Abstract
The antiretroviral protease inhibitor atazanavir inhibits hepatic uridine diphosphate glucuronosyltransferase (UGT) 1A1, thereby preventing the glucuronidation and elimination of bilirubin. Resultant indirect hyperbilirubinemia with jaundice can cause premature discontinuation of atazanavir. Risk for bilirubin‐related discontinuation is highest among individuals who carry two UGT1A1 decreased function alleles (UGT1A1*28 or *37). We summarize published literature that supports this association and provide recommendations for atazanavir prescribing when UGT1A1 genotype is known (updates at www.pharmgkb.org).
The purpose of this guideline is to provide information to allow the interpretation of clinical UGT1A1 genotype tests so that the results can be used to inform the prescribing of atazanavir. Detailed guidelines for the use of atazanavir as well as analyses of cost effectiveness are beyond the scope of this article. CPIC guidelines are periodically updated at http://www.pharmgkb.org.
FOCUSED LITERATURE REVIEW
A systematic literature review focused on UGT1A1 genotype and atazanavir‐related hyperbilirubinemia/jaundice and atazanavir discontinuation (details in Supplement) was conducted.
GENE: UGT1A1
Background
The uridine diphosphate (UDP) glucuronosyltransferases (UGT) are a large family of enzymes (19 in humans) that mediate conjugation of glucuronic acid with lipophilic drugs, xenobiotics, and endogenous substances, thereby increasing their water solubility and enabling efficient elimination in bile and/or urine. Three UGT subfamilies have been identified based on gene sequence similarity: UGT1A, UGT2A, and UGT2B. The UGT1A subfamily of enzymes (nine in humans) are encoded by a single gene locus through differential splicing of unique first exons (exon 1) to shared exons 2 to 5.
The major UGT1A subfamily enzyme, UGT1A1, is expressed primarily in the liver and gastrointestinal tract1 and is essential for the efficient elimination of bilirubin, the main byproduct of heme catabolism.2 Reduced UGT1A1 activity either through developmental delay in neonates,3 genetic variation,4, 5 or catalytic inhibition by drugs6 results in the accumulation of unconjugated (indirect) bilirubin in blood and tissues. When bilirubin elevation is high enough to cause visible yellow discoloration of the skin and eyes it is called jaundice (also known as icterus). In neonates, extreme bilirubin accumulation can lead to severe adverse neurological effects, namely, kernicterus.5
Genetic variants that have been identified in UGT1A1 exon 1, promoter, enhancer, and shared UGT1A exons 2 to 5 as well as the accepted allele nomenclature are listed at http://www.pharmacogenomics.pha.ulaval.ca/files/content/sites/pharmacogenomics/files/Nomenclature/UGT1A/UGT1A1.htm. The most frequent genetic variant that affects UGT1A1 function is a dinucleotide TAn repeat polymorphism (rs8175347) located in a TATAA consensus element in the UGT1A1 promoter at –53 relative to the translation start site. This varies from five to eight TA repeats. In all populations studied to date, TA6 (UGT1A1*1) and TA7 (UGT1A1*28) are most frequent, while TA5 (UGT1A1*36) and TA8 (UGT1A1*37) repeats are infrequent or absent depending on geographic region of ancestry.7, 8
The UGT1A1*28 allele was originally identified as a causative genetic variant of Gilbert syndrome, a form of mild unconjugated hyperbilirubinemia that affects ∼3–9% of individuals of European ancestry.9 Mechanistic studies using promoter‐reporter constructs have shown that the TA7 (UGT1A1*28) allele causes a moderate reduction in gene transcription as compared with the reference TA6 (UGT1A1*1) allele,9 possibly due to reduced binding affinity for transcription factors including TATA‐binding protein.10 The TA8 repeat (UGT1A1*37) appears to cause lower transcription levels compared to TA7, while TA5 (UGT1A1*36) appears to cause higher transcription levels than TA6. In studies of human liver microsomes, the amount of UGT1A1 protein is ∼2‐fold less in UGT1A1*28/*28 donors than in UGT1A1*1/*1 donors.11
Genome‐wide association studies (GWAS) that genotyped single nucleotide polymorphisms but not the TA repeat12, 13, 14 have consistently associated another polymorphism that is within 300 basepairs of the TA repeat, namely, rs887829 (c.‐364C>T; UGT1A1*80), with indirect hyperbilirubinemia in the general population (i.e., Gilbert syndrome). The rs887829 T allele is in very strong linkage disequilibrium (LD) with the TA7 and TA8 alleles, while rs887829 C is in very strong LD with the TA5 and TA6 alleles (r2 ≅ 0.99).7 The rs887829 polymorphism explains ∼15% of interindividual variability in indirect bilirubin concentrations in African‐American and European populations.
Other UGT1A1 single nucleotide polymorphisms (SNP) that have been associated with increased bilirubin concentrations in GWAS include rs11891311 in a Korean population15 and rs6742078 in a Chinese population,16 both of which are in significant LD with rs887829. The SNP rs4148323 (c.211G>A; p.Gly71Arg; UGT1A1*6) was also identified as an independent predictor of bilirubin concentrations in Korean and Chinese populations accounting for about 5% of variability (about 10% total when combined with UGT1A1*28 linked SNPs).15, 16 UGT1A1*6 is relatively common with East Asian (Japanese, Chinese, and Korean) ancestry but is absent in European and African populations.16
A polymorphism affecting a phenobarbital response element in the UGT1A1 enhancer (rs4124874; –3279T>G; UGT1A1*60) has also been associated with Gilbert syndrome,17 likely because this variant is in incomplete LD with UGT1A1*28.18 There is no evidence that UGT1A1*60 alone results in decreased UGT1A1 function. There are also a relatively large number of rare variants that have been discovered through sequencing of the UGT1A1 gene in individuals with Gilbert syndrome (see http://www.pharmacogenomics.pha.ulaval.ca/files/content/sites/pharmacogenomics/files/Nomenclature/UGT1A/UGT1A1.htm).
Genetic test interpretation
Clinical laboratories generally report UGT1A1 genotype assay results for the more frequent alleles, using either the star (*) allele nomenclature and/or the number of TA repeats in the UGT1A1 gene promoter region. Each named * allele is defined by one or more specific polymorphisms (see Supplemental Table S1 online and http://www.pharmacogenomics.pha.ulaval.ca/files/content/sites/pharmacogenomics/files/Nomenclature/UGT1A/UGT1A1.htm). The level of UGT1A1 activity associated with the most frequent allelic variants is summarized in Supplemental Table S2 online. Genotyping rs8175347 for the number of TA repeats allows assignment to UGT1A1*28 (TA7), UGT1A1*36 (TA5), UGT1A1*37 (TA8), or UGT1A1*1 (TA6, reference genotype).19 Because rs887829 is in almost complete linkage with rs8175347 (r2 ≅ 0.99), metabolizer status may also be inferred based on rs887829. Table 1 summarizes the assignment of the likely UGT1A1 phenotype based on * allele and number of TA repeats.
Table 1.
Assignment of likely UGT1A1 phenotypes based on genotypes
| Likely phenotype | Genotypes | Examples of diplotypes |
|---|---|---|
| Extensive metabolizer |
An individual carrying two referenceb function (*1)c and/or increased function alleles (*36). Alternatively identified by homozygosity for rs887829 C/C. |
*1/*1; *1/*36; *36/*36; rs887829 C/C |
| Intermediate metabolizer | An individual carrying one referenceb function (*1)c or increased function allele (*36) plus one decreased function allele (*6, *28, *37). Alternatively identified by heterozygosity for rs887829 C/T. | *1/*28; *1/*37; *36/*28; *36/*37; rs887829 C/T, *1/*6 |
| Poor metabolizer | An individual carrying two decreased function alleles (*6, *28, *37). Alternatively identified by homozygosity for rs887829 T/T (*80/*80) | *28/*28; *28/*37; *37/*37; rs887829 T/T (*80/*80), *6/*6 a |
Homozygosity for UGT1A1*6, which occurs almost exclusively in individuals of Asian descent, is associated with Gilbert's syndrome. However, at this time it is unclear if patients with this diplotype are at increased risk of severe atazanavir‐associated hyperbilirubinemia.
“Reference” function refers to the UGT1A1 alleles to which other alleles are compared.
The reference function *1 allele is fully functional and refers to the rs8175347 TA6 allele.
Alleles of UGT1A1 have been characterized in various geographically, racially, and ethnically diverse populations (Supplemental Table S3). The UGT1A1*6 allele (rs4148323, 211G>A) is associated with reduced UGT1A1 enzyme function and is found almost exclusively among individuals of Asian descent. In general, genotyping tests do not identify very rare or de novo variants.
Available genetic test options
See the Supplementary Material for more information on commercially available clinical testing options.
Incidental findings
Gilbert syndrome
Reduced hepatic UGT1A1 activity to ∼30% of normal is a hallmark of Gilbert syndrome, a benign condition characterized by mild unconjugated hyperbilirubinemia.4 Individuals with Gilbert syndrome may experience transient elevations in unconjugated plasma bilirubin in response to various triggers (e.g., fasting, infection, or medications). Genotypes most commonly implicated in Gilbert syndrome are UGT1A1*28/*28 and UGT1A1*6/*6.
Crigler‐Najjar syndrome
Crigler‐Najjar syndrome is a very rare and severe form of unconjugated hyperbilirubinemia that results from various deleterious UGT1A1 mutations,5, 20 most of which are not tested in commercial genotyping platforms. Crigler‐Najjar syndrome type 1, the severest form of the disease, is characterized by a complete absence of UGT1A1 activity. Without appropriate treatment that includes phototherapy and liver transplantation, patients usually die in childhood. Crigler‐Najjar syndrome type 2 is less severe and is characterized by severely reduced but detectable UGT1A1 activity. Crigler‐Najjar syndrome is generally diagnosed early in life, so is therefore very unlikely to be an incidental finding of genetic screening. However, identification of a heterozygous carrier state for a Crigler‐Najjar mutation may have implications for prenatal genetic counseling. Many rare UGT1A1 mutations have been associated with Crigler‐Najjar syndrome types 1 and 2 (see http://www.pharmacogenomics.pha.ulaval.ca/files/content/sites/pharmacogenomics/files/Nomenclature/UGT1A/UGT1A1.htm).
Implications of UGT1A1 genotype for other drugs
Decreased function genotypes of UGT1A1 may affect toxicity and/or tolerability of other drugs. Such drugs include irinotecan, the active SN‐38 metabolite of which undergoes detoxification to SN‐38‐glucuronide by UGT1A1, belinostat, which is also extensively metabolized by UGT1A1, and nilotinib or pazobapnib, which inhibits UGT1A1.21
Other considerations
None.
DRUG: ATAZANAVIR
Background
Atazanavir efficacy and tolerability
In 2003, atazanavir was US Food and Drug Administration (FDA)‐approved as the first once‐daily human immunodeficiency virus type 1 (HIV‐1) protease inhibitor. To maintain plasma concentrations necessary for optimal antiviral effect, atazanavir is typically prescribed with a pharmacokinetic enhancer, either low‐dose ritonavir or cobicistat. For the past decade, atazanavir, together with low‐dose ritonavir (atazanavir/r) and two nucleoside analogs, had constituted a recommended first‐line regimen for HIV‐1 infection in adults and children 6 years and older and an alternative agent for children 3 months through 5 years of age.22, 23, 24 In April 2015, the US Department of Health and Human Services Panel on Antiretroviral Guidelines for Adults and Adolescents downgraded regimens containing atazanavir/r from recommended to alternative status based on a large comparative trial showing the rate of toxicity‐related discontinuation was greater with the atazanavir/r than with either darunavir/r or raltegravir, each given with tenofovir/emtricitabine.25 In 2015, the FDA approved a coformulated tablet comprising atazanavir with cobicistat,26 which was likewise relegated to alternative status.
Atazanavir/r is generally safe and well tolerated. However, atazanavir inhibits UGT1A1‐mediated glucuronidation of bilirubin, thus increasing plasma indirect bilirubin concentrations.6 Plasma indirect bilirubin increases from baseline in virtually every patient who takes this drug, while direct bilirubin and hepatic transaminase concentrations are unaffected. This indirect hyperbilirubinemia does not indicate hepatic injury.22, 27, 28 With once‐daily atazanavir 300 mg plus ritonavir 100 mg, the frequency of grade 3 or higher bilirubin elevations (at least 2.5 times the upper limit of normal) is ∼40%,22, 28 and of grade 4 (at least 5 times the upper limit of normal) bilirubin elevation is ∼4‐8%.22, 24, 28 Among children and adolescents in a trial that involved atazanavir, 9% had a bilirubin value ≥5.1 times the upper limit of normal and 1.4% experienced jaundice.29
Premature discontinuation of atazanavir and UGT1A1 polymorphisms
Three studies have examined associations between UGT1A1 genotype and premature discontinuation of atazanavir/r.30, 31, 32 Two studies only evaluated all‐cause discontinuation,30, 31 whereas one also evaluated bilirubin‐related discontinuation.32 The latter approach minimizes the impact of factors unrelated to UGT1A1 genotype (e.g., nonadherence) and therefore better reveals genetic associations. Among 121 Swiss HIV Cohort Study participants (80% Caucasian) who had received atazanavir/r, carriage of UGT1A1 decreased function alleles (*28/*28 or *28/*37) was associated with increased risk of all‐cause atazanavir/r discontinuation. Eighteen participants had two decreased function alleles, 48 had one decreased function allele, and 55 participants had none, with estimated first‐year cumulative discontinuation rates of 63%, 24%, and 15%, respectively.30 In contrast, among 646 participants randomized to receive atazanavir/r in the AIDS Clinical Trials Group (ACTG) protocol A5202 there was no significant association between decreased function UGT1A1 genotype (primarily UGT1A1*28) and increased likelihood of all‐cause atazanavir/r discontinuation among either White or Black participants, but there was an association among Hispanic participants.31
Among 481 patients who initiated randomized atazanavir/r with tenofovir disoproxil fumarate (TDF)/emtricitabine in ACTG protocol A5257, bilirubin‐related discontinuation of atazanavir was strongly associated with rs887829 T/T (Figure 1). As discussed earlier, the rs887829 variant is in very high LD with the promoter TA repeat (r2 ≅ 0.99), the C allele being in linkage with (TA)5 and (TA)6, and the T allele with (TA)7 and (TA)8.7, 32 Without T/T homozygosity, bilirubin‐related discontinuation was infrequent regardless of race/ethnicity.32 Positive predictive values of rs887829 T/T for bilirubin‐related discontinuation through 96 weeks of atazanavir (with 95% confidence intervals) were 60% (32–84%) in White, 29% (8–58%) in Hispanic, 20% (9–36%) in Black participants; negative predictive values were 95% (90–98%), 97% (90–100%), and 97% (93–99%), respectively. The authors speculated that the higher discontinuation rate among White participants with rs887829 T/T may have reflected differences in physical manifestations of icterus.
Figure 1.
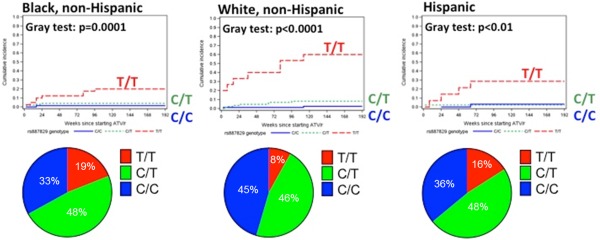
Cumulative incidence of time to bilirubin‐associated discontinuation of atazanavir stratified by UGT1A1 genotype in AIDS Clinical Trials Group protocol A5257 (adapted from ref. 32). Top panels: Lines estimate the cumulative incidence of time to bilirubin‐associated discontinuation of atazanavir, stratified by UGT1A1 rs887829 genotype and self‐reported race/ethnicity. P values are given by Gray's test for testing equality of cumulative incidence functions. Dashed red lines represent rs887829 T/T, dotted green lines rs887829 C/T, and solid blue lines rs887829 C/C. Bottom panels: Proportions of individuals in this analysis with rs887829 T/T, C/T, and C/T genotypes.
Associating genetic variability with variability in drug‐related phenotypes
Substantial evidence associates UGT1A1 genotype with phenotypic variability (see Supplemental Table S4). Most evidence is of high quality based on a standard grading scale (Supplemental Table S4). The evidence in the Supplemental Material and in Supplemental Table S4 provides the basis for the dosing recommendations in Table 2.
Table 2.
Recommended use of atazanavir (boosted with either ritonavir or cobicistat*) by UGT1A1 phenotype
| Phenotype | Implications for phenotypic measures | Dosing recommendations | Classification of recommendationsa |
|---|---|---|---|
| Extensive metabolizer | Referenceb UGT1A1 activity; very low likelihood of bilirubin‐related discontinuation of atazanavir. | There is no need to avoid prescribing of atazanavir based on UGT1A1 genetic test result. Inform the patient that some patients stop atazanavir because of jaundice (yellow eyes and skin), but that this patient's genotype makes this unlikely (less than about a 1 in 20 chance of stopping atazanavir because of jaundice). | Strong |
| Intermediate metabolizer | Somewhat decreased UGT1A1 activity; low likelihood of bilirubin‐related discontinuation of atazanavir. | There is no need to avoid prescribing of atazanavir based on UGT1A1 genetic test result. Inform the patient that some patients stop atazanavir because of jaundice (yellow eyes and skin), but that this patient's genotype makes this unlikely (less than about a 1 in 20 chance of stopping atazanavir because of jaundice). | Strong |
| Poor metabolizer | Markedly decreased UGT1A1 activity; high likelihood of bilirubin‐related discontinuation of atazanavir. |
Consider an alternative agent particularly where jaundice would be of concern to the patient. If atazanavir is to be prescribed, there is a high likelihood of developing jaundice that will result in atazanavir discontinuation (at least 20% and as high as 60%). |
Strong |
*All studies correlating UGT1A1 genotypes with atazanavir adverse events have involved ritonavir boosting. However, concentration‐time profiles are equivalent when boosted with either cobicistat or ritonavir,26 and bilirubin‐related adverse events including discontinuation of atazanavir occur in a similar percentage of patients prescribed atazanavir with cobicistat or ritonavir.26 Associations between UGT1A1 genotype, bilirubin elevations, and atazanavir/r discontinuation therefore almost certainly translate to atazanavir/cobicistat.
Rating scheme is described in Supplementary Data online.
“Reference” function refers to the UGT1A1 allele to which other alleles are compared.
Therapeutic recommendations
Atazanavir‐associated indirect hyperbilirubinemia does not indicate hepatic injury,22, 27, 28 but some patients are not prescribed atazanavir to avoid the possibility of jaundice. Implications of UGT1A1 genotype data for prescribing of atazanavir, boosted with either ritonavir or cobicistat, may be influenced by several factors. These include consequences of jaundice for the particular patient (e.g., workers who frequently interact with the public), access to alternative protease inhibitor antiretrovirals (e.g., darunavir), and whether the provider finds it useful to monitor atazanavir‐induced changes in bilirubin concentrations to assess adherence. Recommendations are provided in Table 2.
A UGT1A1 genotype is most helpful if available before atazanavir is prescribed. If noticeable jaundice does not develop while taking atazanavir chronically (or develops but is not bothersome), then the risk for bilirubin‐related atazanavir discontinuation is probably low regardless of UGT1A1 genotype.
For individuals carrying two UGT1A1 decreased function alleles (i.e., UGT1A1*28/*28, UGT1A1*28/*37, UGT1A1*37/*37, or rs887829 T/T), the likelihood of bilirubin‐related atazanavir discontinuation is substantial.30, 31 Before such individuals are prescribed atazanavir (boosted with either ritonavir or cobicistat), all such patients should be advised about the substantial likelihood of developing jaundice. Prescribing atazanavir to such individuals should generally be avoided unless the patient does not consider jaundice to be a concern, or there are other compelling reasons to prescribe atazanavir.
For individuals carrying fewer than two UGT1A1 decreased function alleles (i.e., *1/*28, *1/*37, *36/*28, *36/*37, rs887829 C/C or rs887829 C/T), the likelihood of bilirubin‐related atazanavir discontinuation is low.31, 33 This risk is extremely low for individuals carrying no UGT1A1 decreased function alleles (i.e., UGT1A1*1/*1, UGT1A1*1/*36, UGT1A1*36/*36, or rs887829 C/C). Among patients with extensive metabolizer UGT1A1 phenotypes it may not be necessary to discuss the possibility of jaundice with atazanavir. This decision about whether to discuss possible jaundice should be based on the clinical situation and provider judgment. If advice is offered, such discussion may note that the likelihood of developing jaundice that would require discontinuation of atazanavir is very low.
Recommendations for pediatrics
At the time of this writing there are no pediatric data regarding associations between UGT1A1 genotypes and likelihood of bilirubin‐related discontinuation of atazanavir. However, UGT1A1 genotypes are expected to affect atazanavir‐related hyperbilirubinemia similarly in adults and children. Therefore, recommendations for adults may be directly adapted to pediatric patients.
Recommendations for incidental findings
Individuals who are homozygous for UGT1A1*28 or UGT1A1*6 are very likely to have Gilbert syndrome. Knowing an individual's UGT1A1 genotype prior to prescribing may have implications for selection and dosing for drugs known to be UGT1A1 substrates or inhibitors, such as irinotecan and nilotinib.
Other considerations
Other UGT1A1 variants
Homozygosity for UGT1A1*6 or *27, which occurs almost exclusively in individuals of Asian descent, is associated with Gilbert syndrome. However, there is a lack of evidence regarding whether patients with these diplotypes are at increased risk of severe atazanavir‐associated hyperbilirubinemia. One study found no association between UGT1A1*6/*6 and the incidence of severe hyperbilirubinemia with atazanavir,34 although the lack of a statistically significant association may reflect the small number of patients with this genotype, with only seven patients homozygous for UGT1A1*6. Therefore, at this time, it is unclear whether UGT1A1*6/*6 or *27/*27 genotypes confer increased risk of severe atazanavir‐associated hyperbilirubinemia.
Higher plasma atazanavir concentrations correlate directly with greater increases in plasma bilirubin concentrations. In a GWAS involving 475 HIV‐infected patients prescribed atazanavir/r, no genetic variant (including candidate pharmacogenetic polymorphisms) was strongly associated with plasma atazanavir clearance.35
Bilirubin as a biomarker of adherence
All HIV‐1 protease inhibitors have relatively high genetic barriers to viral drug resistance and are therefore somewhat forgiving of nonadherence. For this reason, protease inhibitor‐based regimens are sometimes prescribed to patients considered at high risk for nonadherence. With atazanavir/r, failure of plasma bilirubin to increase from baseline (regardless of UGT1A1 genotype) is strong evidence that atazanavir/r was not taken during the prior ∼24 hours.36, 37, 38 This biomarker of adherence, often available from chemistry panels obtained at routine clinic visits, may still be used among UGT1A1 extensive metabolizers who are prescribed atazanavir/r.
Implications of UGT1A1 polymorphisms for atazanavir with cobicistat
All studies correlating UGT1A1 genotypes with atazanavir adverse events have involved ritonavir boosting. Such data are lacking for atazanavir boosted with cobicistat. However, atazanavir plasma concentration–time profiles are equivalent when boosted with either cobicistat or ritonavir.26 In addition, in a double‐blind clinical trial that randomly assigned 692 patients to receive atazanavir with either cobicistat or ritonavir, adverse events related to bilirubin elevations (e.g., hyperbilirubinemia, jaundice, and scleral icterus) occurred in a similar percentage of patients in the cobicistat and ritonavir arms (40.7% and 36.2%, respectively), as did bilirubin‐associated discontinuation of atazanavir (3.5% and 3.2%, respectively).26 Associations between UGT1A1 genotype, bilirubin elevations, and atazanavir/r discontinuation therefore almost certainly translate to atazanavir/cobicistat. These guidelines regarding UGT1A1 genotype do not apply to rare situations where atazanavir is prescribed without either ritonavir or cobicistat.
POTENTIAL BENEFITS AND RISKS FOR THE PATIENT
The benefit of prospective UGT1A1 genotyping would be to determine the individual's likelihood of bilirubin‐related discontinuation of atazanavir prior to beginning therapy.32 This may allow atazanavir to be prescribed to patients at low risk for bilirubin‐related discontinuation and avoided in patients at high risk. Some patients may still develop bilirubin‐related discontinuation of atazanavir despite low‐risk genotypes. There is little apparent risk of UGT1A1 genotyping that results in a recommendation to avoid atazanavir, as alternative protease inhibitor‐containing regimens are comparable in terms of efficacy and pill burden, although costs may vary depending on the payer.
Other possible limitations include laboratory error and an incomplete UGT1A1 genetic profile, as many tests only report results for UGT1A1*28. As individuals' genotypes do not change over time, genotyping errors could remain in the medical record for the lifetime of the patient.
CAVEATS: APPROPRIATE USE AND/OR POTENTIAL MISUSE OF GENETIC TESTS
A systematic review of the literature concluded that homozygosity of UGT1A1*28 is a risk factor for occurrence of severe atazanavir‐associated unconjugated hyperbilirubinemia, with a pooled positive predictive value of 40.3% and a pooled negative predictive value of 88.1%. Bilirubin‐related discontinuation of atazanavir through 96 weeks is strongly associated with rs887829 T/T (in significant LD with UGT1A1*28), with reported positive predictive value ranging from 20% to 60% depending on race/ethnicity.32 Thus, race/ethnicity may modify the genetic effect (Figure 1). Nongenetic factors such as fasting and diet can also affect bilirubin concentrations.39, 40
This CPIC guideline assumes that the UGT1A1 genotype results are already available to the prescriber. It is beyond the scope of this guideline to make recommendations regarding whether or not UGT1A1 genotyping should be performed.
DISCLAIMER
Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines reflect expert consensus based on clinical evidence and peer‐reviewed literature available at the time they are written, and are intended only to assist clinicians in decision‐making, as well as to identify questions for further research. New evidence may have emerged since the time a guideline was submitted for publication. Guidelines are limited in scope and are not applicable to interventions or diseases not specifically identified. Guidelines do not account for all individual variation among patients and cannot be considered inclusive of all proper methods of care or exclusive of other treatments. It remains the responsibility of the healthcare provider to determine the best course of treatment for the patient. Adherence to any guideline is voluntary, with the ultimate determination regarding its application to be solely made by the clinician and the patient. CPIC assumes no responsibility for any injury to persons or damage to property related to any use of CPIC's guidelines, or for any errors or omissions.
CONFLICT OF INTEREST
D.W.H. has been a consultant to Merck. All other authors declare no conflicts.
ACKNOWLEDGMENT
We acknowledge the critical input of members of CPIC of the Pharmacogenomics Research Network (PGRN), particularly MV Relling (St Jude Children's Research Hospital) and the CPIC informatics working group. This work was funded by the National Institutes of Health (NIH) PAAR4Kids (UO1 GM92666), PharmGKB (R24 GM61374), U01 HL0105198, P30 AI110527, R01 AI077505, TR000445 (to D.W.H.), and R01 GM102130 (to M.H.C.).
References
- 1. Court, M.H. et al Quantitative distribution of mRNAs encoding the 19 human UDP‐glucuronosyltransferase enzymes in 26 adult and 3 fetal tissues. Xenobiotica 42, 266–277 (2012). [DOI] [PubMed] [Google Scholar]
- 2. Bosma, P.J. et al Bilirubin UDP‐glucuronosyltransferase 1 is the only relevant bilirubin glucuronidating isoform in man. J. Biol. Chem. 269, 17960–17964 (1994). [PubMed] [Google Scholar]
- 3. Strassburg, C.P. et al Developmental aspects of human hepatic drug glucuronidation in young children and adults. Gut 50, 259–265 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Strassburg, C.P. Pharmacogenetics of Gilbert's syndrome. Pharmacogenomics 9, 703–715 (2008). [DOI] [PubMed] [Google Scholar]
- 5. Kadakol, A. et al Genetic lesions of bilirubin uridine‐diphosphoglucuronate glucuronosyltransferase (UGT1A1) causing Crigler‐Najjar and Gilbert syndromes: correlation of genotype to phenotype. Hum. Mutat. 16, 297–306 (2000). [DOI] [PubMed] [Google Scholar]
- 6. Zhang, D. et al In vitro inhibition of UDP glucuronosyltransferases by atazanavir and other HIV protease inhibitors and the relationship of this property to in vivo bilirubin glucuronidation. Drug Metab. Dispos. 33, 1729–1739 (2005). [DOI] [PubMed] [Google Scholar]
- 7. Horsfall, L.J. et al Prevalence of clinically relevant UGT1A alleles and haplotypes in African populations. Ann. Hum. Genet. 75, 236–246 (2011). [DOI] [PubMed] [Google Scholar]
- 8. Hall, D. , Ybazeta, G. , Destro‐Biso, I G. , Petzl‐Erler, M.L. & Di Rienzo, A. Variability at the uridine diphosphate glucuronosyltransferase 1A1 promoter in human populations and primates. Pharmacogenetics 9, 591–599 (1999). [PubMed] [Google Scholar]
- 9. Bosma, P.J. et al The genetic basis of the reduced expression of bilirubin UDP‐glucuronosyltransferase 1 in Gilbert's syndrome. N. Engl. J. Med. 333, 1171–1175 (1995). [DOI] [PubMed] [Google Scholar]
- 10. Hsieh, T.Y. et al Molecular pathogenesis of Gilbert's syndrome: decreased TATA‐binding protein binding affinity of UGT1A1 gene promoter. Pharmacogenet. Genomics 17, 229–236 (2007). [DOI] [PubMed] [Google Scholar]
- 11. Girard, H. et al UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology 42, 448–457 (2005). [DOI] [PubMed] [Google Scholar]
- 12. Johnson, A.D. et al Genome‐wide association meta‐analysis for total serum bilirubin levels. Hum. Mol. Genet. 18, 2700–2710 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen, G. et al UGT1A1 is a major locus influencing bilirubin levels in African Americans. Eur. J. Hum. Genet. 20, 463–468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sanna, S. et al Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum. Mol. Genet. 18, 2711–2718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kang, T.W. et al Genome‐wide association of serum bilirubin levels in Korean population. Hum. Mol. Genet. 19, 3672–3678 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dai, X. et al A genome‐wide association study for serum bilirubin levels and gene‐environment interaction in a Chinese population. Genet. Epidemiol. 37, 293–300 (2013). [DOI] [PubMed] [Google Scholar]
- 17. Sugatani, J. et al Identification of a defect in the UGT1A1 gene promoter and its association with hyperbilirubinemia. Biochem. Biophys. Res. Commun. 292, 492–497 (2002). [DOI] [PubMed] [Google Scholar]
- 18. Chen, Z. et al UGT1A1 sequence variants associated with risk of adult hyperbilirubinemia: a quantitative analysis. Gene 552, 32–38 (2014). [DOI] [PubMed] [Google Scholar]
- 19. Barbarino, J.M. , Haidar, C.E. , Klein, T.E. & Altman, R.B. PharmGKB summary: very important pharmacogene information for UGT1A1. Pharmacogenet. Genomics 24, 177–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bartlett, M.G. & Gourley, G.R. Assessment of UGT polymorphisms and neonatal jaundice. Semin. Perinatol. 35, 127–133 (2011). [DOI] [PubMed] [Google Scholar]
- 21. Singer, J.B. et al UGT1A1 promoter polymorphism increases risk of nilotinib‐induced hyperbilirubinemia. Leukemia 21, 2311–2315 (2007). [DOI] [PubMed] [Google Scholar]
- 22. Molina, J.M. et al Once‐daily atazanavir/ritonavir versus twice‐daily lopinavir/ritonavir, each in combination with tenofovir and emtricitabine, for management of antiretroviral‐naive HIV‐1‐infected patients: 48 week efficacy and safety results of the CASTLE study. Lancet 372, 646–655 (2008). [DOI] [PubMed] [Google Scholar]
- 23. Daar, E.S. et al Atazanavir plus ritonavir or efavirenz as part of a 3‐drug regimen for initial treatment of HIV‐1. Ann. Intern. Med. 154, 445–456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lennox, J.L. et al Efficacy and tolerability of 3 nonnucleoside reverse transcriptase inhibitor‐sparing antiretroviral regimens for treatment‐naive volunteers infected with HIV‐1: a randomized, controlled equivalence trial. Ann. Intern. Med. 161, 461–471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Panel on Antiretroviral Guidelines for Adults and Adolescents . Guidelines for the Use of Antiretroviral Agents in HIV‐1‐Infected Adults and Adolescents. Department of Health and Human Services. <http://www.aidsinfo.nih.gov/contentfiles/lvguidelines/adultandadolescentgl.pdf>. Accessed on April 14, 2015.
- 26. Gallant, J.E. et al Cobicistat versus ritonavir as a pharmacoenhancer of atazanavir plus emtricitabine/tenofovir disoproxil fumarate in treatment‐naive HIV type 1‐infected patients: week 48 results. J. Infect. Dis. 208, 32–39 (2013). [DOI] [PubMed] [Google Scholar]
- 27. Malan, D.R. et al Efficacy and safety of atazanavir, with or without ritonavir, as part of once‐daily highly active antiretroviral therapy regimens in antiretroviral‐naive patients. J. Acquir. Immune Defic. Syndr. 47, 161–167 (2008). [DOI] [PubMed] [Google Scholar]
- 28. Torti, C. et al Hyperbilirubinemia during atazanavir treatment in 2,404 patients in the Italian atazanavir expanded access program and MASTER Cohorts. Infection 37, 244–249 (2009). [DOI] [PubMed] [Google Scholar]
- 29. Rutstein, R.M. et al Long‐term safety and efficacy of atazanavir‐based therapy in HIV‐infected infants, children and adolescents: the Pediatric AIDS Clinical Trials Group Protocol 1020A. Pediatr. Infect. Dis. J. 34, 162–167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lubomirov, R. et al Association of pharmacogenetic markers with premature discontinuation of first‐line anti‐HIV therapy: an observational cohort study. J. Infect. Dis. 203, 246–257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ribaudo, H.J. et al Impact of UGT1A1 Gilbert variant on discontinuation of ritonavir‐boosted atazanavir in AIDS Clinical Trials Group Study A5202. J. Infect. Dis. 207, 420–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vardhanabhuti, S. et al Screening for UGT1A1 Genotype in Study A5257 would have markedly reduced premature discontinuation of atazanavir for hyperbilirubinemia. Open Forum Infect. Dis. 2, ofv085 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eley, T. et al Clinical and pharmacogenetic factors affecting neonatal bilirubinemia following atazanavir treatment of mothers during pregnancy. AIDS Res. Hum. Retroviruses 29, 1287–1292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park, W.B. et al Genetic factors influencing severe atazanavir‐associated hyperbilirubinemia in a population with low UDP‐glucuronosyltransferase 1A1*28 allele frequency. Clin. Infect. Dis. 51, 101–106 (2010). [DOI] [PubMed] [Google Scholar]
- 35. Johnson, D.H. et al Genomewide association study of atazanavir pharmacokinetics and hyperbilirubinemia in AIDS Clinical Trials Group protocol A5202. Pharmacogenet. Genomics 24, 195–203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petersen, K. et al Use of bilirubin as a marker of adherence to atazanavir‐based antiretroviral therapy. AIDS 19, 1700–1702 (2005). [DOI] [PubMed] [Google Scholar]
- 37. Rekic, D. et al Bilirubin‐a potential marker of drug exposure in atazanavir‐based antiretroviral therapy. AAPS J. 13, 598–605 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morello, J. et al Short communication: use of serum bilirubin levels as surrogate marker of early virological response to atazanavir‐based antiretroviral therapy. AIDS Res. Hum. Retroviruses 27, 1043–1045 (2011). [DOI] [PubMed] [Google Scholar]
- 39. Rodrigues, C. et al Bilirubin dependence on UGT1A1 polymorphisms, hemoglobin, fasting time and body mass index. Am. J. Med. Sci. 343, 114–118 (2012). [DOI] [PubMed] [Google Scholar]
- 40. Navarro, S.L. et al Cruciferous vegetable feeding alters UGT1A1 activity: diet‐ and genotype‐dependent changes in serum bilirubin in a controlled feeding trial. Cancer Prev. Res. (Phila.) 2, 345–352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]


