L., S. Berrón, S. Fernández, M. I. Santiago, A. Malvar, and J. A. Vázquez. 2000. Investigation for a more virulent variant among the C:2b:P1.2,5 Spanish meningococcal epidemic strains by molecular epidemiology. J. Med. Microbiol. 49:1079-1084. [DOI] [PubMed] [Google Scholar]
- 3.Berrón, S., and J. A. Vázquez. 1994. Increase in moderate penicillin resistance and serogroup C in meningococcal strains isolated in Spain. Is there any relationship? Clin. Infect. Dis. 18:161-165. [DOI] [PubMed] [Google Scholar]
- 4.Hartstein, A., C. Phelps, and A. Lemonte. 1995. Typing of sequential bacterial isolates by pulsed-field gel electrophoresis. Diagn. Microbiol. Infect. Dis. 22:309-314. [DOI] [PubMed] [Google Scholar]
- 5.Kyaw, M. H., J. C. Bramley, S. Clarke, et al. 2002. Prevalence of moderate penicillin resistant invasive Neisseria meningitidis infection in Scotland, 1994-9. Epidemiol. Infect. 128:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maiden, M. C. J., J. A. Bygraves, E. Feil, et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mastrantonio, P., P. Stefanelli, C. Fazio, T. Sofia, A. Neri, G. La Rosa, C. Marianelli, M. Muscillo, M. G. Caporali, and S. Salmaso. 2003. Serotype distribution, antibiotic susceptibility, and genetic relatedness of Neisseria meningitidis strains recently isolated in Italy. Clin. Infect. Dis. 36:422-428. [DOI] [PubMed] [Google Scholar]
- 8.Richter, S. S., K. A. Gordon, P. R. Rhomberg, M. A. Pfaller, and R. N. Jones. 2001. Neisseria meningitidis with decreased susceptibility to penicillin: report from the SENTRY antimicrobial surveillance program, North America, 1988-99. Diagn. Microbiol. Infect. Dis. 41:83-88. [DOI] [PubMed] [Google Scholar]
- 9.Spratt, B. G., and K. D. Cromie. 1988. Penicillin-binding proteins of gram-negative bacteria. Rev. Infect. Dis. 10:699-711. [DOI] [PubMed] [Google Scholar]
- 10.Stefanelli, P., A. Carattoli, A. Neri, C. Fazio, and P. Mastrantonio. 2003. Prediction of decreased susceptibility to penicillin of Neisseria meningitidis strains by real-time PCR. J. Clin. Microbiol. 41:4666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefanelli, P., C. Fazio, A. Neri, T. Sofia, and P. Mastrantonio. 2003. First report of capsule replacement among electrophoretic type 37 Neisseria meningitidis strains in Italy. J. Clin. Microbiol. 41:5783-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomasz, A. 1986. Penicillin-binding proteins and the antibacterial effectiveness of beta-lactam antibiotics. Rev. Infect. Dis. 8:S260-S278. [DOI] [PubMed] [Google Scholar]