Abstract
The antibacterial mechanism of the Cu-containing materials has not been fully understood although such understanding is crucial for the sustained clinical use of Cu-containing antibacterial materials such as bone implants. The aim of this study is to investigate the molecular mechanisms by which the Gram-positive Staphylococcus aureus (S.aureus) is inactivated through Cu-bearing titanium alloys (Ti6Al4V5Cu). Cu ions released from the alloys were found to contribute to lethal damage of bacteria. They destroyed the permeability of the bacterial membranes, resulting in the leakage of reducing sugars and proteins from the cells. They also promoted the generation of bacteria-killing reactive oxygen species (ROS). The ROS production was confirmed by several assays including fluorescent staining of intracellular oxidative stress, detection of respiratory chain activity, and measurement of the levels of lipid peroxidation, catalase and glutathione. Furthermore, the released Cu ions showed obvious genetic toxicity by interfering the replication of nuc (species-specific) and 16SrRNA genes, but with no effect on the genome integrity. All of these effects lead to the antibacterial effect of Ti6Al4V5Cu. Collectively, our work reconciles the conflicting antibacterial mechanisms of Cu-bearing metallic materials or nanoparticles reported in the literature, as well as highlight the potential use of Ti6Al4V5Cu alloys in inhibiting bacterial infections.
Keywords: antibacterial mechanism, copper, titanium alloys, bacteria
1. Introduction
Titanium (Ti) and its alloys are widely used in clinics as the preferred metallic materials in the form of human surgical implants, such as the dental implants, bone trauma repair materials and joint prosthesis. [1–3] However, Ti alloys tend to be highly susceptible to bacterial adhesion and infection because of compromised host defense. [4, 5] With sterile operation and perioperative antibiotic defenses, the risk of infection after internal fixation ranges between 0.4% and 16.1% depending on the extent of fracture. [6] To be more precise, the infection rates of periprosthetic joint infections (PJIs) are 0.5–2%, [7] 2–9% [8] and 0.3–1.7% [9] after total joint replacement of the knee, hip and ankle, respectively. Staphylococcus aureus (S.aureus) is one of the most frequent microorganisms causing implant infections, and implant associated infection is considered to be the most serious complications of prosthetic implants. Such infection sometimes results in long-term hospitalization, revision surgeries, and even definitive failure of the implants. [10] Even more, the emergence of drug-resistant bacteria caused by antibiotics abuse brings more obstacles for the treatment of implant associated infections. More seriously, once these infections associated with metal implants occur, implant removals cannot be avoided. Hence, there is a pressing need in the development of antibacterial metallic implant materials. [11]
Several metal ions, like silver (Ag), zinc (Zn), and copper (Cu), have been described as a potent antibacterial agent with a very broad spectrum against numerous bacteria. [12, 13] Compared with Ag and Zn, Cu is an essential element of several enzymes and a metabolizable agent. Furthermore, Cu represents a more promising metal ion to be included in antibacterial metallic implants because of its lower toxicity and higher biocompatibility. [14] Therefore, Cu was employed as the antibacterial agent in the alloys in this study. Previously, we fabricated Cu-bearing medical Ti6Al4V alloys (Ti6Al4V5Cu) and discovered their antibacterial effect for two types of bacteria including Escherichia coli (E.coli; Gram-negative) and S.aureus (Gram-positive). [15] However, we do not know their antibacterial mechanism. We hypothesize that it is the Cu ions released from the alloys that resulted in their antibacterial property. The antibacterial mechanisms of Cu ions have not been elucidated up to date and the molecular mode remains controversial. [16, 17] For example, Santo et al suggested that the metallic Cu was not genotoxic and did not kill bacteria via DNA damage, however, bacterial membranes were was damaged. [18] Keevil and coworkers reported that DNA inside Cu-exposed S.aureus cells was degraded causing cell death. Yet, only little negative effect on cytoplasmic membrane integrity was observed. [19] Hence, we made an attempt to clarify the antibacterial mechanism of Cu ions released from the Ti6Al4V5Cu alloy in this study.
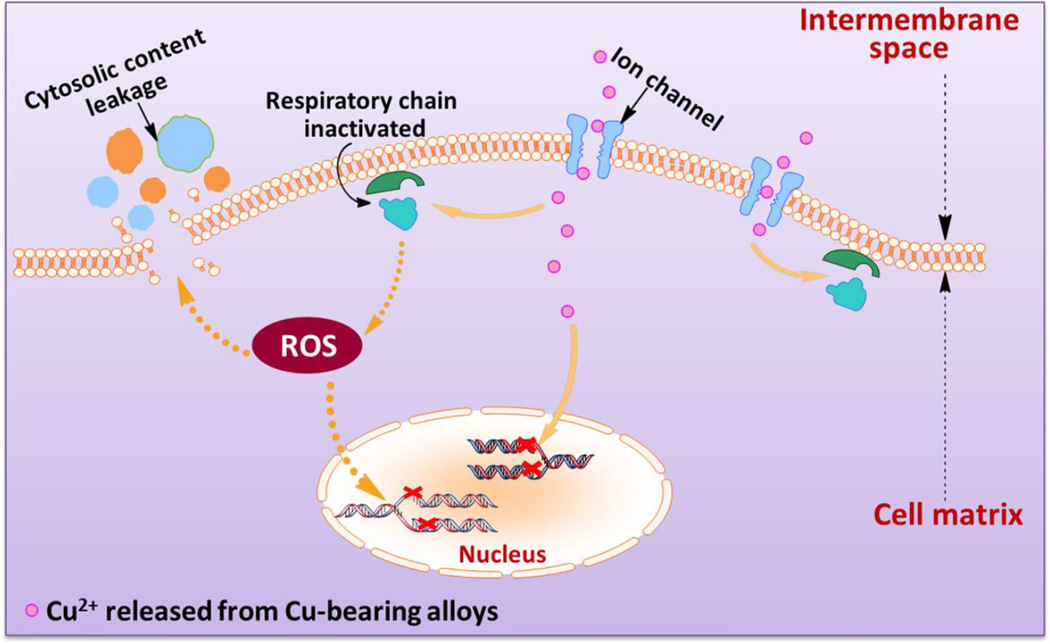
Specifically, in this study we aim to investigate the mechanism of Cu-releasing Ti6Al4V5Cu alloys against S.aureus with a focus on cellular respiration and DNA damage of the bacteria when the bacteria are in contact with experimental samples. We found that the antibacterial activity of Ti6Al4V5Cu alloys was exerted via the destruction of bacterial cell integrity, the production of ROS and genotoxicity (Figure 1), which were all confirmed by a variety of biological assays carried out in this study. This study will also lay foundation for the application of Cu-bearing titanium alloys as a new type of antibacterial orthopedic implants.
Figure 1.
Schematic illustration of the general idea for this study and the possible anti-bacterial mechanisms on the surface of Ti6Al4V5Cu implants. Ti6Al4V5Cu releases Cu ions. The released Cu ions (1) accumulate in the cell membrane affecting membrane permeability; (2) disrupt the activity of respiratory chain; (3) enter bacterial cells to generate ROS; and (4) disturb the gene replication of S.aureus
2. Results
2.1. Characterization studies
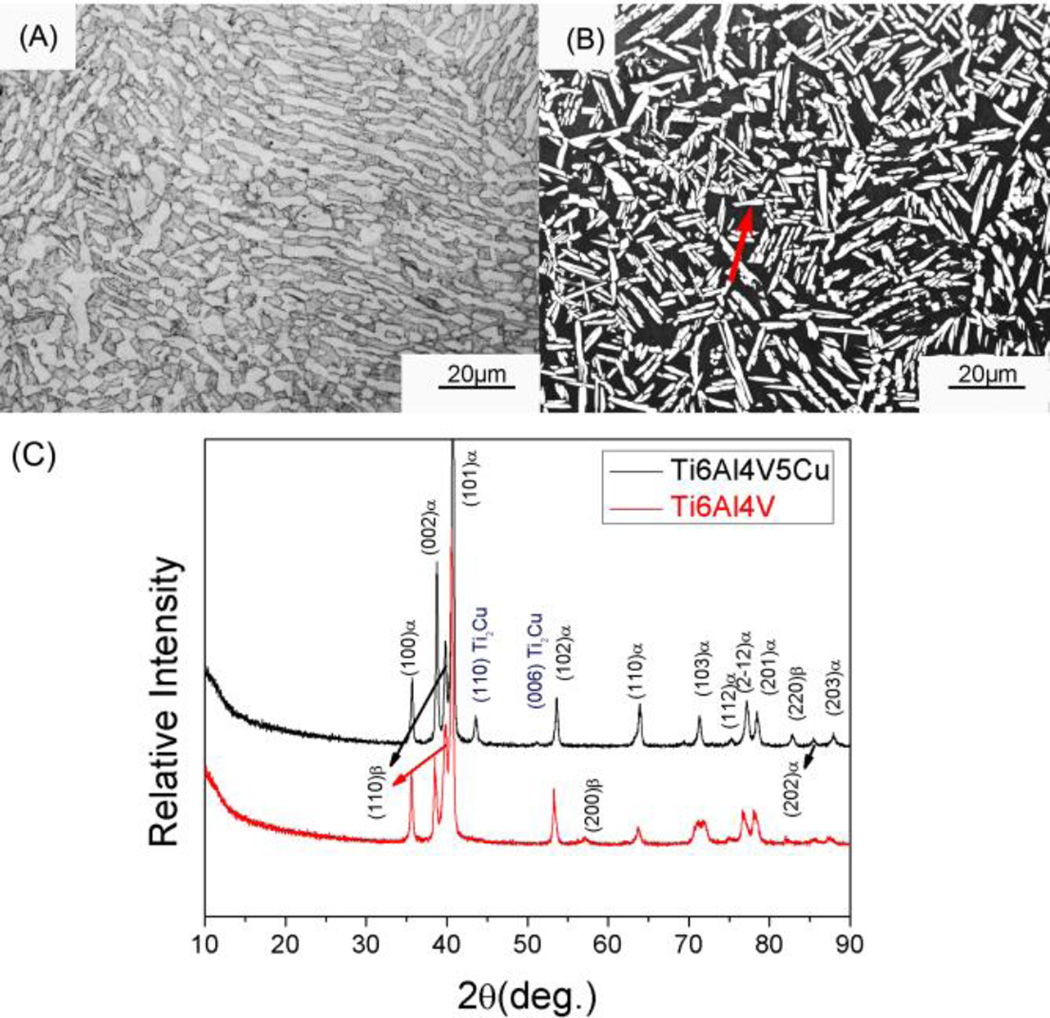
Microstructures of Ti6Al4V and Ti6Al4V5Cu alloys were shown in Figure 2A and 2B, respectively. Metallurgical structures of annealed Ti6Al4V5Cu alloys exhibited the typical biphasic microstructure, which was similar to that of Ti6Al4V alloys. Both Ti6Al4V and Ti6Al4V5Cu alloys were found to show equiaxed α phase uniformly distributed in the matrix of β phase. XRD patterns of Ti6Al4V and Ti-6Al4V5Cu alloys (Figure 2C) indicated that the two profiles were similar and the reflection peaks belonging to α and β phase could be indexed. However, unlike Ti6Al4V alloys, there are two reflection peaks at around 44° and 52.8° in the profile of Ti6Al4V5Cu alloys, which can be identified as (110) and (006) of intermetallic Ti2Cu phase.
Figure 2.
Microstructures (A, B) and XRD patterns (C) of Ti6Al4V and Ti6Al4V5Cu alloys. (B) exhibited metallurgical structures of the annealed Ti6Al4V5Cu alloys, showing that equiaxed α phase (indicated by the red arrow) was distributed in the matrix of β phase. The typical biphasic microstructure of Ti6Al4V5Cu alloys (B) was similar to that of Ti6Al4V alloys (A). (C) exhibited the XRD patterns of Ti6Al4V5Cu and Ti6Al4V alloys. The two profiles were similar and reflection peaks belonging to α and β phase could be clearly indexed. However, an intermetallic phase Ti2Cu was found apparently in the matrix of Ti6Al4V5Cu alloy compared to Ti6Al4V alloy.
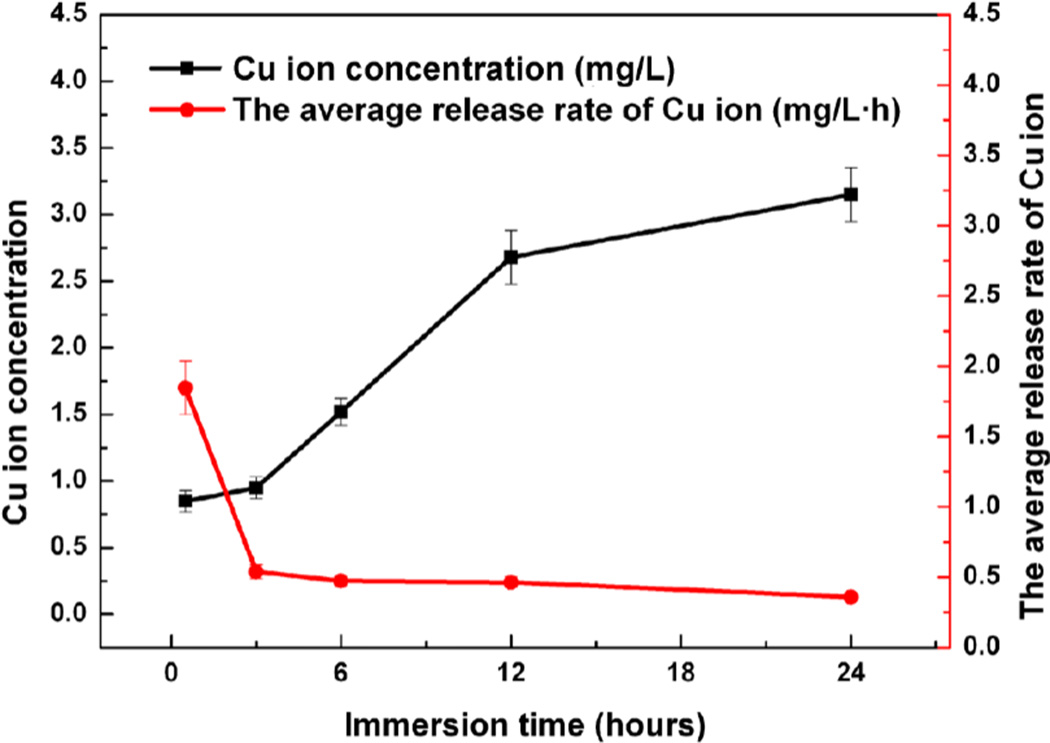
Figure 3 displayed the cumulative concentration of Cu ions released from Ti6Al4V5Cu alloys and the average release rate. The data indicated that the Cu ions were continuously released from Ti6Al4V5Cu alloys in saline and the accumulated concentration was increased with time in 24 h, but not in a linear trend. At the beginning, the Cu ions kept a high releasing rate, however, the rate slowed down with time.
Figure 3.
Cumulative concentration of Cu ions released from Ti6Al4V5Cu alloys and the average release rates. The data indicated that the release of Cu ions was continuous from Ti6Al4V5Cu alloy in saline and the accumulated concentration was increased with time in 24 h, but not in a linear trend. At the beginning, Cu ions kept a high releasing rate, however, the rate slowed down with time. It was these Cu ions released in the early stage that played the antibacterial role.
2.2. Antibacterial effect
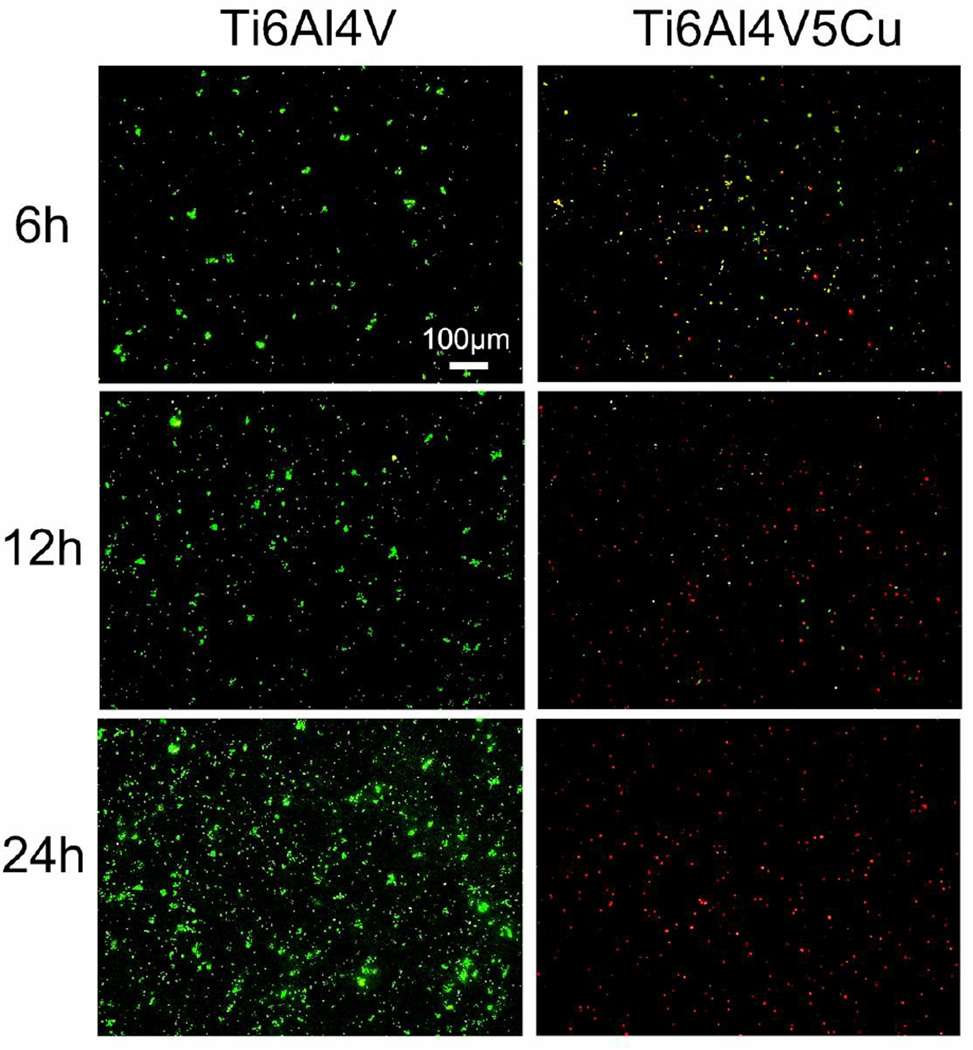
The viable/inviable bacteria after co-cultured with Ti6Al4V or Ti6Al4V5Cu samples for 6 h, 12 h and 24 h were determined by the Live/Dead staining method. Green fluorescent SYTO9 was able to enter viable cells and used for assessing live cell counts, whereas red fluorescent propidium iodide (PI) enters only cells with damaged cytoplasmic membranes. Therefore, the viable and nonviable cells can be distinguished under the fluorescence microscope. Our fluorescence microscopic images (Figure 4) showed that bacterial cells of Ti6Al4V group were intensely stained with SYTO9 and produced green fluorescence, whereas the Ti6Al4V5Cu group was found to be PI positive and produced red fluorescence during the process of co-culture, demonstrating that Ti6Al4V5Cu showed strong antibacterial abilities against S.aureus. With the extension of incubation time, the antibacterial role of Ti6Al4V5Cu was boosted and peaked at 24 h. Previous study also indicated that the Live/Dead staining kit allowed rapid evaluation of the integrity of cell membranes.[20] Therefore, our results also suggested that bacteria cells of Ti6Al4V5Cu group may lose their plasma membrane integrity during the process of co-culture.
Figure 4.
The role of Ti6Al4V5Cu in inhibiting the viability of S.aureus. The live/dead bacterial cells were stained at 6 h, 12 h and 24 h by LIVE/DEAD Baclight assay, and representative images after cell staining were observed by fluorescence microscopy. Live cells were dyed by SYTO9 and showed green fluorescence, whereas dead cells were dyed by PI and showed red fluorescence. The number of live bacteria in Ti6Al4V group with green fluorescence increased significantly in comparison to the Ti6Al4V5Cu group. However, numbers of live cells decreased remarkably in Ti6Al4V5Cu group, almost all bacteria died (red fluorescence) at 24 h.
2.3. Effect of Ti6Al4V5Cu on the membrane leakage of proteins and reducing sugars
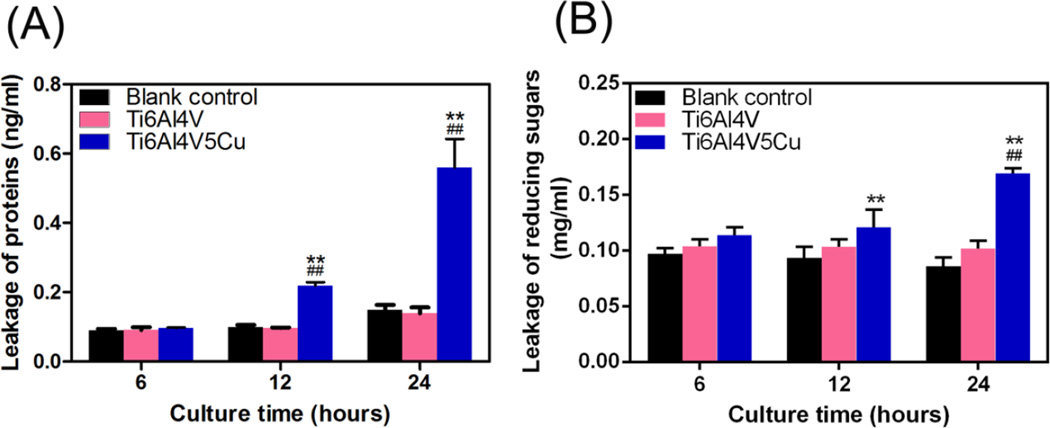
Figure 5A revealed that Ti6Al4V5Cu could enhance the membrane leakage of proteins. Initially, almost no proteins were detected due to the leakage from bacterial cells in blank control group at 6 h, and the differences between Ti6Al4V5Cu group and Ti6Al4V or blank control group were not significant. The leakage amount of proteins from bacterial cells co-cultured with Ti6Al4V5Cu increased significantly (p < 0.01) comparing to those in control and Ti6Al4V group. The leakage amount from Ti6Al4V5Cu samples reached 0.22 ng/mL and 0.56 ng/mL at 12 h and 24 h, respectively. Similarly, Ti6Al4V5Cu also elevated the leakage of reducing sugars through the membrane of S.aureus (Figure 5B). The leakage of reducing sugars in S.aureus treated with Ti6Al4V5Cu for 24 h reached 0.169 mg/mL, which was 97±8.02 % more than the blank control group. No significant differences were identified between the Ti6Al4V group and blank control group. These results showed that Ti6Al4V5Cu accelerated the leakage of the proteins and reducing sugars from bacterial cytoplasm, suggesting that the Cu-bearing titanium alloys could apparently enhance the permeability of membrane.
Figure 5.
Effects of Ti6Al4V5Cu on the membrane leakage of proteins and reducing sugars. (A) Leakage of proteins from S.aureus treated with Ti6Al4V5Cu using BCA assay. The leakage amount of proteins was increased obviously after 12 h co-culturing (p < 0.01). (B) Leakage of reducing sugars from S.aureus co-cultured with Ti6Al4V5Cu. The highest amount of leakage was reached on 24 h, at a level of 97±8.02 % more than the blank control group (p < 0.01). All data represent the mean ± standard deviation of three independent experiments. *: compared to the blank control group, #: compared to the Ti6Al4V group. One symbol, p < 0.05; Two symbols, p < 0.01.
2.4. Oxidative stress markers
2.4.1. ROS generation
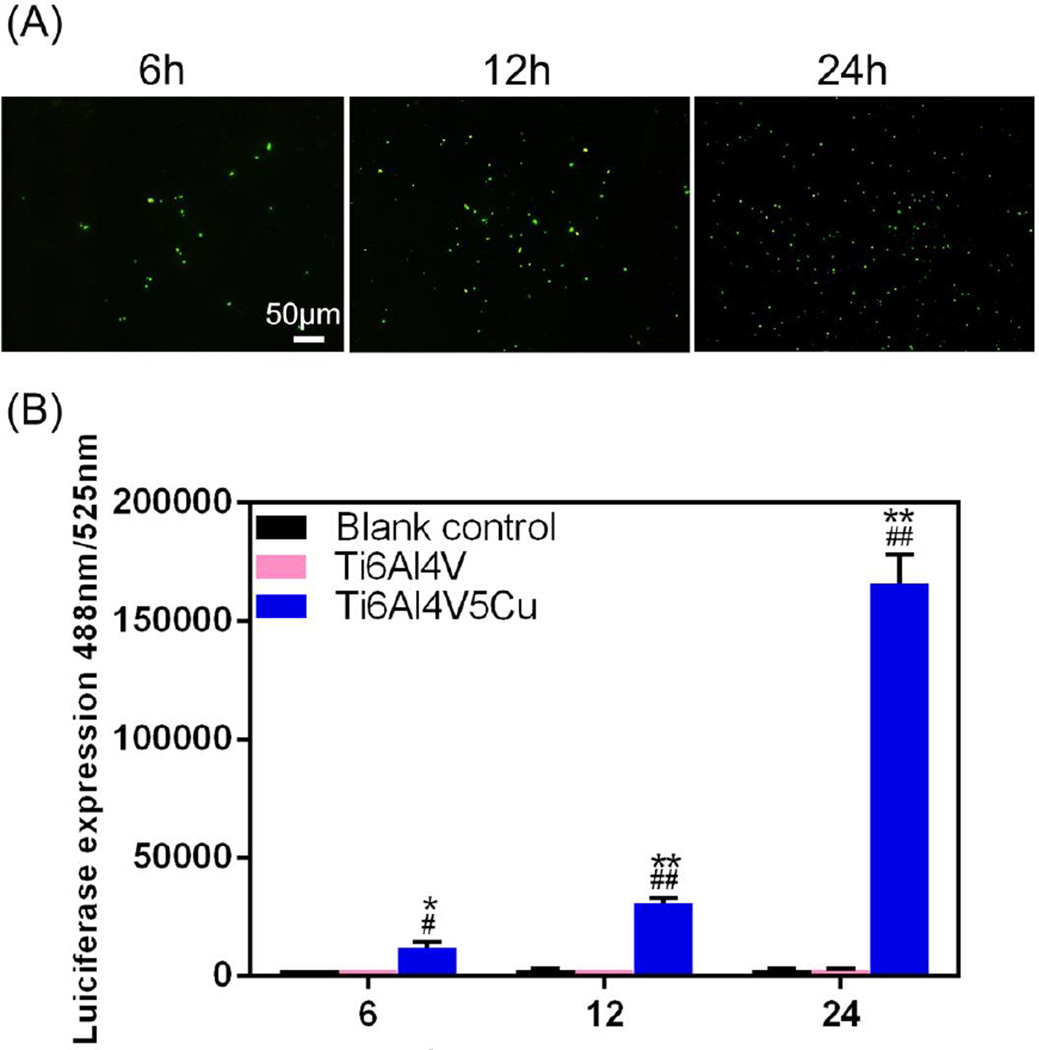
Figure 6A shows the quantification of ROS production through 2’7’ -dichlorofluorescin (DCF) fluorescence intensities after bacteria were co-cultured with Ti6Al4V or Ti6Al4V5Cu samples. No significant change was observed in the probe fluorescence emission from either the adherent bacteria or the suspended bacteria in Ti6Al4V group. On the contrary, the percentage of quantified fluorescence produced as an indicator free radical formation in Ti6Al4V5Cu group was slightly increased, and became more evident with longer exposure times. Besides, the microscopic images of DCF fluorescence in Figure 6B showed noticeable increase in the fluorescence of the ROS generating bacterial cells in the Ti6Al4V5Cu group.
Figure 6.
The role of Ti6Al4V5Cu in ROS generation of S.aureus. (A) Representative micro-photographs of Ti6Al4V5Cu-induced ROS generation in S.aureus adhered on samples. ROS was studied using 2,7-DCFH-DA dye, and images were snapped under fluorescence microscope. The increasing fluorescence value indicated that continuous Cu ions released from Ti6Al4V5Cu accelerated the oxidative damage to S.aureus. (B) ROS fluorescence intensities in the suspended bacteria co-cultured with Ti6Al4V5Cu. ROS generation induced by Cu ions from Ti6Al4V5Cu was elevated remarkably and significantly higher than that from the blank control group and Ti6Al4V group (p < 0.01). All data represent the mean ± standard deviation of three independent experiments. *: compared to the blank control group, #: compared to the Ti6Al4V group. One symbol, p < 0.05; Two symbols, p < 0.01
2.4.2. Respiratory chain dehydrogenases activity
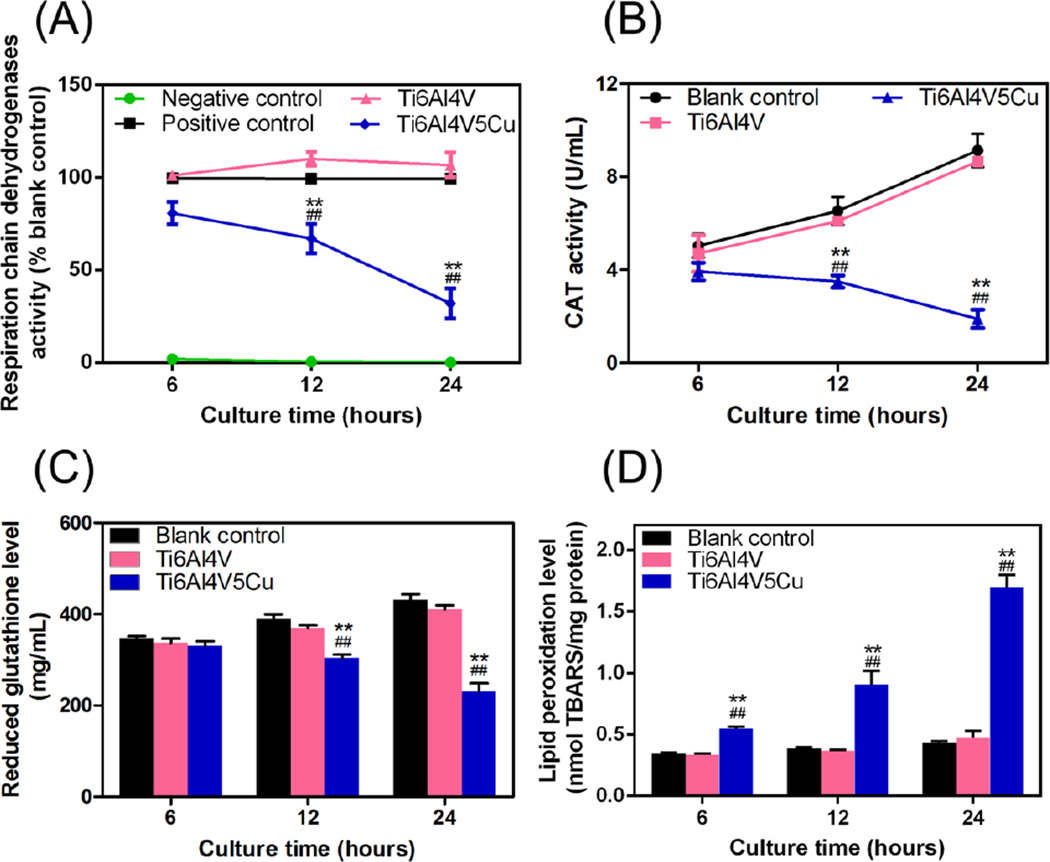
The effects of Cu-bearing titanium alloy samples on respiratory chain dehydrogenases of S.aureus were shown in Figure 7A. The activity of respiratory chain dehydrogenases in the blank control group increased with the extension of incubation time. The differences between Ti6Al4V group and blank control group were not significant and the activity had nearly no change in the negative control group. However, the enzymatic activity of S.aureus treated with Ti6Al4V5Cu fell down rapidly with the increase of cultivation time and reached the lowest level at 24 h. These results indicated that the respiratory chain dehydrogenases activity of S.aureus was inhibited by Ti6Al4V5Cu samples.
Figure 7.
Effects of Ti6Al4V5Cu on the oxidative stress markers in S.aureus. (A) Activity of respiratory chain dehydrogenases analyzed by catalytic reaction using INT reagent. (B) Activity of catalase (CAT) measured by CAT activity kit. Both the two enzyme activity was inhibited in S.aureus treated with Ti6Al4V5Cu obviously with the extension of the culture tim, and reached the lowest level at 24 h. (C) Glutathione (GSH) levels were reduced and (D) Lipid peroxidation (LPO) levels were enhanced significantly in comparison to blank control and Ti6Al4V group (p < 0.01). All data represent the mean ± standard deviation of three independent experiments. *: compared to the blank control group, #: compared to the Ti6Al4V group. One symbol, p < 0.05; Two symbols, p < 0.01.
2.4.3. Catalase (CAT) activity
Significant reduction of the CAT activity was observed in the Ti6Al4V5Cu group at 12 h and 24 h compared with Ti6Al4V group and blank control group (Figure 7B, p < 0.01), suggesting that the Ti6Al4V5Cu group could inhibit the CAT activity of S.aureus. The highest inhibitory rate of the Ti6Al4V5Cu group (59±5.12 %) was reached at 24 h. However, the differences between the Ti6Al4V group and blank control group were no significant.
2.4.4 Glutathione (GSH) levels
GSH levels were compared between S.aureus co-cultured with Ti6Al4V5Cu, Ti6Al4V and blank control group. As shown in Figure 7C, the GSH level of Ti6Al4V5Cu group exhibited a slight downward trend, and was significantly lower than the Ti6Al4V group and blank control group at 12 h and 24 h (p < 0.01). However, there were no significant differences between the Ti6Al4V group and blank control group during the period of incubation, suggesting that an increased level of Cu ions could significantly reduce the GSH level of S.aureus.
2.4.5. Lipid peroxidation (LPO) levels
The levels of LPO serve as another reliable indicator of oxidative stress. Figure 7D showed that the Ti6Al4V5Cu group caused a time-dependent increase in LPO levels, and the highest level was reached at 24 h. However, no significant alteration of LPO levels was seen in the Ti6Al4V and blank control groups.
2.5 Genotoxicity study
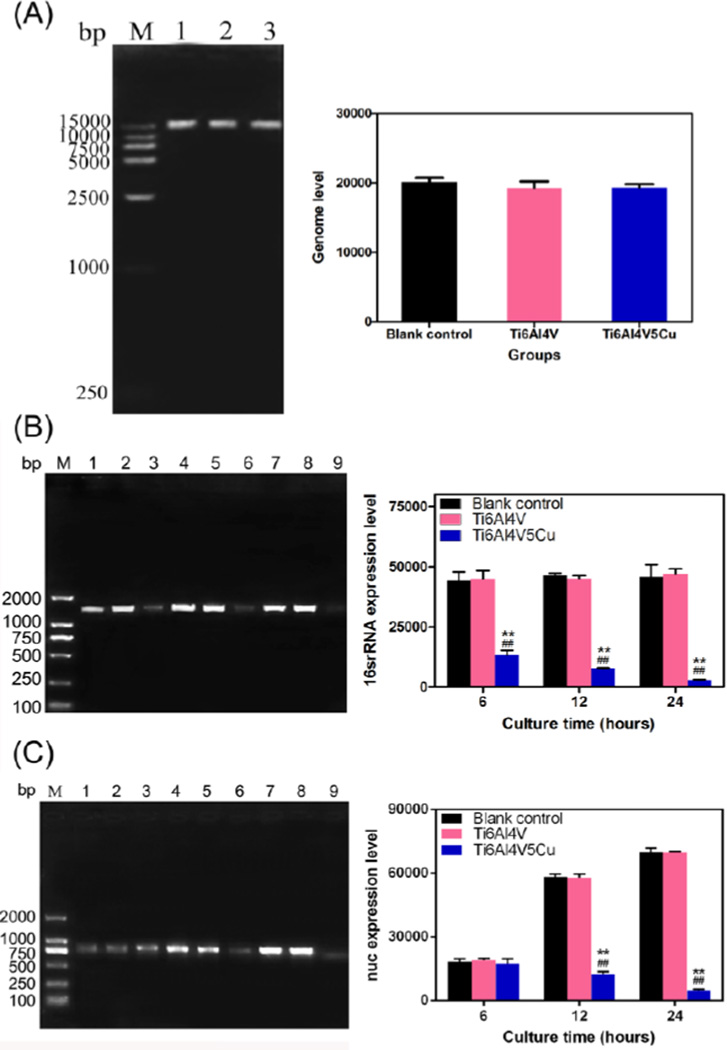
The genome DNA image of agarose gel electrophoresis shown in Figure 8A displayed that the bacterial genome bands were intact. There was no dispersed distribution from the front of electrophoresis strap for the DNA of S.aureus for both the control Ti6Al4V and the antibacterial Ti6Al4V5Cu samples, indicating that antibacterial titanium alloys led to bactericidal effect not through the damage of genomic integrity.
Figure 8.
Effects of Ti6Al4V5Cu on the genotoxicity in S.aureus. (A) Determination of genomic integrity in S.aureus at 24 h. The image of genome DNA obtained by agarose gel electrophoresis and ethidium bromide staining indicated that there were no damages of genomic integrity. Lane M, 1.5 kb DNA ladder; Lane1, Blank control group; Lane 2, Ti6Al4V group; Lane 3, Ti6Al4V5Cu group. (B) Detection of 16srRNA and (C) nuc expression in S.aureus using EMA-PCR method. 16SrRNA and nuc gene expression levels of the bacteria co-cultured with Ti6Al4V5Cu samples decreased constantly within 24 h, and reached the lowest level on 24 h. Lane M, 2000bp DNA ladder; Lane1, 4, 7, Blank control group at 6 h, 12 h and 24 h, respectively; Lane 2, 5, 8, Ti6Al4V group at 6 h, 12 h and 24 h, respectively; Lane 3, 6, 9, Ti6Al4V5Cu group at 6 h, 12 h and 24 h, respectively. Error bars represent standard deviations of the average. All data represent the mean ± standard deviation of three independent experiments. *: compared to the blank control group, #: compared to the Ti6Al4V group. One symbol, p < 0.05; Two symbols, p < 0.01.
Additionally, ethidium bromide monoazide (EMA) selectively enters the membrane-compromised cells and binds to cellular DNA, which subsequently inhibits polymerase chain reaction (PCR) amplification of the DNA. The reduction in PCR amplification could be used as an indicator of cell membrane leakage.[21–24] To assess the membrane integrity and understand the molecular basis of Ti6Al4V5Cu action against S.aureus, a set of S.aureus genes involved in the general and toxin production were selected and EMA-PCR was performed in the gene expression study. Figure 8B showed that the expression level of housekeeping gene (16srRNA) was not significantly up or down-regulated regardless of the cultivation time, while the expression of nuc genes was increased along with the culture time extension both in blank control group and Ti6Al4V group. However, the levels of 16SrRNA and nuc gene expression in the bacteria co-cultured with Ti6Al4V5Cu samples decreased constantly within 24 h, and reached the lowest level at 24 h. These results obviously revealed that the Ti6Al4V5Cu samples and the released Cu ions could suppress the gene replication of S.aureus.
3. Discussion
Ti6Al4V is one of the most commonly used biomaterials for endosseous implants in orthopaedic applications owing to its mechanical properties and biocompatibility. Despite these advantages, a serious post-operative problem leading to the failure of implants is the appearance of implant associated infections. Because systemic antibiotics do not often provide effective treatment for infection, bactericidal or bacteriostatic modifications such as surface treatments have been used to prevent bacterial colonization. [25, 26] However, the problem of the falling off of the antibacterial coating cannot be avoided during long-time sustained antibacterial function. In our previous study, [15, 27] a novel class of Cu-bearing titanium alloys, Ti6Al4V5Cu biomaterials with self-antibacterial function, had been successfully designed and fabricated. We found that this Cu-bearing titanium alloy showed not only intense bactericidal effect, but also good cyto-compatibility with the same level of biomedical Ti6Al4V alloy, suggesting its potential application in clinics.
Cu is a metal ion with well-known antibacterial activities against a broad spectrum of bacteria with a low incidence of resistance. In vitro studies have been carried out to investigate the antibacterial effects of Cu ions from metallic copper surface[28] and Cu-containing nanoparticles. [12] Although many studies have reported the bactericidal mechanism of antibacterial nanomaterials and copolymers in the past few years, [29–31] there have been very few reports about comprehensive mechanisms of the antibacterial effect of Cu ions. In the present study, Ti6Al4V5Cu with antibacterial activity was used to understand the antibacterial mechanism. Staining-based methods had shown that bacteria attached to the alloys present a significant loss of viability progressively, reaching its maximum damage after 24 h of contact (Figure 4), which further demonstrated the bactericidal effect of Ti6Al4V5Cu alloys against the S.aureus. These results were consistent with other different methodologies based on the evaluating of the colony-forming units (CFU) in our earlier study.[15] Ti6Al4V5Cu also apparently accelerated the leakage of membrane, favoring the subsequent leakage of proteins and reducing sugars from bacterial cytoplasm (Figure 5). Hence, it could be conferred that our Cu-bearing alloys would disturb the permeability of the membrane, which was an important factor for antibacterial function. Since the initial time after implantation is crucial for suppressing the formation of biofilms, the reduction of bacteria viability and destruction of bacteria integrity on Ti6Al4V5Cu alloys within 24 h represented a valuable finding in the effort to minimizing the infectious incidences.
Oxygen is essential for aerobic organisms to respire or obtain energy from nutrients, but is also a precursor to ROS, such as superoxide anion radical (O2−), hydrogen peroxide (H2O2), and the highly reactive hydroxyl radicals (·OH). These highly reactive species react with biologically important molecules such as lipids, proteins and nucleic acids, eventually resulting in the oxidative damage or even the death of organisms. [32] Previous reports have pointed out that the generation of intracellular ROS was considered as one of the important factors for the antibacterial activity of silver nanoparticles. [33, 34] 2’,7’- ichlorofluorescin-diacetate (DCFH-DA) probe, which can be oxidized into highly fluorescent DCF by intracellular ROS, was used to indicate the ROS concentration of bacteria on different material samples in our studies. Figure 6 revealed a significant increasing trend of fluorescence intensity produced as an indicator of free radical formation in Ti6Al4V5Cu group compared to the blank control group after 6 h (p < 0.01). Such trend was similar with previous studies on Cu-containing nanoparticles, [35, 36] and became more evident with longer exposure times. These free radicals caused lipid peroxidation damage of cell membrane afterwards, which decreased the fluidity of membrane. Then, the membranous properties and membrane-bound proteins were disrupted continuously. [32] Hence, it can be concluded that the higher amount of cellular ROS, some of which was produced within the cells, was associated with the destruction of the cellular structure of bacteria, as well as the antibacterial effects. Most of the ROS products were derived from the catalysis by several membrane associated respiratory chain enzymes such as nicotinamide adenine dinucleotide (NADH) dehydrogenase in bacteria. [37] Living organisms are able to build balance mechanisms to normally maintain ROS at certain steady levels and protect themselves against oxidative stress, [38] including some enzymes such as CAT. To investigate the major key points in the process of ROS generation in S.aureus under the treatment of different materials, the activity of respiratory chain was detected herein. Our results showed that the activity levels of respiratory chain dehydrogenases (Figure 7A) and CAT (Figure 7B) were all inhibited by Ti6Al4V5Cu alloys. In addition, GSH acts directly as a scavenger of ROS and protects cells from oxidative stress through its ability to bind to and reduce ROS. [39] Our results showed that Ti6Al4V5Cu decreased the GSH levels significantly, and both the disruption of catalytic process and GSH levels could lead to the generation of ROS. This may be due to the interaction between Cu ions released from the alloys and the specific site of enzymes on the respiratory chains. [40]
DNA is also a main target of ROS. [41] However, genotoxicity caused by metallic Cu is controversial, especially in Staphylococci. Quaranta and coworkers clearly reported that metallic Cu did not cause mutation damage to the DNA but membrane damage of Staphylococcus haemolyticus [28]. In contrast, Weaver et al supported that the exposure to copper surfaces rapidly killed S.aureus by damaging genomic DNA. [42] In spite of this, the molecular mode-of-action of copper interaction with genomic DNA of bacteria has not been fully understood. We performed the integrity detection of cellular genomic DNA with agarose gel electrophoresis. Although Figure 8A showed that our Ti6Al4V5Cu alloys had no obvious effect on the genome integrity, we further confirmed the genotoxicity through EMA-PCR method. A number of staphylococcal genes such as 16SrRNA and nuc had been used for specific detection and identification from mixed clinical samples. [43] 16SrRNA was adopted in this study not only because it is a universal target gene conserved among species of bacteria, enabling the identification of the bacteria to species level, but also because rRNA was more stable than mRNA. [44] The nuc gene, which encoded an extracellular thermostable nuclease, have been used to distinguish S.aureus from other Staphlococcus species. [35] The EMA-PCR results (Figure 8B,C) showed that both the relative expression level of 16SrRNA and nuc gene in S.aureus co-cultured with Ti6Al4V5Cu decreased in a time-dependent manner, and reached the minimum expression at 24 h. These results revealed that Cu ions could result in genotoxicity by disturbing gene replication of S.aureus. To the best of our knowledge, this is the first study highlighting the important role of Cu ions in disturbing gene replication. Furthermore, EMA selectively entered membrane-compromised bacteria and bound cellular DNA, which subsequently inhibited PCR amplification of DNA. [45, 46] Hence, the reduction of PCR amplification could also serve as an indicator of cell membrane damage in our experiment.
Taken together, our results indicate that the action model of Ti6Al4V5Cu alloy may be described as follows (Figure 1). First, Cu ions damaged the permeability of outer membrane of the bacteria, resulting in the leakage of cellular materials such as proteins and sugars. Then, Cu ions inactivate the activity of respiratory chains, thus inhibiting cell respiration, producing high level of ROS and suppressing the growth of bacteria. Simultaneously, Ti6Al4V5Cu alloy also causes obvious genetic toxicity by influencing gene replication efficiency. Our work confirms that Ti6Al4V5Cu alloy has unique superiorities over the current materials used for orthopedics due to its excellent antibacterial property.
Our results confirm that Ti6Al4V5Cu alloys outperform the traditional titanium-based orthopedic materials in terms of maintaining the integrated and sustainable antibacterial activity, in addition to their known excellent wearing resistance, strong biomechanical and fine biocompatibility properties. [47, 48] Hence, such biomaterials are especially suitable for use in treating the contaminated wound or cancer. They can decrease the chances of infection in internal fixation, reduce the cost of treatment for outpatients, and then improve the clinical therapy efficacy. Therefore, Ti6Al4V5Cu can minimize the risk of hospital acquired infection and find significant potential clinical applications as surgical implant materials.
4. Conclusions
In summary, Ti6Al4V5Cu alloy was used to understand the antibacterial mechanism of Cu-bearing materials. Characterization of the Ti6Al4V5Cu alloy indicated that the release of Cu ions was continuous from Ti6Al4V5Cu alloy in saline, leading to an increase in the accumulated concentration in 24 h. Therefore, we considered that it was these Cu ions released in the early stage that played the antibacterial role. Excellent bactericidal activity was observed against S.aureus in contact with Ti6Al4V5Cu. The morphology of S.aureus was changed, mainly due to the leakage of membranes, which further resulted in the leakage of the cellular contents such as sugars and proteins in bacteria during the co-culture of the alloys and the bacteria. The mechanisms of antibacterial action were primarily in two folds. Firstly, the activity of respiratory chains was disrupted by Cu ions, generating an increased amount of ROS subsequently. Secondly, the genome integrity was not obviously damaged after contacting with Cu-bearing alloy, but the process of gene replication was disturbed remarkably by the released Cu ions.
5. Experimental Section
Sample preparation and material characterization
Samples were prepared by machining commercial medical grade Ti6Al4V alloys rod into plates of Φ 15mm × 2mm in dimension. In order to obtain antibacterial properties, Ti6Al4V5Cu alloys were manufactured with the addition of high-purity oxygen free copper blocks (99.99%). The Ti6Al4V alloys without Cu served as a control in this study. Before testing, all samples were ground with SiC sand paper up to 2000 grids and sterilized completely (121 °C, 20 min). Microstructures were examined under an Axiovert 200 MAT optical microscope (OM). The samples for OM observations were prepared using the conventional metallographic procedure including grinding, mechanical polishing and then chemical etching in a solution of 13 mL HF, 26 mL HNO3 and 100 mL H2O. The phase compositions were determined by the 2θ/θ coupling method of XRD analysis using a Cu-Kα irradiation at an accelerating voltage of 40 kV and a current of 40 mA. The concentration of Cu ions released from Ti6Al4V5Cu alloys was measured by an inductively coupled plasma emission spectrometer (Thermo Scientific, iCAP Q). The samples were immersed in normal saline (2.86 cm2/mL) at 37 °C according to ISO Standard 10993-12:2002 and 10993-15:2000. An average of three measurements was taken for each group.
Bacterial culture and antibacterial test
S.aureus (ATCC25923) used in the present study was cultured overnight in sterilized Luria-Bertani (LB) broth (1% w/v Tryptone, 0.5% w/v Yeast extract, 0.5%w/v NaCl, pH = 7.0 ~7.2) in a shaking incubator at 37 °C. Bacterial concentration was determined by measuring the optical density at 600 nm (OD600) using a micro-absorbance reader and considering that 0.1 OD600 equals to 108 colony-forming units (cfu/mL). Then, serial dilution of the bacteria was performed in lysogeny broth (LB) medium to achieve a final bacterial suspension of 1 × 105 cfu/mL. Ti6Al4V5Cu and the control sample discs with size of Φ 15mm × 2mm were placed in 24-well plates and 1 mL bacterial suspension was dropped onto each experiment sample. The samples were then incubated at 37 °C for 6, 12, and 24 h. After co-culturing with the samples, the above bacterial suspensions were eliminated and the live and dead bacteria adhered on the samples were doubly stained with SYTO9 (5 µM, green fluorescence) and PI (30 µM, red fluorescence) at 37 °C for 15 min, respectively, and then visualized under the fluorescence microscope (LEICA, DM IRB). The images were acquired from random positions on the sample discs.
Leakage of reducing sugars and protein from S.aureus
The leakage of proteins and reducing sugars through membranes in S.aureus were detected according to previous methods reported by Miller et al[49] and Brown et al,[50] respectively. Similar to the antibacterial test, the sample discs were co-cultured with bacterial suspension at 37 °C for 6, 12, and 24 h. 1 mL co-culture supernatant was then harvested by centrifugation and the precipitate was removed, and then the supernatant extract was frozen at −20 °C immediately. The concentrations of proteins and reducing sugars were determined by bicinchoninic acid (BCA) protein assay kit and dinitrosalicylic acid reagent, respectively. Each experiment was performed in triplicate.
ROS generation
As an oxidation-sensitive fluorescent probe, 2’,7’- ichlorofluorescin-diacetate (DCFH-DA) was used to identify the existence of the overall ROS levels inside the bacterial cells. This non-fluorescent compound DCFH-DA can be oxidized by cellular oxidants into the highly fluorescent compound 2’7’-dichlorofluorescin (DCF).[51] S.aureus strains were prepared and co-cultured with the sample discs at three different time-points as described above for the antibacterial tests. For staining of adherent bacteria on the samples, the discs were removed and gently washed three times with phosphate-buffered saline (PBS). Subsequently, discs were stained in a new 24-well plate with 500 µL DCFH-DA (10 µM) at 37 °C for 30 min and then photographed with a fluorescence microscope. Moreover, for the measurement of ROS level in the suspended bacteria co-cultured with the materials, bacteria solutions were collected and stained with a diluted stock solution of DCFH-DA (10 µM) at 37°C for 30 min. Finally, the fluorescence intensities of the supernatant were detected by means of a Synergy 2 multi-mode microplate reader (BioTek) with an excitation wavelength of 488 nm and emission wavelength of 535 nm.[52]
Respiratory chain dehydrogenases activity
Since colorless Iodonitrotetrazolium chloride (INT) can be catalyzed into dark-red iodo-nitrotetrazolium formazan (INF) by cellular respiratory chain dehydrogenase, the respiratory chain dehydrogenase activity of S.aureus was determined by measuring the formation of formazan through spectrophotometric value.[53, 54] S.aureus strains were co-cultured with the sample discs for 6, 12 and 24 h, and then 500 µL INT solution (0.5% w/v) was added and incubated at 37 °C in dark for 2 h. Finally, the activity of respiratory chain dehydrogenases was calculated according to the OD value of INF at 490 nm. S.aureus cells boiled for 20 min to inactivate the enzyme activity served as a negative control, whereas bacterial cells of blank control group served as a positive control.
Catalase (CAT) activity
CAT is a key antioxidant enzyme in the organism defenses against oxidative stress. The CAT activities in the suspensions of bacterial cells cultured with experiment samples were assayed using CAT assay kit (Nanjing Jiancheng Bioengineering Institute) according to the manufacturer’s instructions. The absorbance at 405 nm was quantitated by a spectrophotometer (Thermo), and the decomposition of 1 µmol H2O2 every second was defined as one unit of activity (U). Data were expressed as U/mL.
Glutathione (GSH) levels
GSH is another antioxidant protecting cells from free radicals. The GSH activities in bacterial cell suspensions were measured using GSH assay kit (Nanjing Jiancheng Bioengineering Institute, China) following the protocol provided by the manufacturer. The colorimetric method was used and the absorbance at 420 nm was measured using a spectrophotometer (Thermo). Data were expressed as mg GSH/mL.
Lipid peroxidation (LPO) levels
LPO is a typical consequence of oxidative stress leading to further cell damages. The experimental setup for LPO level measurement in bacterial cell suspensions was similar to that for the CAT and GSH. The levels were measured in the form of thiobarbituric acid reactive substance (TBARS) by using lipid peroxidation assay kit (Nanjing Jiancheng Bioengineering Institute) according to the manufacturer’s instructions. The absorbance at 532 nm was collected by a spectrophotometer (Thermo). Data were expressed as nmol TBARS/mg protein.
Genome integrity determination
After incubation with different sample discs for 24 h, the co-culture supernatants were collected and subjected to genome DNA extraction using a Bacteria DNA kit (TIANGEN) according to the manufacturer’s instructions. Then, the genome integrity was checked by means of agarose gel electrophoresis, followed by staining with ethidium bromide and visualization through gel imaging analysis system (Bio-rad).[21]
EMA-PCR assay
An improved method using ethidium bromide monoazide (EMA, sigma, USA) in combination with PCR has been developed to detect and differentiate viable and dead bacteria. [55] The EMA selectively penetrates into injured or dead cells with compromised cell membrane/wall systems and binds to DNA molecules, which inhibits PCR amplification of the DNA from dead cells. [56] The present study was designed to optimize EMA-PCR methodology and monitor the behavior of target bacteria. In brief, bacterial cells in the co-culture supernatant was treated with 20 mg/mL EMA in the dark for 5 min and subsequently exposed to a 600 W halogen light for 1 min. Then, the nuc (S.aureus) and 16SrRNA gene was selected as target genes for PCR detection in these bacterial cells. The primers used in the experiments were listed as follows: nuc: F, 5’-TGTC TCGATTGATATC GCAACG-3’ and R, 5’-GATCTAAAAATTATAAAAGTGCCACTAG-3’; 16srDNA: F, 5’-AGATTTATCCTGG CTCAG-3’ and R, 5’-GGTTACCTTGTTACGACTT-3’.
Data analysis and statistical analysis
Data were analyzed by SPSS 13.0. Statistical comparisons among groups were evaluated by One-way ANOVA and Post Hoc multiple comparison LSD. The results were presented as means ± standard deviation (SD). A p-value less than 0.05 and 0.01 was considered statistically significant and highly significant, respectively.
Acknowledgments
We thank the financial support from the National Natural Science Foundation of China (Grant No. 81271957, 81272057) and National Basic Research Program of China (Grant No.2012CB 619106). We also sincerely thank the founding for The key Laboratory of Trauma & Tissue Repair of Tropical Area. PLA and Guangdong Key Lab. of Orthopaedic Technology and Implant Materials. M.Y. acknowledges the support of National Natural Science Foundation of China (21172194), Projects of Zhejiang Provincial Science and Technology Plans (2012C12910), Silkworm Industry Science and Technology Innovation Team (2011R50028), China Agriculture Research System (CARS-22-ZJ0402), and National High Technology Research and Development Program 863 (2013AA102507). Y. Zhu and C.B.M. would like to thank the financial support from National Institutes of Health (EB015190 and CA200504), National Science Foundation (CMMI-1234957 and CBET-1512664), Department of Defense Congressionally Directed Medical Research Program (W81XWH-15-1-0180), Oklahoma Center for Adult Stem Cell Research (434003) and Oklahoma Center for the Advancement of Science and Technology (HR14-160).
Contributor Information
Mei Li, Department of Orthopedics, Guangdong Key Lab of Orthopaedic Technology and Implant Materials, Guangzhou General Hospital of Guangzhou Military Command, Guangzhou, Guangdong 510010, China; State Key Laboratory for Mechanical Behavior of Materials, Xi’an Jiaotong University, Xian, Shaanxi 710049, China.
Zheng Ma, Institute of Metal Research, Chinese Academy of Sciences, Shenyang, Liaoning 110016, China.
Ye Zhu, Department of Chemistry and Biochemistry, Stephenson Life Sciences Research Center, University of Oklahoma, Norman, OK 73019, USA.
Prof. Hong Xia, Department of Orthopedics, Guangdong Key Lab of Orthopaedic Technology and Implant Materials, Guangzhou General Hospital of Guangzhou Military Command, Guangzhou, Guangdong 510010, China
Mengyu Yao, Department of Orthopedics, Guangdong Key Lab of Orthopaedic Technology and Implant Materials, Guangzhou General Hospital of Guangzhou Military Command, Guangzhou, Guangdong 510010, China.
Xiao Chu, Department of Orthopedics, Guangdong Key Lab of Orthopaedic Technology and Implant Materials, Guangzhou General Hospital of Guangzhou Military Command, Guangzhou, Guangdong 510010, China.
Xiaolan Wang, Department of Orthopedics, Guangdong Key Lab of Orthopaedic Technology and Implant Materials, Guangzhou General Hospital of Guangzhou Military Command, Guangzhou, Guangdong 510010, China.
Prof. Ke Yang, Institute of Metal Research, Chinese Academy of Sciences, Shenyang, Liaoning 110016, China
Prof. Mingying Yang, Institute of Applied Bioresource Research, College of Animal Science, Zhejiang University, Yuhangtang Road 866, Hangzhou, 310058, China
Prof. Yu Zhang, Department of Orthopedics, Guangdong Key Lab of Orthopaedic Technology and Implant Materials, Guangzhou General Hospital of Guangzhou Military Command, Guangzhou, Guangdong 510010, China
Prof. Chuanbin Mao, Department of Chemistry and Biochemistry, Stephenson Life Sciences Research Center, University of Oklahoma, Norman, OK 73019, USA School of Materials Science and Engineering, Zhejiang University, Hangzhou, Zhejiang 310027, China.
References
- 1.Mao C, Li H, Cui F, Ma C, Feng Q. J Crystal Growth. 1999;206:308. [Google Scholar]
- 2.Mao C, Li H, Cui F, Feng Q, Ma C. J Mater Chem. 1999;9:2573. [Google Scholar]
- 3.Mao C, Li H, Cui F, Feng Q, Wang H, Ma C. J Mater Chem. 1998;8:2795. [Google Scholar]
- 4.Zimmerli W. J Intern Med. 2014;276:111. doi: 10.1111/joim.12233. [DOI] [PubMed] [Google Scholar]
- 5.Zimmerli W, Moser C. FEMS Immunol Med Microbiol. 2012;65:158. doi: 10.1111/j.1574-695X.2012.00938.x. [DOI] [PubMed] [Google Scholar]
- 6.Helfet DL, Haas NP, Schatzker J, Matter P, Moser R, Hanson B. J Bone Joint Surg Am. 2003;85-A:1156. [PubMed] [Google Scholar]
- 7.Laffer RR, Graber P, Ochsner PE, Zimmerli W. Clin Microbiol Infect. 2006;12:433. doi: 10.1111/j.1469-0691.2006.01378.x. [DOI] [PubMed] [Google Scholar]
- 8.Wu C, Qu X, Liu F, Li H, Mao Y, Zhu Z. PLoS One. 2014;9:e95300. doi: 10.1371/journal.pone.0095300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kessler B, Sendi P, Graber P, Knupp M, Zwicky L, Hintermann B, Zimmerli W. J Bone Joint Surg Am. 2012;94:1871. doi: 10.2106/JBJS.K.00593. [DOI] [PubMed] [Google Scholar]
- 10.Pulido L, Ghanem E, Joshi A, Purtill JJ, Parvizi J. Clin Orthop Relat Res. 2008;466:1710. doi: 10.1007/s11999-008-0209-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury AN, Azam MS, Aktaruzzaman M, Rahim A. J Hazard Mater. 2009;172:1229. doi: 10.1016/j.jhazmat.2009.07.129. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson HL, Cronholm P, Hedberg Y, Tornberg M, De Battice L, Svedhem S, Wallinder IO. Toxicology. 2013;313:59. doi: 10.1016/j.tox.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Cui Z, Quaranta D, Grass G. Microbiologyopen. 2012;1:46. doi: 10.1002/mbo3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirai T, Tsuchiya H, Shimizu T, Ohtani K, Zen Y, Tomita K. J Biomed Mater Res B Appl Biomater. 2009;91:373. doi: 10.1002/jbm.b.31412. [DOI] [PubMed] [Google Scholar]
- 15.Ren L, Ma Z, Li M, Zhang Y, Liu W, Liao Z, Yang K. Journal of Materials Science & Technology. 2014;30:699. [Google Scholar]
- 16.Neal AL. Ecotoxicology. 2008;17:362. doi: 10.1007/s10646-008-0217-x. [DOI] [PubMed] [Google Scholar]
- 17.Damm C, Münstedt H. Applied Physics A. 2008;91:479. [Google Scholar]
- 18.Santo CE, Quaranta D, Grass G. Microbiologyopen. 2012;1:46. doi: 10.1002/mbo3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weaver L, Noyce JO, Michels HT, Keevil CW. J Appl Microbiol. 2010;109:2200. doi: 10.1111/j.1365-2672.2010.04852.x. [DOI] [PubMed] [Google Scholar]
- 20.Soejima T, Minami J, Yaeshima T, Iwatsuki K. Appl Microbiol Biotechnol. 2012;95:485. doi: 10.1007/s00253-012-4086-0. [DOI] [PubMed] [Google Scholar]
- 21.Aitken RJ, Bennetts LE, Sawyer D, Wiklendt AM, King BV. Int J Androl. 2005;28:171. doi: 10.1111/j.1365-2605.2005.00531.x. [DOI] [PubMed] [Google Scholar]
- 22.Kuley R, Smith HE, Frangoulidis D, Smits MA, Jan Roest HI, Bossers A. PLoS One. 2015;10:e0121661. doi: 10.1371/journal.pone.0121661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elizaquivel P, Aznar R, Sanchez G. J Appl Microbiol. 2014;116:1. doi: 10.1111/jam.12365. [DOI] [PubMed] [Google Scholar]
- 24.Liu YH, Wang CH, Wu JJ, Lee GB, Lee GB. Biomicrofluidics. 2012;6:34119. doi: 10.1063/1.4748358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slepicka P, Kasalkova NS, Siegel J, Kolska Z, Bacakova L, Svorcik V. Biotechnol Adv. 2015;33:1120. doi: 10.1016/j.biotechadv.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Qu H, Knabe C, Radin S, Garino J, Ducheyne P. Biomaterials. 2015;62:95. doi: 10.1016/j.biomaterials.2015.05.041. [DOI] [PubMed] [Google Scholar]
- 27.Ma Z, Yao M, Liu R, Yang K, Ren L, Zhang Y, Liao Z, Liu W, Qi M. Materials Technology. 2014;30:B80. [Google Scholar]
- 28.Cui Z, Ibrahim M, Yang C, Fang Y, Annam H, Li B, Wang Y, Xie G, Sun G. Molecules. 2014;19:9975. doi: 10.3390/molecules19079975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Guo Y, Chen S, Ge Z, Yang H, Tang J. J Mater Chem. 2012;22:9092. [Google Scholar]
- 30.Chen S, Guo Y, Zhong H, Chen S, Li J, Ge Z, Tang J. Chemical Engineering Journal. 2014;256:238. [Google Scholar]
- 31.Zhang W, Vinueza NR, Datta P, Michielsen S. Journal of Polymer Science Part A: Polymer Chemistry. 2015;53:1594. [Google Scholar]
- 32.Cabiscol E, Tamarit J, Ros J. Int Microbiol. 2000;3:3. [PubMed] [Google Scholar]
- 33.Ruparelia JP, Chatterjee AK, Duttagupta SP, Mukherji S. Acta Biomater. 2008;4:707. doi: 10.1016/j.actbio.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Bagchi B, Dey S, Bhandary S, Das S, Bhattacharya A, Basu R, Nandy P. Materials Science and Engineering: C. 2012;32:1897. doi: 10.1016/j.msec.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 35.Karlsson HL, Cronholm P, Hedberg Y, Tornberg M, De Battice L, Svedhem S, Wallinder IO. Toxicology. 2013;313:59. doi: 10.1016/j.tox.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 36.Kar S, Bagchi B, Kundu B, Bhandary S, Basu R, Nandy P, Das S. Biochimica et Biophysica Acta (BBA) - General Subjects. 2014;1840:3264. doi: 10.1016/j.bbagen.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 37.Dutta RK, Nenavathu BP, Gangishetty MK, Reddy AVR. Colloids and Surfaces B: Biointerfaces. 2012;94:143. doi: 10.1016/j.colsurfb.2012.01.046. [DOI] [PubMed] [Google Scholar]
- 38.Slauch JM. Molecular Microbiology. 2011;80:580. doi: 10.1111/j.1365-2958.2011.07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piao MJ, Kang KA, Lee IK, Kim HS, Kim S, Choi JY, Choi J, Hyun JW. Toxicology Letters. 2011;201:92. doi: 10.1016/j.toxlet.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 40.Su R, Wang R, Guo S, Cao H, Pan J, Li C, Shi D, Tang Z. Biological Trace Element Research. 2011;144:668. doi: 10.1007/s12011-011-9039-4. [DOI] [PubMed] [Google Scholar]
- 41.Sharma V, Anderson D, Dhawan A. Apoptosis. 2012;17:852. doi: 10.1007/s10495-012-0705-6. [DOI] [PubMed] [Google Scholar]
- 42.Weaver L, Noyce JO, Michels HT, Keevil CW. Journal of Applied Microbiology. 2010;109:2200. doi: 10.1111/j.1365-2672.2010.04852.x. [DOI] [PubMed] [Google Scholar]
- 43.Kilic A, Basustaoglu AC. Research in Microbiology. 2011;162:1060. doi: 10.1016/j.resmic.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 44.Aarthi P, Bagyalakshmi R, Therese KL, Madhavan HN. Microbiol Res. 2013;168:497. doi: 10.1016/j.micres.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 45.Nogva HK, Dromtorp SM, Nissen H, Rudi K. Biotechniques. 2003;34:804. doi: 10.2144/03344rr02. [DOI] [PubMed] [Google Scholar]
- 46.Soejima T, Schlitt-Dittrich F, Yoshida S. Analytical Biochemistry. 2011;418:286. doi: 10.1016/j.ab.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 47.Guo S, Meng Q, Zhao X, Wei Q, Xu H. Sci Rep. 2015;5:14688. doi: 10.1038/srep14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banerjee D, Williams JC. Acta Materialia. 2013;61:844. [Google Scholar]
- 49.Teixeira RS. Carbohydr Res. 2012;363:33. doi: 10.1016/j.carres.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 50.Brown RE, Jarvis KL, Hyland KJ. Anal Biochem. 1989;180:136. doi: 10.1016/0003-2697(89)90101-2. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Joseph JA. Free Radic Biol Med. 1999;27:612. doi: 10.1016/s0891-5849(99)00107-0. [DOI] [PubMed] [Google Scholar]
- 52.Jia SJ, Jiang DJ, Hu CP, Zhang XH, Deng HW, Li YJ. Vascul Pharmacol. 2006;44:143. doi: 10.1016/j.vph.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Munujos P, Coll-Canti J, Gonzalez-Sastre F, Gella FJ. Anal Biochem. 1993;212:506. doi: 10.1006/abio.1993.1360. [DOI] [PubMed] [Google Scholar]
- 54.Iturriaga R, Zhang S, Sonek GJ, Stibbs H. J Microbiol Methods. 2001;46:19. doi: 10.1016/s0167-7012(01)00249-4. [DOI] [PubMed] [Google Scholar]
- 55.Lee JL, Levin RE. J Microbiol Methods. 2006;67:456. doi: 10.1016/j.mimet.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 56.He Y, Chen CY. Food Microbiol. 2010;27:439. doi: 10.1016/j.fm.2009.11.017. [DOI] [PubMed] [Google Scholar]